Abstract
Bacterial resistance to antibiotics has become an increasing threat, requiring not only the development of new targets in drug discovery, but more importantly, a better understanding of cellular response. In the current study, three closely related Escherichia coli strains, a wild type (MG1655), and an isogenic pair derived from the wild type (DPB635 and DPB636) are studied following exposure to sub lethal concentrations of antibiotic (norfloxacin) over time. In particular, genotype similarities between the three strains were assessed based on the lipid regulation response (e.g., presence/absence and up/down regulation). Lipid identification was performed using direct surface probe analysis (Matrix-assisted laser desorption/ionization, MALDI), coupled to high-resolution mass spectrometry (Fourier transform ion cyclotron resonance mass spectrometry, FT-ICR MS) followed by statistical analysis of variability and reproducibility across batches using internal standards. Inspection of the lipid profile showed that for the MG1655, DPB635 and DPB636 E. coli strains, a similar distribution of the altered lipids were observed after exposure to norfloxacin antibiotic (e.g., fatty acids and glycerol phospholipids are up and down regulated, respectively). Additionally, variations in the lipid distribution resemble the extent to which each strain can combat the antibiotic exposure. That is, the topA66 topoisomerase I mutation of DPB636 translates into diminished response related to antibiotic sensitivity when compared to MG1655 and the DPB635 strains.
Keywords: lipid profile, MALDI FT-ICR MS, E. coli, profile analysis, cell response
INTRODUCTION
Lipids are an integral component to cells, involved in energy storage processes and in maintaining structural integrity[1]. Biological pathways that utilize lipids have been associated with tumor progression[2], neurological disorders[3] and infections[4]; and more recently, it has been shown that changes in the lipid profiles can be used as markers for cellular regulation processes[5]. In particular, evidence suggests that SOS cell response[6] to external stress conditions (e.g., antibiotic treatment) triggers gene expression changes[7], cell mutations[8] and macromolecular differentiation which results in bacterial resistance. At the cellular level, these changes translate into morphological (e.g., cell membrane[9]) and physiological changes, suggesting that the SOS cell response can be correlated with the lipid composition and localization.
Over the last decade, significant advances in the development of high throughput analytical techniques have permitted the characterization of lipid profiles from complex biological targets[10] and the study of antibiotic response on surfaces using mass spectrometry[11–14]. While classical lipid analysis includes lengthy extraction, pre-fractionation followed by chromatographic separation (e.g., GC and LC) and mass spectrometry identification[15], current efforts progress towards the development of in situ lipid profiling tools combining surface probes (e.g., DESI, MALDI and TOF-SIMS) with fast, gas phase, post-ionization separation techniques[16–26]. In addition, when complemented with high-resolution mass spectrometry, the analyses of biological surfaces can take advantage of the high mass accuracy for direct lipid identification [27–29]. Recent work has shown that lipid quantification is possible when using internal standards[30, 31]
In the present paper, we utilize the advantages of high-resolution mass spectrometry combined with a direct surface probe – matrix-assisted laser desorption ionization coupled to Fourier transform ion cyclotron resonance mass spectrometry (MALDI FT-ICR MS) – for the analysis of the SOS cell response of three Escherichia coli (E. coli) strains as a preliminary step towards single cell in situ lipid profiling. In particular, SOS cell response is studied for the case of a wild type (MG1655) and two isogenic strains derivatives of the wild type (DPB635 and DPB636) as a function of the concentration and the exposure time of the antibiotic norfloxacin, a fluoroquinolone. Bacterial SOS response to DNA damage by quinolone action has been shown to lead to an increase in general antibiotic resistance.[32, 33] When compared to strains MG1655 and DPB635, strain DPB636 exhibits norfloxacin hypersensitivity (manuscript in preparation) as a result of the topA66 mutation in the C-terminal domain of topoisomerase I[34]; the latter will be used to identify specific lipid changes in the isogenic strains that may be correlated with antibiotic sensitivity. Topoisomerase I is present in all bacteria, and has been shown to be a promising target for discovery of novel antibacterial drugs.[35–37]
EXPERIMENTAL SECTION
Cell growth conditions
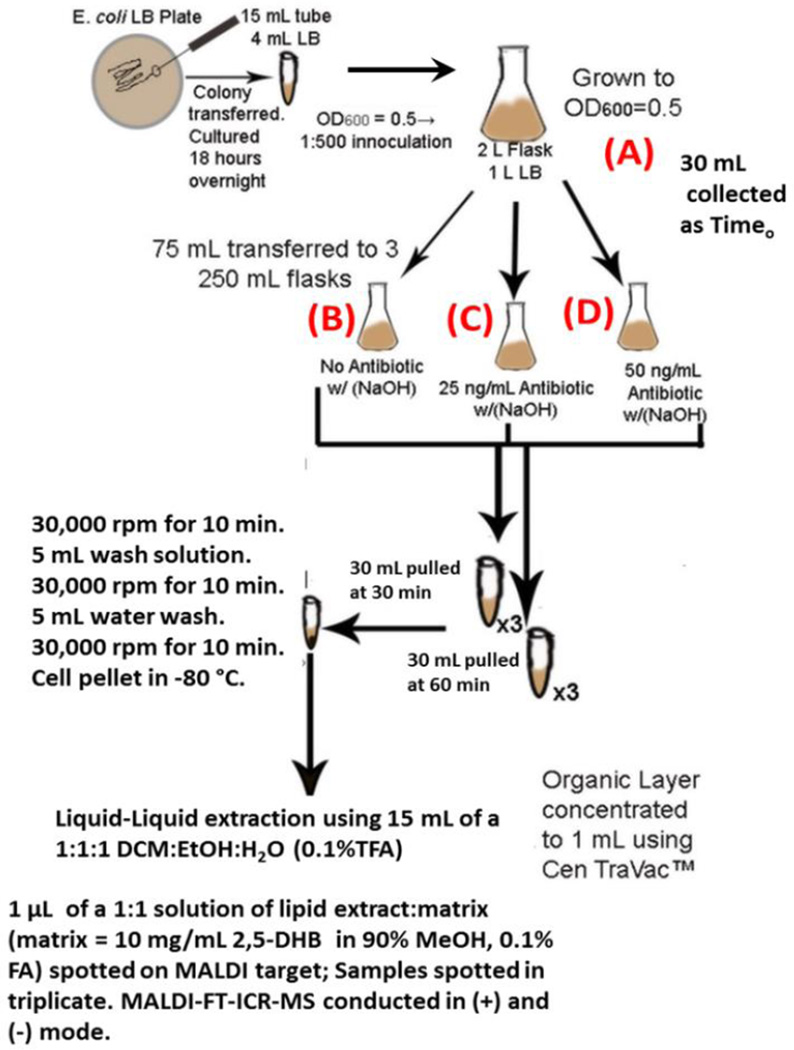
The experimental design used to generate the subset of cultures used in this study has been depicted in Scheme 1. Briefly, E. coli strains MG1655 (K-12 wild-type), DPB635 (MG1655 zci-2250::mini-kan), DPB636 (MG1655 topA66 zci-2250::mini-kan), were obtained from the Yale Coli Genetic Stock Center (New Haven, CT). Cultures of the three E. coli strains were grown overnight in Luria-Bertani (LB) growth medium in a 37 °C shaker at 200 rpms (culture I). A 1:500 inoculation of culture I was added to 500 mL LB in a 1000-mL Erlenmeyer flask (strainstock). Culture A was grown at 37 °C, 200 rpms to an OD600 = 0.5. From culture A, 75 mL aliquots were transferred to three 250-mL Erlenmeyer flasks, denoted culture B, C and D, respectively. The norfloxacin used in the studies was diluted from a 10 µg/mL stock in 0.1 M NaOH in order to obtain sublethal concentrations of 25 ng/mL and 50 ng/mL antibiotic in flasks two and three, respectively. Culture B contained only 0.1M NaOH to account for the presence of this solvent in cultures C and D. Flasks were placed back in the shaker. At 30 and 60 minute time intervals, 30 mL samples were taken from cultures B (straincontrol), C and D (strainstress) in order to evaluate the antibiotic response as a function of time exposure and concentration. Samples were then centrifuged for 10 minutes at 3000 rpms to generate a cell pellet. The cell pellets were washed with 5 mL of 10 mM EDTA (pH 8.0) and 50 mM glucose solution[38], centrifuged and washed with 5 mL autoclaved water. Finally cell pellets were frozen at −80 °C. Growth media was made in large batches and other steps were taken to reduce batch variation in lipid profile that can be influenced by growth conditions [39–41].
Scheme 1.
E. coli growth conditions and antibiotic study preparation and analysis workflow. Sample (A) refers to the strain stock culture, (B) NaOH control culture, (C) low dose antibiotic culture + NaOH and (D) high dose antibiotic culture + NaOH. Cultures (B), (C) and (D) had samples pulled at two time intervals – 30 minutes and 60 minutes following splitting from (A).
MALDI-FTICR-MS preparation and analysis
Frozen cell pellets were reconstituted in 15 mL of a 1:1:1 water (0.1% TFA) : ethanol : dichloromethane solution[39] and left for 18–24 hours to extract the lipids from the total cell material. The lipid layer was concentrated to 1 mL using a CentriVac system. A 10 mg/mL 2,5-dihydroxybenzoic acid (Sigma-Aldrich Co. LLC, St. Louis, MO) in an analytical grade 90 % methanol, 0.1 % FA matrix-solvent system was used as a MALDI matrix. Reproducibility of the analysis was corrected using known concentration of an internal lipid standards mixture of (catalog numbers 840635P, 850745P, 840455P, 840036P, 850141P,700085, 700000P, LM-6002, LM-6000, Avanti Polar Lipids, Inc., Alabaster, Alabama). A 1 µL volume of a 1::1 analyte:internal standard mix solution was spotted on a stainless steel MALDI target and the MALDI matrix was sublimated to guarantee homogeneity across the spot. Each sample was analyzed in triplicate. Analysis was conducted using a Bruker Solarix 7.0T FT-ICR MS equipped with a SmartBeam II laser for MALDI analyses (Bruker Daltonics, Inc., Billerica, MA). The instrument was externally calibrated using a tuning mix standard (Tunemix, G2421A, Agilent Technologies, Santa Clara, CA) under electrospray ionization conditions in both positive and negative modes (< 1 ppm mass accuracy). Internal lipid standards and previously reported lipids were used as MALDI internal calibration post analysis [39, 42] Multiple replica (typically, 5 measurements) of positive and negative mode MALDI spectra were acquired for each experimental condition. The wash solutions, growth media and matrix solutions were run as controls for each batch. Six batches of each E. coli strain were analyzed without antibiotic in order to characterize similarities and differences between the strains. Three batches of the antibiotic study were utilized to evaluate the reproducibility of the stress response of each strain.
Statistical analysis
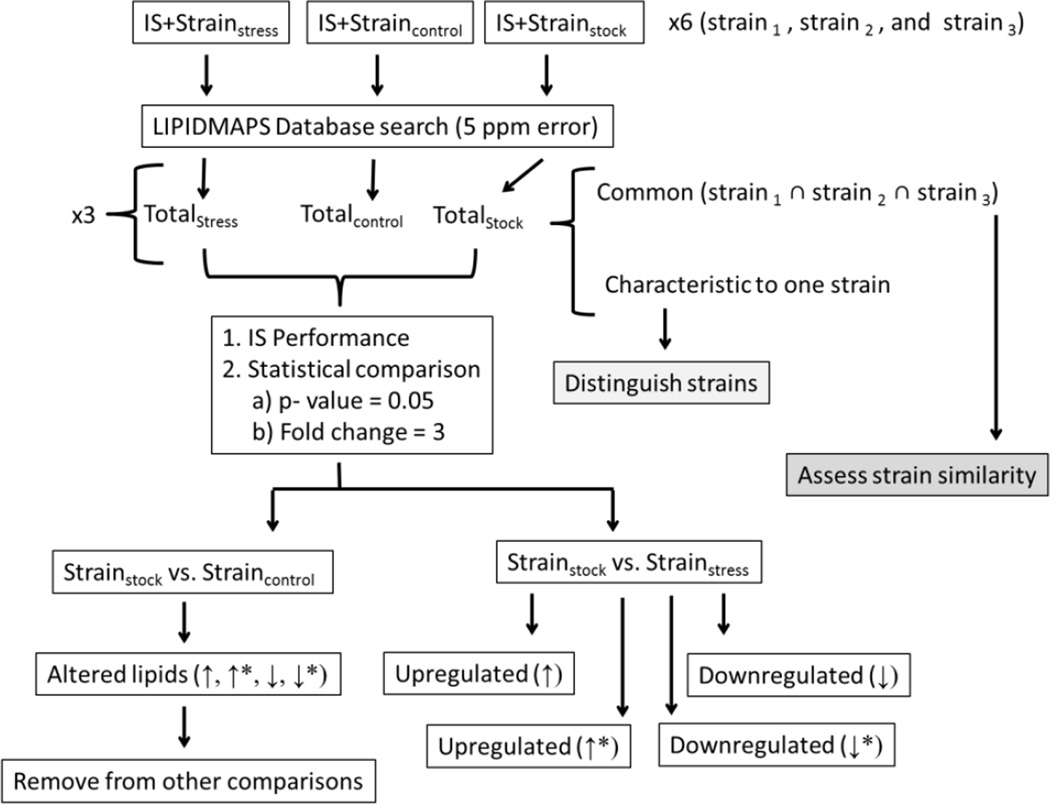
The data processing workflow has been outlined in Scheme 2. In particular, this workflow has been adapted from reference [15] and determines statistically significant lipids using a p-value = 0.05 and an intensity fold change following a student’s t-test. An The criteria for significant fold change was based on the reproducibility of the internal standards as a function of the conditions. In particular, matrix effects observed did not exceed a fold change of two, however to ensure minimal, if any, bias in the trends reported, a fold change of three was utilized to compensate for the variation in lipid standard reproducibility encountered in all samples. A bucketing approach was used to generate the t-test variables by dividing each mass spectrum into 5 mDa intervals to yield a single intensity value paired with a mass-to-charge unit using ProfileAnalysis (Bruker Daltonics, Inc., Billerica, MA). Each bucket list was normalized to the sum of the bucket values in the mass spectrum.
Scheme 2.
Workflow of MALDI-FTICR-MS data processing utilized for E. coli strain comparison using an internal standard (IS). The asterisk (*) corresponds to lipids that are up or down regulated on the basis of an absence (not detection) in the other sample being compared.
Comparisons were conducted based on total differences regardless of the analysis mode in which the lipid molecular ion was detected. Lipids were assigned from the LIPIDMAPS database[43] using a 5 ppm tolerance. Potential adducts were considered in the database search; that is [M+H]+, [M+Na]+ and [M+K]+for positive mode, and [M-H]−, [M-2H+Na]− and [M-2H+K]− for negative mode. The neutral lipid mass was used for trend comparison regardless of the molecular ion detected. All eight lipid categories (as grouped by LIPIDMAPS) were considered in the database search and lipid candidate structures were proposed based on the chemical formula. The complexity of the sample (i.e., multiple lipid signals at the level of nominal) limited the possibility to perform MS/MS experiments for secondary confirmation.
Following the procedure outline in Scheme 2, common and characteristic lipids between the three strains were determined. The extent of lipid differentiation between the three strains were determined from the total lipid assignments (Totalstock) generated for each strain (see supplemental Tables 1–3 for more details). Here after, a characteristic and a common lipid refers to a lipid that was detected in only one strain or in at least two/three strains, respectively. The number of batches (out of 6 total) in which the lipid was encountered can be used to assess the significance of the lipid when determining characteristic lipids among the strains.
For the case of SOS cell response, statistically significant lipid changes associated with the presence of norfloxacin were categorized on the basis of intensity fold change as up (↑) or down (↓) regulated. Lipids that are considered up or down regulated as a result of an absence (not detection either in the initial or final state) have been designated by ↑* or ↓*. All statistical comparisons have been made using the total lipid lists for each strain as described in scheme 2 (i.e., Totalstock, Totalcontrol and Totalstress). Each stress condition is comprised of a time exposure (30 or 60 minutes) and an antibiotic concentration (25 ng/mL or 50 ng/mL) component; these results in four separate stress-related lipid comparisons per strain (see supplemental Tables 5–16 for more detail). In addition, to account for the influence of solvent, in particular the NaOH contribution to changes in the lipid profile following exposure to the norfloxacin antibiotic, lipids up or down regulated as a result of the stock vs. control comparison were removed if encountered in other stock vs. antibiotic comparisons. That is, the listed lipids, either ↑, ↑*, ↓ or ↓* are only representative of the interaction of the antibiotic with the E. coli cell response.
RESULTS AND DISCUSSION
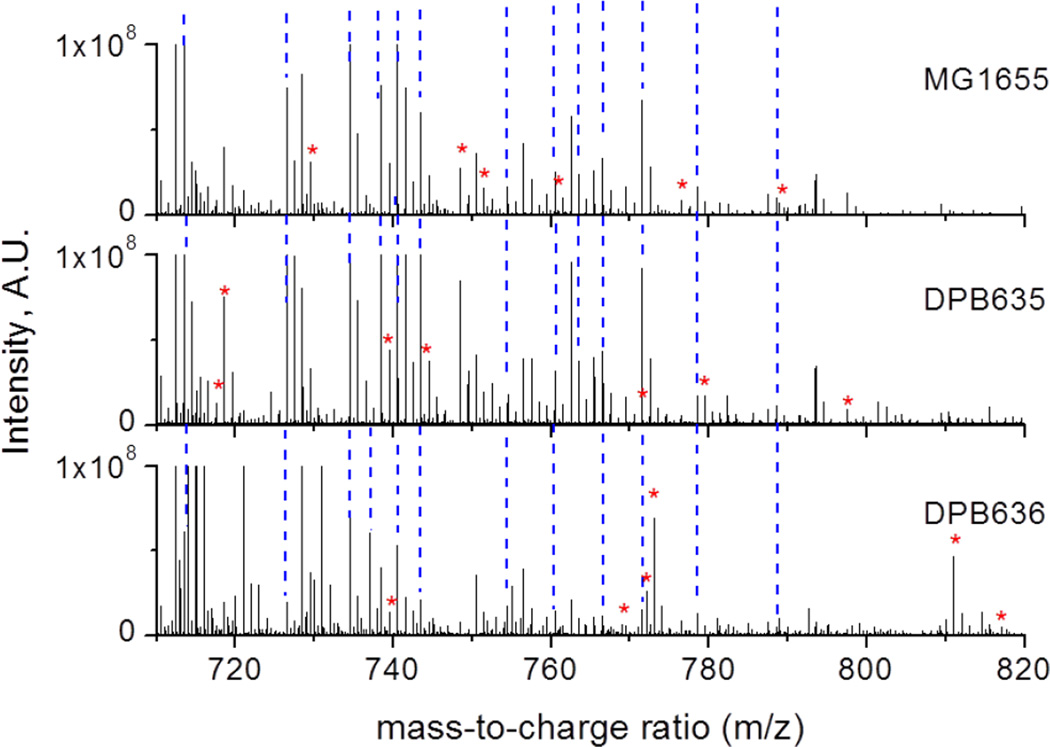
Lipid-related differentiation between the MG1655, DPB635 and DPB636 E. coli strains can be observed from the MALDI FT-ICR MS analysis (see Figure 1). The three strains are closely related which translates into more common lipids than characteristic lipids for the comparison. Inspection of the common lipid IDs show that most of the lipids correspond to fatty acyls, FA (424 of 1364 lipids) and glycerophospholipids, GP (328 of 1364), while other lipid classes are observed in lower abundances (see Table 1 and supplemental Tables 1–4). The higher mass accuracy and resolution of the MALDI FT-ICR MS yielded more lipid IDs than previously reported by MALDI-TOF-MS studies of E.coli identification and/or differentiation. [12,15] This could also be the result of the preconcentration step incorporated into sample preparation, see scheme 1. In particular the current data is in good agreement of previous GP assignments and MALDI-TOF-MS reported lipids were also encountered as common lipids for the three strains studied here[39].
Figure 1.
Typical positive mode MALDI FT-ICR MS spectra for MG1655 (top), DPB635 (middle) and DPB636 (bottom). Peaks labeled with a (*) correspond to characteristic lipids detected for the particular E. coli strain. The blue dashed lines depict lipids common to all three strains.
Table 1.
Distribution of common (∩) and characteristic (Δ) lipids between MG1655, DPB635 and DPB636 strains.
| Strain | PR | FA | ST | GL | GP | SP | SL | PK | Total |
|---|---|---|---|---|---|---|---|---|---|
| MG1655ΔDPB635ΔDPB636 | 121 | 200 | 142 | 168 | 651 | 67 | 7 | 3 | 1359 |
| DPB635ΔMG1655ΔDPB636 | 21 | 49 | 33 | 40 | 175 | 19 | 2 | 339 | |
| DPB636ΔMG1655ΔDPB635 | 28 | 52 | 62 | 18 | 115 | 18 | 1 | 1 | 295 |
| MG1655∩DPB635∩DPB636 | 135 | 436 | 244 | 127 | 745 | 235 | 6 | 11 | 1939 |
| MG1655∩DPB635 | 167 | 496 | 289 | 168 | 932 | 286 | 6 | 11 | 2355 |
| MG1655∩DPB636 | 176 | 497 | 303 | 208 | 1011 | 331 | 10 | 11 | 2547 |
| DPB635∩DPB636 | 147 | 476 | 279 | 155 | 801 | 331 | 8 | 11 | 2208 |
| MG1655∩DPB635ΔDPB636 | 173 | 298 | 228 | 225 | 961 | 160 | 7 | 4 | 2056 |
| MG1655∩DPB636ΔDPB635 | 41 | 61 | 59 | 81 | 266 | 96 | 4 | 0 | 608 |
| DPB635∩DPB636ΔMG1655 | 59 | 124 | 98 | 113 | 406 | 182 | 3 | 2 | 987 |
FA = fatty acyls, PR = Prenol lipids, SP = Sphingolipids, ST = Sterol lipids, GP = Glycerophospholipids, PK = polyketide lipids, GL = Glycerolipids and SL = Saccharolipids
Genotype similarities between the MG1655, DPB635 and DPB636 strains can be observed from the analysis of common and characteristic lipids between two and all three strains (see Table 1). As expected, there is a large number of common lipids between the three strains (over 2000 lipids). An interesting observation comes from the analysis of lipids common to two strains (e.g., MG1655∩DPB635) vs common to two strains and not observed in the third one (e.g., MG1655∩DPB635ΔDPB636). Inspection of Table 1 shows that common lipids are correlated with the genotype similarity; that is, for example, MG1655 has more common lipids with DPB635 (not observed in DPB636) than with DPB636, suggesting that initially, prior to exposure to stress, the topA66 mutation does not play a significant role in lipid expression.
Further inspection of the characteristic lipids for each strain showed that the most abundant classes were common to the three strains (see Table 1). In particular, the larger abundance of fatty acyls (FA), sterols (ST) and glycerophospholipids (GP) relative to polyketides (PK) and saccharolipids (SL) suggesting that these classes may be used as lipid markers for each strain. It should be noted that while common lipids were detected in multiple batches, characteristic lipids were usually detected in low abundance, and encountered in fewer batches (see supplemental Tables 1–3 for details). For instance, at a mid-log point in growth (OD600 = 0.5) under standard laboratory conditions the genetic differences between strains did not yield large changes in the lipid profile obtained with this analysis. The differences in abundance between common and characteristic lipids suggests that common lipids are part of cellular processes, cell membranes and biological pathways that transcend the difference between the E. coli strains studied here. Moreover, characteristic lipids are likely involved in processes that result from genotype expression.
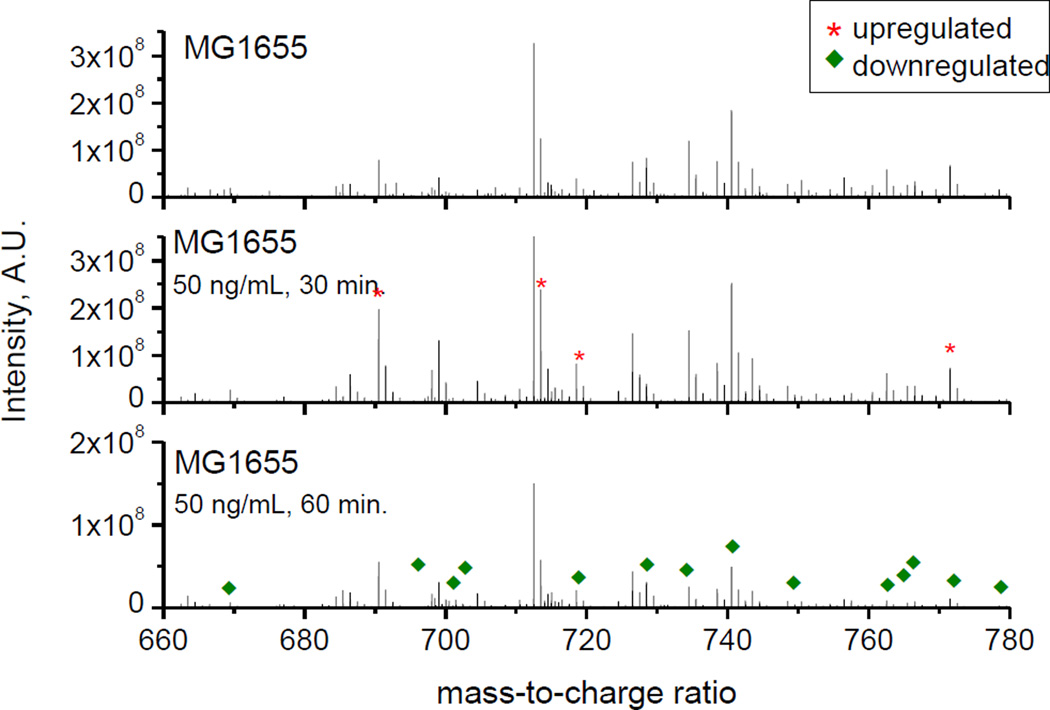
Lipid response resulting from the exposure to norfloxacin was also studied for all three strains (see example in Figure 2 for MG1655). Inspection of Figure 2 shows that following a 30 minute exposure to norfloxacin, the MG1655 strain demonstrates a lipid response to the presence of the antibiotic, resulting in an increase in the number of lipids that are up regulated in comparison to the strain stock (eliminating lipids from the control culture). However, following a 60 minute exposure, the spectra showed a large increase in the number of down regulated lipids (15 vs. 165 lipids at t= 30 and 60 minutes, respectively). Previous work has established that treatment of the MG1655 strain with 50 ng/mL norfloxacin was sufficient to induce multidrug resistance[44], which suggest that the changes in lipid composition are derived from a global cell response mechanism (e.g., cell membrane integrity[45] and cell division mechanism[46]).
Figure 2.
Positive mode MALDI FT-ICR MS spectra for MG1655 stock (top), 50 ng/mL norfloxacin 30 minute exposure (middle) and 50 ng/mL norfloxacin 60 minute exposure (bottom). Peaks labeled with (*) and (♦) correspond to up and down regulated lipids, respectively.
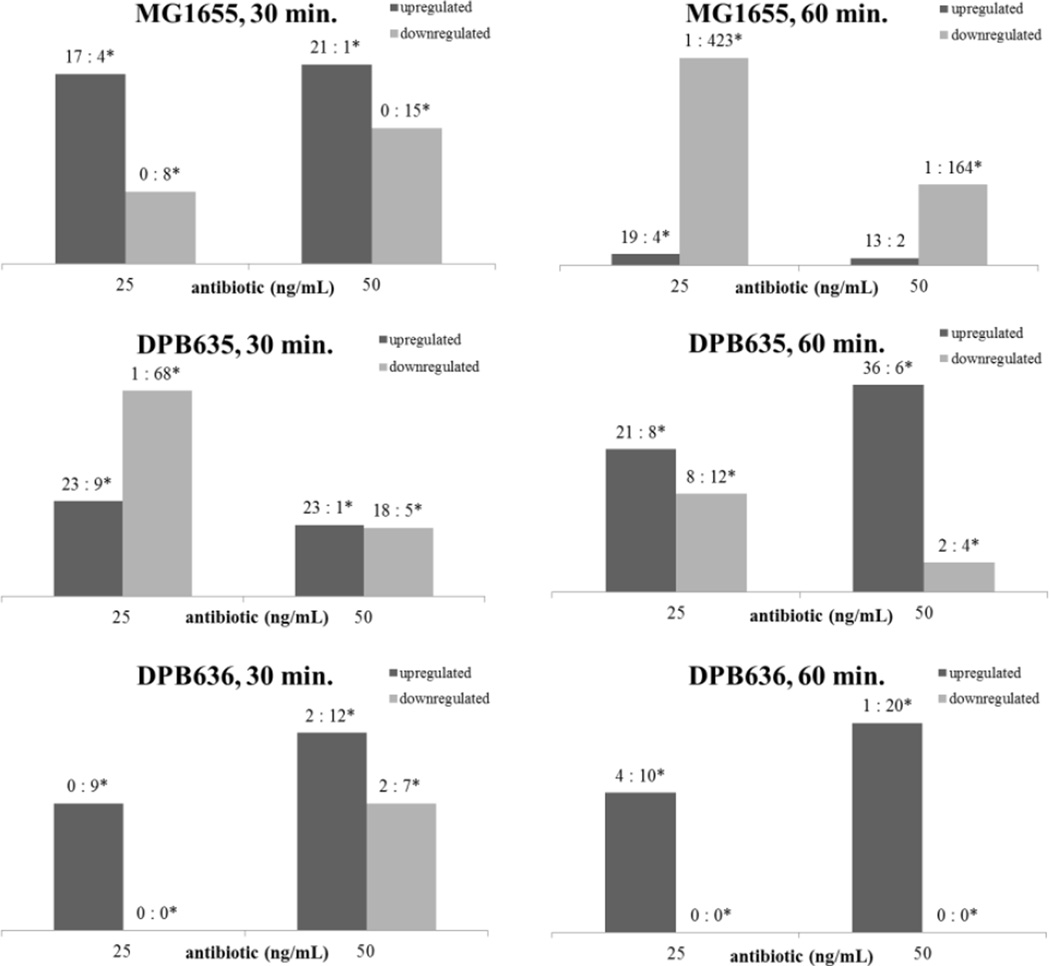
The lipid response as a function of norfloxacin concentration and exposure has been summarized in Figure 3 for all the three strains. For the MG1655 strain, a general trend was observed in that down regulation was amplified at the later time regardless of the antibiotic concentration. However, the isogenic strains responded differently than the wild type (MG1655). For example, if cell response is evaluated by changes in up/down regulated lipids under stress conditions, data presented in Figure 3 suggest that DPB635 is capable of exhibiting a prolonged response (similar to MG1655 at low concentration and early time) by means of the lipid up regulation that extends to higher antibiotic concentrations and the longer duration of exposure. Furthermore, the distribution of the lipid profile for the DPB636 strain suggest that although the cells exhibit a similar trend in up regulation to the DPB635, most likely a result of the mini-kan transposon present in the isogenic strains that is absent in MG1655 [47], the hypersensitivity of DPB636 to norfloxacin is reiterated by the lower number of lipids influenced by the antibiotic (↑, ↑*, ↓, and ↓*) that predominates in the culture. This results in a deficiency in adaptation following norfloxacin exposure. The topA66 mutation in topoisomerase I that differs between the isogenic strains can also be the source for hypersensitivity observed in the DPB636 strain. Detailed information regarding the lipids detected under the stress conditions for each strain can be found in the supplemental tables 5–16.
Figure 3.
Total number of lipids up or downregulated as a function of norfloxacin dose and time exposure for MG1655, DPB635 and DPB636. Values above each column correspond to the number of ↑:↑* lipids or ↓:↓* lipids for each strain and condition. The asterisk (*) corresponds to lipids that are up or down regulated on the basis of an absence (not detection).
Class specific lipid changes were analyzed as a function of the norfloxacin exposure (see Table 2). The fewer number of both up and down regulated lipids observed for the DPB636 strain following norfloxacin exposure can also be attributed to cell death as a result of hypersensitivity of the strain to antibiotics. Inspection of Table 2 shows that for all three strains, changes in the up regulated lipids result in an increase in the number of fatty acyls, whereas down regulated lipids are predominantly glycerophospholipids, two of the most abundance lipid classes detected in E. coli[48, 49]. It is known that fatty acyls serve as building blocks for other more complex lipid groups[50] and their up regulation may be associated to degradation or metabolism of proteins, carbohydrates or sugars[50]. Moreover, when up regulation is coupled with phospholipid depletion, a more complex cell response mechanism may be occurring. For example, fatty acids can be used as starting material in the production of phospholipids[51]and therefore an initial increase in fatty acyl production following GP down regulation could imply a combative response in the cell to compensate for changes in membrane integrity. Phospholipids comprise the inner monolayer of the outer membrane and also the bilayer of the inner membrane[48]. Changes in the phospholipids, in this case an overall down regulation, could imply that cell integrity has been compromised with higher concentration or duration of exposure. Depletion in phospholipids could also imply that the phospholipids have been metabolized or were used as other intermediates to adapt the cellular stress response imposed by the antibiotic as a function of concentration and time. Two main classes of phospholipids, phophatidylglycerols (PG) and phosphatidylethanolamines (PE), have been shown to be involved in the posttranslational modification of lipoproteins[48] that are associated with both the inner monolayer of the outer membrane. This could indicate an additional form of response that lipid down regulation can be correlated to. The two processes in conjunction could imply a change in gene regulation and proteomic composition [49, 52]. Previous work on Canduda albicans fungus[5] has shown that changes in lipid distribution (e.g., PGs in the antibiotic resistant isolates) coincide with the cell combative changes, the onset of antibiotic resistance and a change in membrane integrity[5].
Table 2.
Distribution of up or downregulated lipids as a function of norfloxacin exposure.
| Upregulated lipids, 25 ng/mL | |||||||||
| Strain | Time (min.) | FA | PR | SP | ST | GP | PK | GL | SL |
| MG1655 | 30 | 1 | 0 | 6 | 1 | 12 | 0 | 1 | 0 |
| 60 | 12 | 4 | 2 | 0 | 4 | 0 | 1 | 0 | |
| DPB635 | 30 | 30 | 0 | 3 | 0 | 0 | 0 | 2 | 0 |
| 60 | 20 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| DPB636 | 30 | 1 | 0 | 3 | 0 | 0 | 0 | 5 | 0 |
| 60 | 8 | 1 | 0 | 2 | 3 | 0 | 0 | 0 | |
| Downregulated lipids, 25 ng/mL | |||||||||
| Strain | Time (min.) | FA | PR | SP | ST | GP | PK | GL | SL |
| MG1655 | 30 | 1 | 0 | 1 | 1 | 3 | 0 | 1 | 1 |
| 60 | 114 | 52 | 28 | 72 | 139 | 1 | 18 | 0 | |
| DPB635 | 30 | 0 | 1 | 2 | 1 | 15 | 0 | 1 | 0 |
| 60 | 0 | 1 | 2 | 1 | 15 | 0 | 1 | 1 | |
| DPB636 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Upregulated lipids, 50 ng/mL | |||||||||
| Strain | Time (min.) | FA | PR | SP | ST | GP | PK | GL | SL |
| MG1655 | 30 | 3 | 1 | 1 | 1 | 13 | 0 | 2 | 1 |
| 60 | 7 | 1 | 1 | 0 | 5 | 0 | 1 | 0 | |
| DPB635 | 30 | 19 | 0 | 3 | 1 | 0 | 1 | 0 | 0 |
| 60 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | |
| DPB636 | 30 | 5 | 0 | 1 | 0 | 5 | 5 | 3 | 0 |
| 60 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Downregulated lipids, 50 ng/mL | |||||||||
| Strain | Time (min.) | FA | PR | SP | ST | GP | PK | GL | SL |
| MG1655 | 30 | 2 | 4 | 0 | 3 | 5 | 0 | 1 | 0 |
| 60 | 15 | 11 | 9 | 15 | 104 | 0 | 11 | 0 | |
| DPB635 | 30 | 2 | 0 | 1 | 1 | 18 | 0 | 0 | 1 |
| 60 | 0 | 1 | 0 | 1 | 3 | 0 | 0 | 0 | |
| DPB636 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
CONCLUSIONS
Lipid profiles of three closely related E.coli strains were monitored using high resolution mass spectrometry and statistical analysis using internal lipid standards. Results showed that the present methodology permits the identification of the cell strain as well as insight into SOS response. This was accomplished using both the inspection of common and specific lipids to each strain and by using trends in up/down regulated lipids as a function of the norfloxacin antibiotic exposure. For the MG1655, DPB635 and DPB636 E. coli strains, a similar distribution of the altered lipids, FAs up regulated and GPs down regulated, were observed. The shift in the lipid distribution resembles the extent to which each strain can combat the antibiotic exposure. Results also suggest that the topA66 topoisomerase I mutation of DPB636 translates into diminished response related to antibiotic sensitivity when compared to MG1655 and the DPB635 strains. Despite the complexity of the biological samples, the workflow and methodology presented here permits the analysis of cell strains during the SOS response for lipid differentiation. In particular, high resolution mass spectrometry when coupled to a surface interrogation probe provides in situ detection of lipid differences for closely related E. coli strains.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health, Grant No. R00GM106414 (Fernandez-Lima) and R01GM054226 (Tse-Dinh).
Footnotes
SUPPORTING INFORMATION:
Additional supporting information has been submitted. Supporting information includes: total lipid counts for each strain (supplemental Tables 1–3), common lipid distribution among the three strains (supplemental Table 4), MG1655 lipids as a function of antibiotic exposure (supplemental Tables 5–8), DPB635 lipids as a function of antibiotic exposure (supplemental Tables 9–12), and DPB636 lipids as a function of antibiotic exposure (supplemental Tables 13–16). In addition, the performance of the lipid internal standard has been presented to demonstrate the influence of matrix effects within the E. coli lipid samples.
References
- 1.Wenk MR. Nature Reviews Drug Discovery. 2005;4:594. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 2.Maccarrone M. Curr Pharm Des. 2006;12:759. doi: 10.2174/138161206775474279. [DOI] [PubMed] [Google Scholar]
- 3.Sagin FG, Sozmen EY. Current Alzheimer Research. 2008;5:4. doi: 10.2174/156720508783884648. [DOI] [PubMed] [Google Scholar]
- 4.Marsh M, Helenius A. Cell. 2006;124:729. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh A, Yadav V, Prasad R. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radman M. Basic Life Sci. 1975;5A:355. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- 7.Michel B. PLoS Biol. 2005;3:e255. doi: 10.1371/journal.pbio.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenover FC. The American Journal of Medicine. 2006;119:S3. doi: 10.1016/j.amjmed.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Gode D, Volmer DA. Analyst. 2013;138:1289. doi: 10.1039/c2an36337b. [DOI] [PubMed] [Google Scholar]
- 11.Blaze MT M, Akhmetov A, Aydin B, Edirisinghe PD, Uygur G, Hanley L. Analytical Chemistry. 2012;84:9410. doi: 10.1021/ac302230e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasper GL, Takahashi LK, Zhou J, Ahmed M, Moore JF, Hanley L. Analytical Chemistry. 2010;82:7472. doi: 10.1021/ac101667q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanley L, Zimmermann R. Analytical Chemistry. 2009;81:4174. doi: 10.1021/ac8013675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edirisinghe PD, Moore JF, Skinner-Nemec KA, Lindberg C, Giometti CS, Veryovkin IV, Hunt JE, Pellin MJ, Hanley L. Analytical Chemistry. 2006;79:508. doi: 10.1021/ac0615605. [DOI] [PubMed] [Google Scholar]
- 15.Layre E, Moody DB. Biochimie. 2013;95:109. doi: 10.1016/j.biochi.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cooks RG. Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids. 2011;1811:946. doi: 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Schultz JA, Woods AS. J. Mass Spectrom. 2007;42:1093. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson S, Wang H, Woods A. Journal of the American Society for Mass Spectrometry. 2007;18:17. doi: 10.1016/j.jasms.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Jackson SN, Wang H-YJ, Woods AS, Ugarov M, Egan T, Schultz JA. J. Am. Soc. Mass Spectrom. 2005;16:133. doi: 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Jackson SN, Wang H-YJ, Woods AS. Journal of the American Society for Mass Spectrometry. 2005;16:2052. doi: 10.1016/j.jasms.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Bich C, Touboul D, Brunelle A. Mass Spectrometry Reviews. 2013 doi: 10.1002/mas.21399. n/a. [DOI] [PubMed] [Google Scholar]
- 22.Touboul D, Brunelle A, Laprévote O. Biochimie. 2011;93:113. doi: 10.1016/j.biochi.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Lima FA, DeBord JD, Schweikert EA, Della-Negra S, Kellersberger KA, Smotherman M. Surface and Interface Analysis. 2013;45:294. doi: 10.1002/sia.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Lima FA, Eller MJ, DeBord JD, Verkhoturov SV, Della-Negra S, Schweikert EA. Nucl. Instr. and Meth. in Phys. Res. B. 2012;273:270. doi: 10.1016/j.nimb.2011.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Lima FA, Post J, DeBord JD, Eller MJ, Verkhoturov SV, Della-Negra S, Woods AS, Schweikert EA. Analytical Chemistry. 2011;83:8448. doi: 10.1021/ac201481r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng L, McQuaw CM, Baker MJ, Lockyer NP, Vickerman JC, Ewing AG, Winograd N. Applied Surface Science. 2008;255:1190. doi: 10.1016/j.apsusc.2008.05.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidova V, Pol J, Volny M, Novak P, Havlicek V, Wiedmer SK, Holopainen JM. Journal of Lipid Research. 2010;51:2295. doi: 10.1194/jlr.M005488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith D, Kiss A, Leach F, III, Robinson E, Paša-Tolić L, Heeren RA. Analytical and bioanalytical chemistry. 2013;405:6069. doi: 10.1007/s00216-013-7048-1. [DOI] [PubMed] [Google Scholar]
- 29.Smith DF, Robinson EW, Tolmachev AV, Heeren RMA, Paša-Tolić L. Analytical Chemistry. 2011;83:9552. doi: 10.1021/ac2023348. [DOI] [PubMed] [Google Scholar]
- 30.Angelini R, Vitale R, Patil VA, Cocco T, Ludwig B, Greenberg ML, Corcelli A. Journal of Lipid Research. 2012;53:1417. doi: 10.1194/jlr.D026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landgraf RR, Garrett TJ, Conaway MCP, Calcutt NA, Stacpoole PW, Yost RA. Rapid Communications in Mass Spectrometry. 2011;25:3178. doi: 10.1002/rcm.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elliott TS, Shelton A, Greenwood D. J Med Microbiol. 1987;23:83. doi: 10.1099/00222615-23-1-83. [DOI] [PubMed] [Google Scholar]
- 33.Blondeau JM. Surv Ophthalmol. 2004;49(Suppl 2):S73. doi: 10.1016/j.survophthal.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Valjavec-Gratian M, Henderson TA, Hill TM. Mol Microbiol. 2005;58:758. doi: 10.1111/j.1365-2958.2005.04860.x. [DOI] [PubMed] [Google Scholar]
- 35.Pommier Y. Chem Rev. 2009;109:2894. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng B, Shukla S, Vasunilashorn S, Mukhopadhyay S, Tse-Dinh YC. J Biol Chem. 2005;280:38489. doi: 10.1074/jbc.M509722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tse-Dinh YC. Nucleic Acids Research. 2009;37:731. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold RJ, Karty JA, Ellington AD, Reilly JP. Anal Chem. 1999;71:1990. doi: 10.1021/ac981196c. [DOI] [PubMed] [Google Scholar]
- 39.Gidden J, Denson J, Liyanage R, Ivey DM, Lay JO. Int J Mass Spectrom. 2009;283:178. doi: 10.1016/j.ijms.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams TL, Andrzejewski D, Lay JO, Musser SM. J Am Soc Mass Spectrom. 2003;14:342. doi: 10.1016/S1044-0305(03)00065-5. [DOI] [PubMed] [Google Scholar]
- 41.Arneborg N, Salskoviversen AS, Mathiasen TE. Applied Microbiology and Biotechnology. 1993;39:353. [Google Scholar]
- 42.Oursel D, Loutelier-Bourhis C, Orange N, Chevalier S, Norris V, Lange CM. Rapid Communications in Mass Spectrometry. 2007;21:1721. doi: 10.1002/rcm.3013. [DOI] [PubMed] [Google Scholar]
- 43.Schmelzer K, Fahy E, Subramaniam S, Dennis EA. Methods Enzymol. 2007;432:171. doi: 10.1016/S0076-6879(07)32007-7. [DOI] [PubMed] [Google Scholar]
- 44.Kohanski MA, DePristo MA, Collins JJ. Molecular Cell. 2010;37:311. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez D, Kolter R. Genes Dev. 2010;24:1893. doi: 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barak I, Muchova K. Int J Mol Sci. 2013;14:4050. doi: 10.3390/ijms14024050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biek DP, Cohen SN. J Bacteriol. 1989;171:2056. doi: 10.1128/jb.171.4.2056-2065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dowhan W. Annual Review of Biochemistry. 1997;66:199. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 49.McIntyre TM, Chamberlain BK, Webster RE, Bell RM. J Biol Chem. 1977;252:4487. [PubMed] [Google Scholar]
- 50.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6th ed. W.H. Freemand and Company; 2007. [Google Scholar]
- 51.Polakis SE, Guchhait RB, Lane MD. J Biol Chem. 1973;248:7957. [PubMed] [Google Scholar]
- 52.Bell RM. J Bacteriol. 1974;117:1065. doi: 10.1128/jb.117.3.1065-1076.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.