Abstract
Objectives
To develop the infrastructure and demonstrate the feasibility of conducting microarray-based RNA transcriptional profile analyses for the diagnosis of serious bacterial infections in febrile infants 60 days and younger in a multicenter pediatric emergency research network.
Methods
We designed a prospective multicenter cohort study with the aim of enrolling more than 4000 febrile infants 60 days and younger. To ensure success of conducting complex genomic studies in emergency department (ED) settings, we established an infrastructure within the Pediatric Emergency Care Applied Research Network, including 21 sites, to evaluate RNA transcriptional profiles in young febrile infants. We developed a comprehensive manual of operations and trained site investigators to obtain and process blood samples for RNA extraction and genomic analyses. We created standard operating procedures for blood sample collection, processing, storage, shipping, and analyses. We planned to prospectively identify, enroll, and collect 1 mL blood samples for genomic analyses from eligible patients to identify logistical issues with study procedures. Finally, we planned to batch blood samples and determined RNA quantity and quality at the central microarray laboratory and organized data analysis with the Pediatric Emergency Care Applied Research Network data coordinating center. Below we report on establishment of the infrastructure and the feasibility success in the first year based on the enrollment of a limited number of patients.
Results
We successfully established the infrastructure at 21 EDs. Over the first 5 months we enrolled 79% (74 of 94) of eligible febrile infants. We were able to obtain and ship 1 mL of blood from 74% (55 of 74) of enrolled participants, with at least 1 sample per participating ED. The 55 samples were shipped and evaluated at the microarray laboratory, and 95% (52 of 55) of blood samples were of adequate quality and contained sufficient RNA for expression analysis.
Conclusions
It is possible to create a robust infrastructure to conduct genomic studies in young febrile infants in the context of a multicenter pediatric ED research setting. The sufficient quantity and high quality of RNA obtained suggests that whole blood transcriptional profile analysis for the diagnostic evaluation of young febrile infants can be successfully performed in this setting.
Keywords: febrile infant, serious bacterial infection, RNA transcriptional profiles, microarray
There is a large degree of variability in the evaluation and management of febrile infants younger than 60 days of age in the setting of the emergency department (ED).1 Approximately 6% to 10% of these infants have serious bacterial infections (SBIs) that require prompt antimicrobial therapy,1,2 and many other infants without bacterial infections are hospitalized unnecessarily until culture results are available. Microbiologic culture for identification of bacteria in the relevant body fluids (e.g., blood, urine, cerebrospinal fluid [CSF]) remains the reference standard for diagnosis of SBIs, as it has been for decades.3–6 However, using cultures, and more specifically blood culture, as the gold standard for diagnosis of SBI has important limitations, including a noninconsequential number of false-positive and false-negative results, and lengthy duration (up to 48 to 72 hours) to determine or exclude a positive result.6–8 In great part because of these limitations, many febrile infants are subjected to excessive evaluation procedures, receive empiric antibiotic therapy, and are hospitalized, exposing them to iatrogenic risks.9 Therefore, there is a clear need for developing less invasive, highly accurate, and more timely diagnostic strategies for the evaluation of young febrile infants.
Advances in genomic techniques now allow us to distinguish the types of infections by assessing the host response to infection caused by different microbes. Previous research has shown that host responses can reliably be measured with distinct transcriptional biosignatures in the RNA of host blood leukocytes.6,10,11 Studies in hospitalized children have demonstrated that RNA biosignatures can distinguish patients with bacterial and viral infections with 95% accuracy.10 This transition from bench to clinical research suggests the potential application of transcriptional biosignatures for the evaluation of febrile infants in the ED.
The Pediatric Emergency Care Applied Research Network (PECARN) annually evaluates more than 4000 febrile infants 60 days of age or younger. PECARN, thus, offers an ideal setting to evaluate the application of RNA expression analysis for diagnosis and management of febrile infants in a prospective manner.12 Because there is limited experience in conducting large multi-center studies with this new technology, especially with a young infant population in the setting of the ED, it was necessary to develop an infrastructure to identify and recruit eligible patients in a consistent and reliable fashion. We also sought obtain high-quality blood RNA samples for expression analysis from these patients across multiple sites. Finally, it was important to demonstrate that the RNA samples collected from multiple sites do not substantially degrade during transport, storage, and processing.
We are currently conducting a prospective cross-sectional study of a convenience sample of more than 4000 febrile infants younger than 60 days of age in the PECARN. The objective of the present article is to describe the development of the necessary infrastructure and methods for conducting genomic studies in this population. We also aim to demonstrate the ability to obtain samples in 21 PECARN EDs that yield adequate quantities of high-quality RNA for transcriptional profiles which would be required for identifying the diagnostic bacterial (SBI) and nonbacterial (non-SBI) biosignatures in febrile infants.
METHODS
Study Setting and Population
Febrile infants (fever defined as rectal temperature at triage of ≥38°C, or fever of a similar degree measured at home) evaluated in the ED for SBI with blood cultures were eligible for the study. Infants with obvious clinical sepsis, prematurity, major systemic comorbidities (e.g., serious congenital abnormalities, in-born errors of metabolism), or evidence of focal infections (not including otitis media) were excluded. The laboratory evaluation, beyond the required blood culture and blood sample for RNA biosignatures, was conducted at the discretion of the individual clinician, but typically included a complete blood count, urinalysis and urine culture, and CSF analysis and culture. Many sites also obtained nasopharyngeal samples from these patients for viral diagnostic studies. The study was approved by the Institutional Review Board at each site, and participants were enrolled after written informed consent was obtained from their parents or guardians.
Study Definitions
We classified febrile infants as having SBIs if they were diagnosed with bacteremia, urinary tract infection, or bacterial meningitis. We classified febrile infants as not having SBIs (non-SBIs) if there was no growth or growth of known contaminants in the blood, urine, or CSF cultures. We defined urinary tract infection by the growth of a single bacterial pathogen with colony counts meeting 1 of 3 criteria: (1) ≥1000 cfu/mL for urine cultures obtained by suprapubic aspiration, (2) ≥50,000 cfu/mL from a catheterized specimen, (3) ≥10,000 cfu/mL from a catheterized specimen in association with an abnormal urinalysis. Abnormal urinalysis was defined by a urine dipstick test positive for leukocyte esterase or nitrite, or if more than 5 white cells per high power field were reported on microscopic examination.13,14 These were standard definitions at the time of study initiation.
Development of the Infrastructure
The development of the infrastructure was assisted by the presence of a PECARN Febrile Illness Working Group, which included individuals with extensive experience and expertise in the evaluation and management of young febrile infants. This group helped design the study and reviewed the study protocol. The study infrastructure further included the data coordinating center (DCC), at the University of Utah, which was responsible for ensuring data quality, monitoring data collection, data management, data transfer from the sites to the DCC, and ensuring adherence to study procedures, good clinical practices and compliance with federal and local research regulations. A separate Microarray Analytic Core at the Baylor Institute for Immunology Research, which has substantial expertise in infectious diseases, immunology, and bioinformatics and was responsible for sample processing for RNA quality and RNA expression analyses to define bacterial and nonbacterial biosignatures. By conducting this study in PECARN, we were able to leverage the large and diverse patient population in the network, its experience in conducting large-scale, rigorous clinical research, and the expertise from members of the PECARN Steering Committee and subcommittees on budget and data management, and quality assurance. Before the initiation of the study, we conducted a PECARN-wide survey to determine the willingness and ability of sites to participate, and specifically to determine site capabilities to collect blood samples for RNA expression analyses and facilities to store the samples before shipping to the microarray core. To facilitate the smooth implementation of the study, in addition to identifying investigators responsible for the overall conduct of the study at each site, we designated 4 “study champions” whose role was to oversee the conduct of this study in the EDs of their research nodes.
Development of the Protocol
The protocol included detailed processes for conducting, monitoring, and executing the necessary clinical, regulatory, and analytic aspects of this study. Importantly, the study principal investigators included pediatric emergency medicine and pediatric infectious disease specialists, leveraging the strengths of each. We created a comprehensive manual of operations to describe study procedures with detailed instructions on sample collection, processing, storage at the individual laboratories of each center, and shipping/transport in batches to the microarray core. Site investigators and research coordinators were trained in 2 in-person full-day training sessions, and frequent study updates were provided to sites via a web-based secure portal.
Feasibility of Sample and Data Collection
We asked site investigators to record patient demographics, physical examination findings, and laboratory test results including cultures (blood, urine, and CSF) via an electronic data capturing system. Each site was required to demonstrate ability to identify and enroll patients, and collect, process, and store 1 mL blood samples before shipping to the Microarray Core. Immediate processing of the blood samples at the sites included placing blood in baby tempus tubes (Applied Biosystems Life Technologies, Carlsbad, CA) containing 2 mL of RNA storage solution and ensuring that each tube was shaken vigorously for 20 seconds after placement of blood into the tubes to lyse the cells.15 Although the samples were required to be frozen (−20 to −80°C) to maintain RNA quality, we anticipated enrollment to occur at all times in the ED. Fortunately, samples can remain at room temperature for a period up to 96 hours before being frozen, allowing for significant flexibility for blood sampling while still maintaining RNA quality. Each individual institution shipped batches of collected blood samples via courier to the Microarray Core laboratory. In order to test the quality and quantity of RNA to be extracted, we asked each site to enroll 2 eligible febrile infants after obtaining informed consent.
Determination of Quality of Extracted RNA
The RNA was extracted following established protocols in the Microarray Core, and the quality of the RNA was assessed. In brief, total RNAwas isolated from whole blood followed by depletion of globin mRNA according to the manufacturer's instructions.16 Total and globin-reduced RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and RNA quantity was assessed using the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Dublin, OH). A blood sample was considered satisfactory when it yielded: (a) a minimum of 500 ng of RNA and (b) an RNA integrity number greater than 7 (±S.D <0.3).17 In a second step, 52 samples were run on the gene chips to determine whether they yielded consistent results. The main goal was to demonstrate that more than 90% of samples produced high-quality RNA, which was adequate for microarray analysis. All samples that passed quality control were amplified and labeled using the Illumina TotalPrep-96 RNA amplification kit. Amplified RNA was then hybridized to Illumina HumanHT-12 v4 beadchips (47,231 probes) and scanned on the Illumina Beadstation 500.15
Microarray Data and Statistical Analysis for the Entire Study
Illumina BeadStudio/GenomeStudio software was used to subtract background and scale average samples' signal intensity, and GeneSpring GX 7.3 software (Agilent Technologies) to perform further normalization and analyses.10,15,17 Briefly, RNA transcripts were first selected if they are present in greater than 10% of all samples and had a minimum of 2-fold expression change compared with the median intensity across all samples (quality control transcripts).15 We then followed the strategy outlined below to perform the analysis of the RNA transcripts: (a) supervised analysis (comparative analyses between predefined sample groups) was performed using Mann-Whitney or Kruskal-Wallis testings (P < 0.01) for comparisons of 2 or more study groups, followed by Benjamini-Hochberg false discovery rate correction for multiple testing. Lists of significantly expressed RNA transcripts were filtered to include only those that showed 1.25-fold change or higher in expression level relative to the control group15,17; (b) unsupervised clustering which is unbiased grouping of samples based on their molecular profile without previous knowledge of sample classification; (c) class prediction using the K-Nearest Neighbors algorithm,10 and a P value ratio cutoff of 0.5 was applied to identify the top ranked genes that best discriminated the infants with SBIs from those without SBIs; (d) functional analyses of differentially expressed genes is performed using modular analysis as described15,17 module transcript content and annotations is available online at http://www.biir.net/public_wikis/module_annotation/V2_Trial_8_Modules.
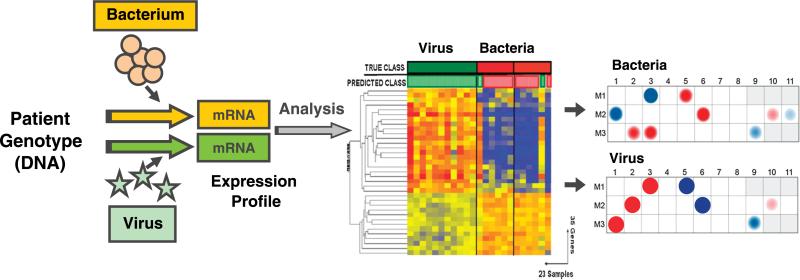
Figure 1 (from a different study conducted by one of the authors [O.R.]) represents an example of how expression patterns (heat maps) will be visualized at the gene level and modular level analyses to display the differences between groups by gene modules (groups of genes with similar biologic function).17
FIGURE 1.
Microarray technology measures the differences in gene transcriptional profiles present in blood immune cells as induced by various types of infectious agents. Expression patterns for individual transcripts are commonly shown in heat maps (middle panel) that show the expression levels of individual genes. The overexpressed genes are represented in red, whereas underexpressed genes are shown in blue. Alternatively, modular analysis is used to display the differences in expression at the modular level (groups of genes that share biological function) between patients with different types of infections (right panel). Red dots indicate overexpressed modules and blue dots under expressed modules.
RESULTS
We successfully established the study infrastructure, and its organization, as described in Figure 2. We tested the quality and quantity of RNA extracted after each site had enrolled 2 eligible febrile infants, as described in Methods. We enrolled 79% (74 of 94) eligible febrile infants across all participating sites in the first few months of the study and were able to obtain 1 mL of blood from 74% (55 of 74) of enrolled participants, with at least 2 samples per participating ED. The blood was of insufficient quantity (less than 0.5 mL) in 15% (11 of 74) of patients, whereas 8 samples had 0.5 mL to 1 mL blood and were not used for RNA extraction.
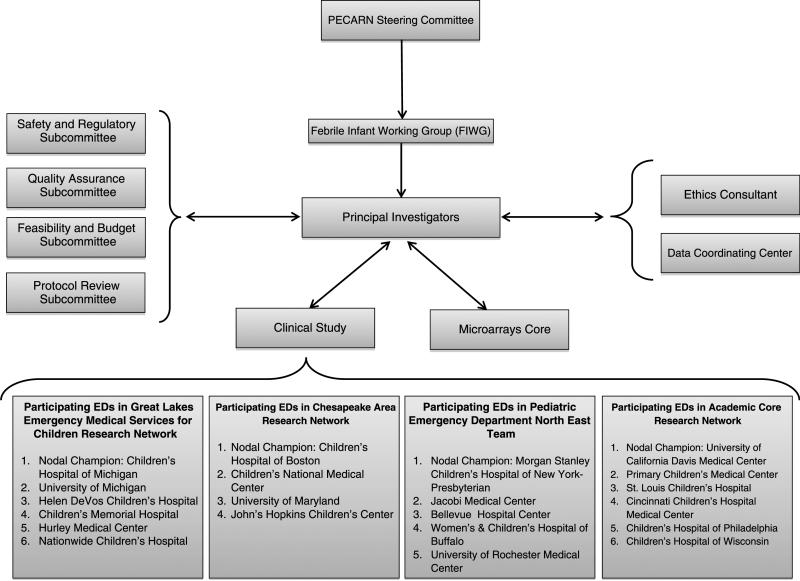
FIGURE 2.
Study Organizational Chart Years 2009 to 2010. GLEMSCRN indicates Great Lakes Emergency Medical Services for Children Research Node; CARN, Chesapeake Area Research Node; PEDNET, Pediatric Emergency Department North East Team; ACORN, Academic Centers Research Note.
Quality of Blood Samples: RNA Extraction and Analysis
The frozen samples were visually inspected to make sure the samples were not thawed, clotted, or the tubes damaged at the laboratory to assure that they arrived frozen and in good condition. After the protocol described above, we documented that 95% (52 of 55) of the samples yielded high-quality RNA as is required for gene expression profiles to evaluate diagnostic biosignatures (note: one of the 55 samples was slightly less than 1 mL; however, the sample still demonstrated sufficient quality for RNA expression upon evaluation).
DISCUSSION
We have demonstrated the successful establishment of a multicenter pediatric ED–based infrastructure that can screen, consent, collect, store, ship, process, and analyze RNA expression profiles from small quantity blood samples from young, febrile infants. The RNA integrity evaluation of these samples demonstrated high quality. Our previous experience in conducting expression analyses along with the high-quality RNA yield from small amounts of blood samples provide confidence in our pursuit of enrolling 4000 febrile infants and defining biosignatures to allow discrimination of young febrile infants with SBIs from those without SBIs.
Conducting comprehensive and rapid RNA expression analysis studies from young infants in a multicenter ED setting poses several challenges including training of personnel with varied skill sets in study procedures, reducing variation in sample collection and processing, and demonstrating the ability to extract adequate quantities of high-quality RNA from small samples of blood. These issues are further accentuated by the implementation of the study in the chaotic setting of the ED, where patient accrual should occur at all times of the day and night, and availability of trained personnel to enroll patients can be challenging. Furthermore, obtaining permission and informed consent from parents of young febrile infants for participating in (complex) genomic studies in the stressful ED environment may also be difficult. Finally, collecting adequate amounts of blood for research purposes in very young infants may be a potential barrier to study enrollment. Despite these challenges, we have demonstrated the feasibility of establishing a research infrastructure necessary to conduct this complicated study in this clinical setting.
The early success in implementing the study protocol resulted from the establishment of a robust and collaborative study infrastructure, and attention to study planning by creation of detailed standard operating procedures to reduce variability in sample collection, processing, and storage. The early implementation of the protocol yielded important information concerning the rates of obtaining informed consent and the success of sample collection from young infants. Finally, this feasibility and planning phase identified barriers and opportunities encountered at multiple sites in patient recruitment and sample processing and demonstrated that the RNA obtained from the samples was of adequate quantity and quality which will allow us to explore RNA expressions of enrolled febrile infants. This initial work was critical as we plan to recruit 4000 young febrile infants into the study, and ultimately identify robust biosignatures for infants with SBIs as well as for those with viral infections. After enrolling the required sample size, we ultimately will define bacterial and nonbacterial biosignatures in these young febrile infants, with the goal of establishing new diagnostic and treatment paradigms for this vulnerable population.
Limitations
With this study, we have successfully established a multicenter ED-based infrastructure and demonstrated the ability to obtain sufficient quantities of high-quality RNA from small blood volumes to conduct microarray analysis for the evaluation of febrile infants. We have yet to demonstrate, however, that this new technology is able to distinguish febrile infants with bacterial infections from those without bacterial infections with sufficient accuracy to change current paradigms, which is the main objective of the study we are undertaking. In addition, in this study, we plan to study the use of RNA biosignatures for a singular purpose. Whether or not the use of this technology can be applied to assess severity of disease, diagnosis of nonbacterial illnesses, and other disease states requires future study.
CONCLUSIONS
We have demonstrated the successful establishment of a multicenter pediatric ED-based research infrastructure to evaluate young febrile infants using a novel genomic technology. Using state-of-the-art molecular assays and bioinformatic analytical tools, we will be able to provide essential information for conducting similar large-scale studies in pediatric acute care settings. This will set the stage for the further study and application of genomic technologies in similar populations, and eventually translate this technology into rapid clinical decision-making at the bedside.
ACKNOWLEDGMENTS
Participating centers and investigators are listed below in alphabetical order: Ann & Robert H. Lurie Children's Hospital (E. Powell); Bellevue Hospital Center (D. Levine, M. Tunik); Boston Children's Hospital (L. Nigrovic); Children's Hospital of Colorado (G. Roosevelt, L. Bjaj); Children's Hospital of Michigan (P. Mahajan); Children's Hospital of Philadelphia (E. Alpern); Children's Hospital of Wisconsin (L. Browne); Children's National Medical Center (S. Atabaki); Cincinnati Children's Hospital Medical Center (R. Ruddy); Helen DeVos Children's Hospital (J. Hoyle, Jr.); Hurley Medical Center (D. Borgialli); Jacobi Medical Center (E. Crain, S. Blumberg); Johns Hopkins Children's Center (J. Anders); Nationwide Children's Hospital (B. Bonsu, D. Cohen); New York Presbyterian-Morgan Stanley Children's Hospital (P. Dayan); Primary Children's Medical Center (R. Greenberg); St. Louis Children's Hospital (D. Jaffe, J. Muenzar); Texas Children's Hospital (A. Cruz); University of California Davis Medical Center (N. Kuppermann, L. Tzimenatos); University of Maryland (R. Gattu); University of Michigan (A. Rodgers); University of Rochester (A. Brayer); Women and Children's Hospital of Buffalo (K. Lillis). We acknowledge the efforts of the following individuals participating in PECARN at the time this study was initiated.
PECARN Steering Committee: N. Kuppermann (Chair), E. Alpern, L. Babcock-Cimpello, S. Blumberg, D. Borgialli, J. Chamberlain, E. Crain, P. Dayan, J. M. Dean, M. Gorelick, J. Hoyle, D. Jaffe, M. Kwok, R. Lichenstein, K. Lillis, P. Mahajan, R. Maio, F. Moler, D. Monroe, D. Nelson,L. Nigrovic, E. Powell, R. Ruddy, R. Stanley, S. Teach, M. Tunik, A. Walker. MCHB/EMSC liaison: D. Kavanaugh.
PECARN Data Coordinating Center: M. Dean (Director), S. Zuspan (Project Manager), J. Burr, K. Call, D. Demarco, A. Donaldson, R. Enriquez, R. Gerard, D. Haward, G. Herron, R. Holubkov, S. Knight, N.C. Mann, C. Torres, B. Yu, J. Zindel.
Feasibility and Budget Subcommittee: E. Kim, D. Nelson, D. Monroe, S. Krug, S. Goldfarb, S. Zuspan.
Grants and Publications Subcommittee: M. Gorelick (Chair), E. Alpern, D. Borgialli, L. Cimpello, A. Donaldson, G. Foltin, F. Moler, L. Nigrovic, S. Teach.
Protocol Concept Review and Development Subcommittee: D. Jaffe (Chair), E. Alpern, K. Brown, J. Chamberlain, P. Dayan, J. M. Dean, R. Holubkov, P. Mahajan, R. Stanley, M. Tunik.
Quality Assurance Subcommittee: K. Lillis (Chair), E. Alessandrini, E. Crain, R. Enriquez, R. Gerard, R. Lichenstein, E. Powell, A. Rogers, R. Ruddy, A. Walker.
Safety and Regulatory Affairs Subcommittee: W. Schalick, J. Hoyle, Co-Chairs; S. Atabaki, J. Burr, K. Call, M. Kwok, R. Maio, R. Ruddy J.
The authors thank the research coordinators in PECARN and the Microarray Core at Baylor Institute for Immunology Research, without whose dedication and hard work this study would not have been possible, and all the clinicians around the PECARN who enrolled children in this study.
Funding: This work was supported in part by H34MCO8509 from Health Resources Administration/Emergency Services for Children and the Pediatric Emergency Care Applied Research Network (PECARN) is supported by cooperative agreements U03MC00001, U03MC00003, U03MC00006, U03MC00007, and U03MC00008 from the Emergency Medical Services for Children (EMSC) program of the Maternal and Child Health Bureau, Health Resources and Services Administration, US Department of Health and Human Services.
Footnotes
N.K. and O.R. served as co-senior authors of this study.
The results of this study were presented in part at the meetings of the Pediatric Academic Societies in Boston, MA in June 2012, and the Society for Academic Emergency Medicine in Chicago, IL in May 2012.
Disclosure: The authors declare no conflict of interest.
REFERENCES
- 1.Harper MB. Update on the management of the febrile infant. Clin Pediatr Emerg Med. 2004;5:5–12. [Google Scholar]
- 2.Bachur RG, Harper MB. Predictive model for serious bacterial infection among infants younger than 3 months of age. Pediatrics. 2001;108:311–316. doi: 10.1542/peds.108.2.311. [DOI] [PubMed] [Google Scholar]
- 3.Korppi M, Heiskenan-Kosma T, Euro Leinonen M. White blood cells, C-reactive protein and erythrocyte sedimentation rate in pneumococcal pneumonia in children. Eur Respir J. 1997;10:1125–1129. doi: 10.1183/09031936.97.10051125. [DOI] [PubMed] [Google Scholar]
- 4.Prilutsky D, Shneider E, Shefer A, et al. Differentiation between viral and bacterial acute infections using chemiluminescent signatures of circulating phagocytes. Anal Chem. 2011;83:4258–4265. doi: 10.1021/ac200596f. [DOI] [PubMed] [Google Scholar]
- 5.Nuutilla J, Lilius EM. Distinguishing between bacterial and viral infections. Curr Opin Infect Dis. 2007;20:304–310. doi: 10.1097/QCO.0b013e3280964db4. [DOI] [PubMed] [Google Scholar]
- 6.Relman D. New technologies, human-microbe interactions, and the search for previously unrecognized pathogens. J Infect Dis. 2002;186(suppl 2):S254–S258. doi: 10.1086/344935. [DOI] [PubMed] [Google Scholar]
- 7.Sard B, Bailey MC, Vinci R. An analysis of pediatric blood cultures in the postpneumococcal conjugate vaccine era in a community hospital emergency department. Pediatr Emerg Care. 2006;22:295–300. doi: 10.1097/01.pec.0000215137.51909.16. [DOI] [PubMed] [Google Scholar]
- 8.McGowan KL, Foster JA, Coffin SE. Outpatient pediatric blood cultures: time to positivity. Pediatrics. 2000;106(2 Pt 1):251–255. doi: 10.1542/peds.106.2.251. [DOI] [PubMed] [Google Scholar]
- 9.DeAngelis C, Joffe A, Wilson M, et al. Iatrogenic risks and financial costs of hospitalizing febrile infants. Am J Dis Child. 1983;137:1146–1149. doi: 10.1001/archpedi.1983.02140380006003. [DOI] [PubMed] [Google Scholar]
- 10.Ramilo O, Allman W, Chung W, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummings CA, Relman DA. Genomics and microbiology. Microbial forensics: “cross-examining pathogens”. Science. 2002;296:1976–1979. doi: 10.1126/science.1073125. [DOI] [PubMed] [Google Scholar]
- 12.The Pediatric Emergency Care Applied Research Network (PECARN) The Pediatric Emergency Care Applied Research Network (PECARN): rationale, development, and first steps. Acad Emerg Med. 2003;10:661–668. [PubMed] [Google Scholar]
- 13.Hoberman A, Wald ER. Urinary tract infections in young febrile children. Pediatr Infect Dis J. 1997;16:11–17. doi: 10.1097/00006454-199701000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Hoberman A, Wald ER, Reynolds EA, et al. Is urine culture necessary to rule out urinary tract infection in young febrile children?. Pediatr Infect Dis J. 1996;15:304–309. doi: 10.1097/00006454-199604000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitley P, Moturi S, Santiago J, et al. Ambion, Inc. Improved microarray sensitivity using whole blood RNA samples. Appl Biosyst Tech Note. 2005 [Google Scholar]
- 17.Ardura MI, Banchereau R, Mejias A, et al. Enhanced monocyte response and decreased central memory T cells in children with invasive Staphylococcus aureus infections. PLoS One. 2009;4:e5446. doi: 10.1371/journal.pone.0005446. [DOI] [PMC free article] [PubMed] [Google Scholar]