Abstract
Background:
Type 2 diabetes (T2D) is a multifactorial disease with susceptibility of several genes that are related to T2D. Insulin secretion pathway starts with potassium channels in pancreatic beta cells. KCNJ11 gene encodes ATP-sensitive potassium channel subunits. Some studies suggested that KCNJ11 (E23K) mutation increases the risk of T2D. Therefore, present study was designed to investigate the association between E23K polymorphism of KCNJ11 gene and type 2 diabetes mellitus (T2DM) in the Iranian population.
Materials and Methods:
The type of study was case-control and 40 unrelated subjects, including 20 healthy controls and 20 diabetic patients were recruited (diagnosed based on American Diabetes Association criteria). Blood samples were used for isolation of genomic deoxyribonucleic acid (DNA). Having extracted the genomic DNA from human blood leukocytes by means of High Pure PCR Template Preparation Kit, PCR-restriction fragment length polymorphism method was used to detect KCNJ11 E23K gene polymorphism. BanII restriction enzyme was used for digestion. Data were analyzed using Chi-square or Fisher exact test or independent t-test, as appropriate. P < 0.05 was considered.
Results:
We found that the carrier homozygous for KK genotype are susceptible to T2D (0.049) and in patients the frequency of K allele was higher than control subjects (0.048).
Conclusion:
The present study suggests that KCNJ11 (E23K) gene polymorphism is associated with T2DM.
Keywords: Iranian population, KCNJ11, polymorphism, potassium channel, restriction fragment length polymorphism, type 2 diabetes
INTRODUCTION
Type 2 diabetes (T2D) is a multifactorial disease that the body cannot control the glucose homeostasis and in many cases this disease is resulted by deficiency of insulin secretion and insulin activity. In general, T2D diagnosed in adults and is related to obesity, life style, age, family history and genetics. Several genes have been reported to be related of T2D susceptibility.[1,2] Prevalence of T2D has been increased in the last decades and relatively 250 million people are suffering from this disease world-wide. T2D is seventh reason of lethality in America.[2]
Insulin secretion pathway starts with inhibition of ATP sensitive potassium channel (KATP) channels by glucose and then by depolarization of the β-cell membrane, influx of calcium ions increased and consequently intracellular calcium stimulates the exocytosis of granules containing insulin. This channel consists of 2 subunits, region of sulfonylurea receptor and K+ subunit (Kir 6.2). ABCC8 and KCNJ11 genes encoding these subunits respectively.[3]
KCNJ11 gene located on chromosome 11 with 4 Kb, encoding a protein with 390 amino acids. Mutation in KCNJ11 can significantly influence KATP channel activity.[4] Several mutations have been reported for KCNJ11 that two of them related to increasing risk of T2D: rs5219 for E23K mutation and rs5215 for I337V mutation. Direct effects of polymorphisms for ABCC8 have not been demonstrated.[5] Investigations have indicated substitution of glutamate (E) to lysine (K) amino acid in codon 23 of KCNJ11 gene being responsible for sensitivity reduction of KATP thus the channel is open for more time and insulin secretion is inhibited.[6,7] Therefore some studies showed correlation between E23K mutation with T2D.[8,9,10] Several studies showed KK homozygote genotype has most relation with T2D in Caucasian population.[3,11,12] Many studies have shown EK heterozygous have great incidence in patients and healthy population.[7,13,14] However frequency of EK in Ragia et al. study has not been greater than others.[15]
Therefore, evidences suggest that E23K polymorphism could be responsible for T2D. The genetic background for KCNJ11 gene for T2D has not been investigated in Iran and therefore the current study was performed to evaluate the incidence of E23K polymorphism and associations between this genetic variation and diabetes susceptibility in Iranian population.
MATERIALS AND METHODS
Subject
The type of study was case-control and was performed on 40 unrelated people that consisted of 20 healthy control and 20 diabetic patients. The subjects were recruited from Sedighe Tahere Research Center (Isfahan, Iran). Criteria for type 2 diabetes mellitus (T2DM) are based on clinical diagnoses (presence of polyuria, polydipsia and polyphagia) and were confirmed by American Diabetes Association criteria.[16] T2DM was determined by a fasting plasma glucose level of more than 126 mg/dl after a minimum fast of 12 h or a 2-h post glucose level (2-h oral glucose tolerance test) of more than 200 mg/dl or an hemoglobin A1c (HbA1c) level of >7%. Control individuals were diagnosed by a fasting plasma glucose of <100 mg/dl or a 2-h glucose of <140 mg/dl or an HbA1c level of <6.4%. Individuals with impaired glucose tolerance and positive family history of T2D were excluded from the study for control group. In order to eliminate interferences with biological variables people with pervious diagnosis of type 1 diabetes, malignity, systematic inflammation and endocrine diseases, impaired renal and hepatic function, those receiving hypercholesterolemia, hypertension or corticosteroids medications were excluded from the study. All participants signed a written consent before their enrolment for the study. The study protocol was approved by the Ethics Committee of Isfahan University of Medical Sciences.
Biochemical measurements
Chemical and biochemical characteristics were performed in Sedighe Tahere Research Center. Cholesterol, triglycerides, high-density lipoprotein (HDL) and plasma glucose were measured by standard enzymatic assays. Low-density lipoprotein (LDL) cholesterol was derived using Friedewald equation.[17] HbA1c was measured by means of ion-exchange high performance liquid chromatography.
Deoxyribonucleic acid analysis
Whole blood sample was used for isolation of genomic DNA. High Pure PCR Template Preparation Kit (Roche, Germany) was employed for extracting genomic DNA from human blood leukocytes. Polymerase chain reaction (PCR) restriction fragment length polymorphism method was used for detecting KCNJ11 E23K gene polymorphism. The sequences of forward and reverse primers were: 5’-GACTCTGCAGTGAGGCCCTA-3’ and 5’ ACGTTGCAGTTGCCTTTCTT-3’, respectively. PCR amplification was performed using the following reagents: 1X PCR reaction buffer (16 mmol/l (NH4) 2SO4, 67 mmol/l TrisHCL and pH 8.8), 10 pmoles of each primer, 2 mmol/l MgCl2, 200 μmol/l of each deoxyribonucleotide triphosphates, 100 ng of genomic DNA, 1 U Taq polymerase (Bioron, Germany) and 0.56 mmol/l DMSO. The final PCR reaction was 25 μl and PCR cycles were as follows: Initial denaturation step at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 60 s, annealing at 60°C for 30 s and extension at 72°C for 60 s and a final extension 72°C for 9 min. The PCR amplification was confirmed by gel electrophoresis (8% acrylamide gel and Ethidium bromide staining). For digestion 8 μl of PCR product (210 bp) was digested by 3U of a restriction enzyme BanII (New England, BioLabs. England) at 37°C for 1 h in 10 μl reaction mixture that contained 1X NE buffer (50 mmol/l potassium acetat 2, 20 mmol/l Tris-acetate, 10 mmol/l magnesium acetate, 1 mmol/l DDT and pH 7.9). The digestion products were analyzed by gel electrophoresis (12% polyacrylamide gel and ethidium bromide staining) with restriction pattern.
Statistical analysis
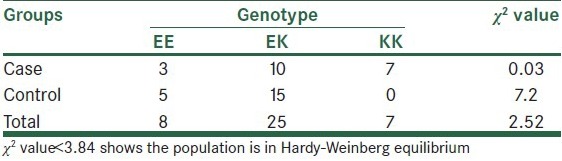
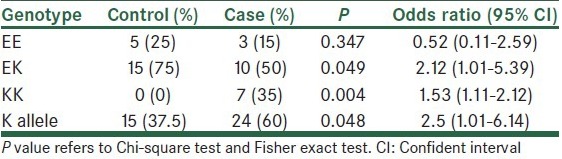
Statistical analyzes were performed using the SPSS version 21 for Windows (SPSS Inc., Chicago, IL, USA). Results are given as mean ± SD or percentage. Data were analyzed by Chi-square or Fisher exact test or independent t-test, as appropriate. P < 0.05 was considered to be significant. Chi-square test was used to test for Hardy–Weinberg equilibrium and comparison of genotype and allele frequencies in the diabetic and non-diabetic subjects [Table 1].
Table 1.
Hardy-Weinberg equilibrium for comparison of genotype and allele frequencies in the case and control subjects
RESULTS
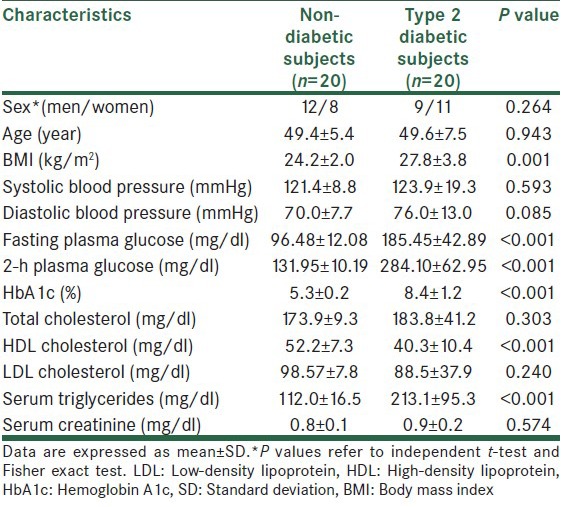
Clinical and biochemical characteristics of participants are summarized in Table 2. Statistically significant differences were observed for HDL, serum triglycerides and diabetes criteria between control and T2D patients. There were no statistically significant difference between control and T2D patient for sex, age, blood pressure, total cholesterol, LDL cholesterol and serum creatinine [Table 2].
Table 2.
Clinical and biochemical characteristics of type 2 diabetic and non-diabetic subjects
PCR amplification of KCNJ11 gene was performed and produced a 210 bp DNA band [Figure 1]. This product digested and gave different patterns as follows: E: 150 bp, 32 bp and 28 bp; K: 178 bp, 32 bp and EK: 178 bp, 150 bp, 32 bp and 28 bp [Figure 2].
Figure 1.
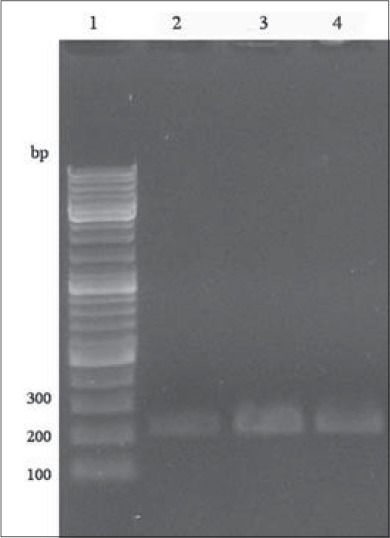
Confirmation polymerase chain reaction (PCR) amplification of KCNJ11 gene using gel electrophoresis. Lane 1:100 bp deoxyribonucleic acid ladder; lane 2, 3 and 4: PCR products (210 bp)
Figure 2.
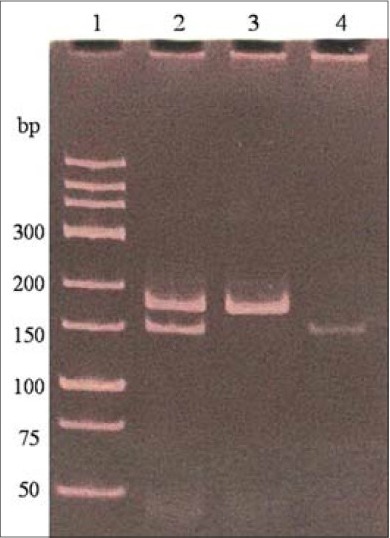
Polymerase chain reaction products digested by BanII restriction enzyme for the detection of E23K polymorphism of KANJ11 gene. Samples were electrophoresed using a 12% polyacrylamide gel and subsequent staining with ethidium bromide. Lane 1: Deoxyribonucleic acid ladder; lane 2: E23K heterozygotes; lane 3: K23 homozygotes; lane 4: E23 homozygotes. Small bands of 32 bp and 28 bp are not visible in this figure
The association between E23K polymorphism and T2D risk was analyzed. As shown in Table 3, carrier homozygous for KK genotype is susceptible to T2D and in patients the frequency of K allele was higher than control subjects. In addition, heterozygous carriers for EK are more in non-diabetic patients (P = 0.049).
Table 3.
Association of KCNJ11 (E23K) with type 2 diabetes
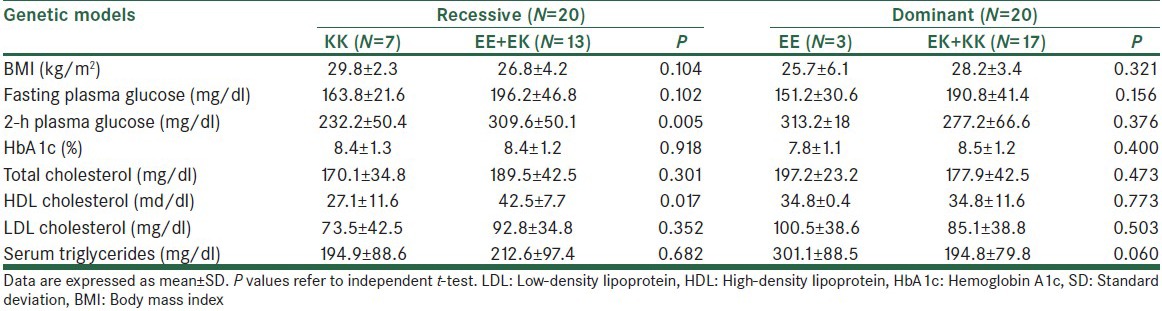
Association of different genetic models with control diabetes criteria in T2D patients according to E23K KCNJ11 genotype was analyzed in Table 4. According to the results there was significant difference (P = 0.005) between KK and combined genotype (EK + EE) for 2-h plasma glucose. Furthermore, a significant difference was shown between HDL of non-diabetic subject and T2D patients (P = 0.017).
Table 4.
Association of different genetic model with diabetes control criteria in T2D patient according to E23K KCNJ11 genotype
DISCUSSION
Prevalence of T2D has increased in the last decades and some genes have been associated with T2D.[2] In our study, KCNJ11 (rs5219) gene was analyzed for investigation of the association between E23K polymorphism with prevalence of T2D. As you can see in Table 3, statistical analysis showed people with KK homozygous genotype have more susceptibility for this disease (P = 0.004). A study in Russia also found KK homozygous genotype had more risk for T2D (P = 0.016).[12] Moreover, we found that carriers of K allele have more risk for T2D (P = 0.048), Chistiakov et al. also found similar results for Russian population (P = 0.023).[12] In Li study also found E23K contributed to T2D susceptibility.[18] In a study published in 2013, Asaf et al. had showed the inheritance of the K allele predisposes for T2D.[19] However in Wang et al. study relationship between the E23K and T2D not seen in Han people in Qingdao area.[20]
We found that frequency of EK genotype is more in non-diabetic patients (P = 0.049). In U.K. prospective diabetes study frequency of EK in T2D was a little more than glucose tolerated people.[21]
Frequency of E allele in healthy people is more prevalent, thus E allele carries probably have lower risk for T2D when compared with carries of K allele (P = 0.048).
In our study, we did not observe a significant association between dominant genetic models with diabetes control criteria [Table 4]. However in recessive model, a significant correlation with HDL cholesterol was found (P = 0.017), so carriers of dominant E allele have more HDL in comparison with KK genotype (P = 0.017). In biochemical characteristics of T2D and non-diabetic people a significant association between HDL and serum triglycerides with two groups was found, so non-diabetic people have more HDL and low serum triglycerides in comparison to T2D patients. This finding could be due dyslipidemia in diabetic patients.[22] In a study by Riedel et al., a mechanistic link was proposed between increased body mass index and plasma free fatty acid levels with high prevalence of T2D by the KATP channel homozygous polymorphic.[23] We also found that population with KK homozygous genotype has more susceptibility to low HDL (P = 0.017).
CONCLUSION
KCNJ11 (E23K) polymorphism has relationship with susceptibility to T2D in Iranian population and carriers of K allele are more susceptible to this disease. Additional studies with larger sample size will be required to confirm this marker in Iranian population.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
REFERENCES
- 1.Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: New genes, new understanding. Trends Genet. 2008;24:613–21. doi: 10.1016/j.tig.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Distefano JK, Watanabe RM. Pharmacogenetics of anti-diabetes drugs. Pharmaceuticals (Basel) 2010;3:2610–46. doi: 10.3390/ph3082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean L, McEntyre J. CH. 3. Bethesda, Md: NCBI : NIDDK; 2004. [Last accessed on 2013 May 27]. The genetic landscape of diabetes (electronic resource) Available from: http://www.ncbi.nlm.nih.gov/books/NBK1667/ [Google Scholar]
- 4.Thomas PM, Cote GJ, Wohllk N, Haddad B, Mathew PM, Rabl W, et al. Mutations in the sulfonylurea receptor gene in familial persistent hyperinsulinemic hypoglycemia of infancy. Science. 1995;268:426–9. doi: 10.1126/science.7716548. [DOI] [PubMed] [Google Scholar]
- 5.Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, Hitman G, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–72. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 6.Samadikuchaksaraei A, Ramandi MF, Khatami S, Hashemi MJ, Haqparast S, Fard-Esfahani P. E23K polymorphism in Iranian patients with coronary heart disease. ARYA Atheroscler. 2010;5:55–60. [Google Scholar]
- 7.He YY, Zhang R, Shao XY, Hu C, Wang CR, Lu JX, et al. Association of KCNJ11 and ABCC8 genetic polymorphisms with response to repaglinide in Chinese diabetic patients. Acta Pharmacol Sin. 2008;29:983–9. doi: 10.1111/j.1745-7254.2008.00840.x. [DOI] [PubMed] [Google Scholar]
- 8.Shaat N, Ekelund M, Lernmark A, Ivarsson S, Almgren P, Berntorp K, et al. Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia. 2005;48:2544–51. doi: 10.1007/s00125-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 9.Schwanstecher C, Meyer U, Schwanstecher M. K (IR) 6.2 polymorphism predisposes to type 2 diabetes by inducing overactivity of pancreatic beta-cell ATP-sensitive K(+) channels. Diabetes. 2002;51:875–9. doi: 10.2337/diabetes.51.3.875. [DOI] [PubMed] [Google Scholar]
- 10.Florez JC, Jablonski KA, Kahn SE, Franks PW, Dabelea D, Hamman RF, et al. Type 2 diabetes-associated missense polymorphisms KCNJ11 E23K and ABCC8 A1369S influence progression to diabetes and response to interventions in the Diabetes Prevention Program. Diabetes. 2007;56:531–6. doi: 10.2337/db06-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hani EH, Boutin P, Durand E, Inoue H, Permutt MA, Velho G, et al. Missense mutations in the pancreatic islet beta cell inwardly rectifying K+channel gene (KIR6.2/BIR): A meta-analysis suggests a role in the polygenic basis of Type II diabetes mellitus in Caucasians. Diabetologia. 1998;41:1511–5. doi: 10.1007/s001250051098. [DOI] [PubMed] [Google Scholar]
- 12.Chistiakov DA, Potapov VA, Khodirev DS, Shamkhalova MS, Shestakova MV, Nosikov VV. The KCNJ11 E23K and ABCC8 exon 31 variants contribute to susceptibility to type 2 diabetes, glucose intolerance and altered insulin secretion in a Russian population. Diabetes Metab Syndr Clin Res Rev. 2008;2:185–91. [Google Scholar]
- 13.Sesti G, Laratta E, Cardellini M, Andreozzi F, Del Guerra S, Irace C, et al. The E23K variant of KCNJ11 encoding the pancreatic beta-cell adenosine 5’-triphosphate-sensitive potassium channel subunit Kir6.2 is associated with an increased risk of secondary failure to sulfonylurea in patients with type 2 diabetes. J Clin Endocrinol Metab. 2006;91:2334–9. doi: 10.1210/jc.2005-2323. [DOI] [PubMed] [Google Scholar]
- 14.Alsmadi O, Al-Rubeaan K, Wakil SM, Imtiaz F, Mohamed G, Al-Saud H, et al. Genetic study of Saudi diabetes (GSSD): Significant association of the KCNJ11 E23K polymorphism with type 2 diabetes. Diabetes Metab Res Rev. 2008;24:137–40. doi: 10.1002/dmrr.777. [DOI] [PubMed] [Google Scholar]
- 15.Ragia G, Tavridou A, Petridis I, Manolopoulos VG. Association of KCNJ11 E23K gene polymorphism with hypoglycemia in sulfonylurea-treated type 2 diabetic patients. Diabetes Res Clin Pract. 2012;98:119–24. doi: 10.1016/j.diabres.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 16.DeFronzo RA, Devjit T, Dawn CS, MaryAnn B, George AB, Thomas AB, et al. Prediction of diabetes based on baseline metabolic characteristics in individuals at high risk. Diabetes Care. 2013;36:3607–12. doi: 10.2337/dc13-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 18.Li YY. The KCNJ11 E23K gene polymorphism and type 2 diabetes mellitus in the Chinese Han population: A meta-analysis of 6,109 subjects. Mol Biol Rep. 2013;40:141–6. doi: 10.1007/s11033-012-2042-9. [DOI] [PubMed] [Google Scholar]
- 19.Asaf A, Ayesh BM, Hamdona OM. Single nucleotide polymorphism E23K of KCNJ11 gene and other risk factors associated with type-2 diabetes mellitus in Gaza. Int J Chem Life Sci. 2013;2:1146–52. [Google Scholar]
- 20.Wang GF, Jiang WJ, Jiang XB, Wu YL, Zhang DF. Correlation study of KCNJ11 gene E23K polymorphism with type 2 diabetes mellitus. Acta Acad Med. 2011;2:4–8. [Google Scholar]
- 21.Gloyn AL, Hashim Y, Ashcroft SJ, Ashfield R, Wiltshire S, Turner RC, et al. Association studies of variants in promoter and coding regions of beta-cell ATP-sensitive K-channel genes SUR1 and Kir6.2 with Type 2 diabetes mellitus (UKPDS 53) Diabet Med. 2001;18:206–12. doi: 10.1046/j.1464-5491.2001.00449.x. [DOI] [PubMed] [Google Scholar]
- 22.Longo DL, Fauci AS, Kasper DL, Hauser SL, Jameson JL, Loscalzo J. 18th ed. Ch. 344. New York: McGraw-Hill Professional; 2011. Harrison's Principles of Internal Medicine. [Google Scholar]
- 23.Riedel MJ, Boora P, Steckley D, de Vries G, Light PE. Kir6.2 polymorphisms sensitize beta-cell ATP-sensitive potassium channels to activation by acyl CoAs: A possible cellular mechanism for increased susceptibility to type 2 diabetes? Diabetes. 2003;52:2630–5. doi: 10.2337/diabetes.52.10.2630. [DOI] [PubMed] [Google Scholar]


