Abstract
We have previously shown that ablation of the three N-linked glycosylation sites in the West Nile virus NS1 protein completely attenuates mouse neuroinvasiveness (≥1,000,000 PFU). Here, we compared the replication of the NS1130-132QQA/175A/207A mutant to that of the parental NY99 strain in monkey kidney Vero cells. The results suggest that the mechanism of attenuation is a lack of NS1 glycosylation, which blocks efficient replication, maturation, and NS1 secretion from the endoplasmic reticulum and results in changes to the virus-induced ultrastructure.
TEXT
West Nile virus (WNV) is a mosquito-borne flavivirus and is distributed in many parts of the world. The WNV genome consists of a 5′ and 3′ untranslated region (UTR), three structural protein (capsid, premembrane/membrane, and envelope [E]) genes, and seven nonstructural protein (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) genes. Using site-directed mutagenesis, we have shown that the ablation of the NS1 N-linked glycosylation (N-X-S/T) sites at residues 130 to 132, 175 to 177, and 207 to 209 of the WNV NY99 infectious clone attenuates mouse neuroinvasiveness and neurovirulence phenotypes (1, 2). The nonstructural NS1 protein is a multifunctional glycoprotein found in the cytoplasm in association with the cell membrane that is secreted, necessary for replication, and involved in virus maturation (3–10). Because this protein is critical in many aspects of the replication cycle, we investigated the mechanism of attenuation of the NS1130-132QQA/175A/207A mutant strain by comparison to the NY99 strain in cell culture. This mutant virus was chosen as it was previously shown to be stable in vitro and in vivo (i.e., no reversion) and was fully attenuated for neuroinvasiveness (>6 log10 PFU).
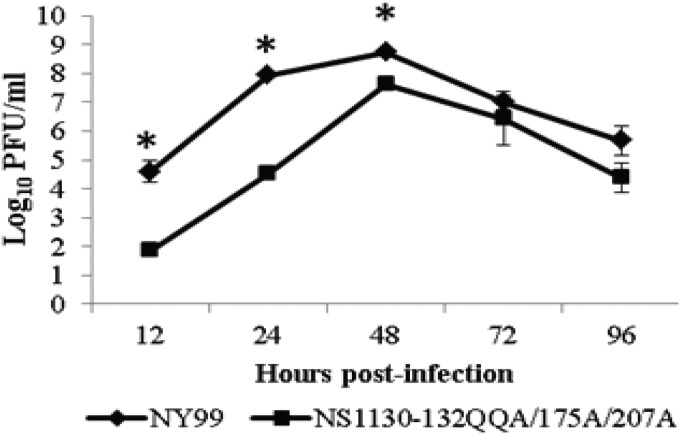
The multiplication kinetics of the parental NY99 and the NS1130-132QQA/175A/207A mutant were compared (in triplicate) in Vero cells at a multiplicity of infection (MOI) of 0.1 (Fig. 1). Significant differences in multiplication kinetics were observed for the NS1130-132QQA/175A/207A mutant compared to those of the parental NY99 virus at 12, 24, and 48 h postinfection, with the greatest differences (4 log10 PFU/ml) at 24 h postinfection. The peak infectivity titer was at 48 h postinfection for both viruses (Fig. 1).
FIG 1.
Multiplication kinetics of parental NY99 and mutant NS1130-132QQA/175A/207A viruses in Vero cells. Supernatants were collected for both viruses in triplicate, and infectivity titers were determined by plaque titration. Statistically significant differences as determined by Student's t test (P < 0.01) occurred at 12, 24, and 48 h p.i. and are noted by asterisks above the time points. Several 48-h time point samples were analyzed for sequencing, and no mutations were detected in the NS1 gene.
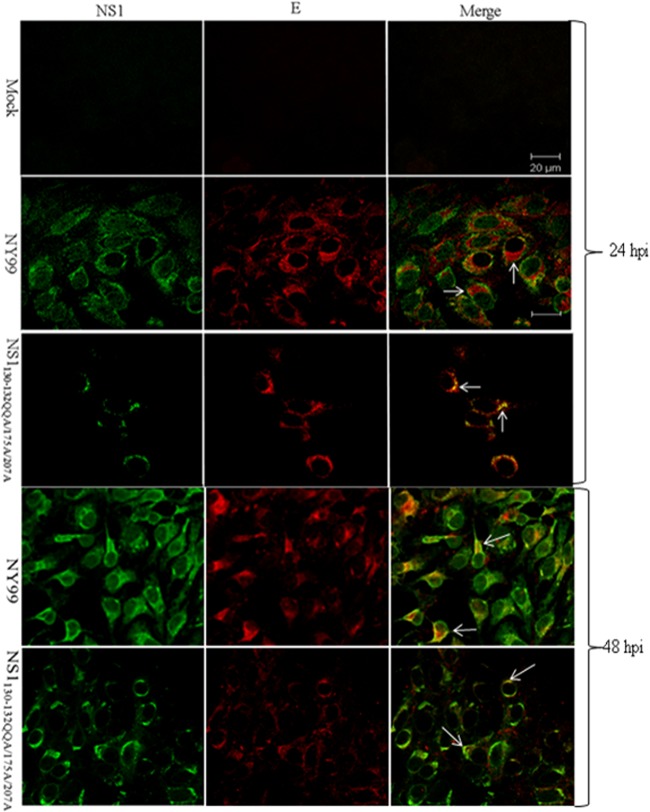
Protein localization was determined for the E and NS1 proteins. Confluent monolayers of Vero cells were infected with either the NS1130-132QQA/175A/207A mutant or the parental NY99 strain at an MOI of 0.1 and fixed at 24 or 48 h postinfection (p.i.) (Fig. 2). Confocal microscopy showed that the E (rabbit anti-envelope domain III [EDIII] antibody; 1:500 [11]) and NS1 (monoclonal antibody [MAb] 8152, 1:100; Millipore) proteins were colocalized in NY99 virus-infected Vero cells. At 24 h p.i., both E and NS1 proteins displayed diffuse cytoplasmic staining with areas of intense fluorescence in apparent vesiclelike structures, and the intensity of the E protein fluorescence correlated with the intensity of the NS1 fluorescence. The NS1130-132QQA/175A/207A mutant virus, however, showed major differences in the staining patterns of both the E and NS1 proteins compared to those of the parental NY99 strain. Specifically, the NS1130-132QQA/175A/207A mutant showed less intense vesiclelike NS1 protein and E protein staining, with an absence of diffuse cytoplasmic staining for NS1. Furthermore, there was a significant lack of colocalization of the NS1 and E proteins in the NS1130-132QQA/175A/207A mutant-infected cells due to the intense diffuse cytoplasmic staining of the E protein but reduced, focal staining of the NS1 protein. At 48 h p.i., the staining patterns were similar for both viruses except that, for the NS1130-132QQA/175A/207A mutant, more cells showed NS1 and E staining.
FIG 2.
Localization of NS1 and E protein in mock-, NY99-, and NS1130-132QQA/175A/207A-infected Vero cells (MOI of 0.1) at 24 h p.i. and 48 h p.i. Images were captured with a Zeiss confocal microscope at ×100 magnification. NS1 was detected by anti-mouse Alexa Fluor 488-conjugated antibody, and E was detected by anti-rabbit Alexa Fluor 568-conjugated antibody. In the images obtained at 24 h p.i., arrows show areas of little NS1/E colocalization in the NY99-infected cells, whereas in mutant NS1130-132QQA/175A/207A-infected cells, NS1 colocalizes to E but not exclusively. At 48 h p.i., E and NS1 colocalize in most NY99-infected cells; however, less colocalization is seen in NS1130-132QQA/175A/207A-infected cells, as indicated by arrows.
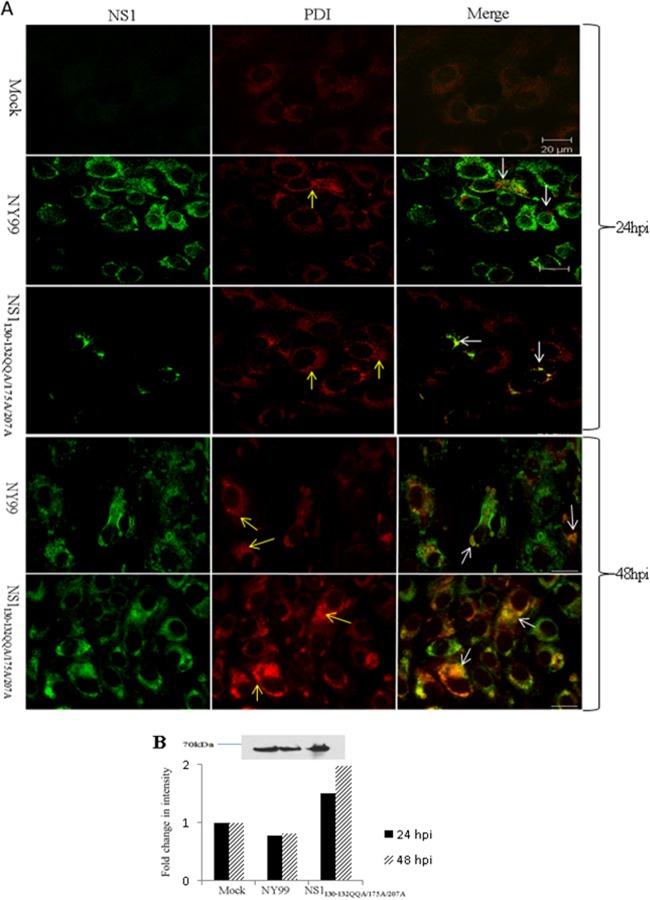
The NS1 protein has been previously demonstrated to have a role in flavivirus RNA replication in association with the endoplasmic reticulum (ER) (12–14). Therefore, virus-infected Vero cells (as described for E/NS1 staining above) were probed with anti-PDI (protein disulfide isomerase) (C81H6, 1:200; Cell Signaling) and anti-WNV NS1 antibodies. Mock-infected cells showed a diffuse staining pattern with anti-PDI antibody, indicating that ER was located throughout the cell. Anti-PDI staining of both the NY99 and NS1130-132QQA/175A/207A virus-infected cells at 24 and 48 h p.i. showed many areas of vesiclelike staining patterns, suggesting that WNV infection induced large ER vesicle formations, consistent with previous reports (Fig. 3A) (14, 15). Colocalization of PDI and NS1 was observed in both NY99- and NS1130-132QQA/175A/207A-infected cells at 24 and 48 h p.i.; however, the NS1130-132QQA/175A/207A-infected cells showed increases in colocalization and PDI staining intensity at 24 h p.i. and, more dramatically, at 48 h p.i., indicating an accumulation of NS1 in the ER compared to its localization in the parental NY99 strain, which showed minimal colocalization of NS1 and PDI. This relative shift in the intensity of PDI staining was confirmed by Western blotting in a separate experiment, in which the PDI protein density was greater in the whole-cell lysates of NS1130-132QQA/175A/207A-infected cells than in those of mock- and NY99-infected cells (MOI of 0.1) at 24 h p.i. (Fig. 3B). Also, the PDI level did not increase over time for NY99. Previous studies have shown that the NS1 protein localizes with the virus replication complex within the ER (14, 16). Thus, an increase in ER in NS1 glycoylation mutant-infected cells may indicate a block at this stage of the replication cycle.
FIG 3.
ER modifications in WNV-infected (MOI of 0.1) Vero cells at 24 and 48 h p.i. (A) Localization of NS1 and PDI (which recognizes ER) in cells infected with NY99 or NS1130-132QQA/175A/207A or uninfected (mock). A vesiclelike pattern of staining is seen in NY99-and NS1130-132QQA/175A/207A-infected cells (yellow arrows) probed with PDI. An apparent lack of colocalization between NS1 and PDI is seen in the NY99-infected cells, while in cells infected with NS1130-132QQA/175A/207A, NS1 staining colocalizes with intense PDI staining (white arrows) at 24 h p.i. At 48 h p.i., NS1 and PDI colocalize for both viruses, most dramatically in NS1130-132QQA/175A/207A-infected cells, which display intense staining for both proteins (white arrows). (B) Western blots were used to verify an increase in PDI staining in the mutant sample compared to the levels in the parental (NY99) and mock-infected cells. The graph shows fold changes in PDI antibody staining intensity compared to that in mock-infected cells for 24 and 48 h p.i., and a representative Western blot from 24 h p.i. is shown.
Following virus replication in the ER, virus particles are then secreted through the Golgi apparatus. Since NS1 is a secreted protein, a block in the ER would impede its subsequent secretion. Thus, we propose that reduced replication at early time points and the accumulation of NS1 in the ER contributes to the mechanism of attenuation of the NS1130-132QQA/175A/207A mutant. Despite increased replication at the 48-h time point, there was a dramatic difference in PDI levels between the NY99- and NS1130-132QQA/187A/207A-infected cells, including increased PDI levels and PDI/NS1 colocalization in NS1130-132QQA/175A/207A-infected cells. We previously showed that there is reduced NS1 protein secretion from cells at 48 h p.i. (2) and hypothesize that the block of NS1 protein in the ER prevents efficient secretion of this protein.
Transmission electron microscopy (TEM) has implicated the ER in the biogenesis of virus-induced ultrastructures in WNV-infected cells (14). The ultrastructure of flavivirus-infected mammalian cells is distinct from that of uninfected cells, with the formation of virus-induced structures, including paracrystalline arrays (PC), convoluted membranes (CM), vesicle packets, and vesicles (also referred to as smooth membrane vesicles in previous publications) (17–20). Using the same experimental conditions as described above for the confocal studies, the ultrastructural differences of cells infected with the NS1130-132QQA/175A/207A mutant and the parental NY99 virus were compared using TEM (Fig. 4A, B, E, and F).
FIG 4.
TEM images and tomograms of a WNV-infected (MOI of 0.1) Vero cell. (A) NY99-infected cell at 24 h p.i. showing convoluted membranes (CM) surrounded by vesicles (V) within vesicle packets (VP) and virus (Vi). Inlay shows a 10% larger VP with vesicles of uniform size and shape. (B) In contrast, an NS1130-132QQA/175A/207A-infected Vero cell at 24 h p.i. has polymorphic vesicles and VP, represented in the 10% larger inlay, and paracrystalline arrays (PC) with a lack of CM localized to the perinuclear region. The large diameters of the vesicles were determined by measuring 100 vesicles from several cells for both NY99- and NS1130-132QQA/175A/207A-infected cells; they averaged 90 nm and 120 nm (P = 0.01), respectively. (C, D) Tomogram sections of NY99- and NS1130-132QQA/175A/207A-infected cells, respectively, at 24 h p.i. Three hundred-nanometer-thick sections were analyzed using tilt series in the −65° to +65° angular range with 1° increments using the SerialEM program (21). Three-dimensional tomograms were reconstructed using IMOD software (22). Scale bar = 500 nm. (E, F) The 48-h p.i. time point was also observed by TEM for NY99-infected cells (E) and NS1130-132/175A/207A-infected cells (F). (Nu, nucleus; Mi, mitochondria).
The ultrastructure of the NS1130-132QQA/175A/207A-infected cells was distinct from that of parental NY99-infected cells. Most mutant-infected cells did not contain CM structures at 24 h p.i., whereas these structures were found in almost every NY99-infected cell at the same time point. Interestingly, the vesicles were frequently more abundant than the CM structures and vesicles were commonly found without the presence of CM, and yet virus particles were often identified in these cells. PC were seen in both NY99 and NS1130-132QQA/175A/207A-infected cells at 24 h p.i. At 48 h p.i., when both viruses were at peak titers, CM were abundant in cells infected with either virus (Fig. 4E and F), indicating that CM form with high-titer virus replication, whereas PC form either at earlier time points or with lower virus titers or both.
The most striking difference between the ultrastructures of the parental NY99- and NS1130-132QQA/175A/207A-infected Vero cells was in the size and shape of the vesicles. The polymorphic vesicles in the mutant NS1130-132QQA/175A/207-infected cells varied from a small round shape to a larger tubular shape (120 ± 20 nm), whereas the parental NY99-infected cells showed uniform round vesicles (90 ± 10 nm). Tomograms confirmed that most vesicles in NS1130-132QQA/175A/207A-infected cells appeared to be polymorphic, long, and tubular (Fig. 4D), and although some vesicles appear in the images to be small and spherical in shape and similar to the vesicles in NY99-infected cells, this may be due to the orientation of the vesicle within the section, where a top view appears round but the side view is tubular in shape (Fig. 4C). The size of the vesicles in NS1130-132QQA/175A/207A-infected cells was calculated from several different TEM images (slices), but because two-dimensional images were used for this measurement, the higher standard deviation may reflect the inability to measure the longest dimension for some vesicles. The tomograms of NY99-infected cells confirmed that vesicles in different cells were similar in size and shape.
Overall, these data suggest that the mechanism of attenuation for the NS1130-132QQA/175A/207A mutant is the focal perinuclear localization of the NS1 protein and its block in the ER, resulting in reduced replication at early time points. This result may also be the result of malformed vesicles that consist of double-stranded RNA and the replication complex, including NS1.
ACKNOWLEDGMENTS
We thank Adriana Paulucci and Leoncio Vergara in the Optical Microscopy Core at UTMB for their technical assistance with confocal microscopy.
M.C.W. was supported by National Institutes of Health T32 training grant AI007536.
REFERENCES
- 1.Whiteman MC, Li L, Wicker JA, Kinney RM, Huang C, Beasley DW, Chung KM, Diamond MS, Solomon T, Barrett AD. 2010. Development and characterization of non-glycosylated E and NS1 mutant viruses as a potential candidate vaccine for West Nile virus. Vaccine 28:1075–1083. doi: 10.1016/j.vaccine.2009.10.112. [DOI] [PubMed] [Google Scholar]
- 2.Whiteman MC, Wicker JA, Kinney RM, Huang CY, Solomon T, Barrett AD. 2011. Multiple amino acid changes at the first glycosylation motif in NS1 protein of West Nile virus are necessary for complete attenuation for mouse neuroinvasiveness. Vaccine 29:9702–9710. doi: 10.1016/j.vaccine.2011.09.036. [DOI] [PubMed] [Google Scholar]
- 3.Blitvich BJ, Scanlon D, Shiell BJ, Mackenzie JS, Pham K, Hall RA. 2001. Determination of the intramolecular disulfide bond arrangement and biochemical identification of the glycosylation sites of the nonstructural protein NS1 of Murray Valley encephalitis virus. J Gen Virol 82(Pt 9):2251–2256. [DOI] [PubMed] [Google Scholar]
- 4.Flamand M, Megret F, Mathieu M, Lepault J, Rey FA, Deubel V. 1999. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J Virol 73:6104–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mason PW. 1989. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology 169:354–364. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlesinger JJ, Brandriss MW, Putnak JR, Walsh EE. 1990. Cell surface expression of yellow fever virus non-structural glycoprotein NS1: consequences of interaction with antibody. J Gen Virol 71(Pt 3):593–599. doi: 10.1099/0022-1317-71-3-593. [DOI] [PubMed] [Google Scholar]
- 7.Winkler G, Randolph VB, Cleaves GR, Ryan TE, Stollar V. 1988. Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology 162:187–196. doi: 10.1016/0042-6822(88)90408-4. [DOI] [PubMed] [Google Scholar]
- 8.Winkler G, Maxwell SE, Ruemmler C, Stollar V. 1989. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology 171:302–305. doi: 10.1016/0042-6822(89)90544-8. [DOI] [PubMed] [Google Scholar]
- 9.Crooks AJ, Lee JM, Easterbrook LM, Timofeev AV, Stephenson JR. 1994. The NS1 protein of tick-borne encephalitis virus forms multimeric species upon secretion from the host cell. J Gen Virol 75(Pt 12):3453–3460. doi: 10.1099/0022-1317-75-12-3453. [DOI] [PubMed] [Google Scholar]
- 10.Post PR, Carvalho R, Galler R. 1991. Glycosylation and secretion of yellow fever virus nonstructural protein NS1. Virus Res 18:291–302. doi: 10.1016/0168-1702(91)90025-Q. [DOI] [PubMed] [Google Scholar]
- 11.Beasley DW, Holbrook MR, Travassos Da Rosa AP, Coffey L, Carrara AS, Phillippi-Falkenstein K, Bohm RP, Ratterree MS, Lillibridge KM, Ludwig GV, Estrada-Franco J, Weaver SC, Tesh RB, Shope RE, Barrett AD. 2004. Use of a recombinant envelope protein subunit antigen for specific serological diagnosis of West Nile virus infection. J Clin Microbiol 42:2759–2765. doi: 10.1128/JCM.42.6.2759-2765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindenbach BD, Rice CM. 1997. trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol 71:9608–9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindenbach BD, Rice CM. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol 73:4611–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol 84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackenzie JM, Jones MK, Westaway EG. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J Virol 73:9555–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufusi PH, Kelley JF, Yanagihara R, Nerurkar VR. 2014. Induction of endoplasmic reticulum-derived replication-competent membrane structures by West Nile virus non-structural protein 4B. PLoS One 9:e84040. doi: 10.1371/journal.pone.0084040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol 71:6650–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Offerdahl DK, Dorward DW, Hansen BT, Bloom ME. 2012. A three-dimensional comparison of tick-borne flavivirus infection in mammalian and tick cell lines. PLoS One 7:e47912. doi: 10.1371/journal.pone.0047912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leary K, Blair CD. 1980. Sequential events in the morphogenesis of Japanese encephalitis virus. J Ultrastruct Res 72:123–129. doi: 10.1016/S0022-5320(80)90050-7. [DOI] [PubMed] [Google Scholar]
- 20.den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 21.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]