ABSTRACT
Virus capsids provide genome protection from environmental challenges but are also poised to execute a program of compositional and conformational changes to facilitate virion entry and infection. The most abundant adenovirus serotype 5 (AdV5) capsid protein, hexon, directly recruits the motor protein cytoplasmic dynein following virion entry. Dynein recruitment is crucial for capsid transport to the nucleus and requires the transient exposure of AdV5 hexon to low pH, presumably mimicking passage through the endosomal compartment. These results suggest a pH-dependent capsid modification during early infection. The changes to hexon structure controlling this behavior have not been explored. We report that hexon remains trimeric at low pH but undergoes more subtle conformational changes. These changes are indicated by increased sensitivities to SDS-mediated dissociation and dispase proteolysis. Both effects are reversed at neutral pH, as is dynein binding by low-pH-treated hexon. Dispase cleavage, which we find maps to a specific site within hypervariable region 1 (HVR1) of AdV5 hexon, has no apparent effect on virion entry but completely inhibits its transport to the nucleus. In addition, an AdV5 mutant containing HVR1 of AdV48 is unable to bind dynein and is strongly inhibited in the postentry transport step. These results reveal that conformational changes involving hexon HVR1 are the basis for a novel viral mechanism controlling capsid transport to the nucleus.
IMPORTANCE The adenovirus serotype 5 (AdV5) capsid protein hexon recruits the molecular motor protein cytoplasmic dynein in a pH-dependent manner, a function critical for efficient transport toward the nucleus and AdV5 infectivity. In this work, we describe how low-pH exposure induces reversible structural changes in AdV5 hexon and how these changes affect dynein binding. In addition, we identified a pH-sensitive dispase cleavage site in hexon HVR1, which depends on the same structural changes and furthermore regulates dynein recruitment and capsid redistribution in infected cells. These data provide the first evidence relating long-known but subtle pH-dependent structural changes in hexon to a more recently established essential but poorly understood role in virus transport. These results have broad implications for understanding virus infectivity in general, and our ability to block the recruitment mechanism has potential therapeutic implications as well.
INTRODUCTION
Adenovirus (AdV) is one of many viruses that require motor proteins for capsid transport to the nucleus early in infection (1–4). AdV serotype 5 (AdV5) enters the cell by receptor-mediated endocytosis followed by rapid escape from the endosome. In the cytoplasm, AdV5 capsids recruit cytoplasmic dynein for transport along microtubules (MTs) (5–7). We previously reported that cytoplasmic dynein is directly recruited by hexon, the major AdV5 capsid subunit (5). Binding was greatly enhanced by prior, transient exposure of capsids or isolated hexon to low-pH conditions. These observations suggested a novel, virus-specific mechanism for the activation of microtubule-mediated transport specifically during the earliest postendosomal phase of infection (5).
The interaction with hexon is mediated by two classes of dynein subunits involved in cargo recruitment. Individually overexpressed intermediate chains (ICs) (both IC1 and IC2) and light intermediate chain 1 (LIC1) can bind hexon (5). However, LIC1 alone appears to have a predominant role in controlling the recruitment of the native dynein complex (8). LIC1 binding to hexon requires phosphorylation by protein kinase A (PKA) (8), which is activated by the initial binding of AdV to α5 integrins at the cell surface (9, 10).
How low-pH exposure activates hexon for dynein recruitment is unknown. Hexon is a stable homotrimeric structure, with each subunit consisting of a conserved core and seven immunogenic hypervariable regions (HVRs), which contribute to serospecificity (11, 12). Exposure of hexon to acidic pH was found to increase its hydrophobicity, as evidenced by detergent partitioning assays (13) and susceptibility to limited dispase proteolysis (14). Whether these effects are associated with the stability of the hexon trimer or other aspects of the hexon structure is unknown but might be relevant to an understanding of motor protein recruitment.
Here, we examine the effects of pH on hexon self-association and conformational behavior. We identify reversible changes in hexon conformation but not self-association correlated with the gain and loss of dynein recruitment activity. Limited dispase proteolysis of hexon HVR1, or mutation of this sequence, abolishes dynein binding and inhibits capsid transport in the cell. These results implicate HVR1 in these critical aspects of adenovirus biology.
MATERIALS AND METHODS
Cells, virus, and antibodies.
293A and A549 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Replication-deficient AdV5 engineered to express green fluorescent protein (GFP) (AdV5-GFP, plaque purified; gift of Hamish Young, Columbia University), a chimeric AdV5 with the AdV48 hexon HVR1 sequence [AdV5HVR48(1)] (15), and replication-deficient wild-type (WT) AdV48 (both generous gifts of Dan Barouch, Harvard Medical School) were propagated in 293A cells, purified by banding on two successive linear CsCl gradients (16), and dialyzed against 10% glycerol in phosphate-buffered saline (PBS) before cryostorage. The particle concentration was determined by spectrophotometry (17) and, similar to previously reported results (15), was ∼3-fold lower for AdV5HVR48(1) than for wild-type AdV5. The viral titer was determined by using fluorescent-focus assays for AdV5-GFP (18).
Antibodies used included mouse monoclonal antihexon (Novocastra), antitubulin (Sigma), and anti-dynein IC (clone 74.1; Chemicon); chicken anti-LIC1 (19); and rabbit anti-adenovirus serotype 5 (Abcam), anti-early endosome antigen 1 (EEA1), and antihemagglutinin (Sigma).
Proteins and biochemical analyses.
Adenovirus hexon was recovered from virus-depleted supernatants of the first CsCl gradient by either immunoisolation using a hexon-specific monoclonal antibody linked to protein A-Sepharose beads (GE Bioscience) (5) or anion-exchange chromatography (20). Monomeric hexon was obtained by exposing chromatographically purified hexon to 1.0% lithium dodecyl sulfate (LDS; Sigma-Aldrich) at 100°C for 5 min.
Purified rat cytoplasmic dynein was prepared by using phosphate-glutamate buffer (pH 7.0) (21) from MTs assembled in rat brain lysates with the aid of 30 μM paclitaxel (Sigma-Aldrich). Cytoplasmic dynein was released from MTs with 10 mM ATP (Sigma-Aldrich) and further purified on a linear sucrose gradient.
Hexon binding assays were described previously (5). Briefly, immunoisolated hexon was incubated for 30 min in Tris-maleate buffer (50 mM Trizma-maleate, 10 mM NaCl, 1 mM EDTA, and 0.1% Tween 20 [pH 4.4]), resuspended in the same buffer at pH 7.4 for the indicated amounts of time, and then exposed to purified rat brain cytoplasmic dynein at 4°C for 1.5 h. Hexon-bound dynein was assayed by immunoblotting.
For controlled proteolysis, hexon or AdV capsids were incubated with a 6-fold molar excess of dispase (neutral protease; Worthington Biochemicals, Freehold, NJ) over hexon in dispase buffer (100 mM Trizma-maleate, 120 mM NaCl, 2.5 mM CaCl2), and the pH was adjusted to either pH 7.4 (neutral) or pH 4.4 (acidic) for 5 min at 37°C until the reaction was stopped by the addition of either protein sample buffer (PSB) or 5 mM EDTA to the mixture.
Sedimentation analysis of hexon was performed on linear 5 to 20% sucrose gradients of a volume of 1.4 ml in Tris-maleate buffer (50 mM Trizma-maleate, 10 mM NaCl, 1 mM EDTA, and 0.1% Tween 20) at the indicated pH values. Samples (100 μl) were centrifuged for 3 h at 200,000 × g in a TLS-55 rotor at 4°C and fractionated into 13 fractions of equal volumes. Dynein (20S) and bovine serum albumin (BSA) (4.4S) were used as calibration standards.
For time-dependent SDS sensitivity assays, hexon was exposed to 250 mM Trizma-maleate buffer at pH 4.4 for 30 min on ice. PSB (60 mM Tris-HCl [pH 6.8], 1.5% SDS, 5% glycerol, 100 mM dithiothreitol [DTT], and 20 μg/ml bromphenol blue [final concentrations]) was added for incubation at room temperature for the indicated time periods.
For time-dependent pH sensitivity assays, hexon was exposed to low pH for the indicated time periods, and for time-dependent reversibility assays, the protein was transferred to neutral-pH buffer for the indicated time periods, followed by the addition of PSB and incubation for 30 min at room temperature.
Virus infection and other cellular methods.
For fixed-cell analysis, A549 cells were infected with AdV at a multiplicity of infection (MOI) of 20 in a small volume of DMEM lacking FBS at 4°C for 30 min to allow virus attachment. The cells were washed twice in cold PBS and incubated in fresh, warm DMEM–10% FBS for 60 min at 37°C to allow virion internalization and intracellular transport. Cells were grown on glass coverslips and fixed by exposure to methanol at −20°C for 5 min or to 4% paraformaldehyde (PFA) in PBS at room temperature for 15 min. Coverslips were blocked for >60 min in 0.5% donkey serum–PBS, incubated with primary antibody at 37°C for 1 h, washed, incubated for 1 h at 37°C with Cy2- and Cy3-conjugated secondary antibody, and then mounted by using ProLong Gold antifade mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). For live-cell analysis of GFP expression, A549 cells were infected with AdV5-GFP as described above but at an MOI of 1, placed inside a stage-top incubator (Live Cell Instrument) at 37°C with 5% CO2, and immediately imaged every 10 min for 15 h. Similarly, for live-cell analysis of capsid motility, A549 cells were infected with Alexa 555 dye (Invitrogen)-labeled capsids (5) at an MOI of 20, placed inside a stage-top incubator, and immediately imaged at video frame rate (16.5 frames per second [fps]) for 1 min at between 15 and 60 min postinfection (p.i.).
For imaging, either a confocal microscope (IX-81 with an FV100 spectral confocal system; Olympus, Center Valley, PA) equipped with a 63× (1.42-numerical-aperture [NA]) oil immersion objective and operated with FluoView imaging software (Olympus) or an epifluorescence inverted microscope (Nikon Ti-Eclipse; Nikon Instruments, Melville, NY) coupled with a CoolSnap ES2 charge-coupled device (CCD) camera (Photometrics, Tucson, AZ), an Andor electron-multiplying (EM)-CCD camera (Andor Technology, South Windsor, CT), and a 4× Plan Fluor or a 100× Plan Apo VC objective operated by NIS-Elements imaging software (Nikon) was used. For evaluation of virus redistribution, virus particles were scored as reaching the nucleus if they were <1 μm from the nuclear surface.
Statistical analysis.
Two-sample comparisons were performed by using Student's t test with Prism (GraphPad, La Jolla, CA). Statistical significance was indicated by P values of <0.05.
RESULTS
Reversible effects of low pH on hexon-dynein binding and structure.
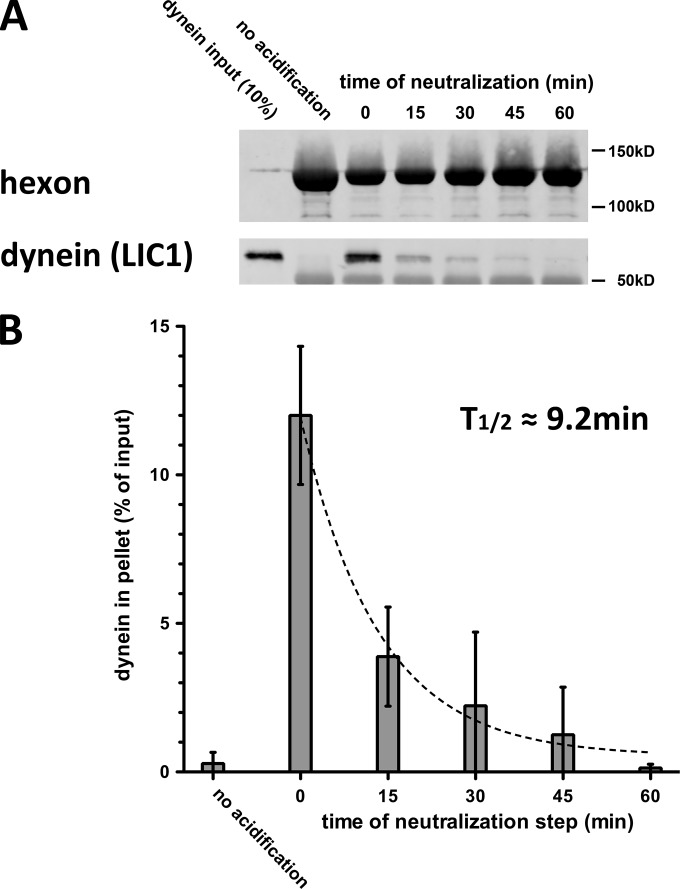
In our previous analysis, we found that transient exposure of hexon to a series of decreasing pH levels increased its ability to bind dynein (5). Optimal dynein binding was seen after hexon was exposed to pH 4.4 buffer (“acidification”), restored to pH 7.4 (“neutralization”), and immediately incubated with dynein. Whether acidification resulted in a stable or transient change in hexon binding activity was not explored. As a basis for further kinetic analysis of hexon conformational behavior, we now evaluated dynein binding as a function of time after hexon neutralization (Fig. 1A). Whereas hexon pulled down high levels of dynein immediately after neutralization, this activity gradually declined and was reduced by 60 min to levels comparable to those observed for the unprimed hexon control, following exponential decay with a half-life (t1/2) of 9.2 min (Fig. 1B).
FIG 1.
Low-pH priming of hexon for dynein binding is reversible. (A) Immunoisolated hexon was acidified, returned to neutral buffer, and kept at neutrality for the indicated periods of time on ice before it was incubated with purified dynein for 1.5 h at 4°C. Pellets from hexon pulldowns were subjected to SDS-PAGE and Western blotting with antihexon and anti-LIC1 antibodies. (B) Quantification of hexon pulldowns of dynein shows an exponential decrease (t1/2 = 9.23 min; R2 = 0.9) with the time that hexon is returned to neutral pH after acid priming. Dynein binding declines to control levels (no acid priming) by 60 min (n = 3; means ± standard deviations).
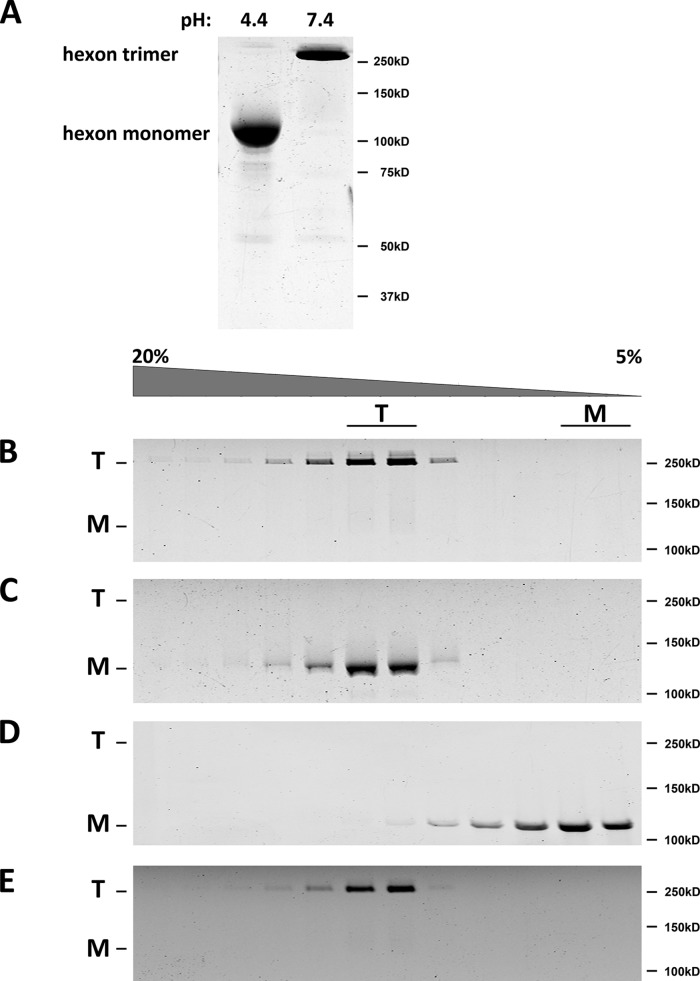
Hexon has been reported to remain trimeric at pH 5.0 (22). It also resists SDS-mediated dissociation unless boiled (23). To test for pH-dependent conformational changes, we first performed SDS-polyacrylamide gel electrophoresis (PAGE) on unboiled hexon samples prepared at pH 7.4 or pH 4.4 (Fig. 2A). Hexon exposed to pH 7.4 ran as a trimer (23), whereas hexon exposed to pH 4.4 ran as monomer (Fig. 2A), indicating low-pH- and/or SDS-dependent disruption of the trimeric structure. To distinguish between these two possibilities, we performed sucrose density gradient centrifugation at pH 7.4 and pH 4.4 (Fig. 2B and C). We found that hexon still sedimented at the hexon trimer position at each pH level, although the pH 4.4 sample again exhibited dissociation on SDS-PAGE gels (Fig. 2B and C). Monomeric hexon produced by a combination of high temperature and LDS treatment ran, as expected, as a low-S peak (Fig. 2D). To test for reversibility of the low-pH-induced structural changes, we exposed hexon to pH 4.4 for 30 min prior to centrifugation on a pH 7.4 sucrose density gradient. Hexon sedimented as a trimer (Fig. 2E) and became insensitive to SDS during SDS-PAGE of unboiled samples. These results together suggest that the acidification of hexon leads to a subtle structural change with no apparent effect on the native hexon trimer but which sensitizes it to SDS dissociation. The effect is completely reversible at neutral pH.
FIG 2.
Hexon dissociation effects of low pH and SDS. (A) Purified AdV5 hexon trimers were tested for low-pH dissociation. Following exposure to pH 7.4 or 4.4, hexon samples were incubated in protein sample buffer overnight, subjected to SDS-PAGE without boiling, and stained by using Coomassie blue. Dissociation into monomers was detected under pH 4.4 conditions but not at pH 7.4. (B and C) Purified hexon was tested for pH-dependent dissociation by centrifugation on linear sucrose density gradients (5 to 20%) at pH 7.4 (B) and pH 4.4 (C). SDS-PAGE of unboiled samples was performed, followed by Coomassie blue staining. Hexon sediments as a trimer at either pH 7.4 or 4.4, although the pH 4.4 sample was dissociated to monomers by SDS-PAGE after exposure to protein sample buffer (without boiling). (D) Dissociation of monomeric hexon (see Materials and Methods) was tested by sucrose density gradient centrifugation at pH 7.4. The sedimentation coefficient and SDS-PAGE indicate a single low-S peak of monomeric hexon. (E) Purified hexon was tested for reversibility of SDS sensitivity for dissociation by pH 4.4 treatment before sedimentation on linear sucrose gradients (5 to 20%) at pH 7.4. SDS-PAGE of samples of the unboiled fraction and Coomassie blue-stained gels show the sedimentation coefficient and SDS-PAGE sedimentation of the trimeric hexon molecule, indicating full reversibility of the low-pH effects. T, trimer; M, monomer.
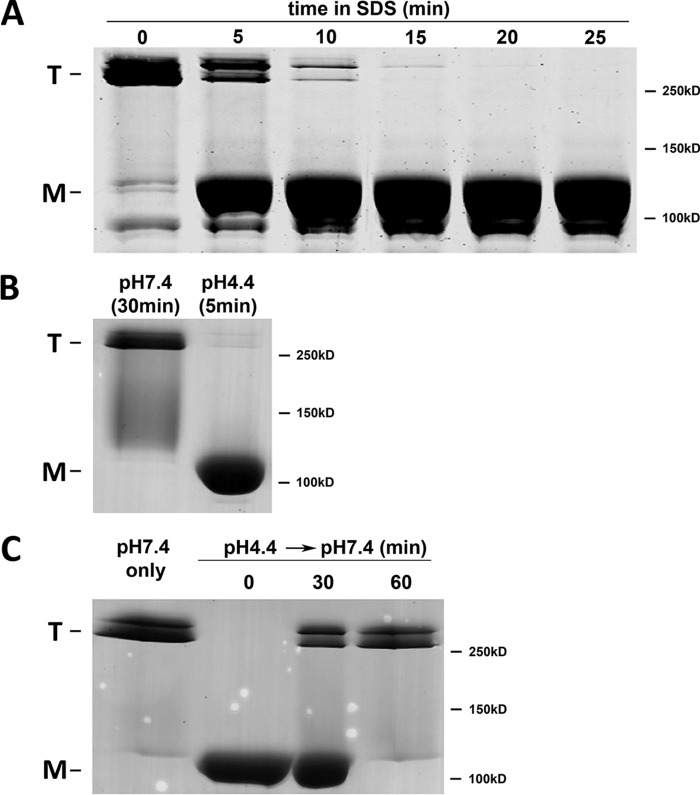
Kinetics of pH-induced hexon structural changes.
In order to assess how well the time course of hexon conformational changes corresponds to that of dynein binding, we first monitored how quickly SDS dissociates low-pH-treated hexon trimers (Fig. 3A). Hexon was incubated at pH 4.4 for 30 min. SDS-containing protein sample buffer was then added at a series of times, and SDS-PAGE was quickly initiated without boiling. SDS-mediated dissociation of the hexon trimers was observed to increase with time and was complete by 20 min (Fig. 3A). We then used SDS sensitization to estimate the time course of the low-pH-induced hexon conformational change and its reversal at neutral pH (Fig. 3B and C). For this purpose, we treated purified hexon at pH 4.4 for 0 to 30 min, followed by uptake in protein sample buffer and SDS-PAGE without boiling. Exposure of hexon to low pH induced SDS sensitivity in <5 min (Fig. 3B). We then examined whether SDS sensitivity was reversed following the return of hexon to neutral pH and the time course thereof. Partial reversal of SDS sensitivity was observed within 30 min, and virtually complete reversal was observed by ∼60 min (Fig. 3C).
FIG 3.
Kinetics of SDS-mediated hexon dissociation and reassociation. (A) To test the kinetics of SDS sensitivity, purified hexon was incubated at pH 4.4 for 30 min and then exposed to PSB for the indicated time periods. Coomassie blue-stained SDS-PAGE gels of unboiled samples show detectable hexon dissociation 5 min after the addition of PSB and a complete effect after 20 min, indicating a gradual SDS effect. (B) To test the kinetics of pH sensitivity, purified hexon was incubated either at pH 7.4 for 30 min or at pH 4.4 for 5 min, followed by 20 min of incubation with PSB and SDS-PAGE of unboiled samples. Hexon sensitivity to dissociation was clearly established within 5 min. (C) To test the kinetics of reassociation, purified hexon was incubated at pH 4.4 for 5 min and neutralized for the indicated time periods, followed by 20 min of incubation with PSB and SDS-PAGE of unboiled samples. Full reassociation occurred within 30 to 60 min.
Together, our results indicate that hexon dissociation into monomers can be induced by acid priming and additional exposure to SDS. For a complete effect, SDS treatment must be >20 min. We also conclude that SDS sensitivity is established rapidly after exposure to pH 4.4, whereas the kinetics of the reversibility of the low-pH effects are substantially slower.
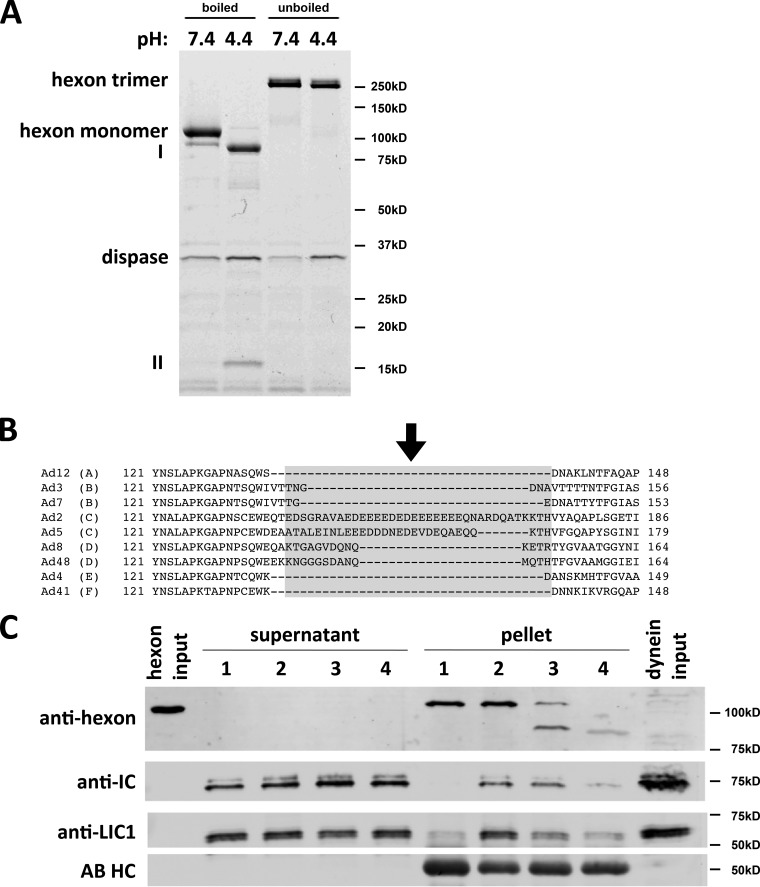
Effects of dispase hydrolysis on the hexon-dynein interaction.
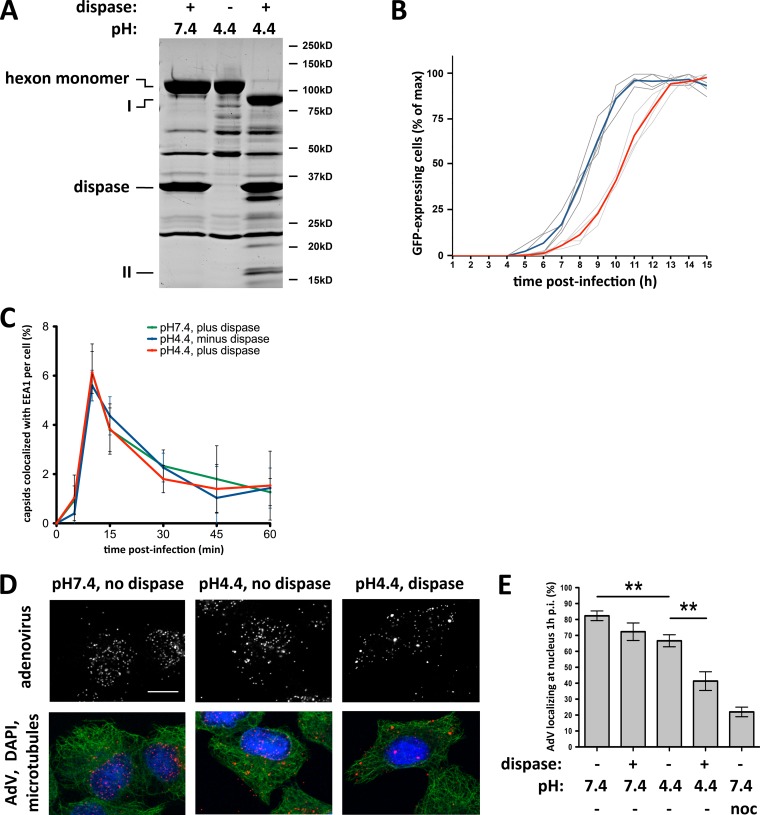
Hexon has been reported to be insensitive to dispase hydrolysis at neutral pH. Below pH 6.5, however, dispase cleaves hexon into a 15-kDa N-terminal fragment and an 85-kDa C-terminal fragment (14, 22). We confirmed these observations and the persistence of the hexon trimer by SDS-PAGE of boiled and unboiled samples (Fig. 4A) (22). These results together are consistent with a single, minimally disruptive nick within the hexon polypeptide. For precise determination of the hydrolysis site, we compared the full-length and 85-kDa dispase digestion products by tandem mass spectrometry (MS/MS) of tryptic subfragments. A single partial tryptic fragment corresponding to the amino acid sequence EVDEQAEQQK was identified, representing the N terminus of the 85-kDa dispase product. This sequence matches E156 to K165 in the AdV5 hexon sequence and is part of hexon hypervariable region 1 (HVR1), responsible for serospecificity and thought to be conformationally flexible (Fig. 4B) (11, 12, 24). To test if dispase hydrolysis affects dynein binding, we exposed hexon to dispase at pH 4.4 for partial or complete digestion and then tested for dynein binding shortly after neutralization at pH 7.4 (Fig. 4C). Partial hexon proteolysis decreased dynein binding, especially as indicated using anti-LIC1 antibody (Fig. 4C, lane 3), and complete proteolysis had a more potent effect (lane 4). These results are consistent with hexon HVR1 mediating the pH-induced structural change and containing the dispase proteolysis site (Fig. 4B; see also Fig. 7B). Interestingly, dynein binding is reduced after dispase treatment, indicating the requirement for an intact polypeptide backbone in this region (Fig. 4C).
FIG 4.
Dispase hydrolysis inhibits low-pH effects on hexon structure and dynein binding. (A) Purified hexon was tested for dissociation after dispase treatment. Dispase exposure of hexon at low pH resulted in two major digestion products (85 kDa [I] and 15 kDa [II]) visible by Coomassie blue staining of SDS-PAGE gels of boiled samples. Unboiled samples show only hexon trimers under digested and undigested conditions, indicating that the hexon trimer remains intact even after complete dispase hydrolysis at low pH. (B) Sequence alignment of HVR1 of hexon from adenovirus subgroups A (serotype 12), B (serotypes 3 and 7), C (serotypes 2 and 5), D (serotypes 8 and 48), E (serotype 4), and F (serotype 41). HVR1 is marked in light gray (37), and the dispase cleavage site in AdV5 hexon is marked by an arrow. (C) Effects of dispase treatment on dynein binding. Hexon was treated with dispase at pH 4.4 and incubated with the purified dynein complex at pH 7.4. Lane 1, nonacidified hexon; lane 2, acidified hexon; lane 3, incompletely dispase-digested hexon; lane 4, fully dispase-digested hexon. Inputs as well as supernatants and pellets from hexon pulldowns were subjected to SDS-PAGE and Western blotting with antihexon, anti-LIC1, and anti-IC antibodies. Dispase pretreatment reduced the hexon affinity for the dynein complex. No proteolysis of dynein subunits was detectable. AB HC, antibody heavy chain.
FIG 7.
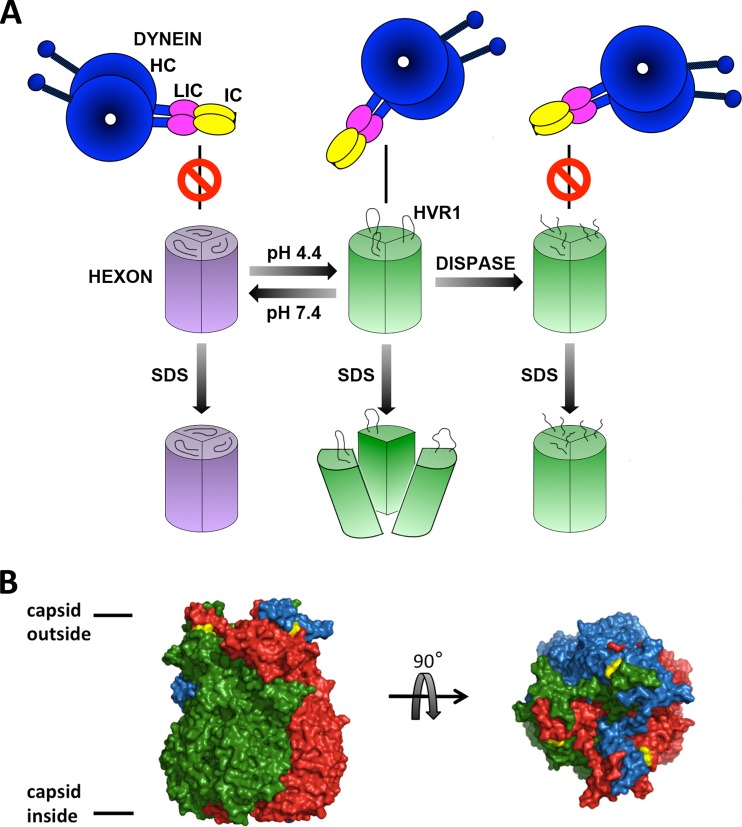
Model of pH-sensitive hexon dissociation upon SDS treatment. (A) Diagram illustrating different structural states of the hexon trimer dependent on pH leading to SDS sensitivity. The hexon trimer at neutral pH (purple) remains monomeric after the addition of SDS. Acidified hexon (green) maintains the trimeric state. Low-pH treatment induces subtle structural changes, which strongly increase dynein affinity (5) and SDS sensitivity, which can be fully reversed by a return to neutral pH. Dispase cleaves only acidified hexon, which is then blocked in its dynein binding activity and SDS sensitivity. (B) X-ray crystal structure of the hexon trimer (PDB accession number 1P30) (37), with monomers shown in red, green, and blue and resolved amino acid residues surrounding the dispase proteolysis region HVR1 shown in yellow (D135 and T165). Side and top views are shown. The illustration was generated with PyMOL (PyMOL molecular graphics system, version 1.5.0.4; Schrödinger LLC).
It was previously reported that dispase treatment of complete AdV5 capsids specifically affects hexon (14). We confirmed this observation and identified the same 85-kDa and 15-kDa products (Fig. 5A). Based on these results, we exposed A549 cells to low-pH/dispase-pretreated capsids and tested the ability of AdV5-GFP to enter the cell, redistribute to the cell center, and induce GFP expression. GFP expression occurred 2 h later for low-pH/dispase-pretreated than for low-pH-treated virions, a delay of ∼25% (Fig. 5B), indicating an attachment, entry, or transport defect. Attachment of virions to the cells appeared to be normal in this assay, as indicated by the number of bound particles at 5 min p.i., and similarly, endosomal escape of pretreated capsids was normal, as judged by colocalization with EEA1 (Fig. 5C). However, redistribution of the digested capsids to the nucleus, which is normally complete by ∼60 min, was significantly reduced (Fig. 5D and E). Exposure of AdV5 capsids to pH 4.4 alone had a minor, though significant, effect.
FIG 5.
Dispase treatment reduces capsid redistribution to the nucleus. (A) pH-dependent dispase cleavage of AdV5 capsid hexon. Purified AdV5 capsids were treated with dispase at pH 7.4 or pH 4.4 or kept at pH 4.4 alone before being boiled in PSB and subjected to SDS-PAGE and Coomassie blue staining to show total capsid proteins. Hexon was hydrolyzed by dispase only at pH 4.4 (85-kDa and 15-kDa products, marked with I and II, respectively), whereas no proteolysis was detectable at pH 7.4 or pH 4.4 without the protease. (B) Effect of dispase cleavage on infectivity. AdV5-GFP capsids were treated with dispase at pH 7.4 or pH 4.4 and used to infect A549 cells, which were continuously imaged for up to 15 h p.i. Graphs represent relative counts of GFP-expressing cells. Dispase-cleaved capsids (light gray lines; the red line is the average) induce GFP expression ∼2 h later than control capsids (dark gray lines; the blue line is the average). (C) Normal cell entry by dispase-treated capsids. A549 cells were infected with untreated or dispase-treated AdV5 capsids, fixed at the indicated times p.i., and processed for immunofluorescence microscopy using antihexon and anti-EEA1 (early endosome antigen 1) antibodies. The graph shows the percentage of colocalization of AdV capsids with EEA1 over time postinfection and indicates similar profiles independent of dispase treatment. (D) Effect of dispase treatment on capsid redistribution to nucleus. A549 cells were infected with untreated or dispase-treated AdV5 capsids, fixed at 60 min p.i., and processed for immunofluorescence microscopy using antihexon antibody to localize capsids within the cells. Cells were also stained for microtubules (antitubulin) and DNA (DAPI). Dispase-treated capsids remained dispersed, whereas control capsids redistributed to the nucleus. Bar, 10 μm. (E) Quantification of virus redistribution. The nuclear localization of AdV5 particles was moderately reduced by pretreatment with dispase at pH 7.4 or without dispase at pH 4.4. More substantial inhibition of virus redistribution resulted from treatment of virus with dispase at pH 4.4 compared to the untreated pH 4.4 control (n = 3; means ± standard deviations). The nocodazole control used to depolymerize microtubules showed near-complete inhibition of virus redistribution to the nucleus. Asterisks mark P values of <0.01.
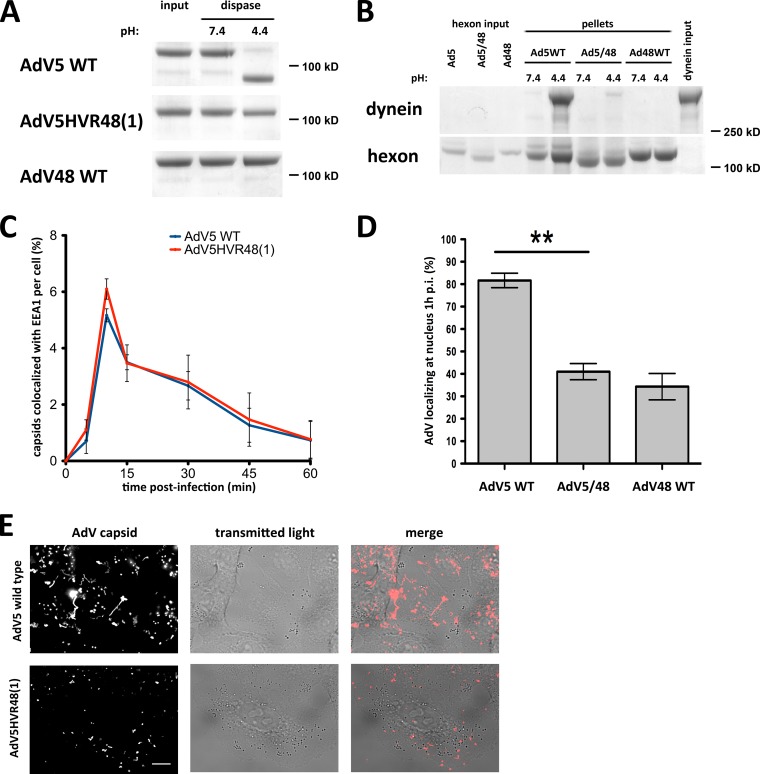
We also used a chimeric AdV5 mutant containing the AdV48 hexon HVR1 sequence [AdV5HVR48(1)] (15) to directly test the role of HVR1 in dynein binding and subcellular transport. AdV48 is a member of AdV serogroup D, some members of which engage with CD46 rather than coxsackie and adenovirus receptor (CAR) as the primary receptor (25–27) and subsequently follow a different uptake pathway than AdV5, remaining in the endolysosomal pathway for up to 8 h (28, 29). We found that the AdV5HVR48(1) mutant hexon cannot be cleaved by dispase (Fig. 6A) and pulls down substantially less dynein than WT AdV5 hexon (Fig. 6B). WT AdV48 hexon is also unable to interact with dynein independent of low-pH priming (Fig. 6B). Furthermore, as for dispase-digested virions, AdV5HVR48(1) capsid redistribution to the nucleus was markedly reduced at 60 min p.i., although endosomal escape occurred normally (Fig. 6C and D). Finally, we compared Alexa-labeled AdV5HVR48(1) and wild-type AdV5 capsids using live-cell microscopy (Fig. 6E; see Movies S1 and S2 in the supplemental material). Whereas wild-type capsids moved over long distances in most infected cells, mutant capsids were highly restricted in their cytoplasmic motility [82.7% versus 27.1% of infected cells showed of capsid movements of >1 μm in a 1-min movie for WT AdV5 and AdV5HVR48(1), respectively], similar to the phenotype observed after microinjection of a function-blocking antidynein antibody (5).
FIG 6.
The AdV5 hexon HVR1 mutant is inhibited in dynein binding and cytoplasmic transport. (A) Dispase insensitivity of mutant hexon. Immunopurified hexons of wild-type (WT) AdV5, AdV5HVR48(1), and WT AdV48 were tested for dissociation after dispase treatment. Dispase hydrolyzed only WT AdV5 hexon at low pH, visible by Coomassie blue staining of SDS-PAGE gels of boiled samples. AdV5HVR48(1) and WT AdV48 samples show hexon trimers independent of pH, indicating that the hexon trimer remains intact even after dispase hydrolysis and low-pH treatment. (B) Effect of mutant HVR1 on dynein binding. Immunopurified hexons from WT AdV5, AdV5HVR48(1) (AdV5/48), and WT AdV48 were exposed to pH 7.4 or pH 4.4 and tested for dynein binding at pH 7.4. Only acid-primed WT AdV5 hexon, but not acid-primed AdV5HVR48(1) or WT AdV48 hexon or hexon kept at pH 7.4, pulls down significant amounts of dynein, as shown by Coomassie blue staining of SDS-PAGE gels of boiled samples. (C) Lack of an effect of mutant HVR1 on virus entry. A549 cells were infected with WT AdV5 or AdV5HVR48(1) capsids, fixed at the indicated times p.i., and processed for immunofluorescence microscopy using antihexon and anti-EEA1 antibodies. The graph shows the percent colocalization of AdV capsids with EEA1 over time and indicates similar profiles for WT and mutant AdV5. (D) Effect of the altered HVR1 sequence on virus redistribution to the nucleus. AdV5HVR48(1) and WT AdV48 each show a significant decrease in nuclear localization at 1 h p.i. relative to WT AdV5 (n = 3; means ± standard deviations). Asterisks mark P values of <0.01. (E) Effect of mutant HVR1 on virus motility. A549 cells were infected with Alexa-labeled WT AdV5 or AdV5HVR48(1) capsids and immediately analyzed by live-cell microscopy (see Materials and Methods). Shown are time stack projections of capsids (AdV capsid) and individual transmitted light micrographs of representative movies for each AdV. WT AdV5 capsids show long-range motility, whereas AdV5HVR48(1) motility was restricted (see Movies S1 and S2 in the supplemental material). Bar, 10 μm.
DISCUSSION
Role of the low-pH effect on capsid transport.
In previous work, we found that following exposure to low pH, hexon specifically and directly recruits cytoplasmic dynein to AdV5 capsids (5). The pH dependence suggested an important new conformational modification required for virus infection. We have now obtained multiple indications for a pH-dependent change in hexon structure, the timing of which correlates well with dynein recruitment activity.
Adenovirus is generally thought to encounter a low-pH environment in the endocytic uptake vesicle (30), although the effects of low pH on infectivity remain under debate (31). The pH of virus-containing organelles was measured to be 4.6 to 6.0 (32), the range in which hexon priming for dynein recruitment occurs (5; this study). We now find furthermore that the pH-priming effect occurs within 5 min of hexon acidification (Fig. 3B), well within the ∼15-min time span reported previously for endosomal passage (33). Conversely, the ability of hexon to bind dynein after neutralization is slowly reversible, with a half-time of ∼9.2 min and a complete loss of activity by about an hour (Fig. 1). These results are consistent with some of the pH-dependent changes in hexon structure we observed (see below). Our data therefore suggest that upon entry into the cytoplasm, virus particles encounter a “window of opportunity” for dynein recruitment. Particles that recruit dynein in a timely manner should be most likely to reach the nuclear envelope to deliver their DNA. Transport of AdV5 to the nucleus is typically complete by ∼60 min p.i., consistent with the lifetime that we determined here for the hexon-dynein binding state.
pH effects on the hexon tertiary structure.
Our data implicate relatively subtle hexon conformational changes in the reversible pH-dependent control of dynein recruitment (Fig. 2). We obtained no evidence for simple dissociation of the hexon trimer to its subunits at reduced pH, arguing against a role for such a major structural change in dynein recruitment. However, we found that the further addition of SDS leads to dissociation specifically of acidified but not untreated hexon, supporting a more subtle change in hexon structure. The area where SDS acts within hexon is unknown, but its effects provide a useful indicator of the low-pH-induced conformational change. Using this behavior as an assay, we determined that the implied conformation change occurs within 5 min of low-pH treatment. However, reversal upon neutralization took between 30 and 60 min, consistent with the low rate of decay for dynein binding activity (Fig. 3B and C).
Another indicator of a low-pH-induced conformational change in hexon is its sensitization to dispase cleavage (14, 22) (Fig. 4 and 7A). We mapped the hydrolysis site between residues D155 and E156. This site is located within the external loop sequence of HVR1 (11, 34–36), which is accessible to dispase at low pH in both individual hexon trimers and the AdV capsid (Fig. 4A, 5A, and 7B).
In view of the increased sensitivity of hexon to dispase at low pH, we suspect that HVR1 becomes more flexible or more exposed under these conditions. HVR1 resides at the hexon surface at the binding interface between adjacent monomers (Fig. 7B) (35, 36). Hence, subtle structural rearrangements of HVR1 might allow access of SDS to more central parts of the hexon trimer, accounting for the specific effect of low pH on the monomer-trimer equilibrium. We note that dispase hydrolysis of hexon at low pH inhibits both SDS sensitivity and dynein binding activity (Fig. 4A and C). Therefore, it appears that a common low-pH-induced molecular rearrangement is involved in increased dynein affinity and SDS and dispase sensitivities.
To test the role of HVR1 in capsid entry further, we used a chimeric AdV5 mutant containing the much shorter HVR1 sequence of AdV48 [AdV5HVR48(1)] (15). Strikingly, dispase hydrolysis, dynein binding of hexon, redistribution of the virus from the cell surface to the nuclear envelope, and postentry capsid motility were all abrogated or reduced for AdV5HVR48(1) (Fig. 6).
Our results are consistent with a direct contribution of HVR1 to dynein recruitment. How serotype-specific sequence conservation of HVR1 correlates with functional activities during entry and pH regulation beyond the current results remains to be explored.
This work further defines the process of dynein recruitment to the AdV capsid and provides mechanistic insight into the low-pH priming of hexon. It also provides a better understanding of AdV capsids as a model dynein cargo. Dynein-mediated transport of the virus and, thus, successful infection depend on a structural change involving the HVR1 hexon loop, alterations which should allow for experimental control of dynein-mediated virus transport. Finally, we note that HVR1 is the first virus recruitment site for dynein to be identified. Similar sites in other viruses could be of further general value in the development of therapeutic strategies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vincent Racaniello, Yinghui Mao, and Chloe Bulinski as well as members of the Vallee laboratory for critical discussions. We also thank Emily Chen of the Proteomics Shared Resource facility at CUMC for her support with the mass spectrometry experiments.
This work was supported by National Institutes of Health grants GM102347 and GM105536.
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02889-14.
REFERENCES
- 1.Dodding MP, Way M. 2011. Coupling viruses to dynein and kinesin-1. EMBO J 30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greber UF, Way M. 2006. A superhighway to virus infection. Cell 124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 3.Radtke K, Dohner K, Sodeik B. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol 8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 4.Scherer J, Vallee RB. 2011. Adenovirus recruits dynein by an evolutionary novel mechanism involving direct binding to pH-primed hexon. Viruses 3:1417–1431. doi: 10.3390/v3081417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bremner KH, Scherer J, Yi J, Vershinin M, Gross SP, Vallee RB. 2009. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 6:523–535. doi: 10.1016/j.chom.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E, Crystal RG. 2000. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther 11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- 7.Suomalainen M, Nakano MY, Keller S, Boucke K, Stidwill RP, Greber UF. 1999. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol 144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scherer J, Yi J, Vallee RB. 2014. PKA-dependent dynein switching from lysosomes to adenovirus: a novel form of host-virus competition. J Cell Biol 205:163–177. doi: 10.1083/jcb.201307116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suomalainen M, Nakano MY, Boucke K, Keller S, Greber UF. 2001. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J 20:1310–1319. doi: 10.1093/emboj/20.6.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309–319. doi: 10.1016/0092-8674(93)90231-E. [DOI] [PubMed] [Google Scholar]
- 11.Rux JJ, Burnett RM. 2000. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol Ther 1:18–30. doi: 10.1006/mthe.1999.0001. [DOI] [PubMed] [Google Scholar]
- 12.Crawford-Miksza L, Schnurr DP. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol 70:1836–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seth P, Willingham MC, Pastan I. 1985. Binding of adenovirus and its external proteins to Triton X-114. Dependence on pH. J Biol Chem 260:14431–14434. [PubMed] [Google Scholar]
- 14.Everitt E, Persson MJ, Wohlfart C. 1988. pH-dependent exposure of endoproteolytic cleavage sites of the adenovirus 2 hexon protein. FEMS Microbiol Lett 49:229–233. doi: 10.1111/j.1574-6968.1988.tb02721.x. [DOI] [Google Scholar]
- 15.Roberts DM, Nanda A, Havenga MJ, Abbink P, Lynch DM, Ewald BA, Liu J, Thorner AR, Swanson PE, Gorgone DA, Lifton MA, Lemckert AA, Holterman L, Chen B, Dilraj A, Carville A, Mansfield KG, Goudsmit J, Barouch DH. 2006. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441:239–243. doi: 10.1038/nature04721. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence WC, Ginsberg HS. 1967. Intracellular uncoating of type 5 adenovirus deoxyribonucleic acid. J Virol 1:851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maizel JV Jr, White DO, Scharff MD. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115–125. [DOI] [PubMed] [Google Scholar]
- 18.Thiel JF, Smith KO. 1967. Fluorescent focus assay of viruses on cell monolayers in plastic petri plates. Proc Soc Exp Biol Med 125:892–895. doi: 10.3181/00379727-125-32232. [DOI] [PubMed] [Google Scholar]
- 19.Tan SC, Scherer J, Vallee RB. 2011. Recruitment of dynein to late endosomes and lysosomes through light intermediate chains. Mol Biol Cell 22:467–477. doi: 10.1091/mbc.E10-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waris M, Halonen P. 1987. Purification of adenovirus hexon protein by high-performance liquid chromatography. J Chromatogr 397:321–325. doi: 10.1016/S0021-9673(01)85015-9. [DOI] [PubMed] [Google Scholar]
- 21.Paschal BM, Shpetner HS, Vallee RB. 1987. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol 105:1273–1282. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga MJ, Bergman T, Everitt E. 1990. Antibodies with specificities against a dispase-produced 15-kilodalton hexon fragment neutralize adenovirus type 2 infectivity. J Virol 64:4217–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fortsas E, Petric M, Brown M. 1994. Electrophoretic migration of adenovirus hexon under non-denaturing conditions. Virus Res 31:57–65. doi: 10.1016/0168-1702(94)90071-X. [DOI] [PubMed] [Google Scholar]
- 24.Bradley RR, Maxfield LF, Lynch DM, Iampietro MJ, Borducchi EN, Barouch DH. 2012. Adenovirus serotype 5-specific neutralizing antibodies target multiple hexon hypervariable regions. J Virol 86:1267–1272. doi: 10.1128/JVI.06165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu E, Trauger SA, Pache L, Mullen TM, von Seggern DJ, Siuzdak G, Nemerow GR. 2004. Membrane cofactor protein is a receptor for adenoviruses associated with epidemic keratoconjunctivitis. J Virol 78:3897–3905. doi: 10.1128/JVI.78.8.3897-3905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfrum N, Greber UF. 2013. Adenovirus signalling in entry. Cell Microbiol 15:53–62. doi: 10.1111/cmi.12053. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Rhee EG, Masek-Hammerman K, Teigler JE, Abbink P, Barouch DH. 2012. Adenovirus serotype 26 utilizes CD46 as a primary cellular receptor and only transiently activates T lymphocytes following vaccination of rhesus monkeys. J Virol 86:10862–10865. doi: 10.1128/JVI.00928-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teigler JE, Kagan JC, Barouch DH. 2014. Late endosomal trafficking of alternative serotype adenovirus vaccine vectors augments antiviral innate immunity. J Virol 88:10354–10363. doi: 10.1128/JVI.00936-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teigler JE, Iampietro MJ, Barouch DH. 2012. Vaccination with adenovirus serotypes 35, 26, and 48 elicits higher levels of innate cytokine responses than adenovirus serotype 5 in rhesus monkeys. J Virol 86:9590–9598. doi: 10.1128/JVI.00740-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol 79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith JG, Wiethoff CM, Stewart PL, Nemerow GR. 2010. Adenovirus. Curr Top Microbiol Immunol 343:195–224. doi: 10.1007/82_2010_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Fernandez M, Longshaw SV, Kirby I, Santis G, Tobin MJ, Clarke DT, Jones GR. 2004. Adenovirus type-5 entry and disassembly followed in living cells by FRET, fluorescence anisotropy, and FLIM. Biophys J 87:1316–1327. doi: 10.1529/biophysj.103.035444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greber UF, Willetts M, Webster P, Helenius A. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477–486. doi: 10.1016/0092-8674(93)90382-Z. [DOI] [PubMed] [Google Scholar]
- 34.Khare R, Reddy VS, Nemerow GR, Barry MA. 2012. Identification of adenovirus serotype 5 hexon regions that interact with scavenger receptors. J Virol 86:2293–2301. doi: 10.1128/JVI.05760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. 2010. Crystal structure of human adenovirus at 3.5 A resolution. Science 329:1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rux JJ, Kuser PR, Burnett RM. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J Virol 77:9553–9566. doi: 10.1128/JVI.77.17.9553-9566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.