ABSTRACT
Viruses with approximately 50% homology to human influenza C virus (ICV) have recently been isolated from swine and cattle. The overall low homology to ICV, lack of antibody cross-reactivity to ICV in hemagglutination inhibition (HI) and agar gel immunodiffusion assays, and inability to productively reassort with ICV led to the proposal that these viruses represented a new genus of influenza virus, influenzavirus D (IDV). To further our understanding of the epidemiology of IDV, real-time reverse transcription-PCR was performed on a set of 208 samples from bovines with respiratory disease. Ten samples (4.8%) were positive and six viruses were successfully isolated in vitro. Phylogenetic analysis of full-genome sequences of these six new viruses and four previously reported viruses revealed two distinct cocirculating lineages represented by D/swine/Oklahoma/1334/2011 (D/OK) and D/bovine/Oklahoma/660/2013 (D/660), which frequently reassorted with one another. Antigenic analysis using the HI assay and lineage-representative D/OK and D/660 antiserum found up to an approximate 10-fold loss in cross-reactivity against heterologous clade antiserum. One isolate, D/bovine/Texas/3-13/2011 (D/3-13), clustered with the D/660 lineage, but also had high HI titers to heterologous (D/OK) clade antiserum. Molecular modeling of the hemagglutinin esterase fusion protein of D/3-13 identified a mutation at position 212 as a possible antigenic determinant responsible for the discrepant HI results. These results suggest that IDV is common in bovines with respiratory disease and that at least two genetic and antigenically distinct clades cocirculate.
IMPORTANCE A novel bovine influenza virus was recently identified. Detailed genetic and antigenic studies led to the proposal that this virus represents a new genus of influenza, influenzavirus D (IDV). Here, we show that IDV is common in clinical samples of bovine respiratory disease complex (BRDC), with a prevalence similar to that of other established BRDC etiological agents. These results are in good agreement with the near-ubiquitous seroprevalence of IDV previously found. Phylogenetic analysis of complete genome sequences found evidence for two distinct cocirculating lineages of IDV which freely reassort. Significant antigenic differences, which generally agreed with the surface glycoprotein hemagglutinin esterase phylogeny, were observed between the two lineages. Based on these results, and on the ability of IDV to infect and transmit in multiple mammalian species, additional studies to determine the pathogenic potential of IDV are warranted.
INTRODUCTION
Influenza viruses are single-stranded, negative-sense, segmented RNA viruses belonging to the family Orthomyxoviridae (1). Influenza A virus (IAV) and influenza B virus (IBV) both contain eight genomic segments, including two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), whereas influenza C (ICV) has only seven segments with one surface glycoprotein, the hemagglutinin-esterase-fusion (HEF) protein (2, 3). While the vast genetic diversity of IAV is found in waterfowl, only limited subtypes infect mammals. IBV and ICV are found principally in humans and rarely infect other species. IBV is a component of seasonal influenza epidemics with clinically significant disease, while ICV infects most humans during childhood and typically results in mild respiratory symptoms and fever (1, 4–6).
In 2011, an influenza virus with moderate homology to ICV was isolated from swine in Oklahoma (D/swine/Oklahoma/1334/2011 [D/OK]) exhibiting influenza-like symptoms. Sequence analysis showed approximately 50% homology to human ICVs (7). D/OK did not cross-react with antibodies against human ICV in hemagglutination inhibition (HI) and agar gel immunodiffusion (AGID) assays. Limited seroprevalence in swine and humans to D/OK (9.5% and 1.3%, respectively) suggested that an alternate species was the reservoir of this novel virus (7). HI assays of bovine sera found seven out of eight herds with titers greater than 40 to both D/OK and the bovine D/bovine/Oklahoma/660/2013 (D/660) strain (8). Eighteen percent of bovine respiratory disease samples were positive by reverse transcription-PCR (RT-PCR) assay targeting the PB1 gene of D/OK. Virus isolation, genome sequencing, and phylogenetic analysis showed that D/OK and three bovine isolates were closely related and did not reassort with human ICV. Likewise, in vitro reassortment experiments between D/OK and human ICV failed to identify viable reassortant viruses. Reassortment of viral segments can yield viable progeny within the same genera but not across genera of influenza virus (2, 9, 10). Taken together, these results led to the proposal to classify D/OK-like viruses as a new genus of influenza virus, influenzavirus D (IDV), with bovines as the potential reservoir (8).
While the current three genera of influenza virus, influenza A, B, and C viruses, all share similar genetic ancestry, they have diverged over time (2). ICVs undergo reassortment frequently in nature, which results in greater genetic diversity of the viruses (3, 6, 11, 12). ICV is a product of multiple-lineage evolution, a result of cocirculating strains in the human population (6, 10, 13, 14). As influenza B and C viruses have further diverged from IAV, significant mutations resulted in the lack of viable reassortant viruses between influenza B and C viruses, and they both are thought to be evolutionarily stable (10, 15). The discovery of IDV warrants new research into its evolutionary history as well as its epidemiology and ecology.
Bovine respiratory disease complex (BRDC) is the most economically significant disease of the beef industry with losses due to morbidity, mortality, treatment costs, and reduced carcass value (16, 17). Established viral etiological agents include bovine viral diarrhea virus (BVDV), bovine herpesvirus 1 (BHV-1), bovine respiratory syncytial virus (BRSV), and parainfluenza virus type 3 (PI3). In the past several years, there has been increasing evidence that bovine respiratory coronavirus also contributes to BRDC in feedlot cattle (17, 18). The finding of IDV in cattle warrants further investigation into its possible role as a BRDC etiological agent.
To further investigate the epidemiology of this proposed new genus, a large sample set of BRDC cases were screened by quantitative real-time reverse transcription-PCR (qRT-PCR) to determine the molecular epidemiology of IDV in association with other bovine respiratory disease viral agents. Phylogenetic analyses of full-genome sequences, along with hemagglutination inhibition (HI) assays, were performed to characterize the genetic and antigenic diversity of IDV.
MATERIALS AND METHODS
Molecular screening of bovine viruses.
Clinical samples from bovine respiratory disease submissions to the Kansas State University Veterinary Diagnostic Lab (n = 208) were screened by a BRDC PCR panel which detects BVDV, BHV-1, BRSV, PI3, and bovine coronavirus (BCV). The 208 samples, consisting of nasal and pharyngeal swabs and lung tissue, originated from 12 states, primarily in the Midwest: Nebraska (n = 48), Kansas (n = 116), Colorado (n = 6), Missouri (n = 1), Mississippi (n = 7), Texas (n = 4), Oklahoma (n = 2), Idaho (n = 2), Montana (n = 6), Oregon (n = 4), Washington (n = 11), and Virginia (n = 1). Additionally, samples were analyzed for IDV using qRT-PCR (7).
Cells and viruses.
Human rectal tumor cells (HRT-18G) (ATCC CRL-11663) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS). Swine testicle cells (ST cells) (ATCC CRL-1746) were maintained in the same manner. Both cell lines were grown at 37°C with ~5% CO2 before infection with viruses. IDVs were isolated at Newport Laboratories from PCR-positive field samples. D/swine/Oklahoma/1334/2011 (D/OK) virus was previously isolated from swine exhibiting influenza-like symptoms (7). D/bovine/Minnesota/628/2013 (D/628), D/bovine/Minnesota/729/2013 (D/729), and D/bovine/Oklahoma/660/2013 (D/660) were previously isolated from bovine respiratory disease samples submitted for diagnostic testing (8).
Virus isolation.
Virus isolation was performed with IDV qRT-PCR-positive bovine samples using confluent monolayers of HRT-18G cells in DMEM without FBS with 0.5 μg/ml trypsin, at 37°C with ~5% CO2. Viral growth was confirmed by qRT-PCR and a hemagglutination assay using 0.5% washed turkey red blood cells, as IDV is noncytopathic on HRT-18G cells. Following one passage on HRT-18G cells, viruses were passaged to ST cells in DMEM without trypsin. Growth was confirmed by qRT-PCR, a hemagglutination assay, and cytopathic effects (CPE) after each passage. PCR-positive samples failing to show CPE or an increase in qRT-PCR and HA titers were passaged three times before they were called negative.
Full-genome sequencing and analysis.
A MegaMax viral RNA kit was used to isolate the viral RNA. Full-genome sequencing was performed as previously described (8). In brief, sequencing libraries were prepared from full-genome amplicons using a NEBNext Fast DNA kit for Ion Torrent. D/OK (accession numbers JQ922305 to JQ922311) was used as the template to assemble the contigs using the SegMan NGen module from DNAStar. Phylogenetic analysis was performed using MEGA6.0 software using the maximum-likelihood algorithm with 1,000 bootstrap replicates to verify tree topology.
Serology.
Rabbit polyclonal antisera generated using D/660 and D/OK were used in the hemagglutination inhibition (HI) assay. All viruses were assayed in triplicate. Colostrum-deprived/cesarean-derived (CDCD; Struve Laboratories) serum was used as a negative control. The HI test was performed according to the WHO protocol, using turkey red blood cells (19). Mean HI titers and standard deviations were calculated from triplicate data. Heterologous mean HI titers were normalized to mean homologous HI titers (mean heterologous titers/mean homologous titers). Relative HI titers were reported to account for differences in homologous HI titers between D/OK and D/660.
HEF structure modeling.
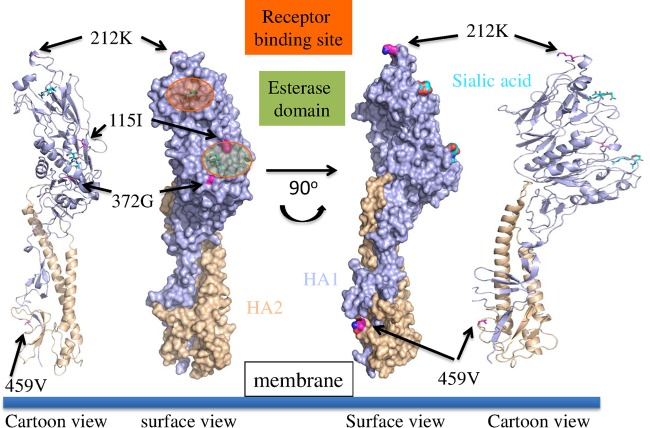
The HEF amino acid sequence of D/bovine/Texas/3-13/2011 (D/3-13) HEF was queried against all sequences in the Protein Data Bank (PDB) structure database using BLAST (20). The HEF structure from ICV C/Johannesburg/1/66 (PDB identifier [ID] 1FLC) (21), which showed the highest sequence identity to D/3-13 HEF (53% sequence identity), was chosen as the template. The sequences of D/3-13 and ICV HEF were then aligned using Muscle (22). The model of D/3-13 HEF was built using Modeler (23). The quality of the structure was checked using Verify 3D (17). For demonstration purposes, the sialic acid binding template is shown on the D/3-13 HEF structure (see Fig. 3).
FIG 3.
Molecular model of D/3-13 HEF depicting cartoon and surface representations. The receptor binding site is in orange, and the esterase domain is in green. Amino acid differences unique to strain D/3-13 are noted.
Nucleotide sequence accession numbers.
Sequences for the six IDVs isolated from clinical samples in this study were deposited in GenBank under accession numbers KM392468 to KM392509.
RESULTS
Molecular epidemiology.
Ten (4.8%) of the 208 samples submitted for BRDC diagnostic testing were positive for IDV by qRT-PCR, with threshold cycle (CT) values ranging from 21 to 29. These samples consisted of nine nasal swabs and one swab of an unknown type. In addition, other viruses associated with BRDC were detected by qRT-PCR, with 36% (n = 75 samples), 7% (n = 15), 5% (n = 11), 3% (n = 7), and <1% (n = 1) of samples positive (CT < 37) for BCV, BHV-1, BVDV, BRSV, and PI3, respectively. Of the 10 samples positive for IDV, one was also positive for BVDV, with a CT of 35. Four of the IDV-positive samples were also positive for BCV, with CT values ranging from 28 to 37. The 10 IDV qRT-PCR-positive samples originated from Kansas, Texas, and Nebraska.
Virus isolation.
Of the 10 samples qRT-PCR positive for IDV, six viruses were isolated in cell culture. All six viruses were initially passaged on HRT-18G cells and were noncytopathic. Viruses were subsequently passaged to ST cells, where CPE was evident. Viral replication was verified after each passage with qRT-PCR and hemagglutination assays. The viruses were designated D/bovine/Kansas/11-8/2012 (D/11-8), D/bovine/Kansas/13-21/2012 (D/13-21), D/bovine/Nebraska/9-5/2012 (D/9-5), D/bovine/Texas/3-13/2011 (D/3-13), D/bovine/Kansas/1-35/2010 (D/1-35), and D/bovine/Kansas/14-22/2012 (D/14-22).
Full-genome sequencing and analysis.
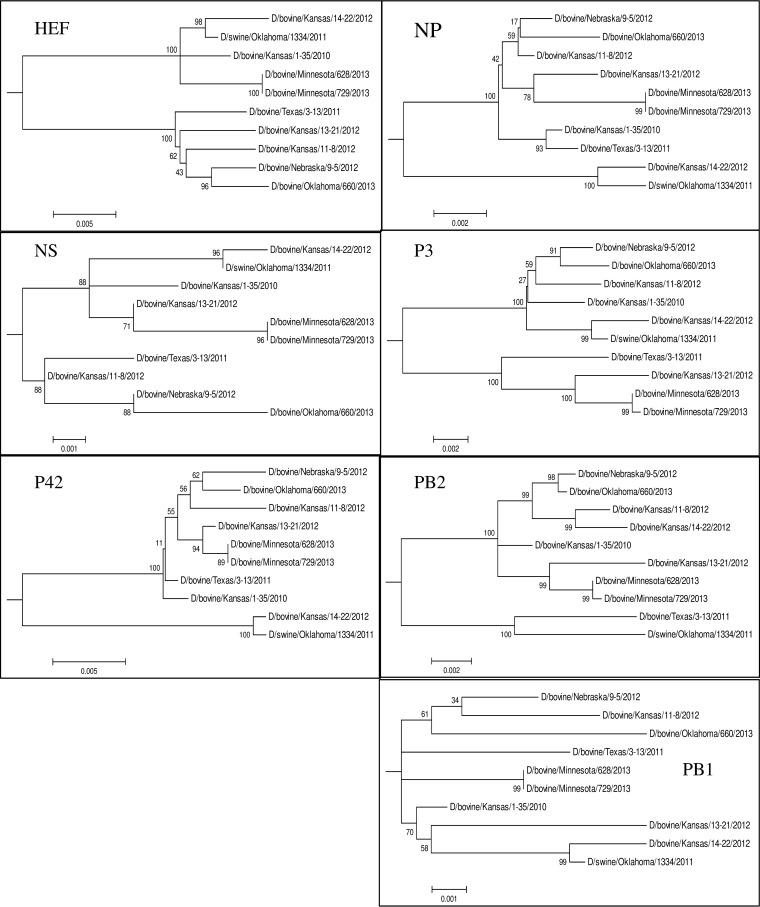
Full-genome sequences were determined for the six IDVs isolated from clinical samples in cell culture. Phylogenetic analysis of each segment was performed for 10 viruses, including nine bovine viruses (D/660, D/628, D/729, D/14-22, D/1-35, D/3-13, D/13-21, D/11-8, and D/9-5) and one porcine virus (D/OK) (Fig. 1). Clustal W alignment showed that HEF was the most divergent segment, with sequence homology ranging from 96.1 to 99.8% among all IDV strains. The phylogenetic analysis showed two distinct lineages of IDV present in cattle, as was evident by two well-defined clades for each segment with the exception of PB1. The PB1 segment showed the highest homology among strains (>99.3% identity), and distinct lineages could not be determined. Incongruency in tree topology for each segment suggests reassortment between the two lineages, resulting in seven distinct genotypes (Table 1). Pairwise amino acid alignments identified residues conserved among bovine viruses compared to the swine D/OK (listed are the D/OK amino acid residue, position, and bovine amino acid): HEF A251T, K252T/A; NP E247D, K381E; P42 C91S; PB2 V521I/M (see Table S1 in the supplemental material).
FIG 1.
Phylogenetic trees of the seven genomic segments of IDV. Maximum-likelihood analysis in combination with 1,000 bootstrap replicates was used to derive trees based on nucleotide sequences of genome segments. Bootstrap values are shown above and to the left of the major nodes. Scale bars indicate the number of substitutions per site.
TABLE 1.
Influenza D virus genotypes determined by phylogenetic analysisa
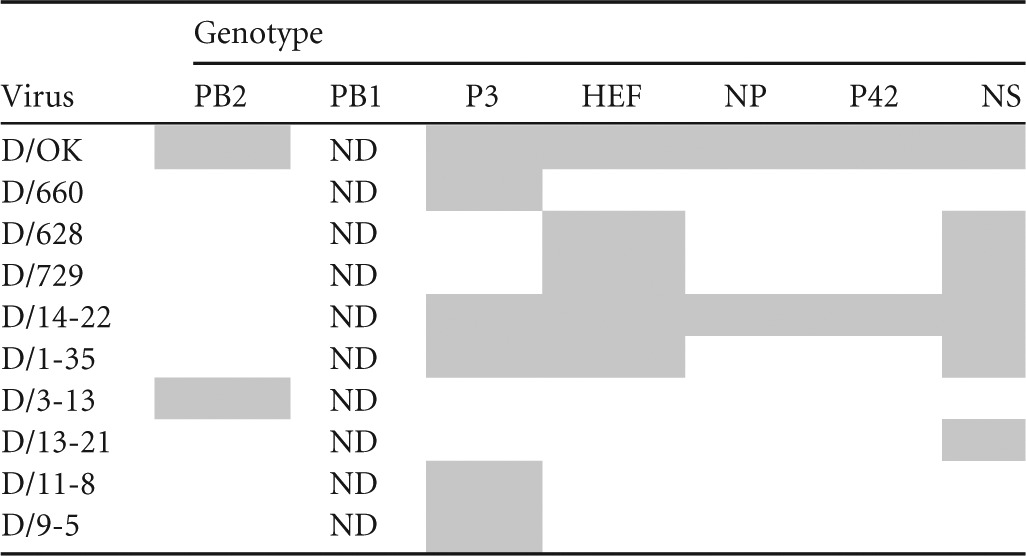
Gray shading indicates segments clustering with D/OK. ND indicates lineage not determined due to high overall homology.
Serology.
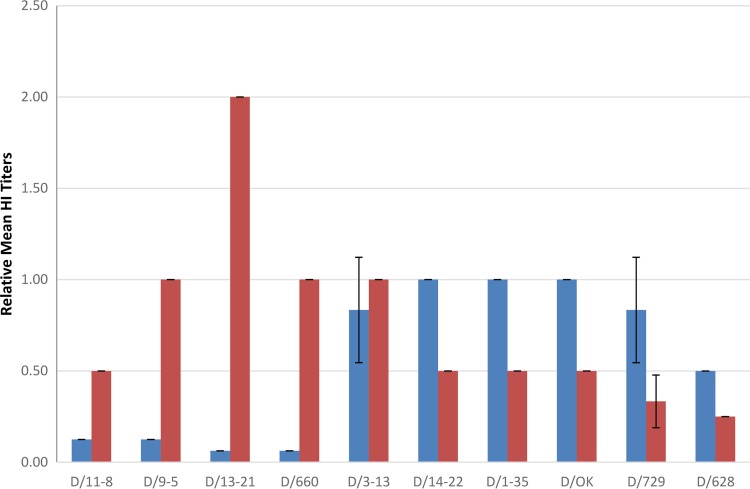
To investigate antigenic differences between the two clades of IDV, HI assays were performed using polyclonal antiserum generated against D/OK and D/660. Homologous HI titers for D/OK and D/660 were 1280 and 320, respectively. Heterologous mean HI titers against D/OK and D/660 antisera were normalized to homologous D/OK and D/660 titers. D/660 clade viruses all reacted most strongly with D/660 antiserum, and D/OK clade viruses reacted most strongly with D/OK antiserum (Fig. 2). The exception was D/3-13, which clustered more closely with the D/660 clade but showed near-equivalent cross-reactivity with D/OK antisera (D/660 relative HI titer, 1.0; D/OK relative HI titer, 0.83). The clade of viruses represented by D/660 all had relative titers to D/660 antiserum ranging from 0.5 to 2.0. The D/660 clade had relative HI titers to the heterologous D/OK antiserum ranging from 0.06 to 0.13, with the exception of D/3-13 (relative HI titer, 0.83). D/OK clade viruses had relative HI titers to homologous D/OK antiserum ranging from 0.5 to 1.0 and heterologous relative HI titers to D/660 ranging from 0.25 to 0.50. While further studies are needed to better evaluate the genetic and antigenic relationships of IDV, these results demonstrate antigenic differences in good agreement with HEF phylogeny.
FIG 2.
Relative mean hemagglutination inhibition titers for 10 influenza D viruses from triplicate data. Blue and red bars represent relative mean HI titers to D/OK and D/660 antisera, respectively. Relative mean HI titers were calculated by normalizing mean heterologous HI titers to mean homologous HI titers (mean heterologous HI titer/mean homologous HI titer). Error bars represent standard deviations.
HEF modeling.
The molecular structure of the HEF protein of D/3-13 (D/660 clade) was modeled to investigate the genetic basis for the cross-reactive mean relative HI titers observed with heterologous antiserum (Fig. 3). One interesting difference is that in the HEF protein of D/3-13, position 212 is occupied by K rather than R, which is present in all other viruses of the D/660 clade. K212 is also found in viruses of the D/OK clade. This position is located at the apex of the HA1 receptor binding domain. Previous antigenic studies with ICV suggested that residues 178 to 284 are likely involved in the binding to cellular receptors. Additionally, amino acid changes at residue 212 were shown to decrease hemagglutinating activity (24, 25). D/3-13 also differs from other IDVs in having the following unique mutations: V115I and D372G in the esterase domain and I459V in the fusion peptide region of HA2 (see Table S1 in the supplemental material).
DISCUSSION
Previous work found that IDV is ubiquitous in cattle, with 87.5% of herds and individuals having HI titers greater than 40 (8). Here, we expand upon this work by showing that IDV is commonly detected in clinical BRDC diagnostic submissions. Aside from BCV, which was detected in 36% of our samples, IDV incidence was similar to that of other recognized BRDC pathogens, such as BHV-1, BVDV, and BRSV (3 to 7%). These results, along with a paucity of titers to IDV in humans and pigs, suggest that bovines are a reservoir for IDV. Further diagnostic testing is needed to determine if other species can serve as IDV maintenance hosts.
Previous phylogenetic analyses found that IDV is most closely related to ICV; the highly conserved PB1 segment has 72% identity to the human C/Ann Arbor/1/1950 reference strain (7), whereas the divergent HEF segment has only 53% identity to this strain. ICV resides in a human reservoir and is comprised of six antigenic and genetic lineages with global distribution (3). It is thought to be evolutionarily stable, with its HEF having an evolutionary rate nine times slower than that of IAV HA (1, 3, 26). Reassortment can occur between different ICVs in nature when different lineages cocirculate in close proximity, giving rise to further genetic variation of ICV (1, 11). Full-genome sequencing was performed for six IDV isolates to further elucidate its genetic diversity and ecology. Phylogenetic analysis found two well-supported clades for all gene segments with the exception of the highly conserved PB1. Reassortment between the two clades is common, as seven distinct genotypes were determined among the 10 genomes analyzed. Antigenic analysis of IDV found a generally good correlation between HEF phylogeny and reactivity in the HI assay. Altogether, these results suggest that at least two lineages of IDV cocirculate in U.S. cattle herds and frequently reassort. IDV, like ICV, consists of a single subtype with cocirculating lineages. IDV is endemic in cattle, as ICV is in humans, though both are capable of infecting other species.
IAV infects multiple species; however, the majority of the genetic diversity is found in its waterfowl reservoir. Reassortment between avian and nonavian viruses sporadically leads to antigenic shift, which can result in IAV epidemics and pandemics. Although ICV has been isolated occasionally from dogs and pigs, it is not found in avian hosts (10). While common in bovines, IDV can also replicate and transmit in ferrets and pigs (7). Importantly, D/OK was originally isolated from pigs with influenza-like illness. The possibility of alternate IDV hosts and interspecies transmission warrants further research. As in swine seroprevalence studies, only low titers to IDV were detected in human sera, suggesting an infection-naive population.
The finding of five of the 10 clinical BRDC samples positive only for IDV suggests a possible primary role for IDV in BRDC, as BRDC pathogenesis is thought to involve a primary viral infection followed by secondary bacterial pneumonia. An unexpected finding was the high incidence of BCV in the BRDC diagnostic submissions. Of the five samples that were positive for both IDV and BRDC pathogens, four were also positive for BCV. BCV can cause respiratory or enteric disease in cattle. Both BCV and ICV utilize N-acetyl-9-O-acetylneuraminic acid (Neu5,9Ac2) as a receptor determinant for attachment (27). Both viruses also possess acetylesterase activity, which releases 9-O-acetyl residues from sialic acids, thereby acting as a receptor-destroying enzyme. Previous work demonstrated that the BCV acetylesterase removed receptors for BCV, but also for ICV, and vice versa (28). A mutant ICV that showed broader cell tropism required less Neu5,9Ac2 on the cell surface receptor (29). Interestingly, D/OK virus demonstrated much broader cell tropism than that of human ICV (7). Further research is needed to study the possible synergy between these viruses and their etiological role in BRDC.
The HI results and molecular model of the D/3-13 HEF suggested that amino acid 212 may play a role in IDV antigenicity. Previous ICV studies have shown that changes in or around the variable region from positions 180 to 214 can result in the loss of recognition by antibodies and that K212 is directly involved in antibody recognition (26). The D/OK HEF clade of viruses possess K212, whereas the D/660 HEF clade has R212. Despite genetically belonging to the D/660 clade, D3/13, which has K replacing R at position 212, had higher titers to heterologous D/660 antiserum. Future research, ideally utilizing reverse genetics analysis, is needed to conclusively demonstrate whether K212 is a determinant of antibody binding in D/OK clade viruses. The amino acid sites specific to bovine strains could also be further evaluated to determine their role in host restriction, though there is only one swine isolate (D/OK) available for comparison.
The presence of cocirculating lineages indicates that IDV is endemic in cattle and that these viruses reassort frequently. The observed antigenic differences between strains could be of importance if IDV's role as a bovine pathogen contributing to BRDC is confirmed. Recently, three IDVs were isolated from bovines in Shandong province in China with 95.35 to 99.22% homology to the IDV strains found in the United States (30). These results suggest that IDV has a global distribution in cattle. Additional studies are needed to investigate the transmissibility and pathogenicity of IDV in bovines, as well as its etiological role in BRDC, both alone and in the presence of other BRDC pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dick Hesse and Barbara Breazeale from the Kansas State University Veterinary Diagnostic Laboratory for providing bovine clinical samples.
Work done in the Feng Li lab was supported in part by South Dakota State University (SDSU) AES 3AH-477 and SD 2010 Research Center (Biological Control and Analysis of Applied Photonics [BCAAP]) Fund SJ163 and NIH/NIAID AI107379. Work performed in the W. Ma lab was partially funded by Kansas State University Startup Funds SRO001, European Commission FP7-GA258084, and the Department of Homeland Security Center of Excellence for Emerging and Zoonotic Animal Diseases, grant 2010-ST061-AG0001.
E.A.C. and B.M.H. were employed by Newport Laboratories, Inc., at the time of this study. Newport Laboratories, Inc., is a for-profit organization that offers diagnostic testing and produces vaccines for the livestock industries, one of the funding sources for this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02718-14.
REFERENCES
- 1.Muraki Y, Hongo S. 2010. The molecular virology and reverse genetics of influenza C virus. Jpn J Infect Dis 63:157–165. [PubMed] [Google Scholar]
- 2.Bouvier NM, Palese P. 2008. The biology of influenza viruses. Vaccine 26:D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speranskaya AS, Melnikova NV, Belenikin MS, Dmitriev AA, Oparina NY, Kudryavtseva AV. 2012. Genetic diversity and evolution of the influenza C virus. Russian J Genet 48:671–678. doi: 10.1134/S1022795412070149. [DOI] [PubMed] [Google Scholar]
- 4.Katagiri S, Ohizumi A, Homma M. 1983. An outbreak of type-C influenza in a childrens home. J Infect Dis 148:51–56. doi: 10.1093/infdis/148.1.51. [DOI] [PubMed] [Google Scholar]
- 5.Yuanji G, Desselberger U. 1984. Genome analysis of influenza C viruses isolated in 1981/82 from pigs in China. J Gen Virol 65:1857–1872. doi: 10.1099/0022-1317-65-11-1857. [DOI] [PubMed] [Google Scholar]
- 6.Matsuzaki Y, Abiko C, Mizuta K, Sugawara K, Takashita E, Muraki Y, Suzuki H, Mikawa M, Shimada S, Sato K, Kuzuya M, Takao S, Wakatsuki K, Itagaki T, Hongo S, Nishimura H. 2007. A nationwide epidemic of influenza C virus infection in Japan in 2004. J Clin Microbiol 45:783–788. doi: 10.1128/JCM.01555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F. 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 9:e1003176. doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, Wang D, Nelson EA, Li F. 2014. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae family. mBio 5(2):e00031-14. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCauley JW, Hongo S, Kaverin NV, Kochs G, Lamb RA, Matrosovich MN, Perez DR, Palese P, Presti RM, Rimstad E. 2012. Orthomyxoviridae, p 749–761 InKing AMQ, Adams MJ, Carstens EB, Lefkowitz E (ed), Virus taxonomy, classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, New York, NY. [Google Scholar]
- 10.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng G, Hongo S, Kimura H, Muraki Y, Sugawara K, Kitame F, Numazaki Y, Suzuki H, Nakamura K. 1996. Frequent occurrence of genetic reassortment between influenza C virus strains in nature. J Gen Virol 77:1489–1492. doi: 10.1099/0022-1317-77-7-1489. [DOI] [PubMed] [Google Scholar]
- 12.Matsuzaki Y, Mizuta K, Sugawara K, Tsuchiya E, Muraki Y, Hongo S, Suzuki H, Nishimura H. 2003. Frequent reassortment among influenza C viruses. J Virol 77:871–881. doi: 10.1128/JVI.77.2.871-881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuzaki Y, Muraki Y, Sugawara K, Hongo S, Nishimura H, Kitame F, Katsushima N, Numazaki Y, Nakamura K. 1994. Cocirculation of two distinct groups of influenza C virus in Yamagata City, Japan. Virology 202:796–802. doi: 10.1006/viro.1994.1401. [DOI] [PubMed] [Google Scholar]
- 14.Matsuzaki Y, Takao S, Shimada S, Mizuta K, Sugawara K, Takashita E, Muraki Y, Hongo S, Mishimura H. 2004. Characterization of antigenically and genetically similar influenza C viruses isolated in Japan during the 1999-2000 season. Epidemiol Infect 132:709–720. doi: 10.1017/S0950268804002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheng Z, Ran Z, Wang D, Hoppe AD, Simonson R, Chakravarty S, Hause BM, Li F. 2013. Genomic and evolutionary characterization of a novel influenza-C-like virus from swine. Arch Virol 159:249–255. doi: 10.1007/s00705-013-1815-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor JD, Fulton RW, Lehenbauer TW, Step DL, Confer AW. 2010. The epidemiology of bovine respiratory disease: what is the evidence for predisposing factors? Can Vet J 51:1095–1102. [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenberg D, Luthy R, Bowie JU. 1997. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404. doi: 10.1016/S0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 18.Saif LJ. 2010. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract 26:349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO manual on animal influenza diagnosis and surveillance. 2002. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 20.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 22.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2007. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci Chapter 2:Unit 2.9. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 24.Sugawara K, Nishimura H, Hongo S, Muraki Y, Kitame F, Nakamura K. 1993. Construction of an antigenic map of the haemagglutinin-esterase protein of influenza C virus. J Gen Virol 72:1661–1666. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki M, Sugawara K, Adachi K, Hongo S, Nishimura H, Kitame F, Nakamura K. 1992. Location of neutralizing epitopes on the hemagglutinin-esterase protein of influenza C virus. Virology 189:79–87. doi: 10.1016/0042-6822(92)90683-G. [DOI] [PubMed] [Google Scholar]
- 26.Muraki Y, Hongo S, Sugawara K, Kitame F, Nakamura K. 1996. Evolution of the haemagglutinin-esterase gene of influenza C virus. J Gen Virol 77:673–679. doi: 10.1099/0022-1317-77-4-673. [DOI] [PubMed] [Google Scholar]
- 27.Schultze B, Gross HJ, Brossmer R, Klenk HD, Herrler G. 1990. Hemagglutinating encephalomyelitis virus attaches to N-acetyl-9-O-acetylneuraminic acid-containing receptors on erythrocytes: comparison with bovine coronavirus and influenza C virus. Virus Res 16:185–194. doi: 10.1016/0168-1702(90)90022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vlasak R, Luytjes W, Spaan W, Palese P. 1988. Human and bovine coronaviruses recognize sialic acid containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci U S A 85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szepanski S, Gros HJ, Brossmer R, Klenk HD, Herrler G. 1992. A single point mutation of the influenza C virus glycoprotein (HEF) changes the viral receptor-binding activity. Virology 188:85–92. doi: 10.1016/0042-6822(92)90737-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang WM, Wang SC, Peng C, Yu JM, Zhuang QY, Hou GY, Liu S, Li JP, Chen JM. 21August2014. Identification of a potential novel type of influenza virus in bovine in China. Virus Genes in press. doi: 10.1007/S11262-014-1107-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.