ABSTRACT
Iron is an essential nutrient for nearly all living organisms, including both hosts and invaders. Proteins such as ferritin regulate the iron levels in a cell, and in the event of a pathogenic invasion, the host can use an iron-withholding mechanism to restrict the availability of this essential nutrient to the invading pathogens. However, pathogens use various strategies to overcome this host defense. In this study, we demonstrated that white spot syndrome virus (WSSV) protein kinase 1 (PK1) interacted with shrimp ferritin in the yeast two-hybrid system. A pulldown assay and 27-MHz quartz crystal microbalance (QCM) analysis confirmed the interaction between PK1 and both ferritin and apoferritin. PK1 did not promote the release of iron ions from ferritin, but it prevented apoferritin from binding ferrous ions. When PK1 was overexpressed in Sf9 cells, the cellular labile iron pool (LIP) levels were elevated significantly. Immunoprecipitation and atomic absorption spectrophotometry (AAS) further showed that the number of iron ions bound by ferritin decreased significantly at 24 h post-WSSV infection. Taken together, these results suggest that PK1 prevents apoferritin from iron loading, and thus stabilizes the cellular LIP levels, and that WSSV uses this novel mechanism to counteract the host cell's iron-withholding defense mechanism.
IMPORTANCE We show here that white spot syndrome virus (WSSV) ensures the availability of iron by using a previously unreported mechanism to defeat the host cell's iron-withholding defense mechanism. This defense is often implemented by ferritin, which can bind up to 4,500 iron atoms and acts to sequester free iron within the cell. WSSV's novel counterstrategy is mediated by a direct protein-protein interaction between viral protein kinase 1 (PK1) and host ferritin. PK1 interacts with both ferritin and apoferritin, suppresses apoferritin's ability to sequester free iron ions, and maintains the intracellular labile iron pool (LIP), and thus the availability of free iron is increased within cells.
INTRODUCTION
Iron is essential for almost all living organisms. In most living cells, the major iron storage protein is ferritin, which acts to modulate the cellular labile iron pool (LIP) and prevents the cell from being damaged by oxidative stress (1, 2). In vertebrates, ferritin is a spherical protein complex composed of 24 subunits of heavy chains and light chains in various different proportions, and one ferritin complex can bind up to 4,500 iron atoms (3). In addition to modulating iron storage, ferritin also acts as part of the host's innate immune system by providing an iron-withholding defense mechanism (4). However, pathogens have been shown to use several different strategies to counter this defense. Bacterial, fungal, or protozoan pathogens can extract iron from the host by (i) binding to ferrated transferrin or lactoferrin on the cell surface to extract the iron content, (ii) synthesizing low-molecular-mass siderophores to extract iron from transferrin or ferritin and then ingesting these ferrated siderophores, (iii) lysing erythrocytes and binding and assimilating heme from digested hemoglobin, (iv) assimilating iron from the host LIP, and (v) accelerating ferritin degradation (5–8). Viral pathogens, on the other hand, secure a source of iron by interfering with host iron homeostasis. For example, the human cytomegalovirus (HCMV) protein US2 alters the iron homeostasis of the host cell by inducing the proteasomal degradation of HFE, a major histocompatibility complex (MHC) class I-like protein which is involved in iron metabolism (9, 10). Another example is the adenovirus protein E1A, which suppresses transcription of the ferritin heavy chain either by targeting FER-1, an enhancer element upstream of the ferritin heavy chain gene (11), or by interfering with the formation of B-box binding factors to reduce cyclic AMP (cAMP) induction (12).
Recently there have been several reports suggesting that ferritin is directly involved in the innate defenses of crustaceans against pathogens and other stressors. For example, the expression level of the ferritin gene was upregulated in Penaeus japonicus shrimps that are resistant to white spot syndrome virus (WSSV) (13), while in Fenneropenaeus chinensis, the ferritin gene expression level was elevated not only by WSSV infection but also after treatment with Zn2+ and Cu2+ (14). The gene expression level of ferritin was also upregulated in WSSV-infected Litopenaeus vannamei and in Penaeus monodon shrimps that were infected with yellow head virus (YHV) (15, 16). Furthermore, injection of ferritin protein into L. vannamei increased the survival rate in shrimps challenged by WSSV (17). Until now, however, it has remained unknown how WSSV interacts with host ferritin.
In the present study, we used a yeast two-hybrid experiment to find that shrimp ferritin interacts with WSSV protein kinase 1 (PK1), which is one of the two serine/threonine protein kinases (PK1 and PK2) expressed by this unique DNA virus (accession number AF440570) (18–20). pk1 is an early gene that encodes an ∼87-kDa polypeptide which contains the major conserved subdomains present in eukaryotic protein kinases (18). Injection of pk1-specific double-stranded RNA (dsRNA) was shown to decrease the cumulative mortality of shrimps after WSSV challenge, leading Kim et al. (21) to suggest that PK1 is essential for WSSV infection. Nevertheless, the role of PK1 in the pathogenesis of WSSV infection remains unknown. Here we used an iron ion binding assay to show that PK1 interferes with apoferritin's ability to incorporate iron. We also showed that overexpression of PK1 in Sf9 cells elevates the level of LIP and that PK1's interaction with ferritin affects host iron homeostasis after WSSV infection.
MATERIALS AND METHODS
Yeast two-hybrid screening for candidate proteins that interact with Lvferritin.
To prepare the bait plasmid, the DNA fragment encoding L. vannamei ferritin (Lvferritin) was amplified by PCR using the specific Lvferritin-F-NdeI/Lvferritin-R-BamHI primer set listed in Table 1. The PCR products were cloned into pGBKT7-1, which is a modified pGBKT7-Rec vector in which the multiple-cloning-site sequence has been replaced by the sequence of pGADT7-Rec (23), together with the GAL4 DNA binding domain (BD). The resulting plasmid, Lvferritin-pGBKT7-1, was then transformed into Saccharomyces cerevisiae strain Y187 (Clontech) to express Lvferritin tagged with c-myc and fused with the BD domain (c-myc-Lvferritin-BD). Transformation was performed in an Eppendorf tube containing competent, freshly prepared yeast strain Y187 cells that had been treated with salmon sperm DNA (Invitrogen). The mixture was heat shocked at 45°C for 30 min, after which the cells were collected by centrifugation, resuspended in sterilized double-distilled water (ddH2O), and spread on an SD/−Trp agar plate.
TABLE 1.
PCR primers used in this studya
| Primer | Sequence (5′–3′) | Usage |
|---|---|---|
| Lvferritin-F-NdeI | GGAATTCCATATGGCCAGCCAAGTCCGC | Plasmid construction (Lvferritin-pGBKT7-1 and Lvferritin-pGADT7-Rec) |
| Lvferritin-R-BamHI | CGCGGATCCTTAGTGGAGTTCCTTATCAAAC | Plasmid construction (Lvferritin-pGBKT7-1 and Lvferritin-pGADT7-Rec) |
| Lvferritin-F-BamHI | CGGGATCCATGGCCAGCCAAGTCCGC | Plasmid construction (Lvferritin-pET28a) |
| Lvferritin-R-NotI | AAGGAAAAAAGCGGCCGCTTAGTGGAGTTCCTTATCAAAC | Plasmid construction (Lvferritin-pET28a) |
| pk1-F-NdeI | CATCCATATGATGGAGGGTGGGGACCAAC | Plasmid construction (pk1-pGBKT7-1 and pk1-pGADT7-Rec) |
| pk1-R-EcoRI | GATCGAATTCCTACTTAACCTTCTTGCTATAC | Plasmid construction (pk1-pGBKT7-1 and pk1-pGADT7-Rec) |
| pk1-F-EcoRI | CATCGAATTCATGGAGGGTGGGGACCAAC | Plasmid construction (pk1-pET28a) |
| pk1-R-XhoI | GATCCTCGAGCTACTTAACCTTCTTGCTATAC | Plasmid construction (pk1-pET28a) |
| pk1-F-BamHI | CGCGGATCCATGGAGGGTGGGGACCAAC | Plasmid construction (pk1-pDHsp70/V5-His) |
| pk1-R-NotI | AAGGAAAAAAGCGGCCGCCTTAACCTTCTTGCTATACAATG | Plasmid construction (pk1- pDHsp70/V5-His) |
| gst-F-EcoRI | CCGAATTCATGTCCCCTATACTAGGTTATTG | Plasmid construction (gst-pET28a) |
| gst-R-XhoI | CCGCTCGAGTTAATCCGATTTTGGAGGATGGTC | Plasmid construction (gst-pET28a) |
A prey library containing WSSV proteins fused with the GAL4 activation domain (AD) was constructed using a Matchmaker Library Construction & Screening kit (Clontech) according to the manufacturer's instructions, with some modifications. Library construction was initially based on the 202 WSSV open reading frames (ORFs) in the Taiwan isolate (GenBank accession no. AF440570) that were predicted to encode functional proteins and which were also present in either the China isolate (GenBank accession no. AF332093) or the Thailand isolate (GenBank accession no. AF369029). ORFs with nucleotide sequences longer than 3 kbp were divided into 2 to 4 subfragments.
The WSSV ORF fragments were amplified by two-step PCR. In the first PCR, the WSSV Taiwan isolate genomic DNA was used as a template, and the ORF fragments were amplified with gene-specific primer sets. The PCR products were then used as templates in the second PCR. The forward (AD-F) and reverse (AD-R) primers used in the second PCR were designed to contain the SMARTIII and CDSIII sequences, respectively, which are homologous to sequences in the prey vector, pGADT7-Rec (Clontech). The SMARTIII/CDSIII-containing PCR products were then cotransformed with SmaI-linearized pGADT7-Rec into S. cerevisiae strain AH109 (Clontech) and spread on SD/−Leu agar plates to select the transformed clones. The prey DNA inserts were checked with colony PCR, and the clones with recombinant plasmids were transferred into 96-well plates containing SD/−Leu medium and then cultured at 30°C. A total of 172 clones were found to contain inserts of the correct size, and these were added to the WSSV AD library.
High-throughput screening was performed by the yeast mating method. First, after the Lvferritin-pGBKT7-1-transformed yeast strain Y187 had grown in 5 ml SD/−Trp medium for 16 h at 30°C, an array of 2-μl spots of this bait culture was dropped on a yeast extract-peptone-dextrose (YPD) agar plate and dried at room temperature. Two microliters of each of the prey cultures in the AD arrays were then dropped on the bait culture spots and again allowed to dry at room temperature. The YPD agar plates were then cultured at 30°C for 24 h to allow the yeast to mate. To select the diploid cells, the yeast colonies that grew on the YPD plates were transferred to SD/−Leu/−Trp agar plates and incubated at 30°C for 3 days. To further screen the positive clones, the diploid cells were progressively transferred to higher-stringency selection medium, first to SD/−Leu/−Trp/−His (SD/−3) agar plates and then to SD/−Leu/−Trp/−His/−Ade/+X-α-gal (SD/−4/+X-α-gal) agar plates. Meanwhile, for the positive and negative controls, the competent cells were transformed with plasmids supplied by the manufacturer (Clontech) and subjected to the same yeast mating procedures.
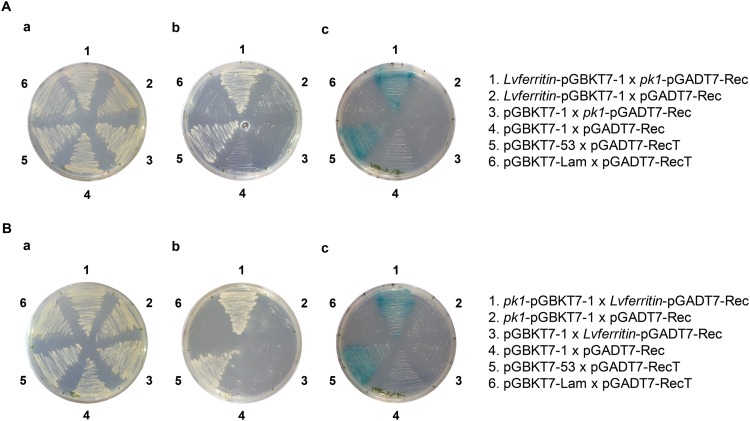
The only prey culture that grew successfully on the SD/−4 plates was WSSV482 (Fig. 1A), which represents the WSSV pk1 gene. This preliminary result was confirmed by domain swapping using a cotransformation-selection procedure. The DNA fragments encoding PK1 and Lvferritin were amplified by PCR using the specific pk1-F-NdeI/pk1-R-EcoRI and Lvferritin-F-NdeI/Lvferritin-R-BamHI primer sets, respectively (Table 1). The resulting PCR products were then cloned into the pGBKT7-1 and pGADT7-Rec vectors, respectively, and the resulting plasmids were named pk1-pGBKT7-1 and Lvferritin-pGADT7-Rec. To cotransform the yeast, equal amounts of bait and prey vectors (pk1-pGBKT7-1 and Lvferritin-pGADT7-Rec) were added to an Eppendorf tube containing freshly prepared competent cells (S. cerevisiae strain AH109). The cells were then heat shocked, collected by centrifugation, and resuspended in sterilized ddH2O as described above. The resuspended cells were then spread on an SD/−Leu/−Trp agar plate. To select the positive clones, the transformed cells that grew on the SD/−Leu/−Trp medium were then transferred to higher-stringency selection medium, first to an SD/−Leu/−Trp/−His agar plate and then to an SD/−Leu/−Trp/−His/−Ade/+X-α-gal agar plate. To exclude the possibility of autonomous activation, the pk1-pGBKT7-1 and Lvferritin-pGADT7-Rec plasmids were cotransformed with the appropriate empty vectors (i.e., pk1-pGBKT7-1 with pGADT7-Rec and pGBKT7-1 with Lvferritin-pGADT7-Rec). For the positive and negative controls, the competent cells were cotransformed with plasmids supplied by the manufacturer, i.e., pGBKT7-53 (p53) and pGADT7-RecT (simian virus 40 large T antigen) for the positive control and pGBKT7-Lam (human lamin C) and pGADT7-RecT for the negative control. The empty vectors pGBKT7-1 and pGADT7-Rec were also cotransformed into the AH109 cells to serve as another negative control.
FIG 1.
WSSV PK1 interacted with host ferritin in the yeast two-hybrid system. (A) Yeast mating. The indicated pairs of plasmid constructs were transformed into two strains of yeast (Y187 and AH109) and grown on SD/−Leu/−Trp (a) and then on agar plates with higher-stringency media, i.e., SD/−Leu/−Trp/−His (b) and SD/−Leu/−Trp/−His/−Ade/+X-α-Gal (c). Growth of yeast on the two high-stringency plates was observed only with Lvferritin-pGBKT7-1/pk1-pGADT7-Rec, pk1-pGBKT7-1/Lvferritin-pGADT7-Rec, and the positive control. (B) To confirm these interactions in a cotransformation domain-swapping experiment, the indicated plasmid constructs were cotransformed into S. cerevisiae strain AH109 and screened with the highest-stringency medium, i.e., SD/−Leu/−Trp/−His/−Ade/+X-α-Gal agar. Growth of cotransformants on this medium was observed only when the yeast was cotransformed with pk1-n-pGBKT7-1/ferritin-pGADT7-Rec or with the positive control.
Expression and purification of His-PK1, His-ferritin, and His-GST.
DNA fragments encoding the full length of WSSV PK1 and L. vannamei ferritin were amplified from the cDNA of WSSV-infected shrimps by using the specific pk1-F-EcoRI/pk1-R-XhoI and Lvferritin-F-BamHI/Lvferritin-R-NotI primer sets, respectively (Table 1). Fragments encoding the glutathione S-transferase (GST) protein were amplified from the vector pGEX4T-1 (GE Healthcare) by using the primer set gst-F-EcoRI/gst-R-XhoI (Table 1). The amplified fragments were cloned into pET28a(+) (Novagen) to generate recombinant plasmids that were named pk1-pET28a, ferritin-pET28a, and gst-pET28a, respectively. The resulting plasmids were transformed into Escherichia coli strain Rosetta 2 cells. The transformants were inoculated into LB broth and incubated at 37°C for 16 h. The cells were further subcultured in fresh LB medium at a ratio of 1:100 and grown at 37°C until the optical density at 600 nm (OD600) of the culture reached 0.4 to 0.6. Expression of the His-PK1, His-ferritin, and His-GST recombinant proteins was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM, followed by incubation at 15°C for 20 h.
For native His-PK1 and His-GST protein purification, the respective IPTG-induced cells were resuspended in lysis buffer (20 mM Tris base, 300 mM NaCl, 10 mM imidazole, pH 7.0) containing 1 mM phenylmethylsulfonyl fluoride (PMSF). After sonication on ice, the cell debris was removed by centrifugation, and the soluble fraction was passed through a column containing Ni-Sepharose 6 Fast Flow beads (GE Healthcare). The beads were then washed several times with wash buffer 1 (20 mM Tris base, 300 mM NaCl, 50 mM imidazole, pH 7.0) and wash buffer 2 (20 mM Tris base, 300 mM NaCl, 100 mM imidazole, pH 7.0). The recombinant protein was eluted with elution buffer (20 mM Tris base, 300 mM NaCl, 250 mM imidazole, pH 7.0), and the protein concentration was measured by the Bradford method, using protein assay reagent dye (Bio-Rad).
His-ferritin was purified under denaturing conditions. Briefly, the IPTG-induced cells were resuspended with 20 mM Tris-HCl containing 8 M urea (pH 8.0) and disrupted by sonication at room temperature. After removing the cell debris, the soluble fraction was mixed with Talon metal-affinity resin (Clontech Laboratories) and constantly inverted at 25°C for 30 min. After binding, the column was washed with 20 mM Tris-HCl containing 8 M urea (pH 8.0) and 20 mM Tris-HCl (pH 8.0), and the recombinant protein was eluted with 200 mM imidazole. This purified, denatured His-ferritin was used as the antigen to raise the anti-Lvferritin antibody in guinea pigs.
Pulldown assays of the interaction between PK1 and ferritin or apoferritin.
Ferritin is a highly conserved protein. Since we could not obtain stable recombinant ferritin under nondenaturing conditions, commercial ferritin and apoferritin were used for the following in vitro analyses. The similarity of amino acid sequences between Lvferritin and commercial ferritin and apoferritin was 72%. For each assay, 40 μg of His-PK1 or His-GST was mixed with 5 μg of ferritin (ferritin from equine spleen; Sigma-Aldrich) or apoferritin (apoferritin from equine spleen; Sigma-Aldrich) in TN buffer (20 mM Tris base, 300 mM NaCl, pH 7.0) containing 1× protease inhibitor cocktail (Roche Diagnostics) and constantly inverted at 4°C for 16 h. After protein binding, Ni-Sepharose 6 Fast Flow beads were added, and the mixture was constantly inverted for another hour at 4°C. The Sepharose beads were then washed with TN buffer four times. The pulldown products were separated by 15% SDS-PAGE in duplicate, transferred to a Hybond-P membrane (GE Healthcare), and subjected to Western blotting. One duplicate was incubated with anti-horse-spleen-ferritin polyclonal antibody (Sigma-Aldrich) and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology) as the primary and secondary antibodies, respectively. The other duplicate was incubated with anti-His monoclonal antibody (Millipore) and HRP-conjugated goat anti-mouse IgG secondary antibody (Santa Cruz Biotechnology). The protein bands were visualized using a chemiluminescence reagent (PerkinElmer) and exposed to Fuji X-ray film.
QCM analysis of the interaction between PK1 and ferritin or apoferritin.
To confirm the interaction between PK1 and ferritin or apoferritin, an Affinix Qμ quartz crystal microbalance (QCM) instrument (Initium Co. Ltd., Tokyo, Japan) was used. Recombinant His-PK1 or His-GST (host protein) was immobilized on the QCM gold electrode in the presence of TN buffer. The excess protein was removed, and the protein-coated electrode in the QCM analysis chamber was covered with fresh TN buffer. When the change in frequency of the quartz crystal resonator had stabilized, commercial ferritin or apoferritin (guest protein) was injected into the QCM analysis chamber. The interaction between host and guest proteins was observed at 25°C.
Effect of PK1 on iron release from ferritin and iron binding to apoferritin.
To determine whether PK1 causes iron-containing ferritin to release iron ions, commercial ferritin was incubated with different amounts of recombinant His-PK1 or His-GST protein. Reactions proceeded in TN buffer, with final concentrations of 0.156 μM ferritin and 0, 0.1, 0.25, 0.5, 1.0, 1.5, and 2.0 μM recombinant protein. The proteins were incubated at 25°C for 1 h and separated in 7.5% native PAGE gels, using the HMW native marker (GE Healthcare) as a standard (440-kDa band for ferritin). The iron-containing ferritin was identified by staining with Ferene S solution (0.75 mM Ferene S, 15 mM thioglycolic acid, and 2% [vol/vol] acetic acid). The same gel was then further stained with Rapid stain (Bioman) to visualize the total ferritin. The signal intensities of the iron-containing ferritin and total ferritin were quantified using the computer software ImageJ (http://imagej.nih.gov/ij/). For each sample, the ratio of iron-containing ferritin to total ferritin was calculated. For each experimental condition, the ratio of the sample containing 0 μM recombinant protein was then set to 1.0, with subsequent values being expressed relative to this initial ratio.
To determine whether PK1 affects the iron ion binding activity of apoferritin, reactions proceeded in TN buffer, with final concentrations of 0.156 μM apoferritin and 0, 0.1, 0.25, 0.5, 1.0, 1.5, and 2.0 μM recombinant protein. After incubation at 25°C for 1 h, ferrous ammonium sulfate (FAS; J. T. Baker) was added to a final concentration of 250 μM, and incubation was continued at 25°C for an additional hour. The subsequent procedures were performed as described above.
Effect of PK1 on cellular LIP.
A DNA fragment that included the PK1 coding region was amplified using the specific pk1-F-BamHI/pk1-R-NotI primer set listed in Table 1 and cloned into the vector pDHsp/V5-His, which contains the Drosophila heat shock protein 70 promoter (24). The resulting plasmid was named pk1-pDHsp/V5-His. For transient expression of this plasmid and of the empty vector, pDHsp/V5-His, 7.5 × 105 Sf9 cells were seeded in 6-well culture plates before transfection, and an Effectene transfection reagent kit (Qiagen) was used for DNA transfection according to the manufacturer's instructions. The cells were incubated at 27°C for 20 h, heat shocked at 42°C for 30 min to induce the Drosophila Hsp70 promoter to drive the downstream gene, and then incubated at 27°C for another 36 h to express the exogenous protein. To confirm the expression of PK1-V5, cells were collected and subjected to a Western blot assay using anti-V5 antibody (Sigma-Aldrich) and HRP-conjugated goat anti-rabbit IgG secondary antibody to recognize the PK1-V5 protein.
At various time points during the experiment, the LIP levels were evaluated using the fluorometric assay described by Espósito et al. (25), with some modifications. Briefly, the cells were rinsed once with PBH buffer (20 mM HEPES, 1 mg/ml bovine serum albumin [BSA] in 1× phosphate-buffered saline [PBS], pH 6.39) and incubated with PBH buffer containing 150 nM calcein-acetomethoxy (CA-AM; Invitrogen) at 27°C for 15 min. CA-AM rapidly permeates the plasma membrane, and once inside the cell, it is hydrolyzed to release the acetomethoxyl group and the fluorescent metal chelator calcein (CA). CA's fluorescence is then quenched immediately by intracellular metals, including iron. Excess CA-AM that did not penetrate into the cells was removed by washing once with PBH buffer and then twice with PBS. The cells were then lysed with 1/3× PBS and scraped with a cell scraper. After incubating on ice for 10 min, the cell lysates were centrifuged at 16,000 × g for 10 min at 4°C to remove cell debris, and the protein content of the supernatants was measured by the Bradford method. CA fluorescence intensity (F0) was measured in an enzyme-linked immunosorbent assay (ELISA) reader (Spectra Max; Molecular Devices) by loading 100 μl supernatant into a 96-well plate and using a filter combination with an excitation wavelength of 488 nm, an emission wavelength of 518 nm, and a cutoff wavelength of 495 nm. After this initial measurement, a strong iron-specific chelator, 2,2′-bipyridine (BIP; Alfa Aesar), was added to a final concentration of 100 μM in order to strip the iron from the chelated CA. The increased CA fluorescence intensity (F1) was then measured in the ELISA reader, using the same filter combination. The change in fluorescence intensity (ΔF = F1 − F0) thus represents the amount of iron that was bound to the CA in the cell (26), i.e., the cellular LIP. In the final calculation, the ΔF value of each sample was normalized against the lysate's protein concentration to eliminate the effects caused by the number of cells. The data are shown as means ± standard deviations (SD). Unpaired Student's t tests were used to check for statistically significant differences (P < 0.05).
Experimental animals, artificial infection, and sample collection.
The Litopenaeus vannamei shrimps (mean body weight, 15 g) used in this study were purchased from the Aquatic Animal Center of National Taiwan Ocean University in Keelung, Taiwan. Before the experiment, the shrimps were reared in a water tank system with a salinity of 32 ppt at 22 to 25°C for 7 days.
The WSSV T-1 isolate (GenBank accession no. AF440570) was used in this study, and the WSSV inoculum stock was prepared as described previously (27). Experimental inoculum was prepared from the viral stock by 1,000× dilution with cold 1× PBS. For artificial infection, the experimental group was injected with 50 μl diluted viral inoculum, while the control group was injected with 50 μl PBS by intramuscular injection. Three pooled samples of stomach were collected from each group at different time points (0, 2, 4, 8, 12, and 24 h postinjection), with each pooled sample being taken from 3 shrimps. Samples were stored in liquid nitrogen until use.
Ethics statement.
All of the shrimps used in this study were supplied by the Aquatic Animal Center of National Taiwan Ocean University. These animals were specifically raised for research purposes. In Taiwan, no specific permits or permissions were required for use of invertebrate experimental animals. However, all of our experimental procedures, including animal sacrifice, were designed to be as humane as possible, and all animals were treated so as to minimize suffering at all times.
Immunoprecipitation of endogenous ferritin and evaluation of iron ion content.
To immunoprecipitate endogenous ferritin, shrimp stomachs were ground to a fine powder in liquid nitrogen and then suspended in 1/3× PBS containing a protease inhibitor cocktail (Roche Diagnostics). After 10 min of incubation on ice, followed by centrifugation (16,000 × g at 4°C for 10 min), the supernatant (tissue lysate) was collected. The tissue lysate (2 mg) was then incubated with protein A agarose (Invitrogen) at 25°C for 1 h to preclean the lysate, and the precleaned lysate was further incubated with or without anti-Lvferritin antibody coupled with protein A agarose at 25°C for 2 h. After incubation, the complex was washed 5 times with 1/3× PBS. One-sixth of the complex was subjected to Western blotting, and the total ferritin (ferritin plus apoferritin) was recognized with anti-Lvferritin antibody and secondary antibody. The signal intensity of the total ferritin was quantified using the computer software ImageJ. The remaining five-sixths of the complex was further treated with 2% HNO3 at 65°C for 16 h and filtered with a 0.22-μm filter. The iron ion content in the filtrate was then quantified by atomic absorption spectrophotometry (AAS) using a Hitachi Z5000 spectrophotometer. To calculate the amount of iron bound by ferritin, the iron ion content determined by AAS was first normalized against the signal intensity of the total ferritin and then expressed relative to the level of the sample from 0 h postinfection for each group.
To evaluate the total ferritin expression levels during WSSV infection, the tissue lysates described above were also subjected to Western blot assay using the anti-Lvferritin antibody and goat anti-rabbit IgG secondary antibody to detect total ferritin. Beta-actin served as an internal control, and this was detected by using anti-beta-actin antibody and goat anti-mouse IgG secondary antibody. The data are shown as means ± SD, and unpaired Student's t test was performed to test for statistically significant differences (P < 0.05).
RESULTS
Interaction of WSSV PK1 with L. vannamei ferritin (Lvferritin).
After a high-throughput preliminary screening using the yeast mating method, we found that Lvferritin-BD interacted with WSSV PK1-AD (Fig. 1A), and a domain-swapping experiment was used to confirm this result. In this experiment, Lvferritin-pGADT7-Rec was cotransformed with pk1-pGBKT7-1 (pk1-pGBKT7-1/Lvferritin-pGADT7-Rec) into yeast strain AH109, while to rule out autonomous activation, pk1-pGBKT7-1 and Lvferritin-pGADT7-Rec were also cotransformed with the appropriate empty vectors (pk1-pGBKT7-1/pGADT7-Rec and pGBKT7-1/Lvferritin-pGADT7-Rec). As shown in Fig. 1B, growth was seen only on the high-stringency selection medium (SD/−Leu/−Trp/−His/−Ade/+X-α-Gal) when the yeast was cotransformed with pk1-pGBKT7-1/Lvferritin-pGADT7-Rec or with the positive control. Neither PK1 nor Lvferritin exhibited any autonomous activation.
The yeast two-hybrid results suggest that PK1 interacts with host ferritin and imply that it might therefore play a role in regulating either the uptake or release of iron. Before investigating these possibilities, we next used two other experimental techniques (i.e., pulldown assay and QCM analysis) to confirm that PK1 interacted not only with ferritin but also with apoferritin.
WSSV PK1 interacts with both ferritin and apoferritin in vitro.
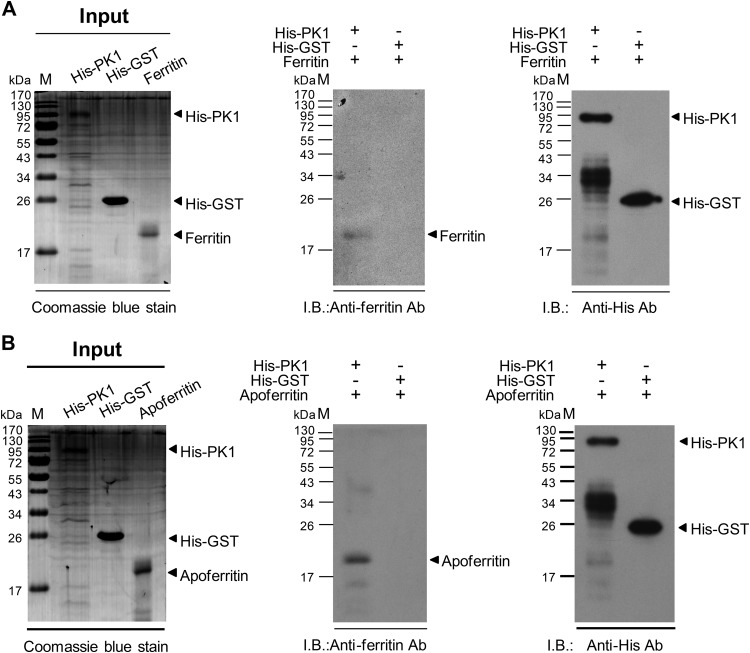
Pulldown assays were used to confirm the interactions between PK1 and ferritin and between PK1 and apoferritin. After incubating ferritin or apoferritin with His-tagged PK1 or His-tagged GST, the resulting protein complexes were pulled down by Ni-Sepharose beads, separated by SDS-PAGE, and subjected to Western blotting. We found that both ferritin and apoferritin interacted with His-PK1 (Fig. 2A and B, middle panels). Neither ferritin nor apoferritin interacted with His-GST.
FIG 2.
Both ferritin and apoferritin were pulled down by WSSV PK1. (A and B) (Left) The input proteins His-PK1 (95 kDa), His-GST (26 kDa), and ferritin (21 kDa) were analyzed by SDS-PAGE and Coomassie blue staining. (Middle) Western blotting (I.B.) of the pulldown products showed that both ferritin and apoferritin interacted with His-PK1 but that neither could be pulled down by His-GST. (Right) The pulldown products were also recognized with anti-His antibody to confirm the presence of His-PK1 or His-GST in these products. (The signal around 34 kDa in the His-PK1 lane was due to a degraded fragment of His-PK1. Even though we used freshly prepared His-PK1, this signal was always present, suggesting that the full-length protein might be unstable.) M, prestained protein marker.
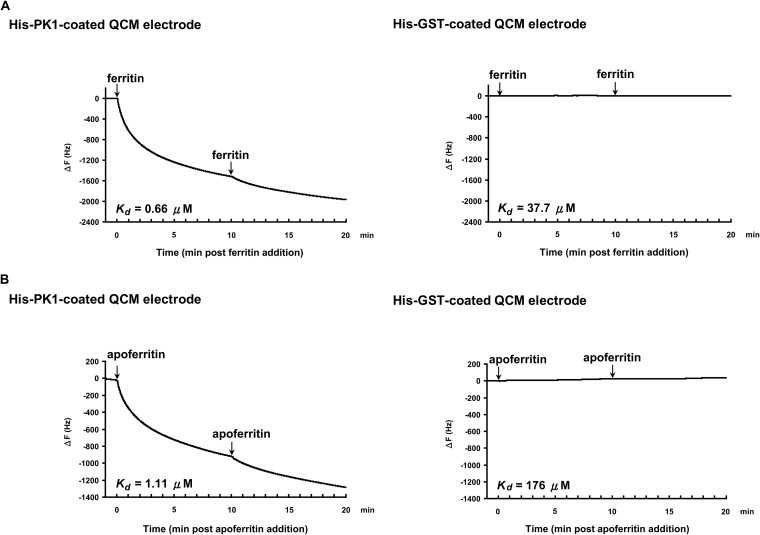
The interactions between PK1 and ferritin and between PK1 and apoferritin were also confirmed by QCM analysis. When purified His-PK1 was used to coat the electrode of a 27-MHz QCM instrument and either ferritin or apoferritin was injected into the analysis chamber, there was a resulting decrease in frequency over time (Fig. 3A and B, left panels). Ferritin bound to His-PK1 with a dissociation constant (Kd) of 0.66 μM, while for apoferritin, the Kd was 1.11 μM. Again, neither ferritin nor apoferritin interacted with His-GST (Fig. 3A and B, right panels).
FIG 3.
QCM analysis of the interactions between PK1 and ferritin or apoferritin. His-PK1 (left panels) and His-GST (right panels) were used as the immobilized host proteins on the QCM electrode of a 27-MHz QCM instrument. The guest protein ferritin (A) or apoferritin (B) was then injected into the analysis chamber at 0 min and 10 min, as indicated. The decrease in frequency (ΔF) is proportional to the interaction between the host and guest proteins. Kd, dissociation constant.
Prevention of apoferritin iron loading by PK1.
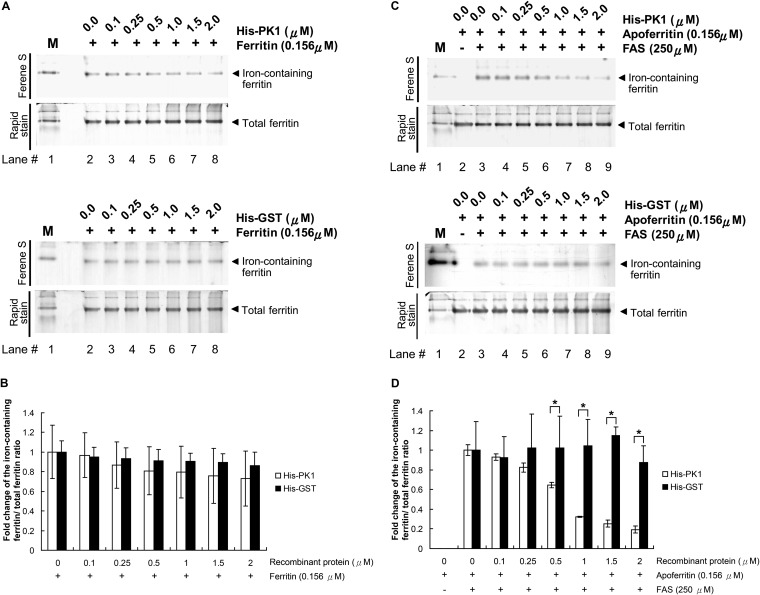
To investigate whether PK1 can regulate the release of iron from ferritin and/or the storage of iron by apoferritin, we conducted two separate assays. In the first assay, we incubated ferritin with His-PK1, and in the second, we preincubated apoferritin with His-PK1 before FAS was added to induce iron loading. The reactants were separated by native PAGE electrophoresis and then stained first with Ferene S, which is a specific stain for iron, and then with Rapid stain solution to detect the total ferritin. It seemed that His-PK1 was no more effective than the His-GST control in promoting the release of iron from ferritin (Fig. 4A, lanes 2 to 8). Quantification of the signal intensities by the computer software ImageJ indicated that there was no significant difference between His-PK1 and His-GST at any of the tested concentrations (Fig. 4B).
FIG 4.
His-PK1 does not promote iron release from ferritin, but it does prevent iron ions from being incorporated by apoferritin. (A) After ferritin was incubated with the indicated amounts of His-PK1 (top) or His-GST control (bottom), the reactants were separated by 7.5% native PAGE and stained with Ferene S solution and Rapid stain to visualize the iron-containing proteins and the total ferritin, respectively. (B) Signal intensities of the iron-containing ferritin and the total ferritin were quantified using the computer software ImageJ. The signal intensity of the iron-containing ferritin was first normalized against the intensity of total ferritin and then expressed relative to the level of the first sample for each group. (C) Apoferritin was preincubated with the indicated amounts of His-PK1 or His-GST before FAS was added. After an additional 1 h of incubation, the proteins were then separated by 7.5% native PAGE and stained with Ferene S solution and Rapid stain. (D) Signal intensities of iron-containing ferritin and total ferritin were quantified using ImageJ. The signal intensity of the iron-containing ferritin was first normalized against the intensity of total ferritin and then expressed relative to that of the first sample containing both apoferritin and FAS. M, native protein marker containing ferritin, which was stained by the Ferene S solution. Bars represent means ± SD for 3 samples. Unpaired Student's t tests were used to test for statistically significant differences (*, P < 0.05).
In contrast, as shown in Fig. 4C, preincubation of apoferritin with His-PK1 prevented binding of the ferrous ion to apoferritin in a dose-dependent manner. Quantification of the signal intensities showed that this effect was statistically significant at His-PK1 concentrations of 0.5 μM and above (Fig. 4D).
Elevation of intracellular LIP by PK1 expression.
Since PK1 blocks the binding of free iron ions to apoferritin in vitro (Fig. 4), we next investigated whether the presence of PK1 affects the level of LIP. This pool consists of the low-molecular-weight transit iron pool and the chelatable iron in the cell, and generally, any increase in LIP levels will stimulate the cell to synthesize ferritin (28–30) that in turn sequesters the excess free iron to maintain iron homeostasis.
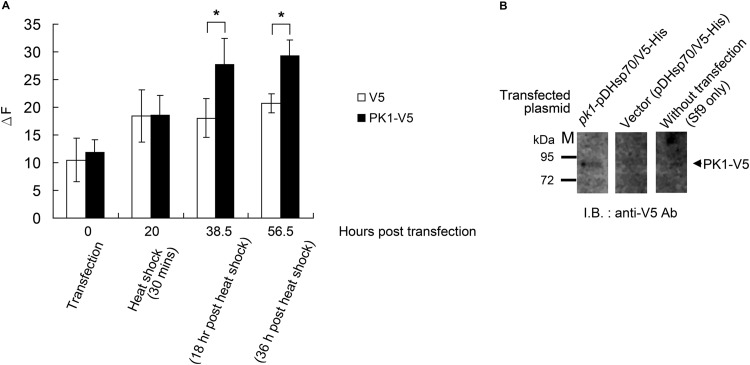
Our experiment showed that overexpression of PK1 in Sf9 cells led to a significantly elevated LIP level 18 h after heat shock and that this elevation continued throughout the experiment, through 56.5 h posttransfection (Fig. 5A). We also used Western blotting to show the expression of PK1-V5 (Fig. 5B). These results suggest that PK1 increases LIP by preventing ferritin or apoferritin from binding additional free iron ions.
FIG 5.
Overexpression of PK1 in Sf9 cells elevates cellular LIP level. (A) When PK1-V5 was overexpressed in Sf9 cells, significant elevations of the LIP level occurred 38.5 h and 56.5 h after transfection. (B) Western blots with anti-V5 antibody were used to detect the PK1-V5 protein expressed in the pk1-pDHsp70/V5-His-transfected cells. The change in fluorescence (ΔF) represents the LIP level. Each bar represents the mean ± SD for 4 samples, and unpaired Student's t tests were used to test for statistically significant differences (*, P < 0.05). M, protein marker.
Reduction of ferritin iron storage after WSSV infection.
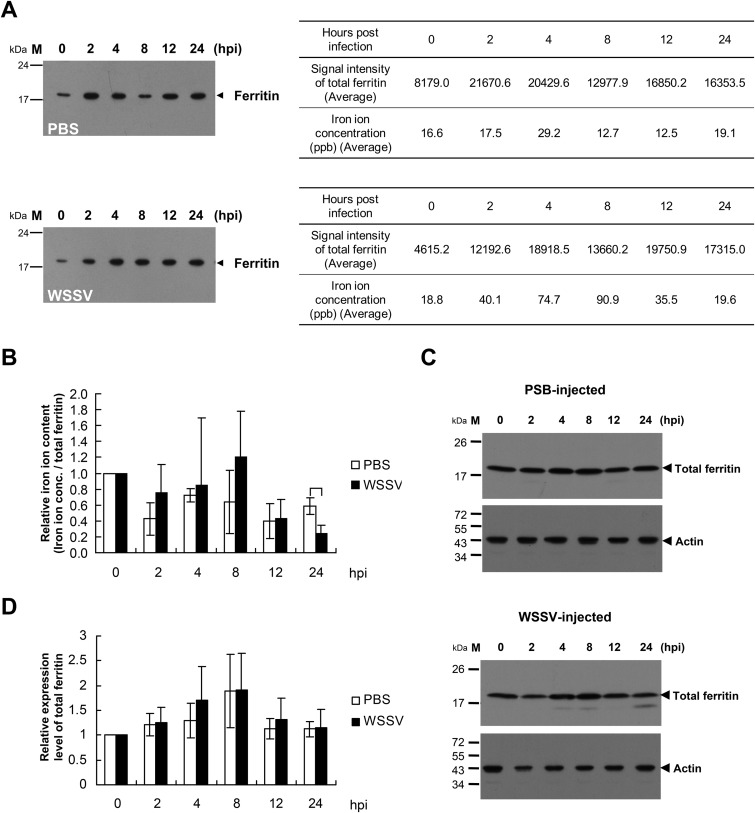
To investigate whether the iron content of shrimp ferritin is affected by WSSV infection, endogenous ferritin from the stomachs of WSSV- or PBS-injected L. vannamei shrimps was immunoprecipitated using anti-Lvferritin antibody coupled with protein A agarose, and the iron content of the immunoprecipitation products was determined using AAS. Figure 6A shows that shrimp ferritin was immunoprecipitated from the stomach lysates. Since the anti-Lvferritin antibody cannot distinguish ferritin from apoferritin, these Western blots represent the total ferritin, i.e., both ferritin and apoferritin. The ratio of iron to total ferritin was then calculated, the initial value was set to 1.0 for each experimental condition, and the subsequent fluctuations in relative iron ion content are shown in Fig. 6B. Although this ratio fluctuated in both the PBS-injected group and the WSSV-injected group, there was a significant relative decrease in the WSSV group at the time that corresponded to approximately the end of the first WSSV replication cycle, i.e., at 24 h postinjection (Fig. 6B). Figures 6C and D further show that although the total ferritin protein expression level fluctuated slightly in both the PBS-injected group and the WSSV-injected group, there was no significant difference between these two groups, nor was there any significant change in expression level within either of the respective groups. Taken together, these results suggest that although WSSV infection does not upregulate ferritin expression, it does reduce shrimp ferritin's ability to bind iron ions.
FIG 6.
The level of iron ions bound by ferritin decreased after WSSV infection. (A) Stomachs excised from L. vannamei shrimps 0, 2, 4, 8, 12, and 24 h after injection with PBS (upper panel) or WSSV (lower panel) were homogenized, and the tissue lysates were mixed with anti-Lvferritin antibody coupled with protein A agarose. The immunoprecipitation products were analyzed by Western blotting to evaluate the amount of total ferritin and by AAS to determine the quantity of iron ions bound by ferritin. (B) The ratio of iron concentration to total ferritin was calculated, and the initial value was set to 1.0 for both the PBS and WSSV injection groups. (C) Stomach tissue lysates prepared from PBS-injected (top) or WSSV-injected (bottom) shrimps were also subjected to Western blot analysis using the anti-Lvferritin antibody or anti-actin antibody to evaluate ferritin expression levels. (D) The ratio of total ferritin to actin was calculated, and the initial value was set to 1.0 for both the PBS and WSSV injection groups. Each bar represents the mean ± SD for 3 samples, where each sample was pooled from 3 shrimps. Unpaired Student's t tests were used to test for statistically significant differences (*, P < 0.05).
DISCUSSION
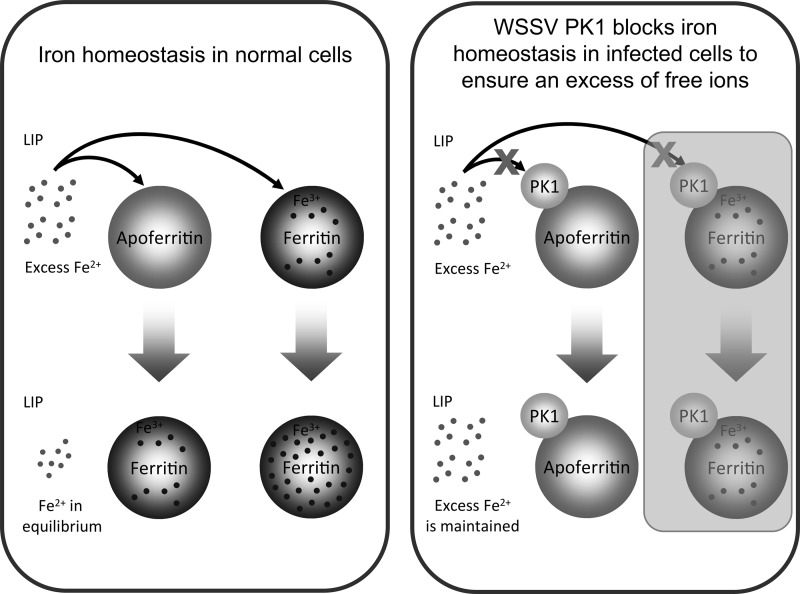
In the cell, iron is important for the activity of many cellular proteins (31), and it is involved in a wide range of physiological reactions (32–36). In particular, iron plays a critical role in ribonucleotide reductase (RR) activity (37–39), and this activity is essential for the replication of at least some DNA viruses. Thus, when the free iron of cells that were infected by vaccinia virus, herpes simplex virus 1, or another DNA virus was chelated with either mimosine or BIP, the resulting suppression of RR activity led to a decrease of both DNA synthesis and virion assembly (40, 41). To fight against pathogenic infection, hosts use a similar iron-withholding mechanism whereby the uptake of iron from outside the cell is reduced, while the free iron ions within the cell are sequestered by ferritin (4). This is potentially an effective antiviral defense strategy regardless of whether the RR is encoded by the virus or the host, because the activity of both types of RR depends on the availability of free iron in the LIP. However, invading pathogens have been shown to use several counterstrategies to defeat this mechanism, including the degradation of iron-containing proteins, such as hemoglobin, the expression of proteins that extract iron from transferrin or ferritin, and the degradation of ferritin to release its store of sequestered iron (5, 7). This is the first study to present evidence that PK1 expressed by the DNA virus WSSV uses a novel mechanism to prevent the host cell's ferritin from sequestering free iron (Fig. 4 to 6). The sequestration of iron is used by the host cells both to maintain iron homeostasis and to support the host cells' iron-withholding defense mechanism. By preventing this sequestration, PK1 thus ensures that WSSV RR activity is maintained for the efficient proliferation of the virus in the host shrimp. This process is represented graphically in Fig. 7.
FIG 7.
Schematic representation of how WSSV PK1 prevents the cell's ferritin from sequestering free iron. This mechanism defeats the cell's iron-withholding defense and ensures that the virus continues to have access to sufficient amounts of this vital nutrient. (The mechanism represented by the shaded part of the schematic has not yet been confirmed experimentally.)
Ferritin is composed of 24 subunits arranged in 432 octahedral symmetry to form a spherical protein cage. Each ferritin molecule has 6 4-fold channels, 8 3-fold channels, and 12 2-fold channels, with the 3-fold channels being used as the pathways for the influx of iron ions (42). The diameter of the 3-fold channel is only a few angstroms, and assuming that all of these properties also apply to shrimp ferritin, the PK1 molecule is too large to enter this channel. We therefore hypothesize that PK1 prevents the sequestration of iron by binding to the ferritin at or near the iron entry site (i.e., the 3-fold channel) and interfering with its conformation and/or hydrophilic properties. We further hypothesize that whatever mechanism PK1 uses to block the entry of iron into apoferritin, it also probably uses the same mechanism to block the further influx of iron ions into ferritin.
After transcription, ferritin translation is regulated by the free iron content in the cell via the iron-regulatory proteins (IRPs) and the iron-responsive elements (IREs) in the 5′-untranslated region (5′ UTR) of ferritin mRNA (43). When PK1 prevents the apoferritin from sequestering free iron and thus elevates the labile iron pool, we would therefore expect an increase in ferritin protein expression. However, as shown in Fig. 6C and D, and contrary to our expectations, there was no significant change in ferritin protein expression levels. The reason for this if presently unclear, but it is possible that WSSV somehow acts to inhibit the usual ferritin translation mechanism. This will need to be investigated in a future study.
In most living cells, ferritin is the main iron storage protein, and one of its major functions is to make iron available to the cell. If a cell does not have access to sufficient amounts of iron, this will lead to mitochondrial ultrastructural aberrations and the release of cytochrome c (44). Meanwhile, by modulating the cellular LIP, ferritin restricts the cytotoxicity of the ferrous ion, Fe(II), which would otherwise generate oxygen radicals, especially the hydroxyl radical (OH·) (45). Oxygen radicals are used as part of the host's innate immune system, but while they can kill some kinds of invaders, they also raise the cell's oxidative stress and can lead to lipid peroxidation, polysaccharide depolymerization, enzyme inactivation, and, ultimately, cell death. Oxidative stress can also be produced by ferritin dysfunction, suggesting that PK1 will also produce these cellular changes when it suppresses ferritin's ability to control iron homeostasis. Some evidence in support of this comes from a recent paper which shows that WSSV infection not only induces changes in metabolism and an increase in the ADP/ATP ratio but also, at the late stage of the infection cycle (24 h), induces several changes associated with cell death, including mitochondrial membrane permeabilization (MMP), elevation of oxidative stress, a decrease in glucose consumption, and disruption of energy production (27).
The peptide sequence of WSSV PK1 contains the major conserved subdomains present in eukaryotic protein kinases (18), which suggests that this protein should be a functional protein kinase. Although the phosphorylation of ferritin has not been studied widely, Ihara et al. (46) found that ferritin can be phosphorylated in vitro by cAMP-dependent protein kinase, and Rama-Kumar (47) reported that plant ferritin can be phosphorylated by both Ca2+/CaM-dependent kinases and calcium-independent kinases in vivo. By producing a net negative charge on the surface of the ferritin, phosphorylation may create putative ionic binding sites for nonferrous metal cations and thus increase ferritin's ability to detoxify heavy metals (47). We were therefore curious to see whether the protein kinase activity of PK1 might also affect ferritin's iron uptake. Since both ATP and Mg2+ are important for PK1 kinase activity, we first tested the effects of various dosages of ATP and MgCl2 on apoferritin's ability to incorporate iron. However, we found that even in the absence of PK1, ATP strongly suppressed the iron uptake in a dose-dependent manner (data not shown). If this suppression was due to the formation of an ATP-Fe complex (48), this would explain why only a few Fe atoms were able to leave this complex to become incorporated into ferritin (49). In any case, unfortunately, this means that this assay could not be used to investigate how the kinase activity of PK1 might affect the iron uptake of apoferritin. Meanwhile, since the absence of any ATP or MgCl2 in the experiments done for Fig. 4 has already shown that kinase activity is not essential for PK1 to suppress the iron ion binding activity of apoferritin, it seems likely that WSSV PK1 uses an alternative mechanism—probably direct protein-protein interaction—to alter ferritin's behavior.
ACKNOWLEDGMENTS
This study was supported financially by the Ministry of Education, Taiwan, Republic of China, under the National Cheng Kung University Aim for the Top University Project Promoting Academic Excellence and Developing World Class Research (grant D103-38A01).
We are indebted to Paul Barlow for his helpful comments on the manuscript.
REFERENCES
- 1.Balla G, Jacob HS, Balla J, Rosenberg M, Nath K, Apple F, Eaton JW, Vercellotti GM. 1992. Ferritin: a cytoprotective antioxidant stratagem of endothelium. J Biol Chem 267:18148–18153. [PubMed] [Google Scholar]
- 2.Epsztejn S, Glickstein H, Picard V, Slotki IN, Breuer W, Beaumont C, Cabantchik ZI. 1999. H-ferritin subunit overexpression in erythroid cells reduces the oxidative stress response and induces multidrug resistance properties. Blood 94:3593–3603. [PubMed] [Google Scholar]
- 3.Theil EC. 1987. Ferritin: structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu Rev Biochem 56:289–315. doi: 10.1146/annurev.bi.56.070187.001445. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg ED, Miklossy J. 2008. Iron withholding: a defense against disease. J Alzheimers Dis 13:451–463. [DOI] [PubMed] [Google Scholar]
- 5.Larson JA, Howie HL, So M. 2004. Neisseria meningitidis accelerates ferritin degradation in host epithelial cells to yield an essential iron source. Mol Microbiol 53:807–820. doi: 10.1111/j.1365-2958.2004.04169.x. [DOI] [PubMed] [Google Scholar]
- 6.Segond D, Abi Khalil E, Buisson C, Daou N, Kallassy M, Lereclus D, Arosio P, Bou-Abdallah F, Nielsen Le Roux C. 2014. Iron acquisition in Bacillus cereus: the roles of IlsA and bacillibactin in exogenous ferritin iron mobilization. PLoS Pathog 10:e1003935. doi: 10.1371/journal.ppat.1003935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg ED. 1999. Iron loading and disease surveillance. Emerg Infect Dis 5:346–352. doi: 10.3201/eid0503.990305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitby PW, Vanwagoner TM, Springer JM, Morton DJ, Seale TW, Stull TL. 2006. Burkholderia cenocepacia utilizes ferritin as an iron source. J Med Microbiol 55:661–668. doi: 10.1099/jmm.0.46199-0. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Arieh SV, Zimerman B, Smorodinsky NI, Yaacubovicz M, Schechter C, Bacik I, Gibbs J, Bennink JR, Yewdell JW, Coligan JE, Firat H, Lemonnier F, Ehrlich R. 2001. Human cytomegalovirus protein US2 interferes with the expression of human HFE, a nonclassical class I major histocompatibility complex molecule that regulates iron homeostasis. J Virol 75:10557–10562. doi: 10.1128/JVI.75.21.10557-10562.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vahdati-Ben Arieh S, Laham N, Schechter C, Yewdell JW, Coligan JE, Ehrlich R. 2003. A single viral protein HCMV US2 affects antigen presentation and intracellular iron homeostasis by degradation of classical HLA class I and HFE molecules. Blood 101:2858–2864. doi: 10.1182/blood-2002-07-2158. [DOI] [PubMed] [Google Scholar]
- 11.Tsuji Y, Akebi N, Lam TK, Nakabeppu Y, Torti SV, Torti FM. 1995. FER-1, an enhancer of the ferritin H gene and a target of E1A-mediated transcriptional repression. Mol Cell Biol 15:5152–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bevilacqua MA, Faniello MC, Quaresima B, Tiano MT, Giuliano P, Feliciello A, Avvedimento VE, Cimino F, Costanzo F. 1997. A common mechanism underlying the E1A repression and the cAMP stimulation of the H ferritin transcription. J Biol Chem 272:20736–20741. doi: 10.1074/jbc.272.33.20736. [DOI] [PubMed] [Google Scholar]
- 13.Pan D, He N, Yang Z, Liu H, Xu X. 2005. Differential gene expression profile in hepatopancreas of WSSV-resistant shrimp (Penaeus japonicus) by suppression subtractive hybridization. Dev Comp Immunol 29:103–112. doi: 10.1016/j.dci.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Li F, Wang Z, Zhang X, Zhou Q, Xiang J. 2006. Cloning, expression and identification of ferritin from Chinese shrimp, Fenneropenaeus chinensis. J Biotechnol 125:173–184. doi: 10.1016/j.jbiotec.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Clavero-Salas A, Sotelo-Mundo RR, Gollas-Galván T, Hernández-López J, Peregrino-Uriarte AB, Muhlia-Almazán A, Yepiz-Plascencia G. 2007. Transcriptome analysis of gills from the white shrimp Litopenaeus vannamei infected with white spot syndrome virus. Fish Shellfish Immunol 23:459–472. doi: 10.1016/j.fsi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Pongsomboon S, Tang S, Boonda S, Aoki T, Hirono I, Yasuike M, Tassanakajon A. 2008. Differentially expressed genes in Penaeus monodon hemocytes following infection with yellow head virus. BMB Rep 41:670–677. doi: 10.5483/BMBRep.2008.41.9.670. [DOI] [PubMed] [Google Scholar]
- 17.Ruan YH, Kuo CM, Lo CF, Lee MH, Lian JL, Hsieh SL. 2010. Ferritin administration effectively enhances immunity, physiological responses, and survival of Pacific white shrimp (Litopenaeus vannamei) challenged with white spot syndrome virus. Fish Shellfish Immunol 28:542–548. doi: 10.1016/j.fsi.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Liu WJ, Yu HT, Peng SE, Chang YS, Pien HW, Lin CJ, Huang CJ, Tsai MF, Huang CJ, Wang CH, Lin JY, Lo CF, Kou GH. 2001. Cloning, characterization, and phylogenetic analysis of a shrimp white spot syndrome virus gene that encodes a protein kinase. Virology 289:362–377. doi: 10.1006/viro.2001.1091. [DOI] [PubMed] [Google Scholar]
- 19.van Hulten MC, Vlak JM. 2001. Identification and phylogeny of a protein kinase gene of white spot syndrome virus. Virus Genes 22:201–207. doi: 10.1023/A:1008127709325. [DOI] [PubMed] [Google Scholar]
- 20.van Hulten MC, Witteveldt J, Peters S, Kloosterboer N, Tarchini R, Fiers M, Sandbrink H, Lankhorst RK, Vlak JM. 2001. The white spot syndrome virus DNA genome sequence. Virology 28:7–22. doi: 10.1006/viro.2001.1002. [DOI] [PubMed] [Google Scholar]
- 21.Kim CS, Kosuke Z, Nam YK, Kim SK, Kim KH. 2007. Protection of shrimp (Penaeus chinensis) against white spot syndrome virus (WSSV) challenge by double-stranded RNA. Fish Shellfish Immunol 23:242–246. doi: 10.1016/j.fsi.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh SL, Chiu YC, Kuo CM. 2006. Molecular cloning and tissue distribution of ferritin in Pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol 21:279–283. doi: 10.1016/j.fsi.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Sangsuriya P, Huang JY, Chu YF, Phiwsaiya K, Leekitcharoenphon P, Meemetta W, Senapin S, Huang WP, Withyachumnarnkul B, Flegel TW, Lo CF. 2014. Construction and application of a protein interaction map for white spot syndrome virus (WSSV). Mol Cell Proteomics 13:269–282. doi: 10.1074/mcp.M113.029199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leu JH, Kuo YC, Kou GH, Lo CF. 2008. Molecular cloning and characterization of an inhibitor of apoptosis protein (IAP) from the tiger shrimp, Penaeus monodon. Dev Comp Immunol 32:121–133. doi: 10.1016/j.dci.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Espósito BP, Epsztejn S, Breuer W, Cabantchik ZI. 2002. A review of fluorescence methods for assessing labile iron in cells and biological fluids. Anal Biochem 304:1–18. doi: 10.1006/abio.2002.5611. [DOI] [PubMed] [Google Scholar]
- 26.Breuer W, Epsztejn S, Millgram P, Cabantchik IZ. 1995. Transport of iron and other transition metals into cells as revealed by a fluorescent probe. Am J Physiol 268:1354–1361. [DOI] [PubMed] [Google Scholar]
- 27.Chen IT, Aoki T, Huang YT, Hirono I, Chen TC, Huang JY, Chang GD, Lo CF, Wang HC. 2011. White spot syndrome virus induces metabolic changes resembling the Warburg effect in shrimp hemocytes in the early stage of infection. J Virol 85:12919–12928. doi: 10.1128/JVI.05385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aziz N, Munro HN. 1987. Iron regulates ferritin mRNA translation through a segment of its 5′ untranslated region. Proc Natl Acad Sci U S A 84:8478–8482. doi: 10.1073/pnas.84.23.8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hentze MW, Rouault TA, Caughman SW, Dancis A, Harford JB, Klausner RD. 1987. A cis-acting element is necessary and sufficient for translational regulation of human ferritin expression in response to iron. Proc Natl Acad Sci U S A 84:6730–6734. doi: 10.1073/pnas.84.19.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisenstein RS. 2000. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr 20:627–662. doi: 10.1146/annurev.nutr.20.1.627. [DOI] [PubMed] [Google Scholar]
- 31.Fraga CG. 2005. Relevance, essentiality and toxicity of trace elements in human health. Mol Aspects Med 26:235–244. doi: 10.1016/j.mam.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Hatefi Y. 1985. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem 54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 33.Jordan A, Reichard P. 1998. Ribonucleotide reductases. Annu Rev Biochem 67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 34.Pang H, Bartlam M, Zeng Q, Miyatake H, Hisano T, Miki K, Wong LL, Gao GF, Rao Z. 2004. Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J Biol Chem 279:1491–1498. doi: 10.1074/jbc.M310022200. [DOI] [PubMed] [Google Scholar]
- 35.Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. 2006. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell 23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 36.Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L. 2007. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat Struct Mol Biol 14:875–877. doi: 10.1038/nsmb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper CE, Lynagh GR, Hoyes KP, Hider RC, Cammack R, Porter JB. 1996. The relationship of intracellular iron chelation to the inhibition and regeneration of human ribonucleotide reductase. J Biol Chem 271:20291–20299. doi: 10.1074/jbc.271.34.20291. [DOI] [PubMed] [Google Scholar]
- 38.Lamarche N, Matton G, Massie B, Fontecave M, Atta M, Dumas F, Gaudreau P, Langelier Y. 1996. Production of the R2 subunit of ribonucleotide reductase from herpes simplex virus with prokaryotic and eukaryotic expression systems: higher activity of R2 produced by eukaryotic cells related to higher iron-binding capacity. Biochem J 320:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chabes A, Domkin V, Larsson G, Liu A, Graslund A, Wijmenga S, Thelander L. 2000. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc Natl Acad Sci U S A 97:2474–2479. doi: 10.1073/pnas.97.6.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai Y, Gold B, Vishwanatha JK, Rhode SL. 1994. Mimosine inhibits viral DNA synthesis through ribonucleotide reductase. Virology 205:210–216. doi: 10.1006/viro.1994.1636. [DOI] [PubMed] [Google Scholar]
- 41.Romeo AM, Christen L, Niles EG, Kosman DJ. 2001. Intracellular chelation of iron by bipyridyl inhibits DNA virus replication: ribonucleotide reductase maturation as a probe of intracellular iron pools. J Biol Chem 276:24301–24308. doi: 10.1074/jbc.M010806200. [DOI] [PubMed] [Google Scholar]
- 42.Laghaei R, Evans DG, Coalson RD. 2013. Metal binding sites of human H-chain ferritin and iron transport mechanism to the ferroxidase sites: a molecular dynamics simulation study. Proteins 81:1042–1050. doi: 10.1002/prot.24251. [DOI] [PubMed] [Google Scholar]
- 43.Durand JP, Goudard F, Pieri J, Escoubas JM, Schreiber N, Cadoret JP. 2004. Crassostrea gigas ferritin: cDNA sequence analysis for two heavy chain type subunits and protein purification. Gene 338:187–195. doi: 10.1016/j.gene.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 44.Dong F, Zhang X, Culver B, Chew HG Jr, Kelley RO, Ren J. 2005. Dietary iron deficiency induces ventricular dilation, mitochondrial ultrastructural aberrations and cytochrome c release: involvement of nitric oxide synthase and protein tyrosine nitration. Clin Sci (Lond) 109:277–286. doi: 10.1042/CS20040278. [DOI] [PubMed] [Google Scholar]
- 45.Halliwell B, Gutteridge JM. 1990. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol 186:1–85. doi: 10.1016/0076-6879(90)86093-B. [DOI] [PubMed] [Google Scholar]
- 46.Ihara K, Maeguchi K, Young CT, Theil EC. 1984. Cell-specific properties of red cell and liver ferritin from bullfrog tadpoles probed by phosphorylation in vitro. J Biol Chem 259:278–283. [PubMed] [Google Scholar]
- 47.Rama-Kumar T. 1998. Ph.D. thesis University of Hyderabad, Hyderabad, India. [Google Scholar]
- 48.Weaver J, Pollack S. 1989. Low-Mr iron isolated from guinea pig reticulocytes as AMP-Fe and ATP-Fe complexes. Biochem J 261:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konopka K, Mareschal JC, Crichton RR. 1981. Iron transfer from transferrin to ferritin mediated by polyphosphate compounds. Biochim Biophys Acta 677:417–423. doi: 10.1016/0304-4165(81)90255-5. [DOI] [PubMed] [Google Scholar]