ABSTRACT
Boid inclusion body disease (BIDB) is a fatal disease of boid snakes, the etiology of which has only recently been revealed following the identification of several novel arenaviruses in diseased snakes. BIBD-associated arenaviruses (BIBDAV) are genetically divergent from the classical Old and New World arenaviruses and also differ substantially from each other. Even though there is convincing evidence that BIBDAV are indeed the etiological agent of BIBD, the BIBDAV reservoir hosts—if any exist besides boid snakes themselves—are not yet known. In this report, we use University of Helsinki virus (UHV; a virus that we isolated from a Boa constrictor with BIBD) to show that BIBDAV can also replicate effectively in mammalian cells, including human cells, provided they are cultured at 30°C. The infection induces the formation of cytoplasmic inclusion bodies (IB), comprised mainly of viral nucleoprotein (NP), similar to those observed in BIBD and in boid cell cultures. Transferring infected cells from 30°C to 37°C ambient temperature resulted in progressive declines in IB formation and in the amounts of viral NP and RNA, suggesting that BIBDAV growth is limited at 37°C. These observations indirectly indicate that IB formation is linked to viral replication. In addition to mammalian and reptilian cells, UHV infected arthropod (tick) cells when grown at 30°C. Even though our findings suggest that BIBDAV have a high potential to cross the species barrier, their inefficient growth at mammalian body temperatures indicates that the reservoir hosts of BIBDAV are likely species with a lower body temperature, such as snakes.
IMPORTANCE The newly discovered boid inclusion body disease-associated arenaviruses (BIBDAV) of reptiles have drastically altered the phylogeny of the family Arenavirus. Prior to their discovery, known arenaviruses were considered mainly rodent-borne viruses, with each arenavirus species having its own reservoir host. BIBDAV have so far been demonstrated in captive boid snakes, but their possible reservoir host(s) have not yet been identified. Here we show, using University of Helsinki virus as a model, that these viruses are able to infect mammalian (including human) and arthropod cells. Our results provide in vitro proof of the considerable ability of arenaviruses to cross species barriers. However, our data indicate that BIBDAV growth occurs at 30°C but is inhibited at 37°C, implying that crossing of the species barrier would be hindered by the body temperature of mammalian species.
INTRODUCTION
Arenavirus is the sole genus in the family Arenaviridae, RNA viruses that have previously been described as almost exclusively rodent borne (1). Several arenaviruses are known to be able to cross the species barrier, for instance to be transmitted from a rodent host to humans (2–4). Such cross-species transmission often leads to severe infections, which manifest as hemorrhagic fever (Lassa, Guanarito, Junin, Lujo, Machupo, Sabia, or Whitewater Arroyo virus) or meningitis (lymphocytic choriomeningitis virus [LCMV]) in humans (1, 5, 6). While these are usually dead-end events for the virus, they can occasionally lead to prolonged chains of transmission between humans (7). More recently, several arenaviruses have been detected in snakes with boid inclusion body disease (BIBD) (8–10), and in vitro experiments, together with statistical associations, provided convincing evidence of an etiological relationship between BIBD and arenavirus infection (10). The pathomorphology of BIBD is manifested by the development of typical eosinophilic intracytoplasmic inclusion bodies (IB) in almost all cell types of affected animals (10–12). The IB consist predominantly, if not entirely, of a 68-kDa protein (11) that has recently been identified as the arenavirus nucleoprotein (NP) (10). They most likely represent complexes required for arenavirus replication (13).
Arenaviruses have a bisegmented genome with an ambisense coding strategy (14). The L segment encodes the RNA-dependent RNA polymerase (RdRp) and the Z protein, and the S segment encodes the glycoprotein precursor (GPC) and the NP (14). Of these, the RdRp is considered the most conserved, whereas all other structural proteins exhibit relatively high levels of variability (14, 15). BIBD-associated arenaviruses (BIBDAV) show very high levels of genome variability, particularly in the GPC region (8–10, 16, 17). This may reflect differences between reservoir hosts of the viruses (17), since the glycoproteins encoded in the GPC mediate binding of the virions to the cellular receptor(s) (18).
To evaluate the potential of BIBDAV to cross species barriers and to shed some light on the potential reservoir hosts of these viruses, we screened a range of vertebrate and arthropod cell lines for their susceptibility to the University of Helsinki virus (UHV), a virus that we isolated from a Boa constrictor with BIBD (10). We also studied the effects of different temperatures on the growth of BIBDAV, since we earlier observed productive UHV infection in Vero E6 cells only when grown at 27 to 30°C (10).
MATERIALS AND METHODS
Viruses and cell lines.
UHV propagated in the boid kidney cell line (named I/1Ki; described in reference 10) used in this study was purified by density gradient ultracentrifugation as described previously (19) and stored at −70°C (supplemented with bovine serum albumin [BSA]) until used for inoculation. The purified UHV (GenBank accession numbers KF297881.1 and KF297880.1) was initially used to infect Vero E6 cells (10), and adaptation to Vero E6 cells was enhanced by three consecutive passages (a fresh batch of Vero E6 cells was infected each time with supernatant collected 12 to 15 days postinfection [p.i.]). A second BIBDAV isolate, T10404 (from snake number 5 in reference 10; GenBank accession number KF564801), was passaged once in the boid kidney cells, concentrated, purified, and stored as described above.
African green monkey kidney cells (Vero and Vero E6; ATCC), human lung adenocarcinoma cells (A549; ATCC), baby hamster kidney cells (BHK-21; ATCC), and Boa constrictor kidney cells (I/1Ki; described in reference 10) were cultured in basal medium Eagle (BME; Biochrom) containing 10% tryptose phosphate broth (TPB) (Difco; Sigma-Aldrich), 15 mM HEPES, 2 mM l-glutamine, 10 μg/ml gentamicin, and 50 IU/ml nystatin (Valeant Pharmaceuticals), pH 7.2 to 7.3, when used for infections with UHV purified from boid cells and in minimal essential medium (MEM) (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS), 25 mM HEPES, 2 mM l-glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin when used for infections with UHV purified from Vero E6 cells and with T10404. When studying the effect of a temperature switch on BIBDAV growth in vertebrate cells, both infected and noninfected cells were maintained at either 30°C or 37°C for 4 to 5 days after virus inoculation, after which half of each group of cells at each temperature, infected and noninfected, were transferred to the opposite temperature, i.e., cells grown at 30°C were maintained at 37°C and vice versa. For Western blotting and real-time PCR, cell samples were collected at 1- to 2-day intervals, and samples for other assays (histology and immunohistochemistry) were collected 4 days after the temperature swap. Cells maintained at 30°C or 37°C for the entire experimental period served as controls.
Three tick embryo-derived cell lines, the Ixodes ricinus cell line IRE/CTVM19 (20, 21), the Rhipicephalus (Boophilus) microplus cell line BME/CTVM2 (22), and the Rhipicephalus appendiculatus cell line RAE/CTVM1 (22) were maintained in 2 ml L-15 (Leibovitz) medium (Sigma-Aldrich) supplemented with 20% FBS, 10% TPB, 2 mM l-glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin in sealed flat-sided culture tubes (Nunc) at 30°C. The medium was changed weekly by the removal and replacement of 3/4 of the medium volume, and resuspended cells were subcultured 1:1 when required (20). Infection of tick cells was done by adding UHV (propagated in boid cells and purified by density gradient ultracentrifugation) to the culture medium, followed by incubation for 14 days at 30°C.
Cloning, production, and purification of recombinant UHV proteins.
A partial S segment of UHV cloned (10) into pGEM-T vector (Promega) was used as the template for PCR cloning of full-length (amino acids [aa] 1 to 582; recombinant nucleoprotein [rNP]) and N-terminal (amino acids 1 to 339; rNP-N) and C-terminal (amino acids 346 to 582; rNP-C) portions of the UHV NP. These regions were chosen based on both the knowledge of functional domains (23) and the structural data of arenavirus NP (24). PCR cloning of the fragments was accomplished using Phusion high-fidelity DNA polymerase (Thermo Scientific) with the following primers: for NP, 5′-GGTACCATGGCTGCACTACAAAGAGC-3′ and 5′-CTCGAGGACCTCCACAGGCC-3′; for N-terminal NP, 5′-GGTACCATGGCTGCACTACAAAGAGC-3′ and 5′-CTCGAGCCTTCTCAAACGGAATACCG-3′; and for C-terminal NP, 5′-CATATGAGGTTATACCCGGACTTGATGGA-3′ and 5′-CTCGAGGACCTCCACAGGCC-3′. The cloning and production of recombinant bacmids were done in accordance with the Bac-to-Bac HBM TOPO secreted expression system manual (Life Technologies). P1 supernatants containing recombinant baculoviruses were harvested 4 to 7 days after transfecting Sf9 cells with purified recombinant bacmids.
For recombinant protein production, confluent High Five cells (75 cm2 bottle) were detached with a cell scraper and pelleted by centrifugation (500 × g for 5 min at room temperature [RT]). The cell pellets were suspended in 1 ml of cell culture supernatant containing recombinant baculoviruses, incubated for 1 h at RT, and transferred into two 175-cm2 bottles to which 25 ml growth medium was added. The cells were collected between 5 and 7 days p.i.
The recombinant proteins were purified from cells infected with recombinant baculovirus as follows: the cell pellet was suspended in 3 ml lysis buffer 1 (50 mM Tris, pH 7.5), and the cell suspension centrifuged (4,000 × g for 10 min at RT). The resultant pellet (containing the insoluble recombinant protein) was suspended in 5 ml lysis buffer 1 and centrifuged as described above. The resultant pellet was suspended in 5 ml lysis buffer 2 (50 mM Tris, 150 mM NaCl, 0.1% Triton X-100, pH 7.5) and centrifuged (4,000 × g for 30 min at 4°C); the procedure was repeated once. After washing with lysis buffer 2, the resultant pellet was suspended in 2 ml lysis buffer 3 (50 mM Tris, 500 mM NaCl, 1% Triton X-100, pH 7.5) and centrifuged (16,000 × g for 5 min at RT). The washing was repeated once using 3 ml lysis buffer 3. The resultant pellet was homogenized in 9 ml binding buffer (50 mM Tris, 500 mM NaCl, 6 M guanidine-HCl, pH 8.0) by being passed several times through an 18-gauge needle on a 5-ml syringe. One ml of Ni-nitrilotriacetic acid (NTA) agarose beads (Invitrogen) equilibrated in binding buffer was added to the cell homogenate, followed by 1 h of incubation at RT. The beads were washed 3 times with binding buffer (centrifuging at 500 × g for 3 min at RT each time), and the recombinant proteins were eluted by the addition of 1 ml elution buffer (50 mM Tris, 500 mM NaCl, 6 M guanidine-HCl, 500 mM imidazole, pH 8.0), followed by centrifugation at 500 × g for 3 min at RT. The elution was repeated three times. Concentration and buffer exchange (to 35 mM HEPES, 135 mM NaCl, 1 M guanidine-HCl, pH 8.2) of the recombinant proteins were done using a centrifugal filter device with a 10-kDa cutoff (Millipore). For SDS-PAGE analysis, guanidine was removed with ethanol (1 part protein solution plus 9 parts ethanol) precipitation (30 min at −70°C), followed by centrifugation at 16,000 × g for 10 min at 4°C.
Antibodies.
Purified rNP-N and rNP-C were used to produce polyclonal antisera by a process similar to that described in reference 10 but with initial injections of 150 μg, 70-μg boosters on days 7 and 14, 150-μg boosters on day 28, and 70-μg boosters on day 42, by BioGenes GmbH. A previously prepared rabbit antiserum against purified and lysed UHV (10) and the antisera produced against rNP-N and rNP-C were affinity purified by rNP coupled to CNBr-activated Sepharose (GE Healthcare) according to the manufacturer's protocol. The affinity-purified rNP-N and rNP-C polyclonal antibodies (PAb) were labeled with horseradish peroxidase (HRP) using the EZ-Link activated peroxidase antibody labeling kit (Thermo Scientific) as instructed by the manual.
SDS-PAGE and immunoblotting.
Protein separations by SDS-PAGE (8 to 12% gels) and wet blotting of proteins onto nitrocellulose (Whatman) were done according to standard protocols. The protein concentrations of cell lysates (in 50 mM Tris, 150 mM NaCl, 1% Triton X-100, pH 8.0, supplemented with EDTA-free protease inhibitor cocktail [Roche]) were determined using the bicinchoninic acid (BCA) protein assay kit (Pierce, Thermo Scientific), and 10 μg of total protein was loaded for each lane. Sample preparation for SDS-PAGE and immunoblotting, as well as the probing and detection of immunoblots using the anti-rNP PAb (affinity purified from anti-UHV serum, used at 0.1 μg/ml concentration), were done as described previously (10). The HRP-labeled anti-rNP-N and anti-rNP-C PAb were used at 0.25 μg/ml to 0.1 μg/ml concentrations, diluted in blocking buffer (50 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 8.0) with 3% skimmed milk powder. The results were recorded on X-ray film (Fuji Medical RX) utilizing in-house enhanced chemiluminescence (ECL) reagents.
qPCR.
Total RNA from UHV-infected boid cells grown at different temperatures was isolated with the RNeasy minikit (Qiagen) according to the manufacturer's protocol. For RNA quantification, 1 μg total RNA was transcribed to cDNA by using RevertAid premium reverse transcriptase (Thermo Scientific) and following the manufacturer's protocol. An equal amount of cDNA from each sample was used as the template for real-time PCR (Stratagene MX3500P) amplification of UHV L segment RNA (primer sequences targeting the Z protein region, originally designed for cloning, were 5′-CATATGAGCGAATCAACCGCAATAGGTC-3′ for the forward direction and 5′-CTCGAGTGGTTCGGGGAGG-3′ for the reverse direction), with Maxima SYBR green quantitative PCR (qPCR) master mix (Thermo Scientific). The relative amounts of viral RNA were estimated based on CT differences compared to the lowest CT value observed (UHV grown at 37°C).
Histological, IHC, immunofluorescence (IF), and transmission electron microscopy (TEM) examinations.
For histology and immunohistochemistry (IHC), cultured cells were detached by trypsinization, pelleted by centrifugation (2,850 × g for 3 min at RT), and then fixed in 2.5% paraformaldehyde (PFA) in 0.2 M PBS (pH 7.4) for 24 h at 5°C and routinely embedded in paraffin wax. Sections (3 to 5 μm) were prepared and either stained with hematoxylin-eosin (HE) and examined for the presence of BIBD IB or used for IHC. Briefly, for IHC, after antigen retrieval with citrate buffer (pH 6.0) in a microwave oven, sections were incubated with affinity-purified rabbit anti-UHV NP antibody (0.25 μg/ml in PBS), followed by HRP-labeled goat anti-rabbit antibody (UltraVision anti-rabbit HRP detection system; Thermo Scientific), with subsequent visualization with diaminobenzidine tetrahydrochloride (DAB) and hematoxylin counterstaining as described previously (10).
Histological and IHC examinations were performed on pellets from two culture flasks for each test in a semiquantitative fashion. In each complete pellet section (×400 magnification), the proportion of cells with one or more intracytoplasmic IB was graded in 10% steps. The IB size was assessed as the diameter in micrometers. Viral NP protein expression was seen in association with IB and as a more diffuse but distinct cytoplasmic reaction. Based on the staining intensity within positive cells and the proportion of positive cells, the IHC reaction was graded on a scale from 0.5 to 3, corresponding to a faint (0.5), weak (1), weak to moderate (1.5), moderate (2), moderate to strong (2.5) or strong (3) overall staining intensity. Subsequently, the IHC reaction strength and histology score were combined as an indicator of the presence and extent of arenavirus infection in the culture. Scoring was undertaken three times by the same investigator (U.H.) in a blinded fashion.
For TEM, cells were scraped from the flasks and pellets were prepared as described above, fixed in 1.5% glutaraldehyde buffered in 0.2 M cacodylic acid buffer, pH 7.3, for 12 h at 5°C, and routinely embedded in epoxy resin. Semithin and thin sections were prepared as previously described (10), and the latter were examined for the presence and morphology of BIBD IB, using a Philips CM 100.
For IF analyses, cells were grown and infected either on diagnostic 10-well slides or in culture vessels, detached by pipetting (tick cells) or trypsinization (vertebrate cells), washed with PBS, diluted in PBS, and dried on slides. After fixation in acetone for 10 min, slides were incubated with the anti-NP and anti-NP-C antibodies diluted in PBS (0.25 μg/ml to 0.5 μg/ml), followed by Alexa Fluor 488-labeled or Alexa Fluor 555-labeled goat anti-rabbit antibody (both diluted 1:1,500 in PBS; Invitrogen) for visualization.
RESULTS
Expression and purification of recombinant UHV NP.
We recently characterized two rabbit antisera raised against UHV purified from cell cultures of boid cells (10). Both antisera also reacted with proteins of boid cells, which prompted us to produce and purify protein-specific antibodies. For this purpose, we produced full-length UHV NP (rNP, aa 1 to 582), the N-terminal portion of NP (rNP-N, aa 1 to 339), and the C-terminal portion of NP (rNP-C, aa 346 to 582) with histidine tags, using a baculovirus expression system. Immobilized-metal affinity chromatography (IMAC) under denaturing conditions served to purify the proteins, and SDS-PAGE and immunoblotting with anti-His antibody were used to estimate the purity of the preparations (Fig. 1, top left and middle). While rNP and rNP-C each yielded a single distinct band, rNP-N produced a triplet band of approximately the expected molecular size. It is likely that the triplet band is the result of either secondary modifications (e.g., phosphorylation) or N-terminally fragmented pieces of the desired product (the His tag is located in the C terminus), since all bands of the triplet were recognized by the anti-His antibody (Fig. 1, top middle).
FIG 1.
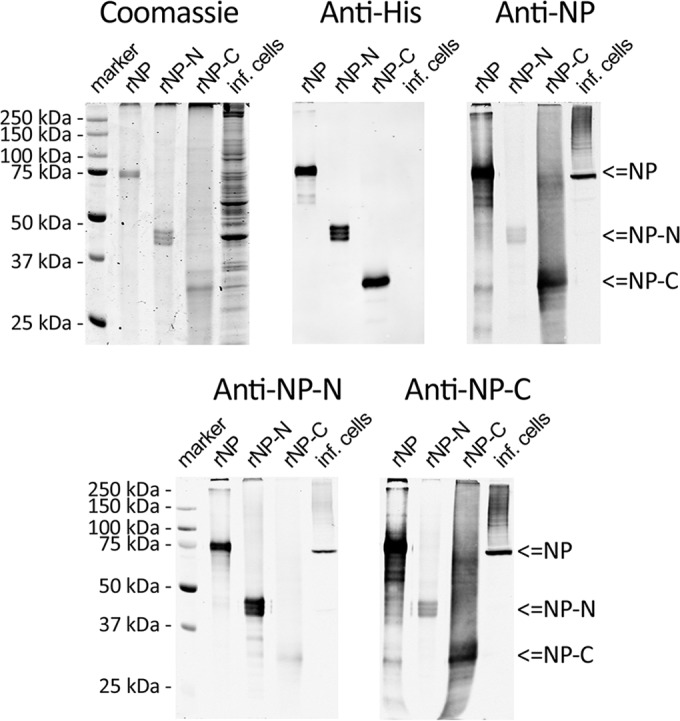
UHV recombinant proteins. (Top) Left, recombinant NP (rNP), rNP-N (N-terminal fragment of NP, amino acids [aa] 1 to 339), and rNP-C (C-terminal fragment of NP, aa 346 to 582) were IMAC purified under denaturing conditions, separated by SDS-PAGE, and visualized by Coomassie brilliant blue staining. Middle and right, immunoblots with anti-His tag and affinity-purified anti-UHV NP antibodies, respectively. (Bottom) Immunoblots with affinity-purified anti-rNP-N (left) and anti-rNP-C (right) antibodies. The immunoblots were visualized by Odyssey Infrared Imaging System (LI-COR).
rNP coupled to CNBr-Sepharose was used to affinity purify anti-NP PAb from antiserum produced against UHV (10) and NP-specific PAbs from antisera produced against rNP-N and rNP-C. The anti-NP antibody reacted strongly with infected cell lysates and rNP, and rNP-C elicited a much stronger immunoreaction than rNP-N (Fig. 1, top right), indicating a predominance of the epitope in the C terminus of NP. The PAbs purified from rNP-N and rNP-C antisera were further labeled with HRP to facilitate their direct use in downstream assays. Both the anti-rNP-N and the anti-tNP-C antibody were found to react with NP from infected cells and with rNP (Fig. 1, bottom). Anti-rNP-N reacted weakly with rNP-C and vice versa (Fig. 1, bottom), most probably due to the presence of the His tag in both proteins.
UHV infects cells at 30°C, but growth is limited or hindered at 37°C.
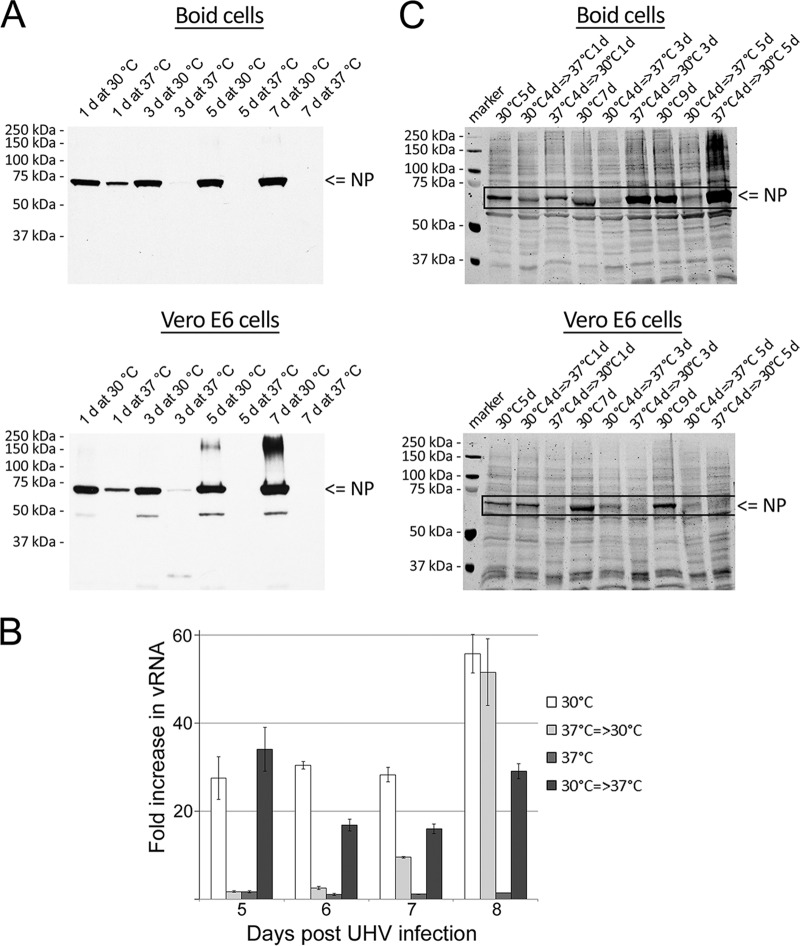
In our recent paper, we demonstrated that UHV not only grows in boid cells but can also infect and be adapted to grow in Vero E6 cells (10). Initial attempts at infecting the cells with UHV under standard culturing conditions (5% CO2 and 37°C) had been unsuccessful, whereas maintenance under conditions similar to those applied to the boid cells (10), i.e., at ambient temperatures between 27°C and 30°C, resulted in the accumulation of UHV antigens. We thus infected both boid kidney and Vero E6 cells with UHV (both Vero E6- and boa cell-adapted UHV), incubated the cells at either 30°C or 37°C, and monitored virus growth by immunoblotting to detect the accumulation of NP. When grown at 30°C, both cell lines already express NP at 2 days p.i., whereas no NP is detected in cells grown at 37°C (Fig. 2), indicating that growth is inhibited or markedly hindered at the higher temperature.
FIG 2.
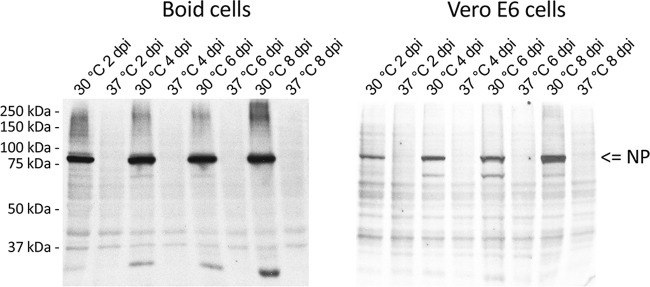
Temperature restricts BIBDAV growth in boid and Vero E6 cells. Boid and Vero E6 cells were infected with Vero-UHV and cultured at 30°C or 37°C. Cells were collected in lysis buffer, and 10 μg of total protein from each time point was analyzed by immunoblotting using HRP-labeled anti-rNP-C antibody. The results were recorded on X-ray film utilizing ECL reagents.
Productive UHV infection is reflected in the presence, size, and quantity of inclusion bodies and viral NP expression.
A specific feature of BIBD is the formation of variably sized eosinophilic intracytoplasmic IB (10). To further characterize the IB, to establish their requirement for active replication, and to assess whether adaptation to Vero E6 cells impairs the ability of UHV to infect boid cells, we collected boid and Vero E6 cells infected with boa- and Vero E6-adapted UHV (boa-UHV and Vero-UHV, respectively) at 8 days p.i. and analyzed them by both light microscopy (histology, IF, and IHC) and ultrastructural microscopy (TEM) methods. The most relevant quantitative results are summarized in Table 1.
TABLE 1.
Results of the semiquantitative assessment of BIBDAV infection of boid kidney and Vero E6 cells with boa-UHV and Vero-UHV maintained at different ambient temperatures for 8 days
| Cell type | Virus | % of cells with IB (range); IHC scorea in cells incubated at: |
|||
|---|---|---|---|---|---|
| 30°C (8 days) | 30°C (4 days) → 37°C (4 days) | 37°C (8 days) | 37°C (4 days) → 30°C (4 days) | ||
| Boid kidney | Boa-UHV | >30–40; 1.5 | >30–40; 1.5 | >30–40; 1.0 | >20–30; 1.0 |
| Vero-UHV | >40–50; 2.0 | >10–20; 1.5 | ≤10; 0.5 | >20–30; 1.0 | |
| Vero E6 | Boa-UHV | >10–20; 1.0 | ≤10; 0.5 | ≤10; 0.5 | ≤10; 0.5 |
| Vero-UHV | >30–40; 2.0 | ≤10; 1.0 | |||
Immunohistochemical expression of BIBDAV NP antigen, graded as faint (0.5), weak (1), weak to moderate (1.5), moderate (2), moderate to strong (2.5), and strong (3) based on the staining intensity within positive cells and the proportion of positive cells.
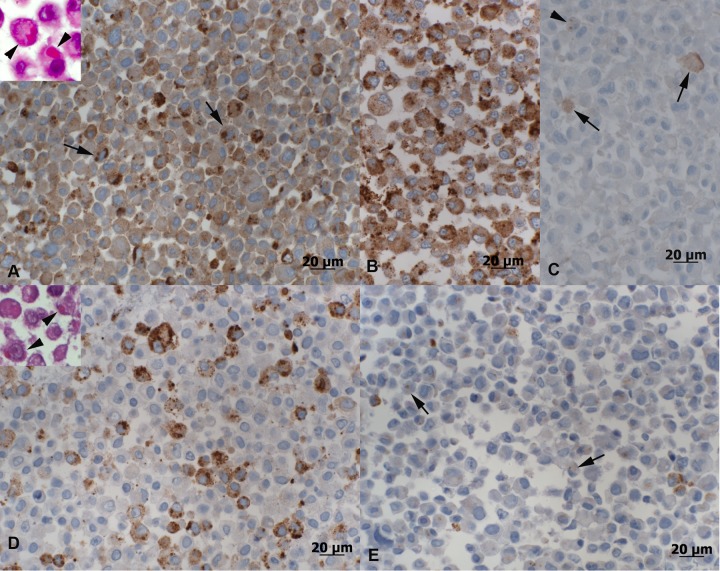
Regardless of the ambient temperature, boid kidney cells infected with boa-UHV exhibited variably sized (0.5 to 3.5 μm) IB in a moderate proportion (up to 40%) of cells, whereas viral NP expression was obvious in more than 50% of the cells, with an overall weak to moderate expression intensity (Fig. 3A). When consistently maintained at 30°C, Vero-UHV induced a more intense infection in the boid cells (up to 80% NP-positive cells and moderate NP expression intensity) (Fig. 3B). In contrast, incubation at 37°C reduced both the proportion of cells with IB and the degree of NP expression. This was most pronounced in cells consistently maintained at 37°C, when IB were often barely visible and only faint NP expression confirmed infection of the cells (Fig. 3C; Table 1). The results show that both boa- and Vero-UHV can productively infect boid cells. However, once the virus is adapted to Vero E6 cells, its replication capacity appears to decline with an increase in ambient temperature.
FIG 3.
Morphological features and BIBDAV NP expression in UHV-infected cell cultures at 8 days p.i. (A to C) Permanent cell line I/1Ki derived from boid kidney cells. (A) Infection with boa-UHV and incubation at 30°C. Viral NP expression is observed in more than 50% of the cells, with an overall weak to moderate staining intensity. NP expression is also seen in the inclusion bodies (arrows); the inset shows variably sized inclusion bodies as evident in HE-stained sections (arrowheads). (B) Infection with Vero-UHV and incubation at 30°C. Around 80% of cells exhibit NP expression, with overall moderate staining intensity. (C) Cells maintained at 37°C for 8 days. NP expression is mainly seen as a faint diffuse cytoplasmic staining in a few cells (arrows) or, rarely, as very small cytoplasmic inclusion bodies (arrowhead); IHC score, 0.5. (D, E). African green monkey (Vero E6) cells after UHV infection and culture at 30°C for 8 days. (D) Infection with Vero-UHV. Viral NP expression is observed in approximately 40% of the cells, with an overall moderate staining intensity; the inset shows variably sized inclusion bodies as evident in HE-stained sections (arrowheads). (E) Infection with boa-UHV results in the formation of small IB that are only visible when stained for NP expression (arrows); NP expression is seen in less than 20% of cells, with generally weak intensity. HRP method, hematoxylin counterstain; insets, HE stain.
At a consistent ambient temperature of 30°C, Vero-UHV in Vero E6 cells and boa-UHV in boid kidney cells yielded similar results (B of 0.5 to 3.0 μm in diameter in up to 40% of cells and moderate NP expression in up to 40% of cells) (Fig. 3D). Boa-UHV infected the Vero E6 cells at 30°C but with low efficiency, since no more than 20% of cells had developed IB at 8 days p.i. and NP was only weakly expressed (Fig. 3E). Incubation at 37°C for any length of time resulted in an even lower efficiency of infection (Table 1). Interestingly, infection with Vero-UHV seemed to fail when the cells were incubated at 37°C immediately after infection (Table 1). These findings suggest that adaptation of UHV to Vero E6 cells is associated with loss of its capacity to infect and grow in these cells at 37°C, perhaps through selection during adaptation.
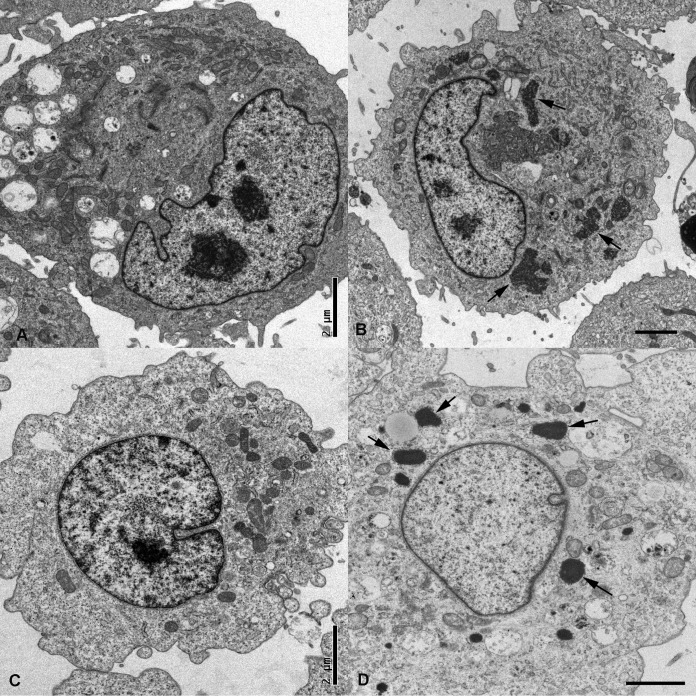
When cells were grown at 37°C, the ultrastructural characteristics of the IB were very similar irrespective of the infected cell line or the virus isolate used (Fig. 4). The kinetics of IB formation appeared to be similar in both cell lines. At 8 days p.i., the majority of IB were of irregular shape. By 12 days p.i., they had acquired the round to ovoid shape of the IB typically seen in cells of boids with BIBD (10). At this stage, the number of IB overall appeared to be lower than at 8 days p.i. (data not shown).
FIG 4.
Ultrastructural features of mock-infected (A) and boa-UHV-infected (B) boid kidney cells and mock-infected (C) and Vero-UHV-infected (D) Vero E6 cells. Cells were maintained at 30°C for 8 days p.i. (B) In boid kidney cells, IB appear as several irregularly shaped intracytoplasmic structures (arrows). (D) In the Vero E6 cells, IB are round to ellipsoid (arrows).
Exposure of infected cells to higher ambient temperatures leads to a decrease in the amount of NP.
Since the growth of UHV was found to be impaired at 37°C, we decided to more closely investigate the effect of a temperature rise to 37°C on cells infected and initially grown at 30°C. We therefore passaged UHV-infected cells at 15 days p.i. (boid cells) and 12 days p.i. (Vero E6 cells) and incubated the new plates at either 30°C or 37°C. UHV replication was monitored at 2-day intervals by quantifying the amount of UHV NP in cells using immunoblotting. The temperature increase to 37°C was associated with a gradual, time-dependent decrease in the amount of UHV NP in cells, indicating that the higher temperature adversely affected the replication of UHV (Fig. 5A). The observed decrease in NP also indicates that the cells lose the accumulated NP deposits, likely through the normal cell turnover, since cytopathic effects were not seen. This would suggest that the IB are indeed dynamic complexes and essential for replication (13).
FIG 5.
The effects of ambient temperature on the growth of UHV and T10404. (A) Boid and Vero E6 cells infected with UHV were transferred to fresh 6-well plates (15 days p.i. and 12 days p.i., respectively) and grown at 30°C or 37°C. Cells were collected at 2-day intervals in lysis buffer, and 10 μg of total protein from each time point was analyzed by immunoblotting using HRP-labeled anti-rNP-C antibody. The results were recorded on X-ray film utilizing ECL reagents. (B) Boid cells were infected with UHV and grown at 30°C or 37°C. At 5 days p.i., a plate of cells grown at 37°C was transferred to 30°C and a plate of cells grown at 30°C to 37°C. Infected cells constantly kept at 30°C or 37°C were used as controls. RNA isolated from cells collected at 5, 6, 7, and 8 days p.i. (x axis) was quantified with qPCR using UHV Z protein-specific primers. The results are shown as the fold increase in comparison to the vRNA level at 2 days p.i. of cells grown at 37°C, the highest CT value measured. (C) Boid and Vero E6 cells were infected with T10404 and cultured at 30°C or 37°C. At 4 days p.i., the cells grown at 37°C were transferred to 30°C (37°C → 30°C), and a plate of cells grown at 30°C was transferred to 37°C (30°C → 37°C). Infected cells kept constantly at 30°C were used as positive controls. Cells were collected in lysis buffer, and 10 μg of total protein from each time point was analyzed by immunoblotting using affinity-purified anti-UHV NP antibody (anti-rNP-N and anti-rNP-C antibodies yielded lower signal intensities). The immunoblot image was recorded using the Odyssey infrared imaging system (LI-COR).
To analyze whether the decrease in NP was due to impaired replication of the virus, we compared the amounts of viral RNA (both genomic and antigenomic) after incubation at different temperatures. We infected boid cells with boa-UHV, incubated them at 30°C or 37°C for 5 days, and then transferred a plate of cells grown at 30°C to 37°C and vice versa. Cells maintained at 30°C or 37°C for the entire examination period served as controls. Samples collected daily until 8 days p.i. were analyzed by qPCR using UHV Z protein-specific primers, which showed that UHV replication was dramatically reduced at 37°C (Fig. 5B); however, when such cells were transferred to 30°C after 5 days, replication restarted. When cells grown at 30°C were transferred to 37°C, the amount of viral RNA decreased; interestingly, the amount of viral RNA started to increase again after 4 days at the higher temperature but at levels approximately 20 to 40 times lower than in cells grown at 30°C. Despite the modest recovery of RNA levels (Fig. 5B), no protein expression was seen in cells grown at 37°C (Fig. 2).
UHV originates from a chronically infected boid bone marrow cell line passaged for more than 10 years at temperatures between 27°C and 30°C (10). Therefore, in order to exclude previous cell culture adaptation to a specific temperature range, we decided to study the temperature preference of a recently isolated, genetically distant BIBDAV (T10404; from snake number 5 in reference 10). We infected both Vero E6 and boid cells with T10404 and maintained the cultures at 30°C or 37°C; at 4 days p.i., we performed temperature swaps and monitored virus growth in samples collected at 5, 7, and 9 days p.i. as described above. The progressive decline in the amount of NP indicates that the replication of T10404 is also impaired in both cell types at 37°C (Fig. 5C, 2nd, 5th, and 8th lanes) in comparison to its replication rate in cells grown at 30°C (Fig. 5C, 1st, 4th, and 7th lanes). Different from the Vero E6 cells, boid cells inoculated with the virus at 37°C started to produce virus once transferred to 30°C (Fig. 5C, 3rd, 6th, and 9th lanes), suggesting that, like UHV, T10404 retains its infectivity and/or replicates at a low level in boid cells at a higher temperature.
UHV infects cells from animals of different classes.
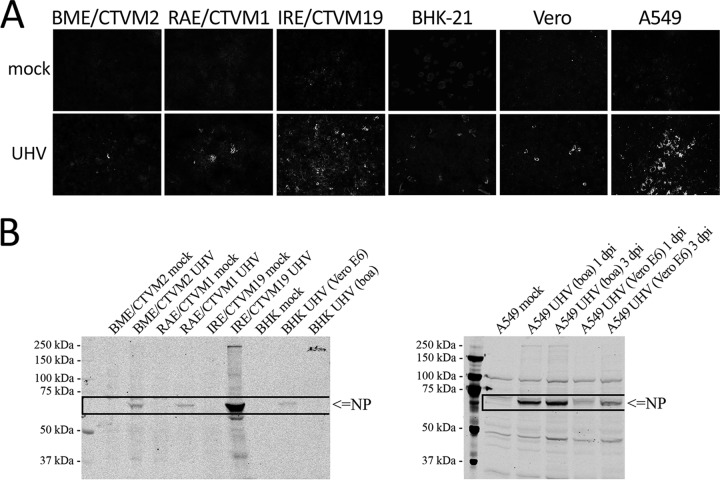
To further investigate the potential of BIBDAV to infect cells other than boid and nonhuman primate cells (Vero E6), we infected a range of cell lines originating from animals of different classes (mammalian and arthropod) with UHV. We wanted to test the replication of BIBDAV in arthropod cells because BIBD epidemics are often concomitant with snake mite infestation (25), and snake mites could act as a vector for BIBDAV. Since mite cells are not available, we used tick cell lines instead. We found evidence of UHV growth (NP production) by both IF (Fig. 6A) and immunoblot (Fig. 6B) assays in both mammalian (Vero, Vero E6, A549, and BHK-21 cell lines) and arthropod [tick cell lines RAE/CTVM1 from R. appendiculatus, IRE/CTVM19 from I. ricinus, and BME/CTVM2 from R. (B.) microplus] cells when they were propagated at 30°C, indicating that these cells are permissive for UHV. Curiously, human cells were found permissive for UHV previously propagated in boid but not in Vero E6 cells. These results highlight the potential of arenaviruses to cross species barriers.
FIG 6.
Susceptibility of different cell lines to UHV infection. BME/CTVM2 [R. (B.) microplus embryo], RAE/CTVM1 (R. appendiculatus embryo), IRE/CTVM19 (I. ricinus embryo) BHK-21 (baby hamster kidney), Vero (African green monkey kidney), and A549 (human lung adenocarcinoma) cells were infected with UHV and grown at 30°C. (A) Infected (UHV) and noninfected (mock) cells were fixed (at 8 days p.i. for BHK and Vero, 14 days p.i. for tick cell lines, and 3 days p.i. for A549) and stained with anti-rNP-C polyclonal antibody in conjugation with anti-rabbit Alexa Fluor 488-labeled secondary antibody. The cells were imaged at ×400 magnification. (B) Immunoblot of UHV-infected tick and BHK-21 cells (left) and A549 cells (right). The cells were collected (at 8 days p.i. for BHK, 14 days p.i. for tick cell lines, and at 1 and 3 days p.i. for A549) in lysis buffer, and 10 μg of total protein analyzed by immunoblotting with affinity-purified anti-UHV NP antibody; the immunoblot image was recorded with the Odyssey infrared imaging system (LI-COR).
DISCUSSION
BIBD is a fatal disease of boid snakes that is morphologically characterized by the formation of typical intracytoplasmic IB. Recent studies have provided convincing evidence that arenaviruses are the causative agents of BIBD; however, the reservoir host of BIBDAV—if any host exists besides boid snakes themselves—is still unknown. For classical arenaviruses, the known reservoir hosts are mammals, i.e., rodents and, for one arenavirus (Tacaribe virus), a bat (4). The isolation of BIBDAV from animals of a different taxonomic class, the Reptilia (i.e., snakes), suggested that arenaviruses might have considerable ability to cross species barriers (10). With this in mind, we tested UHV for its ability to infect a panel of cell lines from mammalian and arthropod species and herein report that BIBDAV can indeed infect cells of various animal classes. Furthermore, we demonstrate that the replication of BIBDAV is temperature sensitive regardless of the cell type and virus strain used.
BIBDAV, like all other arenaviruses in their hosts, cause systemic infections in the snakes; the fact that BIBDAV replicate effectively at 30°C but less or not at all at 37°C indicates that the body temperature of the reservoir host of these viruses is lower than 37°C. Mammals are endothermic, with body temperatures close to 37°C except when hibernating, which suggests that the BIBDAV reservoir hosts might not be mammals. In contrast, snakes are exothermic and can therefore provide a suitable ambient temperature range to promote the effective growth of BIBDAV. Captive boid snakes are commonly housed under controlled-temperature conditions (typically 25°C at night and 27 to 31°C during the day) which we have shown would support the replication of BIBDAV. This could explain the high susceptibility of captive boids to these viruses. In contrast, wild snakes are exposed to a broader range of temperatures, and in the case of boids and other snakes known to develop BIBD, these are often higher. Our findings thus suggest that, in wild snakes, the replication of BIBDAV could be restricted or influenced by changes in the ambient temperature and the susceptibility of a snake species might actually be related to the environment in which it lives. Chronic infection of the reservoir host is seen with classical rodent-borne arenaviruses and is considered almost a hallmark of the family Arenaviridae. Accordingly, BIBDAV could chronically infect boids living in the wild; however, to our knowledge there is so far no report suggesting this.
The occurrence of BIBD in snake collections has been associated with concurrent snake mite (Ophionyssus natricis) infestation (25). Since cell lines of this arthropod species are not available, we tested embryo-derived cell lines of three tick species, I. ricinus, R. appendiculatus, and R. (B.) microplus, for their susceptibility to UHV. Indeed, all three tick cell lines were found to support UHV replication, which provides the first evidence that BIBDAV could indeed be transmitted by an arthropod vector, i.e., be an arbovirus, with arthropods acting as intermediate or even as reservoir hosts for BIBDAV. This hypothesis is supported by the fact that other arthropod-borne viruses, i.e., members of the Togaviridae and Flaviviridae, have been found in snakes (26), and arthropods such as ticks and mosquitoes can harbor persistent arbovirus infections in the absence of obvious deleterious effects (27–29). On the other hand, LCMV, another arenavirus, has been shown to productively infect primary tick cell cultures (30), which indicates that tick cells are in general permissive for arenaviruses. Transmission via an arthropod vector would allow BIBDAV, at least in theory, to cross from one host species to another in the wild. It remains to be studied whether snake mites or other blood-sucking parasites, such as ticks, can indeed transmit the virus.
The facts that UHV was able to infect such a broad range of cell lines and was still able to infect boid cells after it was adapted to Vero E6 cells through cultivation are particularly interesting with regard to its potential receptor usage. Curiously, Vero-UHV infected A549 cells with lower efficacy than boa-UHV. Vero E6 cells are known to secrete interferon-lambda (IFN-λ) in response to infection by New World hantaviruses (31). Thus, the observed growth restriction of Vero-UHV in A549 cells might be due to inhibitory effects of IFN-λ. As few tools are available for detecting boid proteins or genes, the infection of Vero E6 and A549 cells will facilitate studies on, for example, the interaction of BIBDAV with receptors and innate immunity. Rodent-borne arenaviruses use transferrin receptor 1 (TfR1) and α-dystroglycan as their receptors when infecting humans (32, 33). A549 cells are known to express TfR1, and therefore, it is of interest to determine whether BIBDAV could use this receptor for their entry. Thus far, nothing is known about the cellular receptor(s) of BIBDAV. However, the ability of UHV to infect various cell lines and the fact that snakes with BIBD develop IB in basically all cell types suggest that the receptor is more or less ubiquitous and conserved throughout animal classes. This would, again in theory, enable the spread of BIBDAV to different classes of animals. Nevertheless, it appears that the spread of these viruses is limited by their growth temperature requirements.
ACKNOWLEDGMENTS
We are grateful to Kirsi Aaltonen, Irina Suomalainen, and the technicians in the Finnish Centre for Laboratory Animal Pathology, Faculty of Veterinary Medicine, University of Helsinki, as well as Lisbeth Nufer, TEM Unit, Institute of Veterinary Pathology, Vetsuisse Faculty, University of Zurich, for excellent technical assistance. We thank the Tick Cell Biobank, the Pirbright Institute and Anu Hakala for the provision, transfer, and cultivation of tick cell lines.
The study was supported by the Academy of Finland and the Finnish Foundation for Veterinary Research (2011 and 2012 grant rounds).
REFERENCES
- 1.Irwin NR, Bayerlova M, Missa O, Martinkova N. 2012. Complex patterns of host switching in New World arenaviruses. Mol Ecol 21:4137–4150. doi: 10.1111/j.1365-294X.2012.05663.x. [DOI] [PubMed] [Google Scholar]
- 2.Martinez MG, Bialecki MA, Belouzard S, Cordo SM, Candurra NA, Whittaker GR. 2013. Utilization of human DC-SIGN and L-SIGN for entry and infection of host cells by the New World arenavirus, Junin virus. Biochem Biophys Res Commun 441:612–617. doi: 10.1016/j.bbrc.2013.10.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rieger T, Merkler D, Gunther S. 2013. Infection of type I interferon receptor-deficient mice with various old world arenaviruses: a model for studying virulence and host species barriers. PLoS One 8:e72290. doi: 10.1371/journal.pone.0072290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moraz ML, Kunz S. 2011. Pathogenesis of arenavirus hemorrhagic fevers. Expert Rev Anti Infect Ther 9:49–59. doi: 10.1586/eri.10.142. [DOI] [PubMed] [Google Scholar]
- 5.Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M, Nichol ST, Lipkin WI. 2009. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog 5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunberg JH, York J. 2012. The curious case of arenavirus entry, and its inhibition. Viruses 4:83–101. doi: 10.3390/v4010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bausch DG, Hadi CM, Khan SH, Lertora JJ. 2010. Review of the literature and proposed guidelines for the use of oral ribavirin as postexposure prophylaxis for Lassa fever. Clin Infect Dis 51:1435–1441. doi: 10.1086/657315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, Dunker F, Derisi JL. 2012. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio 3(4):e00180-12. doi: 10.1128/mBio.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodewes R, Kik MJ, Raj VS, Schapendonk CM, Haagmans BL, Smits SL, Osterhaus AD. 2013. Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in The Netherlands. J Gen Virol 94:1206–1210. doi: 10.1099/vir.0.051995-0. [DOI] [PubMed] [Google Scholar]
- 10.Hetzel U, Sironen T, Laurinmaki P, Liljeroos L, Patjas A, Henttonen H, Vaheri A, Artelt A, Kipar A, Butcher SJ, Vapalahti O, Hepojoki J. 2013. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J Virol 87:10918–10935. doi: 10.1128/JVI.01123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wozniak E, McBride J, DeNardo D, Tarara R, Wong V, Osburn B. 2000. Isolation and characterization of an antigenically distinct 68-kD protein from nonviral intracytoplasmic inclusions in boa constrictors chronically infected with the inclusion body disease virus (IBDV: Retroviridae). Vet Pathol 37:449–459. doi: 10.1354/vp.37-5-449. [DOI] [PubMed] [Google Scholar]
- 12.Schumacher J, Jacobson ER, Homer BL, Gaskin JM. 1994. Inclusion body disease in boid snakes. J Zoo Wildl Med 25:511–524. [Google Scholar]
- 13.Baird NL, York J, Nunberg JH. 2012. Arenavirus infection induces discrete cytosolic structures for RNA replication. J Virol 86:11301–11310. doi: 10.1128/JVI.01635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charrel RN, Coutard B, Baronti C, Canard B, Nougairede A, Frangeul A, Morin B, Jamal S, Schmidt CL, Hilgenfeld R, Klempa B, de Lamballerie X. 2011. Arenaviruses and hantaviruses: from epidemiology and genomics to antivirals. Antiviral Res 90:102–114. doi: 10.1016/j.antiviral.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Urata S, Yasuda J. 2012. Molecular mechanism of arenavirus assembly and budding. Viruses 4:2049–2079. doi: 10.3390/v4102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodewes R, Raj VS, Kik MJ, Schapendonk CM, Haagmans BL, Smits SL, Osterhaus AD. 2014. Updated phylogenetic analysis of arenaviruses detected in boid snakes. J Virol 88:1399–1400. doi: 10.1128/JVI.02753-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hetzel U, Sironen T, Laurinmaki P, Liljeroos L, Patjas A, Henttonen H, Vaheri A, Artelt A, Kipar A, Butcher SJ, Vapalahti O, Hepojoki J. 2014. Reply to “updated phylogenetic analysis of arenaviruses detected in boid snakes”. J Virol 88:1401. doi: 10.1128/JVI.03044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burri DJ, Palma JR, Kunz S, Pasquato A. 2012. Envelope glycoprotein of arenaviruses. Viruses 4:2162–2181. doi: 10.3390/v4102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huiskonen JT, Hepojoki J, Laurinmaki P, Vaheri A, Lankinen H, Butcher SJ, Grunewald K. 2010. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J Virol 84:4889–4897. doi: 10.1128/JVI.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell-Sakyi L, Zweygarth E, Blouin EF, Gould EA, Jongejan F. 2007. Tick cell lines: tools for tick and tick-borne disease research. Trends Parasitol 23:450–457. doi: 10.1016/j.pt.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Pedra JH, Narasimhan S, Rendic D, DePonte K, Bell-Sakyi L, Wilson IB, Fikrig E. 2010. Fucosylation enhances colonization of ticks by Anaplasma phagocytophilum. Cell Microbiol 12:1222–1234. doi: 10.1111/j.1462-5822.2010.01464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell-Sakyi L. 2004. Ehrlichia ruminantium grows in cell lines from four ixodid tick genera. J Comp Pathol 130:285–293. doi: 10.1016/j.jcpa.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Levingston Macleod JM, D'Antuono A, Loureiro ME, Casabona JC, Gomez GA, Lopez N. 2011. Identification of two functional domains within the arenavirus nucleoprotein. J Virol 85:2012–2023. doi: 10.1128/JVI.01875-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hastie KM, Liu T, Li S, King LB, Ngo N, Zandonatti MA, Woods VL Jr, de la Torre JC, Saphire EO. 2011. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci U S A 108:19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang L, Jacobson ER. 2010. Inclusion body disease, a worldwide infectious disease of boid snakes: a review. J Exotic Pet Med 19:216–225. doi: 10.1053/j.jepm.2010.07.014. [DOI] [Google Scholar]
- 26.Essbauer S, Ahne W. 2001. Viruses of lower vertebrates. J Vet Med B Infect Dis Vet Public Health 48:403–475. doi: 10.1046/j.1439-0450.2001.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labuda M, Nuttall PA. 2004. Tick-borne viruses. Parasitology 129(Suppl):S221–S245. doi: 10.1017/S0031182004005220. [DOI] [PubMed] [Google Scholar]
- 28.Blair CD. 2011. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol 6:265–277. doi: 10.2217/fmb.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballinger MJ, Bruenn JA, Hay J, Czechowski D, Taylor DJ. 2014. Discovery and evolution of bunyavirids in arctic phantom midges and ancient bunyavirid-like sequences in insect genomes. J Virol 88:8783–8794. doi: 10.1128/JVI.00531-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rehacek J. 1965. Cultivation of different viruses in tick tissue cultures. Acta Virol 9:332–337. [PubMed] [Google Scholar]
- 31.Prescott J, Hall P, Acuna-Retamar M, Ye C, Wathelet MG, Ebihara H, Feldmann H, Hjelle B. 2010. New World hantaviruses activate IFNlambda production in type I IFN-deficient Vero E6 cells. PLoS One 5:e11159. doi: 10.1371/journal.pone.0011159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li W, Nagel J, Schmidt PJ, Nunberg JH, Andrews NC, Farzan M, Choe H. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choe H, Jemielity S, Abraham J, Radoshitzky SR, Farzan M. 2011. Transferrin receptor 1 in the zoonosis and pathogenesis of New World hemorrhagic fever arenaviruses. Curr Opin Microbiol 14:476–482. doi: 10.1016/j.mib.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]