ABSTRACT
Giardia lamblia virus (GLV) is a small, nonenveloped, nonsegmented double-stranded RNA (dsRNA) virus infecting Giardia lamblia, the most common protozoan pathogen of the human intestine and a major agent of waterborne diarrheal disease worldwide. GLV (genus Giardiavirus) is a member of family Totiviridae, along with several other groups of protozoal or fungal viruses, including Leishmania RNA viruses and Trichomonas vaginalis viruses. Interestingly, GLV is more closely related than other Totiviridae members to a group of recently discovered metazoan viruses that includes penaeid shrimp infectious myonecrosis virus (IMNV). Moreover, GLV is the only known protozoal dsRNA virus that can transmit efficiently by extracellular means, also like IMNV. In this study, we used transmission electron cryomicroscopy and icosahedral image reconstruction to examine the GLV virion at an estimated resolution of 6.0 Å. Its outermost diameter is 485 Å, making it the largest totivirus capsid analyzed to date. Structural comparisons of GLV and other totiviruses highlighted a related “T=2” capsid organization and a conserved helix-rich fold in the capsid subunits. In agreement with its unique capacity as a protozoal dsRNA virus to survive and transmit through extracellular environments, GLV was found to be more thermoresistant than Trichomonas vaginalis virus 1, but no specific protein machinery to mediate cell entry, such as the fiber complexes in IMNV, could be localized. These and other structural and biochemical findings provide a basis for future work to dissect the cell entry mechanism of GLV into a “primitive” (early-branching) eukaryotic host and an important enteric pathogen of humans.
IMPORTANCE Numerous pathogenic bacteria, including Corynebacterium diphtheriae, Salmonella enterica, and Vibrio cholerae, are infected with lysogenic bacteriophages that contribute significantly to bacterial virulence. In line with this phenomenon, several pathogenic protozoa, including Giardia lamblia, Leishmania species, and Trichomonas vaginalis are persistently infected with dsRNA viruses, and growing evidence indicates that at least some of these protozoal viruses can likewise enhance the pathogenicity of their hosts. Understanding of these protozoal viruses, however, lags far behind that of many bacteriophages. Here, we investigated the dsRNA virus that infects the widespread enteric parasite Giardia lamblia. Using electron cryomicroscopy and icosahedral image reconstruction, we determined the virion structure of Giardia lamblia virus, obtaining new information relating to its assembly, stability, functions in cell entry and transcription, and similarities and differences with other dsRNA viruses. The results of our study set the stage for further mechanistic work on the roles of these viruses in protozoal virulence.
INTRODUCTION
Giardia lamblia (also called G. intestinalis or duodenalis) is a flagellated, parasitic protozoan and an important cause of diarrheal disease worldwide, especially in developing countries. It is a member of the supergroup Excavata, one of the earliest diverging eukaryotic taxa, to which the parasitic protozoa Leishmania, Naegleria, Trichomonas, and Trypanosoma also belong, although in distinct groups or subgroups relative to Giardia (1). Infection by G. lamblia occurs after ingestion of cysts from contaminated water, food, or soil or sometimes by direct contact. After excystation and multiplication, G. lamblia trophozoites adhere to the surface of epithelial cells in the small intestine. Specific pathogenic mechanisms remain poorly characterized, but the resulting diarrheal disease is characterized by malabsorption, maldigestion, chloride hypersecretion, and increased intestinal transit rates (2, 3). Histopathological findings include lymphocyte infiltration and diffuse blunting of the microvilli, enterocyte apoptosis, and disruption of epithelial tight junctions.
Epidemiologically, G. lamblia is the most common protozoan pathogen of the human small intestine, estimated to cause ≥280 million cases yearly (2, 4), and is also common in companion animals and livestock. Most infections are self-limiting, but in immunocompromised persons, including undernourished children and people with AIDS, G. lamblia disease may be more chronic, severe, and atypical and may be associated in children with growth retardation and cognitive impairment (5, 6). Notably, like several other protozoan pathogens, G. lamblia can undergo antigenic variation to evade host adaptive immunity (7), contributing to its capacity for chronic infections. It can also occur in outbreaks due to localized contamination of food or water sources and is therefore recognized by the National Institutes of Health/National Institute of Allergy and Infectious Disease (NIH/NIAID) as a category B priority pathogen (http://www.niaid.nih.gov/topics/biodefenserelated/biodefense/pages/cata.aspx) associated with food and waterborne illness.
G. lamblia itself is susceptible to infection by a small, nonenveloped double-stranded RNA (dsRNA) virus called Giardia lamblia virus (GLV). Discovered in the 1980s (8), GLV represents the prototype and indeed is the sole species in the genus Giardiavirus, family Totiviridae (Fig. 1). It has a nonsegmented, 6.3-kbp genome with two, long, partially overlapping open reading frames (ORFs) on one of the genomic RNA strands, respectively encoding the capsid/coat protein (CP; ORF1) and the RNA-dependent RNA polymerase (RdRp; ORF2) (9). In addition, the ORF1 product undergoes proteolytic cleavage(s) to remove its 32 N-terminal amino acids, which appear not to be present in mature virions (10). The RdRp is expressed in fusion with the CP (i.e., as CP/RdRp) by means of −1 ribosomal frameshifting in the region of ORF1-ORF2 overlap (9, 11). The GLV mRNA, which spans the full length of the genomic plus strand, lacks a 5′ cap and is not polyadenylated; translation is instead initiated by means of an internal ribosomal entry site that partially overlaps ORF1 (12, 13).
FIG 1.
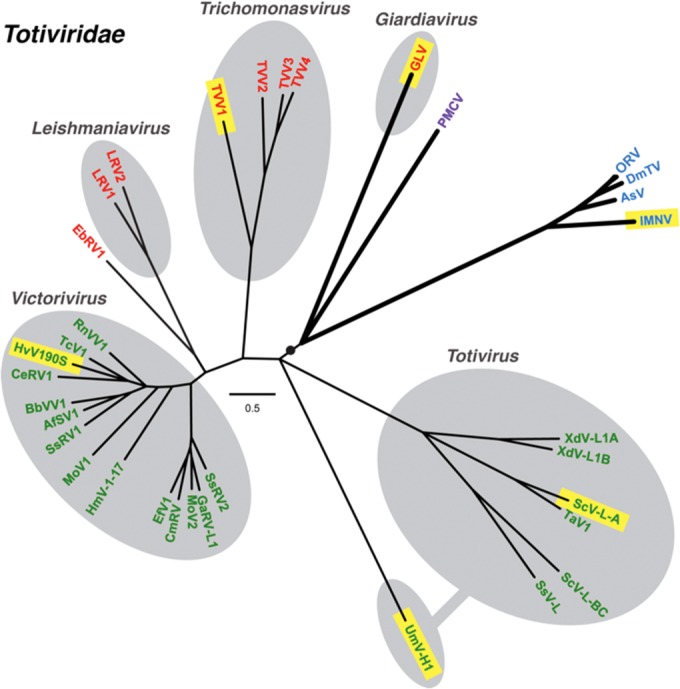
Phylogenetic relationships among taxonomically approved and tentatively assigned members of family Totiviridae. A maximum-likelihood phylogenetic tree was reconstructed from sequences encompassing the predicted CP+RdRp products of each analyzed virus, as described in Materials and Methods. Branches with support values <70% were collapsed to the preceding node, and some terminal branches within the genus Victorivirus are not labeled for simplicity but were all supported at ≥97%. Viruses are labeled with abbreviated names (see Materials Methods for key and GenBank accession numbers), and those for which 3D structures have been determined are highlighted in yellow. Protozoal viruses are labeled in red, fungal viruses are labeled in green, arthropod viruses are labeled in blue, and the one vertebrate virus is labeled in purple. Approved Totiviridae members are grouped within gray ovals according to current genus assignments as labeled; fungal virus UmV-H1 is currently grouped in genus Totivirus as shown but is divergent and appears to warrant reclassification. The black circle and thicker branch lines indicate an apparent subclade of viruses that can undergo efficient extracellular transmission. The scale bar indicates the number of substitutions per aligned position.
Several other parasitic protozoa, including Leishmania, Trichomonas, and Cryptosporidium, are also known to carry small dsRNA viruses: Leishmania RNA viruses (LRVs) 1 and 2 (genus Leishmaniavirus, family Totiviridae); Trichomonas vaginalis viruses (TVVs) 1, 2, 3, and 4 (genus Trichomonasvirus, family Totiviridae); and Cryptosporidium parvum virus 1 (genus Cryspovirus, family Partitiviridae). Moreover, several types of evidence suggest that these other protozoal dsRNA viruses might enhance the pathogenicity of their respective protozoa for causing mammalian disease (14–19). It is not yet clear whether the same may be true for GLV in exacerbating Giardia-associated disease, although an early review by Wang and Wang (20) indicated there is no obvious correlation between GLV and Giardia virulence.
Interestingly, GLV is the only one of the known protozoal dsRNA viruses that can transmit efficiently by extracellular means (8, 21), meaning that GLV virions uniquely include the protein machinery for mediating efficient cell entry. The GLV-Giardia virus-host system thus provides an unusual opportunity to determine the steps involved in cell entry of a “primitive” (early-branching) eukaryote, with a simplified endo/lysosomal vacuolar system (22, 23). Attachment of GLV to the G. lamblia host cell has been shown to involve a cell surface receptor, which is missing from some G. lamblia strains (21, 24), but the identity of this receptor remains unknown. Otherwise, the entry mechanism of GLV is poorly characterized, except for one report suggesting that it is susceptible to inhibition by ammonium chloride and chloroquine and thus may require endocytosis and vacuolar acidification (25).
Resemblance of GLV to other Totiviridae members has not been previously confirmed at the structural level, except by negative-stain transmission electron microscopy (TEM) showing nonenveloped particles with single-layered capsids, appearing to measure 340 to 370 Å in diameter (8, 26). Moreover, in addition to its virions containing the protein machinery for efficient cell entry, which the other Totiviridae members from protozoal or fungal hosts do not, GLV appears to be more closely related to a group of nonsegmented dsRNA viruses isolated in recent years from metazoan hosts, both arthropods (crustaceans and insects) and vertebrates (bony fish) (27–31) and tentatively also assigned to family Totiviridae (Fig. 1). Notably, these metazoan viruses share with GLV the ability to transmit efficiently by extracellular means. The virions of one of these viruses, penaeid shrimp infectious myonecrosis virus (IMNV), have been analyzed by transmission electron cryomicroscopy (cryo-TEM) and icosahedral three-dimensional (3D) image reconstruction, which revealed an unusual protein-fiber complex that projects above the so-called “T=2” capsid at the icosahedral 5-fold axes and is likely to play one or more key roles in cell entry (32, 33). Such “T=2” capsid organization (34) reflects a T=1 structure in which a CP dimer occupies each of the icosahedral asymmetric units (120 CP subunits per capsid) and is common to many dsRNA viruses, including other members of family Totiviridae (35–39).
Considering the variety of interesting biological aspects of GLV and also the importance of its protozoan host G. lamblia as a human disease agent, we undertook this study to determine the virion structure of GLV via cryo-TEM and icosahedral 3D image reconstruction. In parallel, we also investigated several of the in vitro biochemical properties of GLV virions. The results of these studies reveal both similarities and differences between GLV and other Totiviridae members, including TVV1, prototype fungal totivirus Saccharomyces cerevisiae virus L-A (ScV-L-A), and tentatively assigned member IMNV, and set the stage for ongoing studies of cell entry by GLV.
MATERIALS AND METHODS
Protozoan cultures and virion purifications.
A frozen aliquot of G. lamblia strain WBI (GLV-infected WB) cells was kindly provided by C. C. Wang and coworkers (University of California–San Francisco). The batch culture was maintained as described previously (26) and expanded in cell numbers and volume over several days before harvest for virion purification. At harvest time, containers were chilled in an ice bath and swirled to remove attached cells from the glass surface. GLV virions were customarily purified from a batch of 2 × 109 WBI cells, using the same protocol of sonication and centrifugations as previously described for TVV1 virions (18, 39). GLV virions obtained from the last centrifugation step (banding in a CsCl density gradient) were dialyzed overnight against HNM buffer (50 mM HEPES [pH 7.2], 500 mM NaCl, 20 mM MgCl2) and then kept at 4°C for up to 3 weeks or at −80°C for longer periods before use. Concentrations of the dialyzed preparations routinely measured near 200 μg/ml (∼1012 virions/ml) according to a modified Bradford protein assay (Bio-Rad). T. vaginalis isolate UH9 was cultured, and TVV1-UH9 virions were purified as previously described (18, 39). The original T. vaginalis UH9 stock, which was kindly provided by Raina N. Fichorova and coworkers (Brigham and Women's Hospital and Harvard Medical School), had been serially cloned twice in soft agar before use in this study.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Two aliquots of purified GLV virions (each ∼2 × 1010 in 20 μl of HNM buffer) were denatured by heating to 80°C for 5 min and submitted to the Beth Israel Deaconess Medical Center Mass Spectrometry Facility for processing and analysis. Separate digestions were performed with sequencing-grade stocks of trypsin or chymotrypsin. Resulting spectra were analyzed using the Sequest search engine (40) against a customized database of GLV CP and CP/RdRp sequences. Analyses were otherwise performed as described previously (39).
TEM.
Prior to performing cryo-TEM, the integrity and homogeneity of samples were evaluated by using negative-stain TEM. For this, 3.5-μl aliquots of sample were absorbed to a continuous carbon grid that had been glow-discharged for ∼60 s in an Emitech K350 evaporation unit. The absorbed sample was then stained with 1% (unbuffered) uranyl acetate and rinsed with deionized distilled water. Micrographs were recorded on a 4K2 Gatan Ultrascan charge-coupled device camera in an FEI G2 Tecnai (Polara) microscope. Initial experiments performed to judge proper storage conditions for purified GLV virions and to provide an initial gauge of preparation quality were performed similarly, but using uranyl formate as stain and a JEOL 1200EX 80-kV microscope at the Harvard Medical School Department of Cell Biology Conventional EM Facility.
For cryo-TEM, 3.5-μl aliquots of sample were vitrified and examined using established procedures (36). In brief, samples were applied to Quantifoil holey grids that had been glow-discharged for ∼15 s. Grids were then blotted with Whatman filter paper for ∼5 s, plunged into liquid ethane, and transferred into a precooled FEI Polara multispecimen holder, which maintained the specimen at liquid-nitrogen temperature. Micrographs were recorded on Kodak SO-163 electron-image film in the Polara microscope operated at 200 keV and under minimal-dose conditions (∼25 e/A2) at a nominal magnification of ×59,000 and with the objective lens settings ranging between 0.78 and 3.41 μm underfocus.
Icosahedral 3D image reconstruction.
Micrographs exhibiting minimal astigmatism and specimen drift were digitized at 6.35-μm intervals (1.09-Å pixel size at the specimen) on a Nikon Super Coolscan 8000 microdensitometer. RobEM (http://cryoem.ucsd.edu/programs.shtm) was used to extract 14,125 particle images, each 601 × 601 pixels in size, and to preprocess the images as described previously (41). The defocus and astigmatism levels of each micrograph were estimated using the program CTFFIND3 (42). The random-model computation procedure (43) was used to generate an initial 3D density map at an ∼29-Å resolution from the images of 150 particles. This map served as a starting model to initiate full orientation and origin determinations of the entire set of images using AUTO3DEM v4.03.1 (44). After several cycles of orientation and origin refinement with the PO2R and P3DR functions of AUTO3DEM, the resolution converged to ∼6.0 Å (based on the 0.5 threshold of the Fourier shell correlation criterion [45]). The effects of the microscope contrast-transfer function were compensated in part by inverting image phases in spatial frequency bands where contrast reversals occurred (46). An inverse temperature factor of 1/(80 Å2) was applied to the final reconstruction (47).
The Segger v1.7 tool in the Chimera visualization package (48) was used to segment out individual CP-A and CP-B subunits in the final map and to examine interactions between them as described previously (38). Briefly, segmentation is obtained with this tool by means of an iterative procedure, starting from an initial arbitrary assignment of density regions to the two subunits and then refining those assignments in a semiautomatic manner by imposing additional restrictions. The additional restrictions in this case were based on evidence from the estimated capsid volume that the capsid indeed comprises 120 CP subunits, i.e., two CP subunits per icosahedral asymmetric unit, and on the following assumptions: (i) each subunit forms a compact domain, (ii) global morphology and particular features are repeated in CP-A and CP-B, (iii) symmetry constraints of the icosahedral arrangement of subunits within the capsid are maintained, and (iv) no obvious capsid density is left unassigned. Under these constraints, the final result does not depend on the initial arbitrary assignment to the two subunits. GLV subunit A was manually docked into GLV subunit B for comparison and the Chimera “fit-in-map” tool was then used to refine the fit. The normalized correlation value of the final fitting was 0.91 as provided by Chimera (48). Location and length of density features that could be assigned to putative α-helical segments in the segmented A subunits of the different totiviruses were predicted automatically using the VolTrac algorithm (49) implemented in the Sculptor graphics software (50). The procedure was performed as described by Dunn et al. (38).
Temperature stability of virions.
GLV or TVV1 virions (2 × 109 to 5 × 109 in 10 μl) were incubated in HNM buffer for 5 min at the indicated temperatures, using the gradient setting of a thermal cycler (MultiGene; Labnet International). RNase III (New England BioLabs) was then added to a final concentration of 0.02 U/μl, followed by incubation for 5 min at 37°C to digest dsRNA that was released from, or made accessible inside, destabilized virions. RNase III digestion was quenched by addition of EDTA to a final concentration of 100 mM. Protected dsRNA was then released from intact virions by heating the mixture for 5 min at 65–75°C. The genomic dsRNA bands of GLV and TVV1 were visualized on a 0.7% agarose gel, stained with ethidium bromide, and quantitated by using ImageJ software (http://imagej.nih.gov/ij/). The plot and fit for Fig. 6A were generated using DataGraph 3.1.1.
FIG 6.
Different responses of GLV and TVV1 virions to temperature and pH. Experiments were performed as detailed in Materials and Methods: GLV (closed symbols), TVV1 (open symbols). (A) Normalized amount of dsRNA resistant to RNase III as a function of temperature. The data points from three (GLV) or four (TVV) experiments with each virus were superimposed in the graph and used to generate the quadratic fit. (B) RNA transcriptase activity as a function of pH. The data points from two experiments with each virus, using two different buffers (Tris-acetate [diamonds] and Tris-HCl [circles]) were superimposed in the graph and used to generate the polynomial fit.
In vitro transcription.
GLV or TVV1 virions (108 in 20 μl) were incubated in transcription buffer (150 mM NaCl; 2 mM MgCl2; 200 μM concentrations each of ATP, CTP, and UTP; 20 μM [α-32P]GTP; and 50 mM Tris-HCl or Tris/acetate buffer at the indicated pH value) for 30 min at 37°C. The transcription reaction was terminated by addition of formamide and EDTA to final respective concentrations of 30% and 17 mM. Transcripts were separated on an 8% denaturing polyacrylamide gel containing 7 M urea in 0.5× Tris-borate-EDTA buffer. Radiolabeled RNA bands were then visualized by phosphorimaging on a Typhoon 9400 variable mode imager and quantitated using ImageQuant TL software (GE Healthcare). The plot and fit for Fig. 6B were generated using DataGraph 3.1.1.
Phylogenetic tree and polarity plot.
A maximum-likelihood phylogenetic tree was reconstructed from sequences encompassing the predicted CP+RdRp products of each analyzed virus (see Fig. 1). Multiple-sequence alignments were conducted using MAFFT 7.1, as implemented with default settings at http://mafft.cbrc.jp/alignment/software. Model testing was then performed using ProtTest 3.3 (51), which showed the Blosum62 substitution matrix and I+G+F distributions (proportion of invariable sites, 0.01; gamma shape parameter, 2.975; and empirically determined amino acid frequencies) to score most highly according to both Akaike and Bayesian information criteria. Phylogenetic analyses were next performed with these predetermined parameters using PhyML 3.0 (52) as implemented at http://www.hiv.lanl.gov/. The starting tree was optimized by both branch length and tree topology, and tree improvement was performed according to the best of nearest neighbor interchange and subtree pruning and regrafting. Branch support values (%) were estimated by the approximate likelihood ratio test with SH-like supports. The final tree was refined for display using FigTree 1.4.0. The abbreviations (along with the corresponding GenBank accession numbers) for the viruses included in the tree are as follows: GLV (NC_003555); TVV1 (NC_003824); TVV2 (NC_003873); TVV3 (NC_004034); TVV4 (HQ607522); LRV1 (NC_002063); LRV2 (NC_002064); EbRV1, Eimeria brunetti RNA virus 1 (NC_002701); SsRV2, Sphaeropsis sapinea RNA virus 2 (NC_001964); GaRV1-L1, Gremmeniella abietina RNA virus L1 (NC_003876); MoV2, Magnaporthe oryzae virus 2 (NC_010246); CmRV, Coniothyrium minitans RNA virus (NC_007523); EfV1, Epichloe festucae virus 1 (AM261427); HmTV1-17, Helicobasidium mompa totivirus 1-17 (NC_005074); MoV1, Magnaporthe oryzae virus 1 (NC_006367); SsRV1, Sphaeropsis sapinea RNA virus 1 (NC_001963); AfSV1, Aspergillus foetidus slow virus 1 (HE588147); BbVV1, Beauveria bassiana victorivirus 1 (HE572591); RnVV1, Rosellinia necatrix victorivirus 1 (AB698489); CeRV1, Chalara elegans RNA virus 1 (NC_005883); HvV190S, Helminthosporium victoriae virus 190S (NC_003607); TcV1, Tolypocladium cylindrosporum virus 1 (NC_014823); XdV-L1A, Xanthophyllomyces dendrorhous virus L1A (NC_020903); XdV-L1B, Xanthophyllomyces dendrorhous virus L1b (JN997473); TaV1, Tuber aestivum virus 1 (HQ158596); ScV-L-A (L1) (NC_003745); ScV-L-BC (La) (NC_001641); SsV-L, Scheffersomyces segobiensis virus L (KC610514); UmV-H1, Ustilago maydis virus H1 (NC_003823); ORV, Omono River virus (AB555544); DmTV, Drosophila melanogaster totivirus (NC_013499); AsV, Armigeres subalbatus virus (NC_014609); IMNV (NC_007915); and PMCV, piscine myocarditis virus (NC_015639).
A polarity plot was generated for the full-length GLV ORF 1 product (CP precursor) according to the method of Grantham (53) as implemented at http://web.expasy.org/protscale/ with a running window size of 15 (see Fig. 3B).
FIG 3.
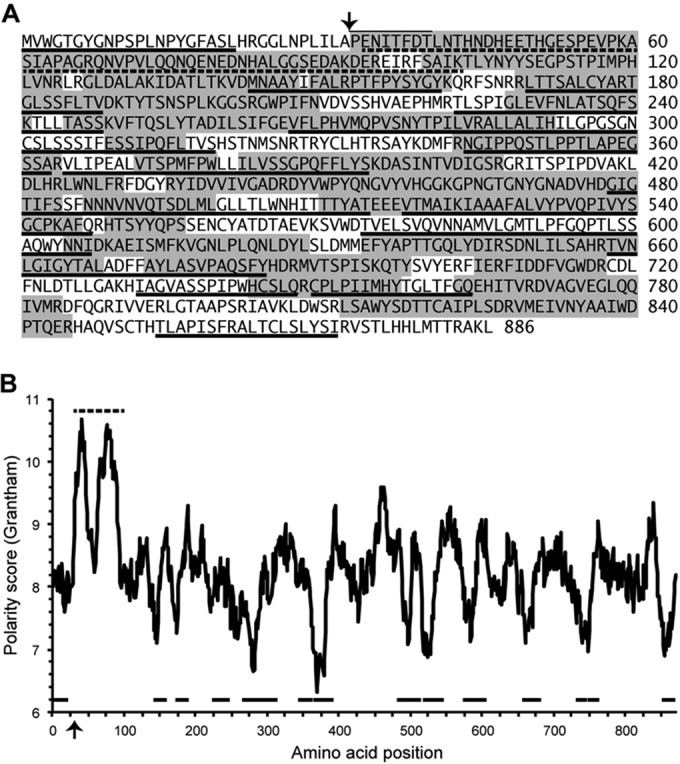
Mass spectrometry to assess protein contents of GLV virions. (A) Sequence of GLV CP precursor is shown. Arrow indicates position of the N-proximal processing cleavage (overlining, N-terminal sequencing results [10]). Sequences represented in the peptides recovered from LC-MS/MS after trypsin or chymotrypsin cleavage of GLV virions are indicated by gray shading. Predominantly uncharged regions as defined in the Discussion are underlined. The highly charged region near the CP N terminus is indicated by dotted underlines. (B) A polarity plot was generated as described in Materials and Methods. Highlighted features (arrow, underlines) are translated from panel A.
Accession number.
The 3D density map of the cryo-reconstruction of the GLV virion has been deposited in the Electron Microscopy Data Bank at the European Bioinformatics Institute with accession code EMD-5948.
RESULTS
Initial characterizations of virions used for cryo-TEM studies.
G. lamblia strain WBI (strain WB infected with GLV strain Portland I) was analyzed, because it has been extensively studied by Wang and coworkers (8–14, 21, 24–26). WBI was serially passaged in liquid batch culture, with a doubling time of ∼24 h. After harvest, cells were sonicated to release virions, centrifuged to deplete debris, and sedimented into a CsCl density gradient. The gradient was fractionated, and fractions containing virions were identified by SDS-PAGE to track GLV CP. Virions consistently concentrated in only one or two consecutive fractions, corresponding to a visible band in the gradient, well separated from any debris. The virion fractions were dialyzed against buffer and stored at either 4°C or −80°C before use.
SDS-PAGE of the purified GLV virions showed a major Coomassie-stained band migrating near 100 kDa, a finding consistent with the sequence-predicted mass of 95 to 98 kDa for GLV CP (95 kDa if missing its N-terminal 32 amino acids [aa] [10]) (Fig. 2A). A minor Coomassie blue-stained band migrating near 200 kDa was also routinely visible, consistent with the lower copy number and sequence-predicted mass of 211 kDa for GLV CP/RdRp (only 1 or 2 copies per virion versus 119 or 118, respectively, for CP). An ethidium-stained band migrating near the top of the SDS-PAGE gel, and consistent with the GLV dsRNA genome, was additionally seen (Fig. 2A).
FIG 2.

Gels and electron micrographs of purified GLV virions. (A) Gel analyses. SDS-PAGE and Coomassie blue staining (left) shows the GLV capsid protein (a) migrating near 100 kDa and the GLV CP/RdRp (b) near 190 kDa. Minor protein bands near 85 and ∼28 kDa are thought to be contaminants or degradation products. Agarose gel electrophoresis and ethidium bromide staining (right) shows a single band for GLV dsRNA (c). (B) Negative-stain TEM. Virions were stained with uranyl acetate to assess particle quality. The scale bar also applies to panel C. (C) Cryo-TEM. Unstained, vitrified virions are shown in one field used for image reconstruction. Examples of full and empty GLV particles are highlighted with black and white arrows, respectively. White dotted arrows point to putative strands of free dsRNA in the solvent background, also seen in the enlarged inset at higher contrast.
To evaluate suitability of the sample for structural analysis, we negatively stained the purified GLV virions and inspected them by TEM. The particles were seen to be well distributed, with minimal breakage and consistent widths near 450 Å (Fig. 2B). Most displayed a regular, circular outline. Essentially all particles appeared to be penetrated by stain, but most had a heterogeneous distribution of interior staining consistent with the presence of centrally packaged GLV genome. A few “empty” particles, with more homogeneous interior staining consistent with the absence of genome, were also observed. Similar findings were obtained for purified virions that had been stored at 4°C or at −80°C for several weeks, with or without addition of glycerol. Storage at −20°C was less reliable, however, as particles in clumped, empty, and broken states were observed in increased numbers.
Purified GLV virions were submitted for LC-MS/MS to assess their protein contents. Parallel analyses were performed: one for peptides generated by trypsin treatment and the other by chymotrypsin treatment. The identified peptides, respectively, covered 56 and 46% of the deduced CP sequence, or 65% coverage by the combined results (Fig. 3A). A notable finding was that no peptides were identified from the N-terminal 32 aa of the ORF1-encoded CP sequence, which have been indicated by Yu et al. (10) to be removed by proteolytic cleavage in infected cells and to be missing from virions. Peptides were also recovered from the RdRp region of CP/RdRp in these analyses, although at lower frequency, 25% coverage of the predicted RdRp sequence by the combination of peptides from either treatment, consistent with the lower copy number of CP/RdRp. Peptides spanning the CP/RdRp fusion junction were not recovered.
Cryo-TEM and 3D reconstruction: surface views and equatorial section.
Purified GLV virions vitrified and visualized by cryo-TEM appeared consistently sized and shaped, with mostly unbroken capsids (Fig. 2C). The genomic dsRNA enclosed within the capsid layer appeared as distributed density, without the “fingerprint” motif that is observed in virions of other totiviruses, including tentatively assigned member IMNV (32, 35, 36, 38, 39). A few particles, however, exhibited lower central densities, and linear strands suggestive of free dsRNA were seen at places in the solvent background, consistent with loss of genome from a few particles in the preparation (Fig. 2C). The capsids in this mixture of mostly “full” and few “empty” particles displayed a very regular, circular outline, without any obvious protrusions. An icosahedral 3D image reconstruction was computed from 14,125 particle images recorded on 101 micrographs. The resolution of the final reconstruction was estimated at 6.0 Å based on a Fourier-shell correlation threshold of 0.5 (45).
As expected for totiviruses (32, 35–39), a surface view of the 3D reconstruction revealed that GLV appears to possess a so-called “T=2” capsid (34) comprising 120 CP subunits in a T=1 lattice (Fig. 4A and C). The estimated volume of the capsid shell is also consistent with this conclusion. Large portions of the CP outer surfaces form a raised but mostly flat “plateau” centered on each of the icosahedral 5-fold axes, with the portion of each subunit contributing to this plateau having the appearance of an elongated, triangular “pizza slice.” The wider (“crust”) end of each slice lies distal to the 5-fold axis. From this perspective, it is notable that each CP subunit occupies either of two nonidentical positions within the capsid (Fig. 4A). In one position (designated “A” as in other “T=2” capsids), the CP subunits approach one another around each 5-fold axis via the narrow (“toppings”) end of the figurative slice, while in the other position (designated “B”), the CP subunits lie wedged between neighboring A subunits, in a similar orientation but more distant from the 5-fold axis. Each plateau thus appears to be constituted by portions of 10 CP subunits: five A and five B. By comparison, each of the icosahedral 3-fold axes, which are low points on the capsid surface, appears to be immediately surrounded by three B subunits, each from a different decameric plateau (Fig. 4A). Each of the icosahedral 2-fold axes, on the other hand, appears to be immediately surrounded by four CP subunits: one A and one B subunit from one decameric plateau and one A and one B subunit from the adjacent decameric plateau (Fig. 4A). Moreover, each of these A subunits appears to make contact in a quasi-2-fold-symmetrical, “tail-to-tail” (or “crust-to-crust”) fashion with the B subunit from the adjacent decamer, thereby forming two elongated A:B ridges that pass alongside the icosahedral 2-fold axis, which is another low point on the capsid surface.
FIG 4.
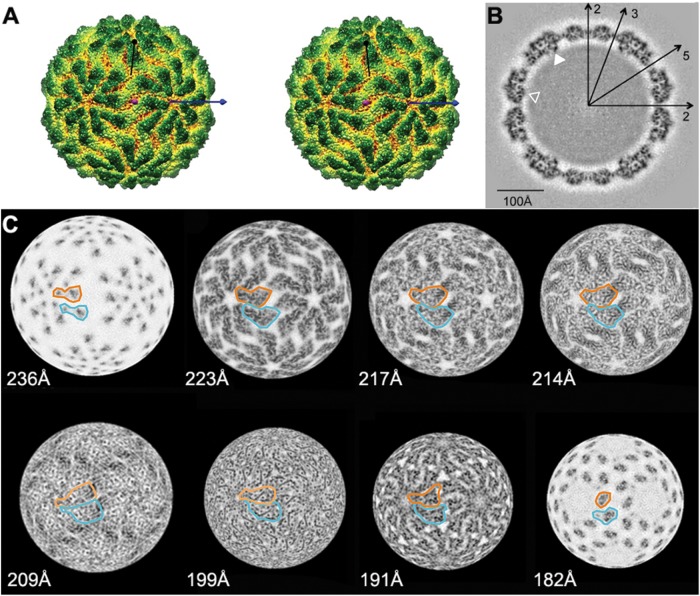
Icosahedral 3D image reconstruction of GLV virions. (A) Space-filling stereo view of the GLV particle surface as viewed down an icosahedral 2-fold axis (magenta). One icosahedral 3-fold axis (black) and one icosahedral 5-fold axis (blue) are marked. The map is color-coded by radius (dark green, outermost; red, innermost). (B) Central (equatorial), 1-pixel (1.09-Å)-thick section through the virion density map shown in grayscale (black and white corresponding to highest and lowest densities, respectively). Symmetry axes (icosahedral two-, three-, and 5-fold) are marked, as well as close contacts between capsid and RNA densities (arrowheads). (C) Radial sections through the capsid, centered at indicated radii and each 1 pixel (1.09 Å) thick. The map is coded in grayscale as in panel B. The outlines of one CP-A (orange) and one CP-B (cyan) subunit are shown in each radial section.
The equatorial section of the 3D reconstruction shows both punctate and linear features, which at this level of resolution suggest a prevalence of α-helical secondary structure in the CP subunits (Fig. 4B). It also shows that the single-layered capsid of GLV has a maximum thickness of ∼60 Å (median, ∼40 Å). The outermost capsid diameter is ∼485 Å, making this the largest totivirus capsid studied to date (35–39), including the capsid (sans spikes) of IMNV (32). The central section also reveals an interior capsid surface that is smoother than the exterior, except for short inward protrusions near the icosahedral 3-fold axes.
Unlike what has been observed to date for other totiviruses (32, 35, 36, 38, 39), the packaged genome of GLV does not appear in the reconstruction as a series of concentric shells of density in the particle interior. This finding is consistent with the lack of central fingerprint densities in the raw cryo-micrographs of GLV (Fig. 2C). Instead, densities attributable to genome are distributed almost homogeneously throughout the interior of the capsid, except for a hint of greater density in some localized regions nearest the shell (Fig. 4B). The most notable contacts between capsid and RNA densities appear to occur near the icosahedral 3-fold axes, as evident in the equatorial section (Fig. 4B, filled white arrowhead). An additional, smaller contact appears to be located between the icosahedral 5-fold and 2-fold axes (Fig. 4B, unfilled white arrowhead).
Subunit arrangements observed in radial sections.
Radial sections of the 3D reconstruction were found to complement surface views for gaining a better understanding of GLV capsid organization. Clearly distinguishable, triangular “pizza-slice” elements fully constitute the sections at higher radii (e.g., 236 to 214 Å in Fig. 4C). Visualized especially well at radius 214 Å, but also at 223 and 217 Å, is how the capsid is organized with four of these elements, representing two A and two B subunits, arrayed around each icosahedral 2-fold axis (Fig. 4C). As also inferred from the surface view, these four subunits around each icosahedral 2-fold axis appear to be arranged as two antiparallel A:B pairs, within each of which the A and B subunits make quasi-2-fold-symmetrical, tail-to-tail contacts across a broad, flat interface. These A:B contacts appear to be the major ones that connect adjacent decamers at these higher radii, although B:B contacts around the icosahedral 3-fold axes appear also to contribute. Progressing to lower radii in the sections, it is nearly impossible to distinguish individual CP subunits at radii 209 and 199 Å (Fig. 4C). Instead, at these radii almost continuous protein densities are seen, representing the core of the capsid shell. At the lowest radii in the capsid (e.g., radii 191 and 182 Å in Fig. 4C), however, the A and B subunits are again distinguishable.
The close correspondence in features between A and B subunits, across the full capsid thickness, is notable in the radial sections (Fig. 4C). This correspondence pertains to not only their similar outlines but also their similar internal features appearing to represent secondary-structure elements. The A and B subunits thus appear to lie at nearly the same radial heights throughout the capsid, although close inspection suggests that the A subunits may lie just a few angstroms (i.e., 1 to 3 Å) higher. In addition, a small clockwise rotation along the longitudinal axis of the A subunits (relative to B subunits; see below) may explain subtle differences observed between the two classes of subunits, especially at lower radii (e.g., see radius 191 Å).
The positions of channels through the GLV capsid are of interest, in particular with regard to RNA transcription and RNA release from particles. Channels 10 to 15 Å wide would be large enough to permit translocation of nucleotide triphosphates and perhaps newly synthesized RNA transcripts, but not the dsRNA genome in the absence of movements of the capsid elements. In the equatorial section, open channels that span the full capsid thickness appear to be present at the icosahedral 5-fold axes (Fig. 4B). Using the radial sections for size estimates, however, we found that this channel narrows in width to only 10 to 11 Å at radius 209 Å and remains nearly as narrow at radius 199Å (Fig. 4C). Open channels are not evident at the icosahedral 3- and 2-fold axes, although the capsid in these locations does appear to be quite thin.
Capsid segmentation.
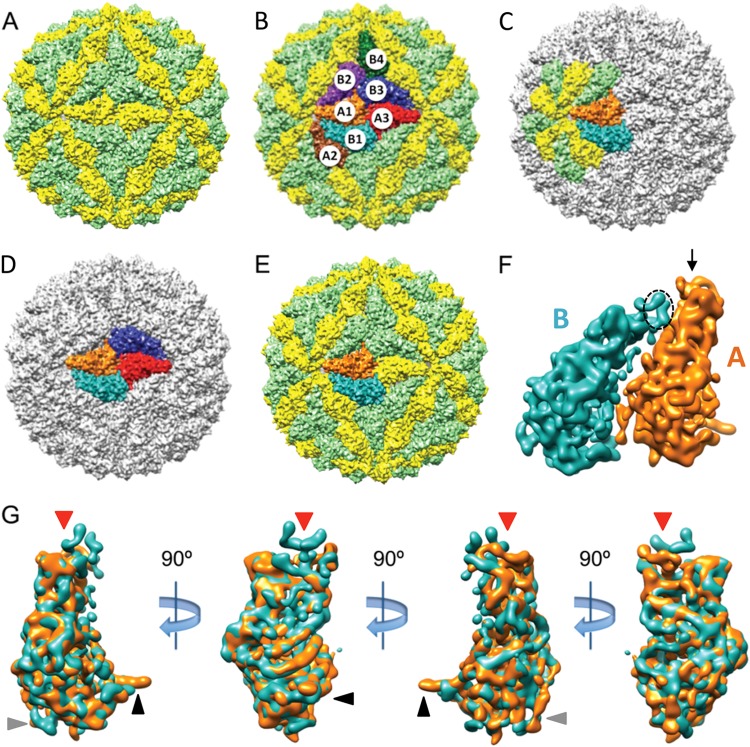
To ascribe specific features in the GLV 3D density map to the A and B subunits (Fig. 5), we used the ab initio approach previously used to segment the maps of HvV190S and TVV1 (38, 39). The segmentation procedure was most difficult to perform near the icosahedral 2-fold axes, where the A and B subunits from adjacent decamers seem to interact extensively in a quasi-2-fold-symmetrical arrangement (see radii 214 and 209 Å in Fig. 4C). Nonetheless, iterative refinement of density assignments, obeying the constraints described previously (39) and carried out independently by two persons, provided consistent identification of the molecular envelopes and boundaries of CP-A and CP-B (Fig. 5). Secondary-structure elements similar in shape and placement were visible as repeating units in both segmented subunits, and no evidence for major domain swapping between them was detected (Fig. 5).
FIG 5.
Segmentation of the GLV virion capsid and possible assembly intermediates. (A) Densities corresponding to CP-A and CP-B subunits according to segmentation analysis are colored yellow and light green, respectively. (B) Subunits are labeled to identify contacts between them. Colors are the same as in panel A, plus subunit A1 in orange, subunit B1 in cyan, subunit A2 in brown, subunit B2 in magenta, subunit A3 in red, subunit B3 in blue, and subunit B4 in green. (C and D) The most plausible assembly intermediates are shown: compact decamer (C) and compact tetramer (D). Colors are the same as in panel B. (E) One of the AB dimers in the GLV capsid in which the two subunits are more parallel in orientation. (F) Rotated 90° clockwise and enlarged view of the AB dimer in panel E. The black arrow indicates the region of CP-A that approaches the icosahedral 5-fold channel, and the dotted ellipse indicates the comparable region of CP-B, which has a different orientation. (G) Superimposition of segmented A and B subunits shown in four different orientations, beginning with a view as seen from outside the capsid (as in panel E) and subsequently rotated three more times, each by 90° clockwise. Predominant differences occur at the tip of each of the two subunits closest to the icosahedral 5-fold axis (red arrow) and at the AB interface near the icosahedral 2-fold axis (black/gray arrows).
Despite having different local environments, CP-A and CP-B as defined by segmentation have very similar envelopes. Superimposition of CP-A and CP-B densities yielded a correlation value of 0.91 (see Materials and Methods), which strongly suggests they share a similar tertiary structure. Indeed, the most prominent tubular density features in each subunit (ascribed to α-helical elements) are nearly identically located and oriented. Each subunit has a length of maximally ∼100 Å, and as viewed radially from inside or outside the capsid, the “crust” region is about twice as wide as the “toppings” region.
CP-A and CP-B as defined by segmentation do differ at their boundaries, however. One major difference occurs at the narrow (toppings) end of each subunit. Here, CP-A contains densities that line the icosahedral 5-fold channel (Fig. 5F, arrow) and contact an adjacent, 5-fold-related A subunit (e.g., A1 contacting A2 in Fig. 5B) but that are not apparent at the comparable locations in CP-B. Instead, features of similar volume and shape in CP-B are oriented toward a neighboring A subunit of the asymmetric A:B pair (e.g., B1 contacting A1 in Fig. 5B and dotted ellipse in Fig. 5F), such that they appear to be extended outward and flipped by 90° from their position in CP-A (Fig. 5G, red arrowhead). Two other notable differences occur near the wider (crust) end of each subunit. One occurs at the outer edge of the quasi-2-fold-symmetrical A:B interaction (e.g., for A1B3 in Fig. 5B and black arrowhead in Fig. 5G), on the side distal to the icosahedral 2-fold axis, where a helix-like density from CP-A extends outward toward two neighboring B subunits (e.g., A1 contacting B2 and B3 in Fig. 5B). In contrast, this same density in CP-B does not protrude away from the subunit but instead extends in a radial direction toward the outside of the capsid. The other notable difference in this domain is a slightly shifted loop of density in CP-B near the icosahedral 3-fold axis, which contacts the other 3-fold-related CP-B subunits (e.g., B2 contacting B3 and B4 in Fig. 5B). This same region in CP-A, though more compact compared to that in CP-B, is the one that contacts another CP-A subunit directly across the icosahedral 2-fold axis (e.g., A1 contacting A3 in Fig. 5B).
Different responses of GLV and TVV1 virions to temperature and pH.
The capacity of GLV to transmit between cells extracellularly (8, 21), whereas other known protozoal or fungal totiviruses cannot, led us to hypothesize that GLV virions may be more stable than many of these other viruses in order to withstand extracellular environments. This idea is consistent with the preceding observation that in our preparations of GLV virions (Fig. 2B and C) only a few particles appeared to have lost their dsRNA genomes after purification, in contrast to the greater number of such particles seen in preparations of TVV1 and ScV-L-A (35, 36, 39). To examine this stability difference more quantitatively, we adapted a previously described assay using RNase III to digest accessible viral genomic dsRNA (36) and used TVV1 virions for comparison with GLV virions. This assay allows for measurement of the amount of genome that becomes exposed from its protected location inside capsids upon preheating virions to different temperatures. The results showed that GLV virions were substantially more thermoresistant than TVV1 virions (Fig. 6A), a finding consistent with the greater stability of GLV.
Optimal growth of T. vaginalis trophozoites occurs at more acidic conditions (pH < 6.5) than does that of G. lamblia trophozoites (pH > 7) (54–56). Moreover, Tritrichomonas foetus, a relative of T. vaginalis, has been shown to possess a more acidic cytoplasm (pH 6.3) than do most eukaryotic cells, including G. lamblia (pH > 7) (55, 57). Hence, we hypothesized that the transcriptase activities of GLV and TVV1 may exhibit different pH optima, reflecting these different cytoplasmic pH values. By adapting in vitro transcriptase assays previously described for these viruses (58, 59), we found that GLV virions mediate transcription across a fairly broad pH range, peaking near pH 8.0, whereas TVV1 virions mediate transcription across a narrower range, peaking near pH 6.5 (Fig. 6B). These results are consistent with the different intracellular conditions in which these two viruses are commonly found in their two distinct hosts.
DISCUSSION
Lack of projecting fibers in GLV.
GLV appears to be more closely related than other taxonomically approved totiviruses to a group of recently discovered, nonsegmented dsRNA viruses from metazoan hosts, including IMNV (Fig. 1). It is thus noteworthy that GLV has no projecting surface fibers, as are found at the 5-fold axes in IMNV (32) (Fig. 7). This absence of fibers is consistent with the lack of coding capacity for a fiber protein in the GLV genome, which IMNV and the related insect “totiviruses” contain upstream of their respective CPs in ORF1 (27–30). Coding for the proteolytically separated, 32-aa N-terminal peptide of the GLV CP precursor (see Fig. 3) is similarly positioned in the genome (i.e., upstream of CP in ORF1), but this peptide would be much too small to form a long fiber. It follows that in GLV, required functions for cell entry and tropism (e.g., receptor binding and membrane penetration) are likely to be mediated by as-yet-unlocalized regions of the capsid, rather than by any surface fibers as in IMNV.
FIG 7.
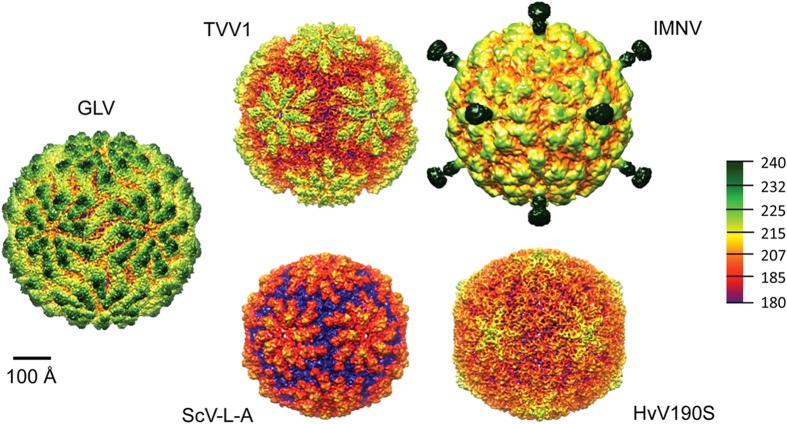
Comparison of GLV to four other taxonomically approved or tentatively assigned totiviruses. All particles are viewed along an icosahedral 2-fold axis, and the surfaces are color coded by radius according to the spectrum shown at right, which lists radii in angstroms. IMNV is tentatively assigned to family Totiviridae.
In the tentatively assigned vertebrate (piscine) totivirus PMCV, a putative receptor-binding protein, containing a chemokine-superfamily motif, is encoded by a unique downstream ORF, designated ORF3 (31). This additional, recently discovered GLV-related virus (see Fig. 1) also transmits efficiently by extracellular means via virion-associated cell entry machinery. Since this property is not shared with other approved totiviruses of protozoal or fungal hosts (see Fig. 1), we suggest that GLV, IMNV and related insect viruses, and PMCV might be usefully classified in either a distinct taxonomic family or, more conservatively, in a distinct subfamily within the family Totiviridae.
Sequence features with potential relevance to RNA interactions and cell entry.
Examination of the GLV CP precursor sequence (GenBank NC_003555) reveals a few, previously unnoted features of interest. Immediately following the 32-aa N-terminal region that is separated by proteolysis from the CP region (10) is a strongly polar, 71-aa region dominated by charged residues (see Fig. 3). This region seems reminiscent to us of the disordered, N-terminally positioned, small R (RNA-interacting) domain of many plus-strand RNA virus CPs (60), although the R domain in those viruses is rich in basic residues, whereas this part of the sequence in GLV is acidic (calculated pI = 4.6 for this region). We hypothesize that this strongly polar region projects into the central cavity enclosed by the GLV capsid, where its negatively charged residues may promote sliding movements of the negatively charged genomic RNA during transcription. The apparent CP contacts with RNA densities reflect where these R-domain-like regions of GLV CP are proposed to project into the interior (see Fig. 4B). These regions might also be involved in interacting with and positioning the RdRp domain of the CP/RdRp subunit(s), as has been suggested, for example, for a similar N-terminal domain in rotaviruses (61, 62).
Given the probable role of GLV CP in membrane penetration during cell entry by GLV virions into G. lamblia cells, it seemed informative to us to identify regions of CP sequence with hydrophobic character. Figure 3 highlights 14 regions of predominantly uncharged residues (regions ≥16 aa in length, including at most one charged residue), 12 of which are spread across the central portions of the CP sequence, suggestive of the folded core of this protein. In addition, there is one such region isolated near the C terminus and another near the N terminus. In fact, the N-terminal 32 aa cleaved from the CP precursor (MVWGTGYGNPSPLNPYGFASLHRGGLNPLILA) include only two charged residues (underlined), as well as a high proportion of small residues (44% Gly, Ala, Pro, and Ser), reminiscent of the membrane-penetration peptides of birnaviruses, orthoreoviruses, and others (63–66). Since only 65% of the GLV CP sequence was covered by the tryptic or chymotryptic peptides identified by LC-MS/MS analysis in this study (see Fig. 3), the possibility remains that this N-terminal peptide, or smaller peptides derived from it, might still be present in the GLV virion.
Conserved capsid elements in GLV and other Totiviridae members.
Despite very little primary-sequence homology among the CPs of GLV, ScV-L-A, HvV190S, TVV1, and IMNV (pairwise-identity scores all <18% using EMBOSS Stretcher [http://www.ebi.ac.uk/Tools/psa/]), their capsid organizations are highly similar (Fig. 7), other than the projecting fibers in IMNV. Although the GLV capsid is somewhat larger than the others (17.5% larger than ScV-L-A; ∼5–7% larger than HvV190S, TVV1, and IMNV), comparable decameric plateaus are seen on the outer surfaces of each, although HvV190S is notably the smoothest. The larger capsid of GLV correlates with it having a larger CP than the other approved Totiviridae members shown here, although the IMNV CP is the largest (IMNV, 99 kDa > GLV, 98 kDa > HvV190S, 81 kDa > ScV-L-A, 76 kDa > TVV1, 75 kDa). The genome size of IMNV is also larger than that of GLV and the other totiviruses (IMNV, 7,561 bp > GLV, 6,277 bp > HvV190S, 5,179 bp > TVV1, 4,647 bp > ScV-L-A, 4,579 bp). Using the inner capsid diameter to estimate the central cavity volume from the density maps of these viruses, however, it turns out that GLV appears to have more space available for its genome than does IMNV (GLV 20,035 to 21,497 nm3 > IMNV 16,362 to 19,858 nm3; since the viral capsids are not perfectly round, the range reflects use of either the smallest or the largest inner diameter of the capsid for calculation). Assuming a single genome molecule per virion, this would result in a lower genome packing density for GLV. By being less densely packed, the GLV genome might be relatively too disorganized to give rise to concentric spherical shells of dsRNA densities in the icosahedral reconstruction as seen in IMNV and other totiviruses (32, 36, 38, 39).
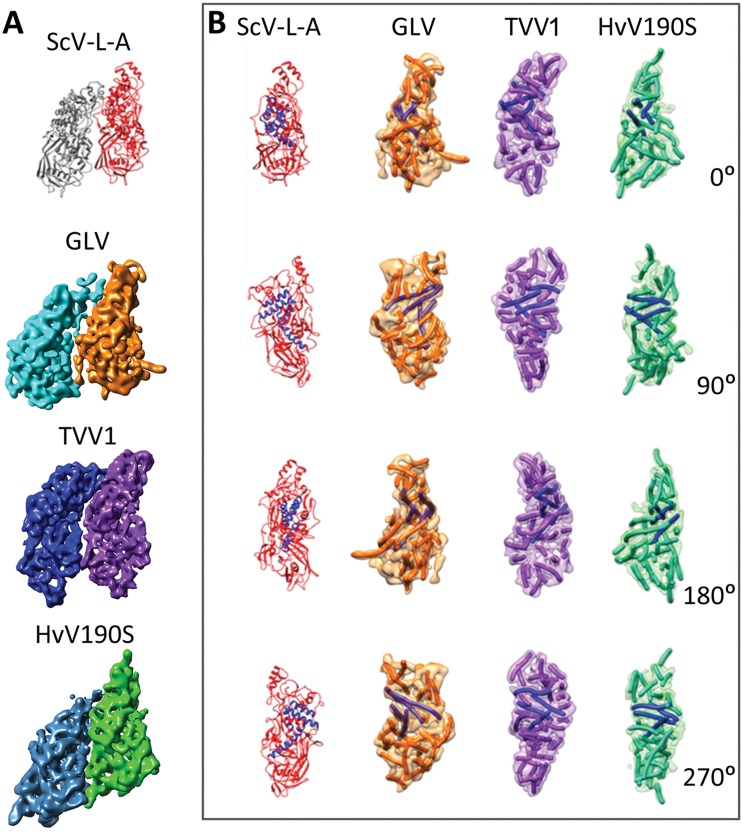
Besides overall capsid organization, the individual GLV A and B subunits show a striking resemblance to those of ScV-L-A, HvV190S, and TVV1 (Fig. 8A). In addition, the orientation of GLV CP-A and CP-B with respect to each other is almost identical to the orientation of these subunits in these three viruses. The resolution of the GLV cryo-reconstruction is too low to permit a reliable trace of the entire CP polypeptide backbone, but it is clear that even at a 6.0-Å resolution the GLV CP is rich in twisted, tubular elements likely to represent α-helices, as are also the CPs of ScV-L-A, HvV190S, and TVV1 (Fig. 8B). Of particular note are four putative, interacting long α-helices (colored blue in Fig. 8B) that are similarly located and oriented in the CP subunits of all four viruses (best seen in the 90° and 180° side views in Fig. 8B). Thus, despite some differences in length, orientation, and angular position in the different viruses, these putative α-helices appear to constitute a conserved core of the CP subunits that might be common to all totiviruses (38), including GLV.
FIG 8.
Comparison of the CP subunits of GLV to those of ScV-L-A, HvV190S, and TVV1. (A) Structure of the A1B1 dimer as defined in Fig. 5. Top to bottom, as seen from outside the particle: ScV-L-A (A, red; B, gray), GLV (A, orange; B, cyan), TVV1 (A, purple; B, blue), and HvV190S (A, green; B, steel blue). The structures shown are from segmentation of cryo-TEM maps except for the X-ray crystal structures shown for ScV-L-A. (B) Structure of the A subunits of ScV-L-A, GLV, TVV1, and HvV190S, viewed from four different orientations each. The rotations indicated are around the y axis. Color scheme for the subunits is translated from panel A. Helices in the A subunit of ScV-L-A are shown as coiled ribbons, whereas putative helices in the A subunits of GLV, HvV190S, and TVV1 are depicted by tubes of similar color as their respective density maps. Four putative, long α-helices, located and oriented similarly in the A subunits of these four viruses, are shown in blue.
Capsid channels and relative stability of GLV and TVV1.
Following entry into the G. lamblia cytoplasm, the particle-enclosed GLV genome is presumably transcribed by the RdRp domain of the CP/RdRp subunit(s) still inside the confines of the “T=2” icosahedral capsid, and the plus-strand transcripts are then released from the particle (e.g., see evidence for this in vitro activity in Fig. 6B), allowing translation of the GLV proteins and packaging into nascent, progeny GLV virions. This strategy for transcription inside the icosahedral capsid is thought to be common to all dsRNA viruses except possibly birnaviruses (67), which lack the “T=2” capsid organization (68). The only solvent-accessible channels that span the GLV capsid appear to occur at each of the 12 5-fold axes, and though somewhat narrow (ca. 10 to 11 Å in width), they are probably wide enough to permit both entry of nucleoside triphosphates and exit of newly synthesized transcripts during transcription. It is also conceivable, and perhaps likely, that conformational changes in the capsid during cell entry and/or in association with transcription may enlarge these channels or perhaps open other ones.
The 5-fold channels in GLV are considerably smaller than those in TVV1, which narrow to only ∼16 Å in the lower portions of the capsid shell (39). Although these 5-fold channels, absent conformational changes, are not wide enough in either virus for genomic dsRNA to pass through, we have found that purified preparations of TVV1 virions contain a substantial fraction of “empty” particles, i.e., particles devoid or depleted of genome, with accompanying evidence for dsRNA release following purification (39). Indeed, we have suggested that the relatively wide 5-fold channels in TVV1 may serve as the conduit for this release of genome upon limited conformational widening of these channels. This may in fact have significant biological relevance as the source of dsRNA for TLR3/TRIF-dependent signaling of proinflammatory responses by mammalian cells that encounter TVV1-positive Trichomonas vaginalis parasites in the human genitourinary tract, thereby contributing to human disease (18, 19). Regarding a similar possible effect of GLV virions in contributing to human enteric disease by G. lamblia, we note that our purified preparations of GLV virions contained a much lower fraction of empty particles or evidence for dsRNA release (see Fig. 2B and C), a finding consistent with our experimental evidence that GLV virions are substantially more thermoresistant than TVV1 virions (Fig. 6A). This greater stability of GLV virions is furthermore consistent with its capacity for transmission between cells by extracellular means, entailing a greater need for it to withstand a wider range of environments but potentially reducing its propensity for proinflammatory signaling as observed for TVV1 (18, 19).
Possible pathways of GLV/totivirus capsid assembly.
Assembly pathways for the capsids of icosahedral viruses often include symmetrical dimers or trimers as initial intermediates (69). Assembly of a “T=2” capsid like that of GLV, however, could not simply involve CP trimers, because only half of the subunits (60 CP-B) are arranged as 20 trimers in the capsid; the other half (60 CP-A) are arranged as 12 pentamers. Although not impossible, the formation of two such notably different intermediates by the chemically identical CP subunits seems unlikely.
“T=2” assembly pathways potentially involve compact decamers as intermediates (69), 12 of which would coalesce to form the complete capsid. Such a pathway involving compact decamers would rule out the participation of either symmetrical trimers or symmetrical dimers as intermediates and would instead rely on the formation of asymmetrical CP dimers (one A and one B subunit arranged in a near-parallel, side-by-side manner). Although this asymmetrical dimer followed by decamer assembly is certainly plausible, inspections of the GLV and other totivirus capsid structures suggest that their “T=2” capsid assembly pathway may instead involve 2-fold-symmetrical, extended CP dimers, two of which could associate to form compact, 2-fold-symmetrical CP tetramers. A related scenario is that these extended CP dimers might be transient and reversible, such that the tetramer represents the first stable intermediate. In any case, 30 such tetramers could then coalesce to form the complete capsid, possibly by first assembling into trimers of tetramers (69), 10 of which would then combine. Yet another, conceivable “T=2” assembly pathway involving a stable tetramer intermediate would have extended CP dimers continuing to accrete to an initial, nucleating tetramer.
Interestingly, studies of Pseudomonas bacteriophages φ6 and φ8 (family Cystoviridae) (70, 71), three fungal partitiviruses (family Partitiviridae) (72, 73), and rabbit picobirnavirus (family Picobirnaviridae) (65) have all identified 2-fold-symmetrical CP tetramers as the most probable assembly intermediates. Possibly, some “T=2” capsids assemble via a decamer pathway (e.g., family Reoviridae), whereas others assemble via a tetramer pathway (e.g., families Cystoviridae, Partitiviridae, and Picobirnaviridae) (69). In the case of family Totiviridae, the greater CP mass and more elongated CP structure of its members (Fig. 8) might suggest these viruses are more like members of family Reoviridae and assemble via the decamer pathway, but we consider the tetramer pathway to remain a viable alternative. In fact, the substantial interactions seen between CP-A and CP-B within each quasi-2-fold-symmetrical dimer in the capsid (segmentation was most difficult to perform at this subunit interface) provide direct evidence in favor of the dimer/tetramer assembly pathway.
Beyond these considerations strictly about CP subunit assembly, other key questions remain about how the CP/RdRp fusion subunit(s) and the genomic RNA assemble into mature virions. For example, regardless of the precise manner by which the GLV/totivirus CP subunits form assembly intermediates, do the one or two CP/RdRp molecules per virion always occupy either an A or a B position, or can the CP/RdRp molecule(s) assemble and function in either of these positions? Also, is the viral RNA recruited by a particular intermediate, presumably one containing a CP/RdRp subunit? These and other challenging questions need to be addressed in future studies.
ACKNOWLEDGMENTS
We are grateful to C. C. Wang and coworkers (University of California, San Francisco) for providing a stock of G. lamblia WBI cells and to Raina N. Fichorova and coworkers (Brigham and Women's Hospital and Harvard Medical School) for providing an original stock of T. vaginalis UH9 cells. We also thank Norman H. Olson for technical assistance with microscopy, Maria Ericsson for negative-stain microscopy of GLV virions that had been stored at different conditions, and John M. Asara and coworkers (Beth Israel Deaconess Medical Center and Harvard Medical School) for LC-MS/MS analyses of GLV virions.
This study was supported in part by NIH grants R37 GM033050 (to T.S.B.) and R56 AI091889 (subcontract to M.L.N.). Support from the University of California San Diego and the Agouron Foundation (all to T.S.B.) was used to establish the cryo-TEM facilities used in this study.
REFERENCES
- 1.Walker G, Dorrell RG, Schlacht A, Dacks JB. 2011. Eukaryotic systematics: a user's guide for cell biologists and parasitologists. Parasitology 138:1638–1663. doi: 10.1017/S0031182010001708. [DOI] [PubMed] [Google Scholar]
- 2.Upcroft P, Upcroft JA. 2001. Drug targets and mechanisms of resistance in the anaerobic protozoa. Clin Microbiol Rev 14:150–164. doi: 10.1128/CMR.14.1.150-164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cotton JA, Beatty JK, Buret AG. 2011. Host parasite interactions and pathophysiology in Giardia infections. Int J Parasitol 41:925–933. doi: 10.1016/j.ijpara.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Lane S, Lloyd D. 2002. Current trends in research into the waterborne parasite Giardia. Crit Rev Microbiol 28:123–147. doi: 10.1080/1040-840291046713. [DOI] [PubMed] [Google Scholar]
- 5.Stark D, Barratt JL, van Hal S, Marriott D, Harkness J, Ellis JT. 2009. Clinical significance of enteric protozoa in the immunosuppressed human population. Clin Microbiol Rev 22:634–650. doi: 10.1128/CMR.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartelt LA, Roche J, Kolling G, Bolick D, Noronha F, Naylor C, Hoffman P, Warren C, Singer S, Guerrant R. 2013. Persistent G. lamblia impairs growth in a murine malnutrition model. J Clin Invest 123:2672–2684. doi: 10.1172/JCI67294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Saraiya AA, Wang CC. 2013. Experimental verification of the identity of variant-specific surface proteins in Giardia lamblia trophozoites. mBio 4:e00321-13. doi: 10.1128/mBio.00321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang AL, Wang CC. 1986. Discovery of a specific double-stranded RNA virus in Giardia lamblia. Mol Biochem Parasitol 21:269–276. doi: 10.1016/0166-6851(86)90132-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang AL, Yang HM, Shen KA, Wang CC. 1993. Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc Natl Acad Sci U S A 90:8595–8599. doi: 10.1073/pnas.90.18.8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu D, Wang CC, Wang AL. 1995. Maturation of giardiavirus capsid protein involves posttranslational proteolytic processing by a cysteine protease. J Virol 69:2825–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Wang AL, Wang CC. 2001. Structural analysis of the −1 ribosomal frameshift elements in giardiavirus mRNA. J Virol 75:10612–10622. doi: 10.1128/JVI.75.22.10612-10622.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garlapati S, Wang CC. 2004. Identification of a novel internal ribosome entry site in giardiavirus that extends to both sides of the initiation codon. J Biol Chem 279:3389–3397. doi: 10.1074/jbc.M307565200. [DOI] [PubMed] [Google Scholar]
- 13.Garlapati S, Wang CC. 2009. Giardiavirus internal ribosome entry site has an apparently unique mechanism of initiating translation. PLoS One 4:e7435. doi: 10.1371/journal.pone.0007435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang A, Wang CC, Alderete JF. 1987. Trichomonas vaginalis phenotypic variation occurs only among trichomonads infected with the double-stranded RNA virus. J Exp Med 166:142–150. doi: 10.1084/jem.166.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogg MM, Carrion R Jr, Botelho AC, Mayrink W, Correa-Oliveira R, Patterson JL. 2003. Short report: quantification of leishmaniavirus RNA in clinical samples and its possible role in pathogenesis. Am J Trop Med Hyg 69:309–313. [PubMed] [Google Scholar]
- 16.Jenkins MC, Higgins J, Abrahante JE, Kniel KE, O'Brien C, Trout J, Lancto CA, Abrahamsen MS, Fayer R. 2008. Fecundity of Cryptosporidium parvum is correlated with intracellular levels of the viral symbiont CPV. Int J Parasitol 38:1051–1055. doi: 10.1016/j.ijpara.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Ives A, Ronet C, Prevel F, Ruzzante G, Fuertes-Marraco S, Schutz F, Zangger H, Revaz-Breton M, Lye LF, Hickerson SM, Beverley SM, Acha-Orbea H, Launois P, Fasel N, Masina S. 2011. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science 331:775–778. doi: 10.1126/science.1199326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fichorova RN, Lee Y, Yamamoto HS, Takagi Y, Hayes GR, Goodman RP, Chepa-Lotrea X, Buck OR, Murray R, Kula T, Beach DH, Singh BN, Nibert ML. 2012. Endobiont viruses sensed by the human host: beyond conventional antiparasitic therapy. PLoS One 7:e48418. doi: 10.1371/journal.pone.0048418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fichorova RN, Buck OR, Yamamoto HS, Fashemi T, Dawood HY, Fashemi B, Hayes GR, Beach DH, Takagi Y, Delaney ML, Nibert ML, Singh BN, Onderdonk AB. 2013. The villain team-up or how Trichomonas vaginalis and bacterial vaginosis alter innate immunity in concert. Sex Transm Infect 89:460–466. doi: 10.1136/sextrans-2013-051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang AL, Wang CC. 1991. Viruses of parasitic protozoa. Parasitol Today 7:76–80. doi: 10.1016/0169-4758(91)90198-W. [DOI] [PubMed] [Google Scholar]
- 21.Miller RL, Wang AL, Wang CC. 1988. Identification of Giardia lamblia isolates susceptible and resistant to infection by the double-stranded RNA virus. Exp Parasitol 66:118–123. doi: 10.1016/0014-4894(88)90056-2. [DOI] [PubMed] [Google Scholar]
- 22.Touz MC, Rivero MR, Miras SL, Bonifacino JS. 2012. Lysosomal protein trafficking in Giardia lamblia: common and distinct features. Front Biosci (Elite Ed) 4:1898–1909. doi: 10.2741/511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivero MR, Jausoro I, Bisbal M, Feliziani C, Lanfredi-Rangel A, Touz MC. 2013. Receptor-mediated endocytosis and trafficking between endosomal-lysosomal vacuoles in Giardia lamblia. Parasitol Res 112:1813–1818. doi: 10.1007/s00436-012-3253-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sepp T, Wang AL, Wang CC. 1994. Giardiavirus-resistant Giardia lamblia lacks a virus receptor on the cell membrane surface. J Virol 68:1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tai JH, Ong SJ, Chang SC, Su HM. 1993. Giardiavirus enters Giardia lamblia WB trophozoite via endocytosis. Exp Parasitol 76:165–174. doi: 10.1006/expr.1993.1019. [DOI] [PubMed] [Google Scholar]
- 26.Miller RL, Wang AL, Wang CC. 1988. Purification and characterization of the Giardia lamblia double-stranded RNA virus. Mol Biochem Parasitol 28:189–195. doi: 10.1016/0166-6851(88)90003-5. [DOI] [PubMed] [Google Scholar]
- 27.Poulos BT, Tang KF, Pantoja CR, Bonami JR, Lightner DV. 2006. Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J Gen Virol 87:987–996. doi: 10.1099/vir.0.81127-0. [DOI] [PubMed] [Google Scholar]
- 28.Wu Q, Luo Y, Lu R, Lau N, Lai EC, Li WX, Ding SW. 2010. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci U S A 107:1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhai Y, Attoui H, Mohd Jaafar F, Wang HQ, Cao YX, Fan SP, Sun YX, Liu LD, Mertens PP, Meng WS, Wang D, Liang G. 2010. Isolation and full-length sequence analysis of Armigeres subalbatus totivirus, the first totivirus isolate from mosquitoes representing a proposed novel genus (Artivirus) of the family Totiviridae. J Gen Virol 91:2836–2845. doi: 10.1099/vir.0.024794-0. [DOI] [PubMed] [Google Scholar]
- 30.Isawa H, Kuwata R, Hoshino K, Tsuda Y, Sakai K, Watanabe S, Nishimura M, Satho T, Kataoka M, Nagata N, Hasegawa H, Bando H, Yano K, Sasaki T, Kobayashi M, Mizutani T, Sawabe K. 2011. Identification and molecular characterization of a new nonsegmented double-stranded RNA virus isolated from Culex mosquitoes in Japan. Virus Res 155:147–155. doi: 10.1016/j.virusres.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Haugland O, Mikalsen AB, Nilsen P, Lindmo K, Thu BJ, Eliassen TM, Roos N, Rode M, Evensen O. 2011. Cardiomyopathy syndrome of Atlantic salmon (Salmo salar L.) is caused by a double-stranded RNA virus of the Totiviridae family. J Virol 85:5275–5286. doi: 10.1128/JVI.02154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang J, Ochoa WF, Sinkovits RS, Poulos BT, Ghabrial SA, Lightner DV, Baker TS, Nibert ML. 2008. Infectious myonecrosis virus has a totivirus-like, 120-subunit capsid, but with fiber complexes at the fivefold axes. Proc Natl Acad Sci U S A 105:17526–17531. doi: 10.1073/pnas.0806724105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nibert ML, Takagi Y. 2013. Fibers come and go: differences in cell-entry components among related dsRNA viruses. Curr Opin Virol 3:20–26. doi: 10.1016/j.coviro.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Ziéntara S, Mertens PPC, Stuart DI. 1998. The atomic structure of the bluetongue virus core. Nature 395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 35.Cheng RH, Caston JR, Wang GJ, Gu F, Smith TJ, Baker TS, Bozarth RF, Trus BL, Cheng N, Wickner RB, Steven AC. 1994. Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric Gag dimers. J Mol Biol 244:255–258. doi: 10.1006/jmbi.1994.1726. [DOI] [PubMed] [Google Scholar]
- 36.Castón JR, Trus BL, Booy FP, Wickner RB, Wall JS, Steven AC. 1997. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J Cell Biol 138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. 2002. L-A virus at 3.4 Å resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol 9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- 38.Dunn SE, Li H, Cardone G, Nibert ML, Ghabrial SA, Baker TS. 2013. Three-dimensional structure of victorivirus HvV190S suggests coat proteins in most totiviruses share a conserved core. PLoS Pathog 9:e1003225. doi: 10.1371/journal.ppat.1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parent KN, Takagi Y, Cardone G, Olson NH, Ericsson M, Yang M, Lee Y, Asara JM, Fichorova RN, Baker TS, Nibert ML. 2013. Structure of a protozoan virus from the human genitourinary parasite Trichomonas vaginalis. mBio 4:e00056-13. doi: 10.1128/mBio.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eng JK, McCormack AL, Yates JR III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 41.Baker TS, Olson NH, Fuller SD. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev 63:862–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142:334–347. doi: 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 43.Yan X, Dryden KA, Tang J, Baker TS. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol 157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM: an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol 157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harauz G, van Heel M. 1986. Exact filters for general geometry three dimensional reconstruction. Optik 73:146–156. [Google Scholar]
- 46.Bowman VD, Chase ES, Franz AW, Chipman PR, Zhang X, Perry KL, Baker TS, Smith TJ. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J Virol 76:12250–12258. doi: 10.1128/JVI.76.23.12250-12258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Havelka WA, Henderson R, Oesterhelt D. 1995. Three-dimensional structure of halorhodopsin at 7 Å resolution. J Mol Biol 247:726–738. doi: 10.1006/jmbi.1995.0176. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Russu M, Wriggers W. 2012. Evolutionary bidirectional expansion for the tracing of alpha helices in cryo-electron microscopy reconstructions. J Struct Biol 177:410–419. doi: 10.1016/j.jsb.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birmanns S, Rusu M, Wriggers W. 2011. Using Sculptor and Situs for simultaneous assembly of atomic components into low-resolution shapes. J Struct Biol 173:428–435. doi: 10.1016/j.jsb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 53.Grantham R. 1974. Amino acid difference formula to help explain protein evolution. Science 185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 54.Diamond LS. 1986. In vitro cultivation of the Trichomonadidae: a state of the art review. Acta Univ Carol Biol 30:221–228. [Google Scholar]
- 55.Hetsko ML, McCaffery JM, Svärd SG, Meng TC, Que X, Gillin FD. 1998. Cellular and transcriptional changes during excystation of Giardia lamblia in vitro. Exp Parasitol 88:172–183. doi: 10.1006/expr.1998.4246. [DOI] [PubMed] [Google Scholar]
- 56.Petrin D, Delgaty K, Bhatt R, Garber G. 1998. Clinical and microbiological aspects of Trichomonas vaginalis. Clin Microbiol Rev 11:300–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregor M, Tachezy J, Slavik J. 1996. The effect of lysosomal pH on lactoferrin-dependent iron uptake in Tritrichomonas foetus, p 95–99 InSlavik J. (ed), Fluorescence microscopy and fluorescent probes. Plenum Press, Inc, New York, NY. [Google Scholar]
- 58.Khoshnan A, Provenzano D, Alderete JF. 1994. Unique double-stranded RNAs associated with the Trichomonas vaginalis virus are synthesized by viral RNA-dependent RNA polymerase. J Virol 68:7108–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White TC, Wang CC. 1990. RNA-dependent RNA polymerase activity associated with the double-stranded RNA virus of Giardia lamblia. Nucleic Acids Res 18:553–559. doi: 10.1093/nar/18.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dolja VV, Koonin EV. 1991. Phylogeny of capsid proteins of small icosahedral RNA plant viruses. J Gen Virol 72:1481–1486. doi: 10.1099/0022-1317-72-7-1481. [DOI] [PubMed] [Google Scholar]
- 61.Lawton JA, Estes MK, Prasad BVV. 2000. Mechanism of genome transcription in segmented dsRNA viruses. Adv Virus Res 55:185–229. doi: 10.1016/S0065-3527(00)55004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Estrozi LF, Settembre EC, Goret G, McClain B, Zhang X, Chen JZ, Grigorieff N, Harrison SC. 2013. Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. J Mol Biol 425:124–132. doi: 10.1016/j.jmb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nibert ML, Schiff LA, Fields BN. 1991. Mammalian reoviruses contain a myristoylated structural protein. J Virol 65:1960–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ivanovic T, Agosto MA, Zhang L, Chandran K, Harrison SC, Nibert ML. 2008. Peptides released from reovirus outer capsid form membrane pores that recruit virus particles. EMBO J 27:1289–1298. doi: 10.1038/emboj.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J, Navaza J, Delmas B, Rey FA. 2009. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J 28:1655–1665. doi: 10.1038/emboj.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galloux M, Libersou S, Alves ID, Marquant R, Salgado GF, Rezaei H, Lepault J, Delmas B, Bouaziz S, Morellet N. 2010. NMR structure of a viral peptide inserted in artificial membranes: a view on the early steps of the birnavirus entry process. J Biol Chem 285:19409–19421. doi: 10.1074/jbc.M109.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luque D, Saugar I, Rejas MT, Carrascosa JL, Rodríguez JF, Castón JR. 2009. Infectious bursal disease virus: ribonucleoprotein complexes of a double-stranded RNA virus. J Mol Biol 386:891–901. doi: 10.1016/j.jmb.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 69.Poranen MM, Bamford DH. 2012. Assembly of large icosahedral double-stranded RNA viruses. Adv Exp Med Biol 726:379–402. doi: 10.1007/978-1-4614-0980-9_17. [DOI] [PubMed] [Google Scholar]
- 70.Poranen MM, Paatero AO, Tuma R, Bamford DH. 2001. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol Cell 7:845–854. doi: 10.1016/S1097-2765(01)00228-3. [DOI] [PubMed] [Google Scholar]
- 71.Kainov DE, Butcher SJ, Bamford DH, Tuma R. 2003. Conserved intermediates on the assembly pathway of double-stranded RNA bacteriophages. J Mol Biol 328:791–804. doi: 10.1016/S0022-2836(03)00322-X. [DOI] [PubMed] [Google Scholar]
- 72.Nibert ML, Tang J, Xie J, Collier AM, Ghabrial SA, Baker TS, Tao YJ. 2013. 3D structures of fungal partitiviruses. Adv Virus Res 86:59–85. doi: 10.1016/B978-0-12-394315-6.00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pan J, Dong L, Lin L, Ochoa WF, Sinkovits RS, Havens WM, Nibert ML, Baker TS, Ghabrial SA, Tao YJ. 2009. Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc Natl Acad Sci U S A 106:4225–4230. doi: 10.1073/pnas.0812071106. [DOI] [PMC free article] [PubMed] [Google Scholar]