ABSTRACT
Cellular immunity is pivotal in HIV-1 pathogenesis but is hampered by viral sequence diversity. An approach to minimize this diversity is to focus immunity on conserved proteome sequences; therefore, we selected four relatively conserved regions (Gag amino acids 148 to 214 and 250 to 335, Env amino acids 521 to 606, and Nef amino acids 106 to 148), each created in three mosaics, to provide better coverage of M-group HIV-1 sequences. A conserved-region vaccine (CRV) delivering genes for these four regions as equal mixtures of three mosaics each (each region at a separate injection site) was compared to a whole-protein vaccine (WPV) delivering equimolar amounts of genes for whole Gag, Env, and Nef as clade B consensus sequences (separate injection sites). Three rhesus macaques were vaccinated via three DNA primes and a recombinant adenovirus type 5 boost (weeks 0, 4, 8, and 24, respectively). Although CRV inserts were about one-fifth that of WPV, the CRV generated comparable-magnitude blood CD4+ and CD8+ T lymphocyte responses against Gag, Env, and Nef. WPV responses preferentially targeted proteome areas outside the selected conserved regions in direct proportion to sequence lengths, indicating similar immunogenicities for the conserved regions and the outside regions. The CRV yielded a conserved-region targeting density that was approximately 5-fold higher than that of the WPV. A similar pattern was seen for bronchoalveolar lymphocytes, but with quadruple the magnitudes seen in blood. Overall, these findings demonstrate that the selected conserved regions are highly immunogenic and that anatomically isolated vaccinations with these regions focus immunodominance compared to the case for full-length protein vaccination.
IMPORTANCE HIV-1 sequence diversity is a major barrier limiting the capability of cellular immunity to contain infection and the ability of vaccines to match circulating viral sequences. To date, vaccines tested in humans have delivered whole proteins or genes for whole proteins, and it is unclear whether including only conserved sequences would yield sufficient cellular immunogenicity. We tested a vaccine delivering genes for four small conserved HIV-1 regions compared to a control vaccine with genes for whole Gag, Env, and Nef. Although the conserved regions ranged from 43 to 86 amino acids and comprised less than one-fifth of the whole Gag/Env/Nef sequence, the vaccines elicited equivalent total magnitudes of both CD4+ and CD8+ T lymphocyte responses. These data demonstrate the immunogenicity of these small conserved regions and the potential for a vaccine to steer immunodominance toward conserved epitopes.
INTRODUCTION
CD8+ T lymphocytes (CTLs) play a key protective role in the pathogenesis of human immunodeficiency virus type 1 (HIV-1) infection. The ontogeny of this immune response is temporally associated with the decline of peak viremia during acute infection (1, 2), mutation in response to CTLs is a major determinant of viral sequence evolution (3–5), and depletion of CD8+ cells in vivo leads to increased simian immunodeficiency virus (SIV) replication in the macaque model of AIDS (6–8). Despite the contribution of this response to slowing disease progression, however, CTLs are ultimately unsuccessful in preventing AIDS in the vast majority of HIV-1-infected persons with untreated infection.
It is likely that ineffective epitope targeting of HIV-1 is a major reason for this failure. The best genetic correlate for the rate of disease progression is human leukocyte antigen class I (HLA-I), which indicates that targeting is a crucial factor in CTL efficacy. Notably, the HLA-I types with the strongest associations with delayed disease, including HLA B*57, B*27, and B*13, have all been associated with highly immunodominant targeting of epitopes that are highly sequence conserved and associated with limited options for epitope escape mutations that are associated with a significant loss of replicative capacity. It further appears that protective CTL targeting against conserved epitopes occurs early in acute infection, which is a period that is usually marked by targeting of variable epitopes (9, 10), with rapid viral escape and CTL retargeting eventually leading to targeting of relatively conserved epitopes in chronic infection (11, 12), by which time severe depletion of total body and HIV-1-specific CD4+ helper T lymphocytes has occurred (13, 14) and CTLs become hypofunctional (15, 16). Thus, an early focus of CTL targeting on highly sequence-constrained epitopes appears to be important for immune containment of HIV-1 (17, 18), and this is a process that typically occurs too late for most HLA-I types.
CTL-based preventive vaccines have been proposed as a means to attenuate infection if not to block it. In addition to the above-mentioned importance of targeting for immune control, the remarkable sequence plasticity of HIV-1 is an additional rationale for targeting of CTLs to highly conserved regions, to maximize the likelihood that vaccine and incoming HIV-1 epitope sequences match. With these goals in mind, several investigators have proposed vaccines consisting of highly conserved epitopes in concatenated sequences, with mixed success in animal models (19–21). In this proof-of-concept study, we demonstrated a strategy for vaccination with short, highly conserved regions of HIV-1 and found them to be highly immunogenic despite their limited immunogenicity in the context of whole-protein gene vaccination, showing that selective vaccination with genes for highly conserved sequences to maximize vaccine sequence matching and exclude easily escaped variable regions is feasible without concatenating these short regions.
MATERIALS AND METHODS
Selection of conserved HIV-1 sequences and mosaic variants.
The regions of conserved sequences were selected as previously described in detail (22), including two stretches within Gag (“conserved Gag-1 and Gag-2”), one within Env (“conserved Env”), and one within Nef (“conserved Nef”). The Mosaic Vaccine Designer tool at the Los Alamos National Laboratory (LANL) HIV Database website (23, 24) was run to analyze the LANL HIV Database curated sequence collections of 2008 for the M group with recombinant sequences, using the following parameters: cocktail size = 1 or 2 or 3 or 4, epitope length = 9, rare threshold = 2, max runtime = 100, population size = 200, stall factor = 10, and internal crossover probability = 0.5. For each conserved region, the Mosaic Vaccine Designer was run on the full set of amino acid sequences from that region within the entire collection. The output mosaic sequences were then tested for coverage of all 9-mer stretches within the entire collection, using the LANL HIV Database Epicover tool (23, 24). For reference, the coverage provided was compared to the combination of the four consensus sequences for clades A1, B, C, and D from the LANL HIV Sequence Database. The Epicover tool was also utilized to assess coverage of the vaccine sequences with the screening M-group consensus sequence peptides (for the final vaccine sequences, see Sequences S1 and S2 in the supplemental material).
DNA vaccine construction and production.
Insert sequences corresponding to the four conserved regions, each in three mosaic versions, and clade B consensus sequences for whole gag, env, and nef (with a glycine-to-alanine mutation at the second amino acid to disrupt the myristoylation signal) were synthesized as codon-optimized genes (GeneArt/Life Technologies, Grand Island, NY) and cloned into the DNA vaccine vector VRC 4401 (25–27) (kindly provided by Gary Nabel) through replacement of the insert after restriction digestion with SalI and BamHI, and the clones were confirmed by sequencing. Plasmid preparation for the DNA vaccine was performed by culturing transformed Escherichia coli in 15-liter fermentation chambers at the UCLA Fermentation Core Facility. After overnight culture, bacterial pellets were obtained by centrifugation and used for plasmid preparation with an endonuclease-free PureLink HiPure Plasmid Gigaprep kit (Invitrogen, Grand Island, NY) according to the manufacturer's protocol. The concentration and purity of the plasmid preparations were checked by Nanodrop 2000c spectrophotometry (Thermo Fisher Scientific, Wilmington, DE); each 20-g bacterial pellet generally yielded 8 to 10 mg plasmid on a single isolation column.
rAd5 construction and production.
The rAd5 vaccine component was produced using the AdEasy XL adenoviral vector system (Agilent Technologies, Santa Clara, CA) according to the manufacturer's protocol. Briefly, the same inserts used for the plasmid DNA vaccine were spliced into the pShuttle vector. Due to Env-mediated toxicity during the virus production stage, the env gene was further modified by in-frame deletions of V2, V3, and both heptad repeat sequences. BJ5183-AD-1 cells (bacteria transformed with pAdEasy-1 vector) were electroporated with PmeI-linearized recombinant pShuttle vector (for homologous recombination), followed by plating and incubation overnight at 37°C. The presence of recombinants was initially screened by PacI digestion of the plasmids isolated from the selected colonies. When restriction yielded expected band patterns, recombination was further confirmed by nucleotide sequence analysis. Recombinant viral genomes were linearized with PacI, transfected by use of PolyFect transfection reagent (Qiagen, Chatsworth, CA) into 1.2 × 106 AD-293 cells (derived from HEK293 cells) in a 2- by 60-mm2 dish in Dulbecco's modified Eagle's medium (DMEM) with 10% heat-inactivated fetal calf serum (FCS), and grown at 37°C with 5% CO2 for 10 to 14 days, until most cells showed cytopathic effects. The supernatant was harvested, and three cycles of freeze-thawing in liquid nitrogen were carried out to release virus particles. The virus was further propagated in progressively larger-scale AD-293 cell cultures. The purification and concentration of adenovirus were performed using an AdEasy virus purification kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's protocol. Briefly, Benzonase nuclease was added for 30 min at 37°C before binding to a Sartobind syringe filter containing an ion-exchange membrane absorber that selectively binds adenoviral particles. After washing of the membrane, the viral particles were eluted in a storage/physiological buffer (20 mM Tris-HCl, pH 8.0, 25 mM NaCl, 2.5% glycerol). The viral particle concentration was determined by absorbance measurement at 260 nm, using a conversion factor of 1.1 × 1012 particles per absorbance unit. The virus vaccine stocks were then diluted and stored in single-use aliquots at −80°C until injection.
Vaccination of rhesus macaques.
The 6 female adult (12 to 15 years old) Indian-origin rhesus macaques (Macaca mulatta) used in this study were housed at the Oregon National Primate Research Center (ONPRC) in accordance with the guidelines of the Institutional Animal Care and Use Committee for Oregon Health & Science University/Oregon National Primate Research Center and the Guide for the Care and Use of Laboratory Animals (31). All macaques were free of cercopithecine herpesvirus 1, simian type D retrovirus, and simian T-lymphotropic virus type 1. Two groups of three macaques each received plasmid DNA as a prime, at weeks 0, 4, and 8, and the replication-defective rAd5 as a boost, at week 24. One group received vaccine constructs expressing the full-length HIV-1 proteins Gag, Env, and Nef (clade B consensus) injected individually into one biceps and both quadriceps, and the other group received the constructs expressing the four conserved regions, Gag-1, Gag-2, Env, and Nef (mosaic mixtures of each), injected individually into both biceps and both quadriceps. The DNA (1.25 mg per construct in 1 ml) was administered three times intramuscularly (i.m.) using Bioject injectors (Bioject Medical Technologies Inc., Tigard, OR), at weeks 0, 4, and 8, and rAd5 (6.25 × 1010 particles per construct in 1 ml) was administered via standard i.m. injection at week 24. Each control vaccination (full-length Gag, Env, and Nef) or conserved-region mixture (Gag-1, Gag-2, Env, and Nef conserved-region mosaics) was given consistently in the same limb.
Assessment of cellular immune responses.
Peripheral blood mononuclear cells (PBMCs) isolated from whole blood and bronchoalveolar lavage (BAL) fluid cells were collected and stimulated with peptide antigens. The responding T cells secreting cytokines were measured by intracellular cytokine staining as previously described (32). The cells were stimulated in R-10 medium (RPMI 1640 supplemented with 10% FCS, 2 mM l-glutamine, 25 mM HEPES, and penicillin-streptomycin) with 5 μg/ml anti-CD28 (28.2; BD Biosciences, San Jose, CA) and 5 μg/ml anti-CD49d (9F10; BD Biosciences, San Jose, CA), with or without pools of 15-mer peptides (2 μg/ml). Peptides spanning M-group consensus Gag, Env, and Nef sequences were obtained from the NIH AIDS Reagent Repository. The Gag peptides were divided into three pools, spanning the conserved Gag-1 region (peptides 39 to 54), the conserved Gag-2 region (peptides 66 to 88), and the remainder of Gag (peptides 1 to 38, 55 to 65, and 89 to 129). The Env peptides were divided into two pools, spanning the conserved Env region (peptides 9122 to 9184) and the remainder of Env (peptides 8974 to 9100 and 9101 to 9121). The Nef peptides were also divided into two pools, spanning the conserved Nef region (peptides 27 to 38) and the remainder of Nef (peptides 1 to 26 and 39 to 53).
Cells were incubated at 37°C for 1 h, followed by an additional 5-h incubation in the presence of 5 μg/ml brefeldin A (Sigma-Aldrich, St. Louis, MO). Cells were then washed, stained for CD4 and CD45 for 30 min at room temperature in the dark, fixed, permeabilized, and stained for CD8, CD3, CD69, gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin-2 (IL-2) for 45 min at room temperature in the dark. The following antibodies were used: CD3-Pacific Blue (SP34-2; BD Biosciences, San Jose, CA), CD4-peridinin chlorophyll protein (PerCP)-Cy5.5 (L200; BD Biosciences), CD8a-phycoerythrin (PE) (RPA-T8; BD Biosciences), CD69-ECD (TP1.55.3; Beckman Coulter, Brea, CA), IFN-γ–fluorescein isothiocyanate (FITC) (B27; BD Biosciences), TNF-α–PE–Cy7 (MAB11; BD Biosciences), and IL-2–allophycocyanin (APC) (MQ1-17H12; BD Biosciences). In addition, CD45 antibodies (DO58-1283; BD Biosciences) were custom conjugated with Q585. Cells were acquired by LSRII flow cytometry (BD Biosciences) with DiVa software. Postacquisition analyses were performed with the FlowJo software program (version 8.8.6; Tree Star, Inc., Ashland, OR). Data shown on the graphs represent response frequencies from which background values (no peptides) have been subtracted.
Macaque MHC typing.
Rhesus major histocompatibility complex (MHC) alleles were identified by cDNA PCR amplification of a 297-nucleotide fragment spanning the highly polymorphic peptide binding domains of MHC class I molecules, as previously described (33).
Ethics statement.
The Laboratory Animal Care and Use Program at the Oregon National Primate Research Center (ONPRC) (Beaverton, OR) is fully accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC), with approved assurances (no. A3304-01) for the care and use of animals on file with the Office for Protection from Research Risks at the NIH. This study protocol (protocol IS0882) was approved by the ONPRC's Institutional Animal Care and Use Committee (IACUC) (with membership constituted to comply with NIH policy and Animal Welfare Act regulations) under the NIH Office for Laboratory Animal Welfare (OLAW).
The rhesus macaques were housed at the ONPRC, a category I facility, in accordance with the standards of the Institutional Animal Care and Use Committee for Oregon Health & Science University/ONPRC and the Guide for the Care and Use of Laboratory Animals (31). They were housed in indoor or indoor-outdoor facilities with twice-daily collection of waste pans, in cages that were sanitized in a central cage washer at least every 2 weeks. They were fed commercially prepared primate feed milled within the past 90 days, supplemented daily with fruit and special diets prepared in the ONPRC diet kitchen. Fresh potable water was provided by the municipal water district via automatic water systems.
Ketamine HCl (5 to 10 mg/kg of body weight given i.m.) was used to induce anesthesia for all routine noninvasive clinical procedures associated with the study protocols (blood sampling, vaccine administration, and clinical examinations) and as a preoperative induction anesthetic. Isoflurane gas vaporized in 100% oxygen was used for maintenance anesthesia. Telazol was used as an anesthetic for minimally invasive clinical/surgical procedures, such as bronchoalveolar lavage. Postoperative and postprocedure analgesia was provided through administration of buprenorphine (0.03 mg/kg i.m.) every 6 to 8 h, with veterinary staff assessment of pain relief according to animal care guideline GL-016. The macaques were observed for species-specific behaviors, food and water consumption, and urine and feces production for reporting of abnormalities to the attending veterinarian. All animals were evaluated for clinical signs of disease on a daily basis by the Pathobiology and Immunology Division Animal Core and Department of Comparative Medicine animal care staff, and a clinical veterinarian was responsible for determining if an animal was experiencing any pain or suffering. This study did not involve euthanasia of the animals.
RESULTS
Four conserved regions from Gag, Env, and Nef in three-mosaic-sequence sets have broad coverage of HIV-1 variability.
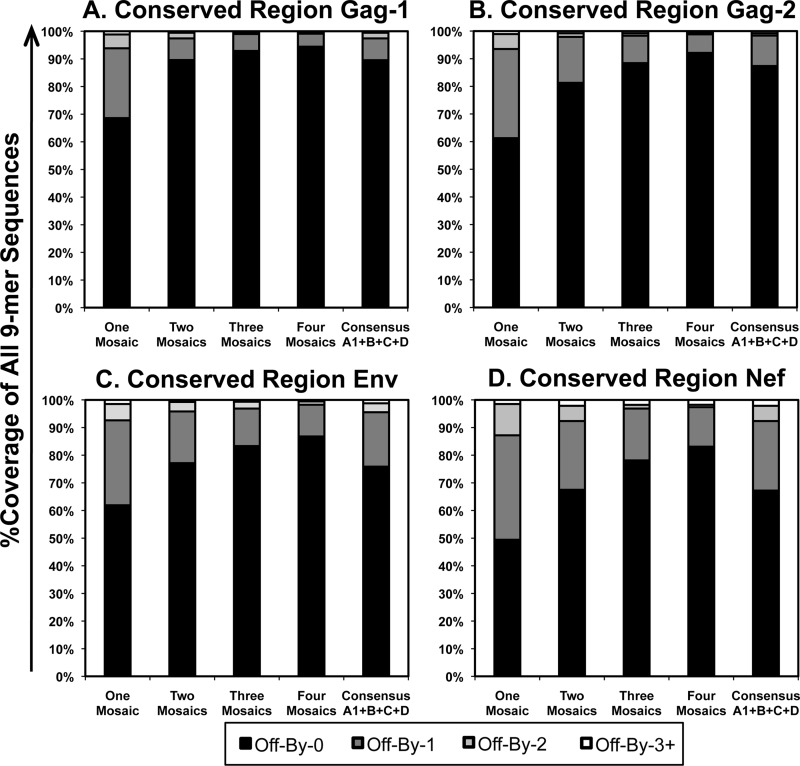
Using the LANL HIV Database Mosaic Vaccine Designer tool, combinations of one, two, three, or four mosaics were designed to maximize coverage of all 9-mer amino acid sequences from the LANL curated collection of all M-group HIV-1 sequences (including recombinants) for the regions of Gag amino acids 148 to 214, Gag amino acids 250 to 335, Env amino acids 521 to 606, and Nef amino acids 106 to 148. The coverage of all 9-amino-acid stretches within the entire M-group sequence collection was then assessed for the generated mosaic sets in comparison to consensus sequences for clades A1, B, C, and D (Fig. 1). Single mosaic sequences achieved 49 to 69% coverage of all 9-mer stretches, which increased to 79 to 93% coverage for three-mosaic-sequence sets. Increasing to four mosaic sequences further increased coverage only to 83 to 94%. In comparison, sets of four clade consensus sequences (clades A1, B, C, and D) provided 67 to 89% coverage, which was consistently less than that with three-mosaic-sequence sets.
FIG 1.
Coverage of selected conserved regions of the HIV-1 proteome by mosaic and clade consensus sequences. Mosaic sequences optimized to cover HIV-1 M-group Gag, Env, and Nef 9-mer stretches were assessed for coverage of all 9-mers in comparison to a combination of clades A1, B, C, and D for the four conserved regions. (A) Gag-1 (Gag amino acids 148 to 214); (B) Gag-2 (Gag amino acids 250 to 335); (C) Env (Env amino acids 521 to 606); (D) Nef (Nef amino acids 106 to 148). “Off-By-0” refers to the percentage of all possible 9-mer sequences within the Los Alamos HIV Database curated M-group sequences that were perfectly represented within the mosaic sequence(s), “Off-By-1” refers to the percentage that were represented by a one-amino-acid mismatch within the mosaic sequence(s), etc.
Despite lesser antigen content, a CRV elicited cellular immune responses with magnitudes similar to those obtained with a WPV containing Gag, Env, and Nef.
The four sets of three mosaic sequences covering the two conserved regions in Gag and single conserved regions in Env and Nef (see Fig. S1 in the supplemental material) were selected for further study as a conserved-region gene vaccine (CRV). This vaccine consisted of four separate mixtures of each mosaic set in equal amounts, both in plasmid DNA expression vectors and in rAd5 vectors from the NIH Vaccine Research Center. A control whole-protein gene vaccine (WPV) had three separate components: full-length clade B consensus gag, env, and nef. Both vaccines were administered as three intramuscular injections for DNA priming (weeks 0, 4, and 8) followed by a rAd5 boost (week 24). Each conserved-region gene mixture of mosaics or whole-protein gene component was injected at a separate site: the four conserved-region mosaic gene mixtures were given in both biceps and both quadriceps, while the three whole-protein genes were given in one biceps and both quadriceps. The total amounts of DNA and rAd5 were equal between CRV and WPV vaccinations. Groups of three rhesus macaques received the CRV or control WPV.
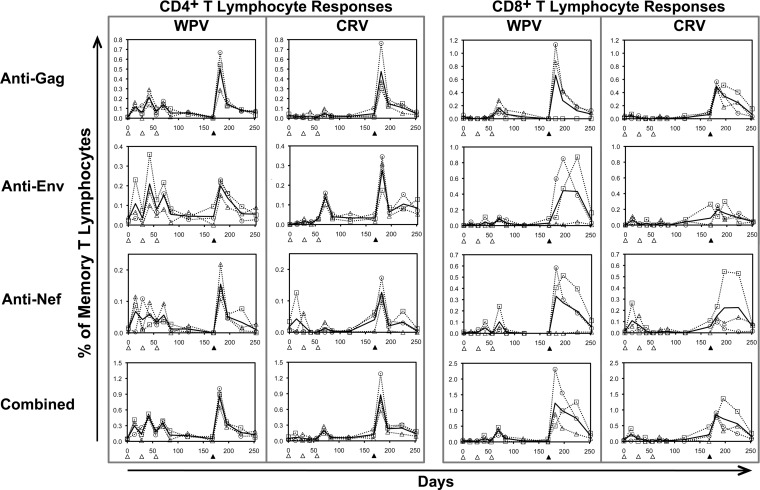
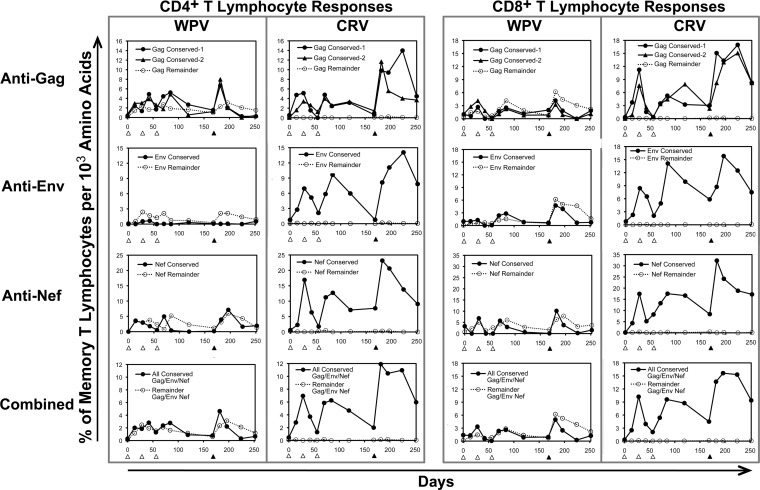
Cellular immunity in blood was assessed using overlapping peptides spanning the whole HIV-1 Gag, Env, and Nef proteins. These peptides were pooled to correspond to the four conserved regions or the remainder of each protein (conserved Gag-1, conserved Gag-2, remainder of Gag, conserved Env, remainder of Env, conserved Nef, and remainder of Nef). All macaques in both vaccine groups developed both CD4+ and CD8+ T lymphocyte responses against all three proteins, generally peaking after the rAd5 booster vaccination (Fig. 2). For both vaccines, Gag responses were highest in magnitude for both CD4+ and CD8+ T lymphocytes. Despite the sizeable discrepancies in CRV versus WPV inserts (153 versus 500 Gag amino acids, 86 versus 723 Env amino acids, and 43 versus 206 Nef amino acids, for a total of 282 versus 1,197 amino acids), responses against each protein peaked after rAd5 boosting at similar magnitudes between vaccines, although there were statistically nonsignificant trends for higher-magnitude responses in the WPV group than in the CRV group during the DNA priming vaccinations. Overall, these results demonstrated that the selected conserved regions are consistently immunogenic.
FIG 2.
Overall immunogenicity of vaccination with whole-protein versus conserved-region sequences. The frequencies of HIV-1-specific responses in the memory CD4+ and CD8+ T lymphocyte compartments were plotted for WPV and CRV. The open triangles at the x axis indicate times of DNA prime vaccinations, and the closed triangles indicate times of rAd5 booster vaccinations. For the WPV, open triangles show data for animal 19798, open squares are for animal 20252, and open circles are for animal 21894. For the CRV, open triangles show data for animal 19825, open squares are for animal 20175, and open circles are for animal 21927. The solid lines indicate means for the three animals in each vaccine group.
Whole-protein gene vaccination predominately yielded responses against sequences outside the conserved vaccine regions.
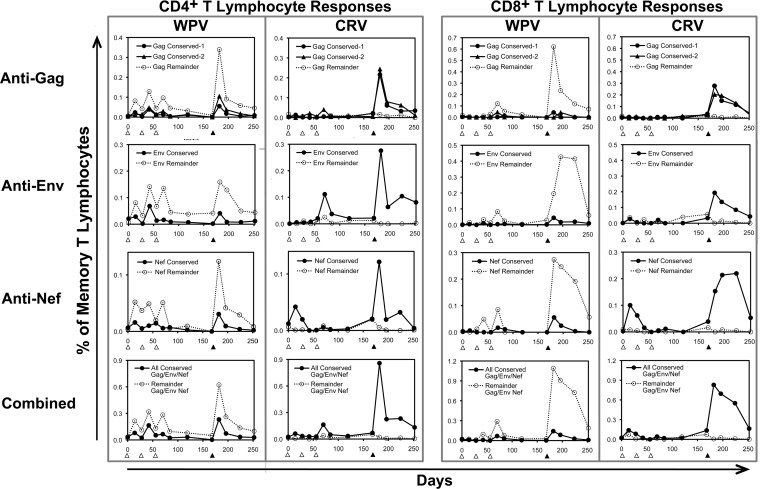
When the blood CD4+ and CD8+ T lymphocyte responses were examined according to targeting of the selected conserved vaccine regions versus sequences outside those areas within the Gag, Env, and Nef proteins, it was apparent that the WPV predominately yielded responses against the latter (Fig. 3). Despite having much smaller inserts, the CRV produced a comparable magnitude of cellular immune targeting against each protein, although it targeted only the selected conserved regions of the proteins. Thus, while the WPV also elicited responses against the conserved regions, they were much lower in magnitude than those with the CRV. These findings indicated that isolated vaccination with the selected conserved sequences sharply focused responses on these regions compared to vaccination with the full-length source proteins, which had a predominance of responses against the remaining regions of the proteins.
FIG 3.
Cellular immune targeting elicited by WPV versus CRV against the conserved regions in the CRV. The responses shown in Fig. 2 were broken down according to recognition of the four conserved regions in the CRV (solid lines and filled symbols) versus the areas outside those regions in Gag, Env, and Nef (dotted lines and open symbols), with plotting of the means for responses of the three animals in each group.
The selected conserved regions of Gag, Env, and Nef were comparably immunogenic to the sequences outside those regions, but the intensity of targeting of those regions was markedly augmented when they were given in isolation.
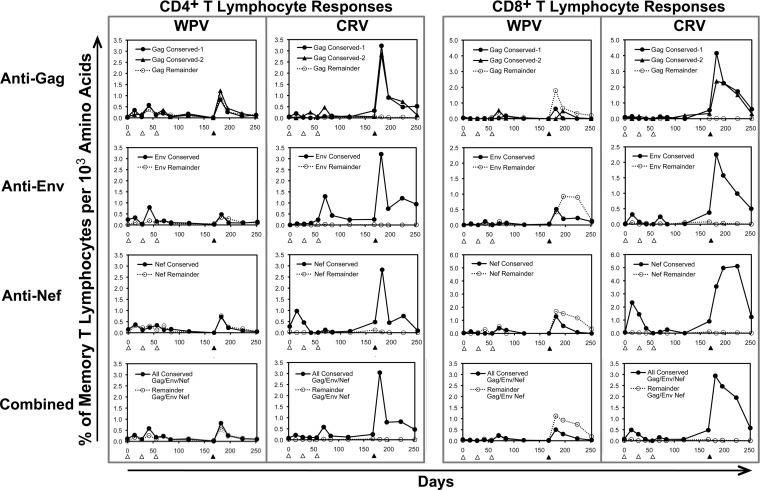
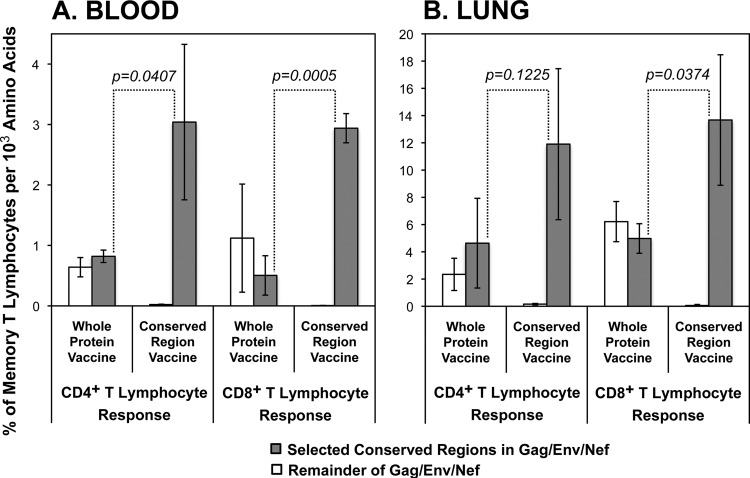
To examine whether the WPV preferential targeting of the areas outside the selected conserved regions was due to an intrinsic difference in immunogenicity, we next considered the density of blood cellular immune responses (frequency per targeted protein size) for the selected conserved regions versus the nonselected regions of Gag, Env, and Nef. For each protein, the WPV yielded consistently similar densities of targeting of the conserved regions versus the remainder of the protein (Fig. 4), suggesting that the lower magnitudes of responses against the conserved regions were due to different sequence lengths rather than differing immunogenicities. As expected, the CRV yielded only targeting of the conserved regions, but at a much higher density than that with the WPV for all four conserved regions. At the peak of cellular immune responses occurring after the rAd5 boosts, the density of overall targeting of the conserved regions elicited by the CRV was about 5-fold higher than that with the WPV for both CD4+ and CD8+ T lymphocytes in both the blood and lung compartments (Fig. 5A). These results suggested that vaccination with the smaller conserved-region genes yielded proportionally more intense targeting against those regions than that with whole-protein gene vaccination and that conserved-region immunogenicity is similar to that for other regions of the proteins.
FIG 4.
Intensities of targeting against selected conserved regions of Gag, Env, and Nef versus outside regions after vaccination with WPV versus CRV. The data shown in Fig. 3 were replotted in terms of targeting density for the vaccine regions, as percentages of HIV-1-specific responses in the memory T lymphocyte compartment per 1,000 amino acids of the vaccine target protein. For example, a vaccination with a protein region of 550 amino acids that yielded a response of 0.12% of memory CD4+ T lymphocytes would have a targeting density of 1,000 × (0.12 ÷ 550) = 2.18.
FIG 5.
Comparison of targeting against selected conserved regions of Gag, Env, and Nef versus outside regions after vaccination with WPV versus CRV at the peak after rAd5 boost. The peak responses at day 182 were plotted as means ± standard deviations for each group of three animals, according to the targeting densities plotted in Fig. 4. The significances of differences between targeting levels of conserved regions between the two vaccines are indicated (Student's t test).
Similarly targeted but higher-magnitude cellular immune responses were observed in the lung.
As with the blood, the density of CD4+ and CD8+ T lymphocyte targeting in relationship to protein size was assessed for lung T lymphocytes obtained from bronchoalveolar lavage fluid. Overall, the lung responses were about 4-fold higher than those seen in the blood (Fig. 5B) and were notable during the DNA vaccinations (Fig. 6). However, the general pattern of cellular immune responses was similar (compare Fig. 5B and 6 to Fig. 5A and 4, respectively), with the WPV showing similar densities of targeting of the selected conserved regions of Gag, Env, and Nef and the remainders of those proteins and the CRV eliciting an about 3-fold more intense targeting of the conserved regions. These results indicated that the immunodominance patterns seen in blood likely mirrored those at effector sites, such as the mucosa.
FIG 6.
Cellular immune response densities against the WPV and CRV in the lung mucosal compartment. Mononuclear cells from bronchoalveolar lavage fluid were assessed for targeting density, as shown for blood in Fig. 4.
DISCUSSION
Although the diversity of HIV-1 sequences is noted as a significant problem in HIV-1 vaccine development, it remains unclear how best to cope with this barrier. An effective vaccine must be able to generate immune responses that cover this diversity. One approach considered is the use of multivalent vaccines, based on the hope that presenting diverse antigens will allow immune responses that match that diversity. However, the extreme variability of HIV-1 precludes adequate representation of the diversity of any of its proteins with a small number of variants, even with optimization methods such as the LANL mosaic program. Although mosaic-mixture vaccines appear to give broader cellular immune responses than those with clonal vaccines, it is also unclear whether cellular immunity is capable of matching the diversity of the virus in general.
The other major vaccine strategy to deal with HIV-1 diversity has been to deliver gene sequences corresponding to relatively more conserved whole viral proteins, regions of proteins, or individual epitopes. For some immunodominant epitopes associated with protective HLA-I types, such as B*57, it appears that viral fitness limitations constrain variability, allowing CD8+ T lymphocytes to provide effective coverage. Whether this phenomenon can be generalized to the whole human population is unclear given the dependence of epitope targeting on HLA types. Furthermore, the optimal approach to focus immunity on conserved sequences has not been determined. Because the most highly conserved stretches of the HIV-1 proteome are short, most delivery attempts have concatenated genes for several short segments into a single construct. Results have been mixed for eliciting cellular immune responses against such constructs (19–21).
This study was a proof-of-concept examination of vaccination with short, highly conserved regions of the HIV-1 proteome, using DNA priming and rAd5 boosting vaccination of macaques as a model system. Our results directly contradict a similar study by Stephenson et al. (21), who recently compared full-length HIV-1 Gag, Pol, and Env to concatenated conserved regions (totaling 247 amino acids) delivered by prime-boost vaccination of macaques with heterologous rAd vectors. Their six conserved regions (three in Gag, one in Pol, and two in Env) ranged from about 30 to 70 amino acids and included three that mostly overlapped three of our conserved regions: Gag residues 148 to 217 (versus our conserved Gag-1 region, spanning Gag residues 148 to 214), Gag residues 257 to 311 (versus our conserved Gag-2 region, spanning Gag residues 250 to 335), and Env residues 537 to 566 (versus our conserved Env region, spanning Env residues 521 to 606). In contrast to our results, they found that their whole-protein gene vaccine elicited >2-fold higher responses than those with the conserved protein gene vaccine against their selected conserved regions, as well as having a greater breadth of response. Although there were a few technical differences (such as the vaccine vectors used, use of two mosaics for the whole proteins, and use of two mosaics for the conserved regions), it is possible that a factor in this discrepancy was the concatenation of their conserved regions versus our separate administration of each conserved region.
Supporting this possibility, our data suggest that targeting density is directly proportional to protein size and not sequence conservation (Fig. 5). Thus, a concatenated combination of six conserved regions totaling 247 amino acids would be targeted as a single small protein, in contrast to the separation of the six regions in three whole proteins in individual vectors. This is further supported by controlled experimental observations in murine vaccination models showing that epitopes codelivered within a single protein compete to yield immunodominance and subdominance of targeting, whereas the same epitopes delivered concurrently in separate proteins can be codominately targeted (34, 35). Our findings thus raise concerns about strategies that concatenate conserved regions, although another study using this strategy with a complex regimen of five vaccinations with three types of vectors demonstrated broad responses against multiple regions (20), as did another recent study with a simpler DNA vaccination protocol (19).
Our observation regarding an increased overall magnitude and density of targeting of the selected conserved regions after vaccination with the CRV versus WPV shows a difference in immunodominance between these vaccination strategies. While targeting was shifted toward conserved regions with the CRV, the data did not address an increased breadth of targeting within these conserved regions (due to cell limitations preventing epitope mapping), which may be an important parameter for containment of HIV-1 infection. It is more likely that the increased magnitude also reflects a greater breadth of targeting of these regions, but this will require formal demonstration.
Some questions remain about conserved-region HIV-1 vaccine strategies as a whole. It is not clear whether sequence conservation necessarily corresponds to sequence constraint. Although the degree of protection associated with CD8+ T lymphocyte responses against specific epitopes is associated with a degree of sequence conservation (36, 37), highly conserved sequences can contain relatively mutable residues (38). More importantly, although conserved regions of the HIV-1 proteome do contain known epitopes recognized in infected persons (22) and are immunogenic in macaques, their immunogenicity in human vaccination is unknown. It has been suggested that HIV-1 has undergone selection in the human population that has depleted common epitopes through fixed escape mutations (39), leading to regions being conserved due to a lack of immunogenicity and immune pressure. This is further suggested by the “rare allele advantage” for class I human leukocyte antigens (40, 41).
An additional finding of our study was vigorous T lymphocyte responses in the lung (measured in bronchoalveolar lavage fluid), which had frequencies about quadruple those in the blood that were demonstrable during the DNA prime vaccinations. In contrast, a prior macaque study of HIV-1 gag DNA vaccination showed lower responses in the lung than in blood that increased to comparable levels with the addition of a CCL27 adjuvant (42). The major technical difference was the mode of delivery (Bioject injectors in our study), suggesting that this may be a factor in determining responses at mucosal effector sites. Enhanced vaccine-elicited responses in the lung have been noted using other vaccine vectors (43, 44). Our data confirm that this also occurs with our DNA prime and rAd5 boost regimen. Although rAd5-based regimens are unlikely to be pursued for preventive vaccines, this approach may be considered for therapeutic vaccination.
There are some technical caveats related to the measurement of cellular immunity in this study. We cannot exclude that our results were biased by mismatch between the M-group consensus sequence peptides used for detection of vaccine-elicited cellular immune responses and the vaccine sequences of whole proteins versus conserved regions. However, such bias would favor detection of conserved sequence targeting given the greater likelihood of sequence matching, leading to overestimation rather than underestimation of responses against the conserved regions compared to other protein regions in the macaques that received the WPV. This was supported by analyzing the coverage of all 9-mer amino acid stretches in the full-length clade B consensus sequences of Gag, Env, and Nef versus the conserved-region mosaic sequences (see Fig. S2 in the supplemental material), which in fact demonstrated that the M-group consensus peptides used for cellular immune detection provided better coverage of the conserved regions in the clade B consensus sequences than in the mosaic sequences. Another caveat is that quantitation of responses was performed with relatively large pools of peptides, which might have led to a reduced sensitivity of detection, and thus the comparisons of targeting of different regions may be somewhat biased by differences in the sizes of peptide pools used for detection.
Another potential confounder may have been MHC differences between macaques, particularly given the small numbers of animals in these groups. Comparison of Mamu class I alleles known to be associated with protective cellular immunity in SIV infection showed similar frequencies between groups (see Table S1 in the supplemental material). While it is unclear that immunogenicity against SIV can be extrapolated to that against HIV-1, there were no clear differences in Mamu class I alleles between the WPV and CRV groups to explain the differences seen in this study.
In summary, presentation of conserved regions of HIV-1 by a vaccine demonstrated their immunogenicity and greater intensity of targeting than that with whole-protein gene vaccination in macaques. These results suggested an advantage of giving these sequences individually and, furthermore, demonstrated the potential utility for keeping such regions in separate vectors. Whether targeting of conserved regions will be similar in human vaccination and protective against HIV-1 remains to be determined.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andrew Sylwester and Shoko Hagen for their technical and administrative assistance, Alfred W. Legasse and Shannon Planer for their expert animal care, Michael K. Axthelm for management of the animal protocols, and Janet Treger, Makoto Sato, and Brooke Bogan for their assistance with recombinant adenovirus production.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02370-14.
REFERENCES
- 1.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol 68:6103–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol 68:4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O'Sullivan MK, Desouza I, Feeney ME, Eldridge RL, Maier EL, Kaufmann DE, Lahaie MP, Reyor L, Tanzi G, Johnston MN, Brander C, Draenert R, Rockstroh JK, Jessen H, Rosenberg ES, Mallal SA, Walker BD. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol 79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, McNevin J, Cao J, Zhao H, Genowati I, Wong K, McLaughlin S, McSweyn MD, Diem K, Stevens CE, Maenza J, He H, Nickle DC, Shriner D, Holte SE, Collier AC, Corey L, McElrath MJ, Mullins JI. 2006. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J Virol 80:9519–9529. doi: 10.1128/JVI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 6.Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med 189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matano T, Shibata R, Siemon C, Connors M, Lane HC, Martin MA. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol 72:164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 9.Goulder PJ, Altfeld MA, Rosenberg ES, Nguyen T, Tang Y, Eldridge RL, Addo MM, He S, Mukherjee JS, Phillips MN, Bunce M, Kalams SA, Sekaly RP, Walker BD, Brander C. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J Exp Med 193:181–194. doi: 10.1084/jem.193.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones NA, Wei X, Flower DR, Wong M, Michor F, Saag MS, Hahn BH, Nowak MA, Shaw GM, Borrow P. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J Exp Med 200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frahm N, Korber BT, Adams CM, Szinger JJ, Draenert R, Addo MM, Feeney ME, Yusim K, Sango K, Brown NV, SenGupta D, Piechocka-Trocha A, Simonis T, Marincola FM, Wurcel AG, Stone DR, Russell CJ, Adolf P, Cohen D, Roach T, St John A, Khatri A, Davis K, Mullins J, Goulder PJ, Walker BD, Brander C. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol 78:2187–2200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koibuchi T, Allen TM, Lichterfeld M, Mui SK, O'Sullivan KM, Trocha A, Kalams SA, Johnson RP, Walker BD. 2005. Limited sequence evolution within persistently targeted CD8 epitopes in chronic human immunodeficiency virus type 1 infection. J Virol 79:8171–8181. doi: 10.1128/JVI.79.13.8171-8181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol 77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg ES, LaRosa L, Flynn T, Robbins G, Walker BD. 1999. Characterization of HIV-1-specific T-helper cells in acute and chronic infection. Immunol Lett 66:89–93. doi: 10.1016/S0165-2478(98)00165-5. [DOI] [PubMed] [Google Scholar]
- 15.Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, Little S, Havlir DV, Richman DD, Gruener N, Pape G, Waters A, Easterbrook P, Salio M, Cerundolo V, McMichael AJ, Rowland-Jones SL. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med 8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 16.Kalams SA, Buchbinder SP, Rosenberg ES, Billingsley JM, Colbert DS, Jones NG, Shea AK, Trocha AK, Walker BD. 1999. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J Virol 73:6715–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, Yu XG, Draenert R, Johnston MN, Strick D, Allen TM, Feeney ME, Kahn JO, Sekaly RP, Levy JA, Rockstroh JK, Goulder PJ, Walker BD. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581–2591. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- 18.Streeck H, Lichterfeld M, Alter G, Meier A, Teigen N, Yassine-Diab B, Sidhu HK, Little S, Kelleher A, Routy JP, Rosenberg ES, Sekaly RP, Walker BD, Altfeld M. 2007. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J Virol 81:7725–7731. doi: 10.1128/JVI.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni V, Valentin A, Rosati M, Alicea C, Singh AK, Jalah R, Broderick KE, Sardesai NY, Le Gall S, Mothe B, Brander C, Rolland M, Mullins JI, Pavlakis GN, Felber BK. 2014. Altered response hierarchy and increased T-cell breadth upon HIV-1 conserved element DNA vaccination in macaques. PLoS One 9:e86254. doi: 10.1371/journal.pone.0086254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosario M, Bridgeman A, Quakkelaar ED, Quigley MF, Hill BJ, Knudsen ML, Ammendola V, Ljungberg K, Borthwick N, Im EJ, McMichael AJ, Drijfhout JW, Greenaway HY, Venturi V, Douek DC, Colloca S, Liljestrom P, Nicosia A, Price DA, Melief CJ, Hanke T. 2010. Long peptides induce polyfunctional T cells against conserved regions of HIV-1 with superior breadth to single-gene vaccines in macaques. Eur J Immunol 40:1973–1984. doi: 10.1002/eji.201040344. [DOI] [PubMed] [Google Scholar]
- 21.Stephenson KE, SanMiguel A, Simmons NL, Smith K, Lewis MG, Szinger JJ, Korber B, Barouch DH. 2012. Full-length HIV-1 immunogens induce greater magnitude and comparable breadth of T lymphocyte responses to conserved HIV-1 regions compared with conserved-region-only HIV-1 immunogens in rhesus monkeys. J Virol 86:11434–11440. doi: 10.1128/JVI.01779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang OO. 2009. Candidate vaccine sequences to represent intra- and inter-clade HIV-1 variation. PLoS One 4:e7388. doi: 10.1371/journal.pone.0007388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, Kuiken C, Haynes B, Letvin NL, Walker BD, Hahn BH, Korber BT. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med 13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 24.Thurmond J, Yoon H, Kuiken C, Yusim K, Perkins S, Theiler J, Bhattacharya T, Korber B, Fischer W. 2008. Web-based design and evaluation of T-cell vaccine candidates. Bioinformatics 24:1639–1640. doi: 10.1093/bioinformatics/btn251. [DOI] [PubMed] [Google Scholar]
- 25.Catanzaro AT, Roederer M, Koup RA, Bailer RT, Enama ME, Nason MC, Martin JE, Rucker S, Andrews CA, Gomez PL, Mascola JR, Nabel GJ, Graham BS. 2007. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine 25:4085–4092. doi: 10.1016/j.vaccine.2007.02.050. [DOI] [PubMed] [Google Scholar]
- 26.Graham BS, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, Martin JE, McCluskey MM, Chakrabarti BK, Lamoreaux L, Andrews CA, Gomez PL, Mascola JR, Nabel GJ. 2006. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 DNA candidate vaccine. J Infect Dis 194:1650–1660. doi: 10.1086/509259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabel GJ, Huang Y, Xu L, Chakrabarti B, Wu L, Yang Z-Y, Gall JD, King R. 23February2006. Vaccines against AIDS comprising cmv/r nucleic acid constructs. US patent WO 2006020071 A2.
- 28.Reference deleted.
- 29.Reference deleted.
- 30.Reference deleted.
- 31.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed.National Academies Press, Washington, DC. [Google Scholar]
- 32.Walker JM, Maecker HT, Maino VC, Picker LJ. 2004. Multicolor flow cytometric analysis in SIV-infected rhesus macaque. Methods Cell Biol 75:535–557. doi: 10.1016/S0091-679X(04)75022-0. [DOI] [PubMed] [Google Scholar]
- 33.Wiseman RW, Karl JA, Bimber BN, O'Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E Jr, Wright C, Harkins T, O'Connor DH. 2009. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med 15:1322–1326. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmowski MJ, Choi EM, Hermans IF, Gilbert SC, Chen JL, Gileadi U, Salio M, Van Pel A, Man S, Bonin E, Liljestrom P, Dunbar PR, Cerundolo V. 2002. Competition between CTL narrows the immune response induced by prime-boost vaccination protocols. J Immunol 168:4391–4398. doi: 10.4049/jimmunol.168.9.4391. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez F, Harkins S, Slifka MK, Whitton JL. 2002. Immunodominance in virus-induced CD8(+) T-cell responses is dramatically modified by DNA immunization and is regulated by gamma interferon. J Virol 76:4251–4259. doi: 10.1128/JVI.76.9.4251-4259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunwar P, Hawkins N, Dinges WL, Liu Y, Gabriel EE, Swan DA, Stevens CE, Maenza J, Collier AC, Mullins JI, Hertz T, Yu X, Horton H. 2013. Superior control of HIV-1 replication by CD8+ T cells targeting conserved epitopes: implications for HIV vaccine design. PLoS One 8:e64405. doi: 10.1371/journal.pone.0064405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mothe B, Llano A, Ibarrondo J, Daniels M, Miranda C, Zamarreno J, Bach V, Zuniga R, Perez-Alvarez S, Berger CT, Puertas MC, Martinez-Picado J, Rolland M, Farfan M, Szinger JJ, Hildebrand WH, Yang OO, Sanchez-Merino V, Brumme CJ, Brumme ZL, Heckerman D, Allen TM, Mullins JI, Gomez G, Goulder PJ, Walker BD, Gatell JM, Clotet B, Korber BT, Sanchez J, Brander C. 2011. Definition of the viral targets of protective HIV-1-specific T cell responses. J Transl Med 9:208. doi: 10.1186/1479-5876-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rolland M, Manocheewa S, Swain JV, Lanxon-Cookson EC, Kim M, Westfall DH, Larsen BB, Gilbert PB, Mullins JI. 2013. HIV-1 conserved-element vaccines: relationship between sequence conservation and replicative capacity. J Virol 87:5461–5467. doi: 10.1128/JVI.03033-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yusim K, Kesmir C, Gaschen B, Addo MM, Altfeld M, Brunak S, Chigaev A, Detours V, Korber BT. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J Virol 76:8757–8768. doi: 10.1128/JVI.76.17.8757-8768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, Kuse N, Oka S, Duda A, Prendergast A, Crawford H, Leslie A, Brumme Z, Brumme C, Allen T, Brander C, Kaslow R, Tang J, Hunter E, Allen S, Mulenga J, Branch S, Roach T, John M, Mallal S, Ogwu A, Shapiro R, Prado JG, Fidler S, Weber J, Pybus OG, Klenerman P, Ndung'u T, Phillips R, Heckerman D, Harrigan PR, Walker BD, Takiguchi M, Goulder P. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trachtenberg E, Korber B, Sollars C, Kepler TB, Hraber PT, Hayes E, Funkhouser R, Fugate M, Theiler J, Hsu YS, Kunstman K, Wu S, Phair J, Erlich H, Wolinsky S. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat Med 9:928–935. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- 42.Kraynyak KA, Kutzler MA, Cisper NJ, Khan AS, Draghia-Akli R, Sardesal NY, Lewis MG, Yan J, Weiner DB. 2010. Systemic immunization with CCL27/CTACK modulates immune responses at mucosal sites in mice and macaques. Vaccine 28:1942–1951. doi: 10.1016/j.vaccine.2009.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M Jr, Lifson JD, Nelson JA, Jarvis MA, Picker LJ. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park H, Adamson L, Ha T, Mullen K, Hagen SI, Nogueron A, Sylwester AW, Axthelm MK, Legasse A, Piatak M Jr, Lifson JD, McElrath JM, Picker LJ, Seder RA. 2013. Polyinosinic-polycytidylic acid is the most effective TLR adjuvant for SIV Gag protein-induced T cell responses in nonhuman primates. J Immunol 190:4103–4115. doi: 10.4049/jimmunol.1202958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.