ABSTRACT
All members of the Caliciviridae family of viruses produce a subgenomic RNA during infection. The subgenomic RNA typically encodes only the major and minor capsid proteins, but in murine norovirus (MNV), the subgenomic RNA also encodes the VF1 protein, which functions to suppress host innate immune responses. To date, the mechanism of norovirus subgenomic RNA synthesis has not been characterized. We have previously described the presence of an evolutionarily conserved RNA stem-loop structure on the negative-sense RNA, the complementary sequence of which codes for the viral RNA-dependent RNA polymerase (NS7). The conserved stem-loop is positioned 6 nucleotides 3′ of the start site of the subgenomic RNA in all caliciviruses. We demonstrate that the conserved stem-loop is essential for MNV viability. Mutant MNV RNAs with substitutions in the stem-loop replicated poorly until they accumulated mutations that revert to restore the stem-loop sequence and/or structure. The stem-loop sequence functions in a noncoding context, as it was possible to restore the replication of an MNV mutant by introducing an additional copy of the stem-loop between the NS7- and VP1-coding regions. Finally, in vitro biochemical data suggest that the stem-loop sequence is sufficient for the initiation of viral RNA synthesis by the recombinant MNV RNA-dependent RNA polymerase, confirming that the stem-loop forms the core of the norovirus subgenomic promoter.
IMPORTANCE Noroviruses are a significant cause of viral gastroenteritis, and it is important to understand the mechanism of norovirus RNA synthesis. Here we describe the identification of an RNA stem-loop structure that functions as the core of the norovirus subgenomic RNA promoter in cells and in vitro. This work provides new insights into the molecular mechanisms of norovirus RNA synthesis and the sequences that determine the recognition of viral RNA by the RNA-dependent RNA polymerase.
INTRODUCTION
Noroviruses, members of the Caliciviridae family of small positive-sense RNA viruses, are a major cause of viral gastroenteritis in the developed world (1, 2). Despite their impact, noroviruses remain poorly characterized, as an in-depth understanding of the molecular mechanisms of norovirus genome translation and replication has been hampered by the inability to culture human norovirus (3). Murine norovirus (MNV) is a model system with which to understand the norovirus life cycle (4), as it replicates in cultured cells (5) and a number of tractable reverse genetics systems are available (6–9). Studies of MNV have therefore led to a number of significant advances in the understanding of the molecular mechanism of norovirus genome translation and replication (reviewed in reference 10).
The norovirus RNA genome typically carries three open reading frames (ORFs) (11). MNV also has an additional ORF in the region overlapping the VP1-coding region (12, 13) (Fig. 1A). All members of the Caliciviridae family of small positive-sense RNA viruses synthesize a shorter-than-genome-length, subgenomic RNA (sgRNA) which directs the translation of the major and minor structural proteins, VP1 and VP2, respectively (10). The MNV sgRNA also encodes the VF1 protein, an antagonist of the innate immune response (12). Production of an sgRNA during the viral life cycle is a common feature of many positive-sense RNA viruses (14), and it is often used to control the expression of the viral proteins.
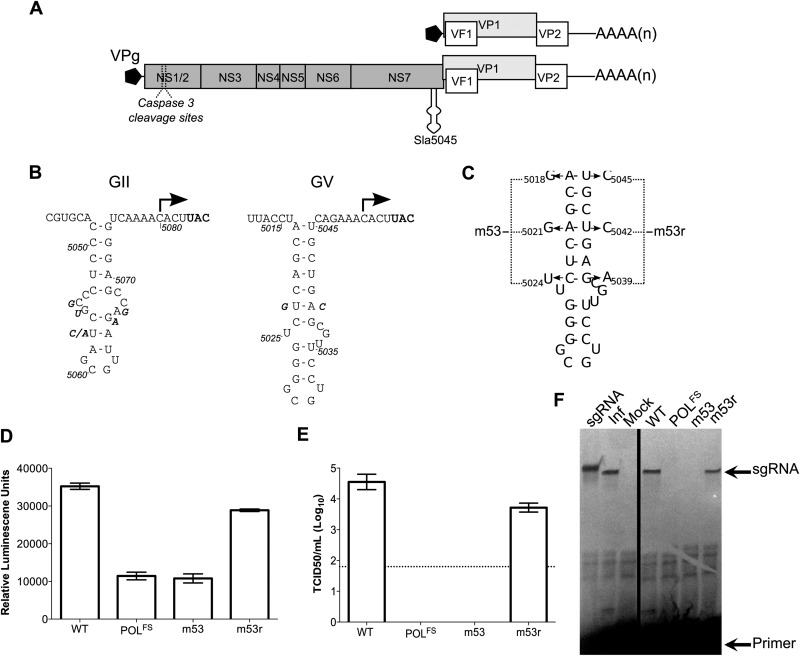
FIG 1.
The stability of Sla5045 is required for norovirus infectivity, replication, and sgRNA synthesis. (A) A schematic of the murine norovirus (MNV) genome, highlighting the four open reading frames and Sla5045. (B) Genetic conservation of the Sla5045 sequences on the norovirus negative-sense viral RNA. The position of the potential norovirus subgenomic RNA initiation site is indicated with an arrow. The position of the VP1 initiation codon on the positive-sense RNA is highlighted in bold, and sequence variation observed in different isolates is shown as bold italic. For genogroup II (GII) noroviruses, the sequence of the GII isolate Hu/GII.4/MD-2004/2004/US is shown (DQ658413), with variation between the following isolates shown: FJ595907.1, AB447425.1, HM635151, HM635164, JF262610.1, and JF262592. For genogroup V (GV) murine norovirus, the sequence of the CW1 isolate (DQ285629) is shown, with variation between the following isolates displayed: JN975491 and JN975492. Note that for simplicity, only sequences showing variation in the stem-loop sequence were used in the analysis and shown in the figure. (C) Structure of Sla5045 and the mutations introduced to generate the mutants m53 and m53r as described previously (16). In the case of m53r, the mutations shown are in addition to those present in m53. (D) Luciferase replicon-based analysis of the effect of Sla5045 disruption. Luciferase levels represent the mean for triplicate independent samples. (E) Virus yield after reverse genetics recovery of full-length cDNA constructs containing either wild-type (WT) MNV, a polymerase frameshift in the NS7 (POLFS), m53, or m53r. The virus titers represent the mean values obtained at 24 h posttransfection of viral cDNA expression constructs. The detection limit of the assay is indicated with a dotted line. In all cases where shown, error bars represent the standard error of the mean (SEM). (F) Primer extension-mediated detection of sgRNA synthesis in MNV-infected cells (Inf) or cells transfected with capped RNAs of WT, POLFS, m53, or m53r clones. In vitro-transcribed sgRNA was used as a positive control (sgRNA) and produces a product 3 nt longer due to the addition of three 5′ G nucleotides by T7 RNA polymerase. Note that 1,000-fold less RNA was used for the reaction using RNA from infected cells.
The mechanism of calicivirus sgRNA synthesis is not well understood. Initial in vitro biochemical approaches using rabbit hemorrhagic disease virus (RHDV) suggested that a sequence upstream of the first nucleotide of the subgenomic RNA is required for sgRNA synthesis (15). This sequence acts in the complement of the RHDV genomic RNA. Bioinformatic analysis also confirmed the presence of evolutionarily conserved and stable RNA stem-loop structures upstream of the starts of the sgRNAs of caliciviruses (16). Within the region coding for the viral RNA-dependent RNA polymerase (NS7, RdRp) in all caliciviruses analyzed, an ∼24- to 50-nucleotide (nt) stem-loop structure on the negative-sense genomic RNA could be predicted, precisely 6 nt 3′ of the sgRNA start site. This stem-loop structure in the MNV minus-strand RNA will be referred to as Sla5045 (for stem-loop anti-sense 5045). We previously observed that the stable structure of Sla5045 is essential for the recovery of viable MNV in cultured cells (16). In the current study, we present results consistent with the hypothesis that Sla5045 forms the core of the norovirus sgRNA promoter.
MATERIALS AND METHODS
Cell lines and plasmid constructs.
The murine leukemia macrophage cell line RAW264.7 was grown and maintained in Dulbecco modified Eagle medium (DMEM) (Gibco) with 10% (vol/vol) fetal calf serum (FCS), penicillin (100 SI units/ml), streptomycin (100 μg/ml), and 10 mM HEPES buffer (pH 7.6). Baby hamster kidney cells (BHK-21) engineered to express T7 RNA polymerase (BSR-T7 cells, obtained from Karl-Klaus Conzelmann, Ludwig Maximillian University, Munich, Germany) were maintained in DMEM containing 10% FCS, penicillin (100 SI units/ml), streptomycin (100 μg/ml), and 1.0 mg/ml Geneticin (G418). All cells were maintained at 37°C with 10% CO2.
The full-length MNV-1 cDNA clone pT7:MNV3′Rz contains the MNV-1 genome under the control of a truncated T7 polymerase promoter (7). Mutant derivatives of pT7:MNV3′RZ were made as previously describe (7, 16). These mutations include (i) a frameshift in the NS7 region of ORF1 (pT7:MNV 3′Rz F/S), (ii) the m53 mutations to destabilize the Sla5045 (pT7:MNVm53 3′Rz), and (iii) the m53r mutations to restore the Sla5045 (pT7:MNVm53r 3′Rz). Other full-length MNV-1 cDNA clones carrying m53 mutant suppressors (m53SupA, m53SupG, and m53SupH) in the m53 backbone construct were generated by overlapping PCR mutagenesis using pT7:MNVm53 3′Rz as a template. All primer sequences used in this work and protocols will be made available upon request. Mutant suppressors were also engineered into pT7:MNV3′Rz, producing WTSupA, WTSupB, and WTSupC cDNA constructs. Additional synonymous mutations at position 4922 in the wild-type (WT) and m53 cDNA constructs with adenylate and guanylate substitutions were also generated by overlapping mutagenesis PCR. The luciferase reporter-expressing MNV replicons used in this study were as reported previously (17). Briefly, the luciferase gene was fused to VP2 in the WT, POLF, m53, and m53r constructs and separated by the foot-and-mouth disease virus 2A protease (FMDV 2A). The expression of the luciferase reporter gene in these constructs is under the control of the MNV TURBS sequence (18, 19). Translation of the luciferase reporter protein occurs as a VP2 fusion protein, and the protein is cotranslationally cleaved at the specific FMDV 2A cleavage site to release both luciferase-2A and VP2.
Sla5045Dup contains a second copy of the WT or m53 stem-loop (Sla2) inserted outside the NS7-coding region in the intergenic region between Orf1 and Orf2 and was used to generate constructs WT/WT, WT/m53, m53/WT, and m53/m53 by overlapping PCR mutagenesis. A second panel of Sla5045Dup mutant constructs was also generated based on the m53/WT construct as detailed below.
Reverse genetics and virus yield determination.
Recoveries of full-length infectious MNV-1 cDNA clones were performed using the established reverse genetics system (7). Typically, BSR-T7 cells were infected with fowlpox virus expressing T7 RNA polymerase at a multiplicity of infection (MOI) of 0.5 PFU per cell before transfection with 1 μg of cDNA was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Note that the levels of T7 RNA polymerase normally expressed in BSR-T7 cells are not sufficient to drive MNV recovery. Where indicated, the RNA-mediated reverse genetics system was used. In this case, RNA transcripts were produced using in vitro transcription reactions with reaction mixtures consisting of 200 mM HEPES (pH 7.5), 32 mM magnesium acetate, 40 mM dithiothreitol (DTT), 2 mM spermidine, 7.5 mM each nucleoside triphosphate (NTP) (ATP, UTP, GTP, and CTP), 40 units of RNase inhibitor (Promega), 250 ng of linearized DNA template, and 50 μg/ml of T7 RNA polymerase. The reaction mixtures were incubated at 37°C for 2 to 7 h and treated with DNase I (New England BioLabs), and the RNA was purified by precipitation using lithium chloride and resuspended in RNA storage solution (Ambion) (9). Prior to transfection into BSR-T7 cells, posttranscriptional enzymatic capping was performed on the purified RNA transcripts using a ScriptCap system (Epicentre), according to the manufacturer's instructions. Typically, one microgram of capped RNA was transfected using Lipofectamine 2000 according to the manufacturer's instructions. The yield of infectious virus was determined at 24 h posttransfection of cDNA or capped RNA, using 50% tissue culture infectious dose (TCID50). Note that BSR-T7 cells, a BHK cell derivative, can support only a single cycle of virus replication, possibly due to the lack of a suitable receptor for virus reinfection.
RNA purification and genome copy determination.
All RNA purifications from infected and transfected cells were performed using the GenElute mammalian total RNA miniprep kit (Sigma-Aldrich) according to the manufacturer's instructions. RNA was quantified by spectrophotometry and qualitatively inspected by on agarose gels stained with ethidium bromide.
The expression levels of genomic RNA for WT and m53SupA MNVs were quantified using one step reverse transcription-quantitative PCR (RT-qPCR). Viral genome copies were determined using the MESA Blue qPCR assay (Eurogentec) performed in parallel with control RNAs of known concentration using the primers 404F (GGAGCCTGTGATCGGCTCTATCTTGGAGCAGG) and 491R (GCCTGGCAGACCGCAGCACTGGGGTTGTTGACC) to amplify the viral genomic RNA.
Where described, strand-specific RT-qPCR was performed using tagged RT primers to detect either positive- or negative-sense genomic RNA as previously described (20). In addition, a set of primers was designed to amplify nucleotides 5473 to 5422 to facilitate the simultaneous quantification of both genomic and subgenomic RNAs. Control RNAs for the genomic or subgenomic RNAs, of positive- or negative-sense polarity, were generated by in vitro transcription and used as standards to facilitate quantification.
Western blotting.
Samples for Western blot analysis were obtained by lysing cells in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% SDS). Protein concentration in the lysates were quantified using the bicinchoninic acid (BCA) protein assay (Pierce). Equal amounts of protein per sample were separated by SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) Immobilon-P transfer membrane (Millipore) for detection using enhanced chemiluminescence (ECL) reagents by semidry transfer. The blocking buffer (PBST) contained 0.1% Tween 20 in phosphate-buffered saline (PBS) containing 5% (wt/vol) milk powder and the rabbit anti-MNV NS7 or VP1 serum. Antibody binding was detected using a horseradish peroxidase-conjugated secondary antibody (source). The signal was detected with an enhanced chemiluminescence kit (ECL; Amersham Biosciences).
Luciferase reporter assay.
Cell lysates used to detect luciferase activity were prepared by lysing the transfected cells in 1× reporter lysis buffer (Promega) according to the manufacturer's instructions. The protein concentration in each lysate was standardized, and total protein (30 μg) from each cell lysate was analyzed. The luciferase assays used a 1:500 dilution of coelenterazine (Promega) in PBS as a substrate. Luciferase activity was measured in an autoinjector luminometer (FLUOstar Omega; BMG Labtech).
Northern blot assay.
Total RNAs were purified from infected cells and denatured by glyoxylation according to the manufacturer's instructions (Ambion). Denatured RNAs along with the relevant control RNA generated by in vitro transcription to produce full-length MNV genomic and subgenomic RNAs of both polarities were separated by agarose electrophoresis. Denatured RNAs were transferred to Hybond N+ nylon membranes using mild alkaline conditions and the viral RNA detected using the NorthernMax Northern blotting system (Life Technologies). RNA probes used to detect the positive- and negative-sense viral RNAs were generated by incorporating [32P]UTP or CTP using the Maxiscript in vitro labeling system according to the manufacturer's instructions (Ambion). The RNA probe used to detect the negative-sense viral RNA consisted of nt 1 to 200 of the positive-sense sgRNA, whereas the positive-sense viral RNA was detected using a probe complementary to nt 7161 to 7382.
5′ RACE.
Total RNA preparations were subjected to cDNA synthesis using a reverse primer complementary to sequences ∼700 bases downstream (on positive polarity) of Sla5045. The resulting cDNAs were treated with RNase to degrade the viral RNA template. The purified cDNAs were then used as a template for rapid amplification of cDNA ends (RACE) assay using 5′ RACE according to the manufacturer's instructions (Life Technologies). Purified cDNAs were first subjected to the 3′-end poly(C) tailing reaction catalyzed by terminal transferase, followed by PCR using an anchor primer that anneals to the poly(C) 3′ end of cDNA and the MNV-specific reverse primer. Sequences of the PCR products were determined using the BigDye Terminator kit and analyzed using the Vector NTI software (Invitrogen).
Primer extension analysis.
Primer extension analysis was performed on RNA isolated from either infected cells or cells transfected with in vitro-transcribed capped RNAs generated from MNV cDNA-containing clones. The [γ-33P]ATP-labeled primer 5129R (GGTTGAATGGGGACGGCCTGTTCAACGG) was annealed to 10 μg of total RNA and extended using Superscript III according to the manufacturer's instructions (Life Technologies). Reactions were stopped by the addition of formamide-containing dye, followed by separation on 6% urea-PAGE. The extended products were visualized following exposure to autoradiography film and compared to samples isolated from infected cells and control RNA produced by in vitro transcription.
Recombinant RdRp expression and purification.
The cDNA encoding MNV NS7 (nt 3537 to 5069) was subcloned in pET-15b. The construct contains an N-terminal six-histidine tag to facilitate protein purification. The plasmid expressing MNV NS7 was transformed into Escherichia coli Rosetta cells, which were grown at 37°C until the optical density at 600 nm (OD600) was between 0.6 and 0.8. Isopropylthiogalactoside was added to a final concentration of 1 mM to induce protein expression for 16 to 18 h at 16°C. The cells were harvested by centrifugation and resuspended in 40 ml lysis buffer (100 mM Tris-Cl [pH 7.9], 300 mM NaCl, 10% glycerol, 15 mM imidazole, 5 mM β-mercaptoethanol, 0.1% Triton X-100, protease inhibitor cocktail [Sigma]). The recombinant protein was purified with Ni-nitrilotriacetic acid (NTA) (Qiagen) according to the manufacturer's instructions. The eluted protein was dialyzed in buffer containing 100 mM Tris-Cl (pH 7.9), 300 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol and stored in aliquots at −80°C.
Recombinant GII.4 NS7 was expressed and purified as previously described (17). Briefly, E. coli Top 10 cells transformed with pBAD GII.4NS7 plasmid were cultured at 37°C until an OD600 of around 0.6 was reached. l-Arabinose was added to a final concentration of 0.02% to induce protein expression. The cells were grown at 37°C for 5 h and then harvested and suspended in lysis buffer (50 mM HEPES [pH 7.9], 150 mM NaCl, 10% glycerol, 10 mM imidazole, proteinase inhibitor cocktail [Sigma]). The protein was purified by Talon metal affinity resin (Clontech Laboratories) and the eluted protein dialyzed in buffer containing 100 mM Tris-Cl (pH 7.9), 300 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol and stored in aliquots at −80°C. All recombinant proteins were quantified by using the Bradford method followed SDS-PAGE and staining with Coomassie brilliant blue to examine their purity.
RNA synthesis assay.
RNA synthesis by the recombinant RdRps was assayed as previously described (21) with minor modifications. The 20-μl reaction mixture contained 20 mM sodium glutamate (pH 7.4), 12.5 mM dithiothreitol, 4 mM MgCl2, 1 mM MnCl2, 0.5% (vol/vol) Triton X-100, 0.2 mM GTP, 0.1 mM ATP and UTP, 3.3 nM [α-32P]CTP (MP Biomedicals), 50 nM MNV RNA template, 250 nM recombinant RdRp. The reaction mixture was incubated at 30°C for 2 h, and then the reaction was stopped by the addition of EDTA (pH 8.0) to final concentration of 10 mM. The RdRp products were subjected to electrophoresis in a 24% polyacrylamide gel containing 7.5 M urea and 0.5× Tris-borate-EDTA (TBE). The radiolabeled RNA products were visualized and quantified by using a PhosphorImager (Typhoon 9210; Amersham Biosciences) and ImageQuant software.
RESULTS
The stability of Sla5045 contributes to norovirus infectivity and subgenomic RNA synthesis.
Sequence analyses of published noroviruses genomes indicate that the stem-loop sequence of Sla5045 is highly conserved (Fig. 1B and data not shown). Several positions in genogroup II (GII) or GV, representing human and murine noroviruses, respectively, appear to show some variation in sequence, although in all cases a complex stem-loop structure is retained (Fig. 1B). We initially sought to test the functional significance of the stability of Sla5045. Three nucleotide substitutions were introduced into the MNV genome, resulting in a construct named m53 (Fig. 1C). m53 was previously shown to have a severely reduced recovery of infectious virus (16). However, it was not determined whether the inability to produce detectable virus was due to a defect in RNA encapsidation, in sgRNA production, or both. To further validate the role of Sla5045 in the norovirus life cycle and to examine whether the m53 mutations affect viral RNA synthesis, we inserted the previously described m53 substitutions into the MNV luciferase replicon Mflc-R (17). The replicon consists of a full-length MNV genome expressing a Renilla luciferase-FMDV 2A peptide-VP2 fusion protein in the VP2-coding region. Transfection of BSRT7 cells with enzymatically capped, in vitro-transcribed RNA from the Mflc-R cDNA leads to luciferase expression in the absence of infectious virus production. As luciferase is expressed from ORF3 and expression occurs via a conserved termination-reinitiation mechanism of translation at the translational termination signal of ORF2 (18, 19), it functions as a highly sensitive marker for the production of sgRNA. The introduction of the m53 mutations resulted in luciferase levels similar to those for a replication-defective replicon containing a frameshift in the NS7-coding region (Fig. 1D). A construct containing mutations compensatory to m53, named m53r (Fig. 1C), that restored the stability of Sla5045 was found to restore luciferase expression levels to nearly the level of the WT MNV replicon (Fig. 1D). The introduction of m53r mutations also resulted in the recovery of infectious virus (Fig. 1E). These data are consistent with the hypothesis that Sla5045 plays a role in viral RNA synthesis and does not simply affect a late stage in the viral life cycle such as encapsidation.
We analyzed the production of sgRNA in cells transfected with capped RNAs of m53 and m53r using a primer extension assay. A radiolabeled primer complementary to nucleotides 5121 to 5129 was annealed to RNA isolated from transfected cells and extended using reverse transcriptase, generating a product of 105 nt corresponding to the start of the sgRNA. While sgRNA was detected in infected or cells transfected with WT RNA, it was absent in m53 but restored to almost wild-type levels in m53r (Fig. 1F). Taken together, these data confirm that the stability of Sla5045 contributes to MNV sgRNA synthesis, as well as that the production of luciferase and the presence of infectious virus correlate with sgRNA synthesis.
Rapid reversion and suppression of Sla5045 disruption occur in cell culture.
The replication cycles of many positive-sense RNA viruses are error prone, and the reversion or phenotypic suppression of the introduced mutations can occur even with low levels of viral replication. Such reversion or suppression can become apparent after repeated passage of samples obtained by reverse genetics recovery. To examine whether it was possible to isolate revertants or suppressors of MNV containing the m53 mutations, named MNVm53, clarified tissue culture supernatants from three independent reverse genetics recoveries were serially passaged in RAW264.7 cells until cytopathic effect (CPE) was observed. In all cases CPE was observed after 3 passages in permissive cells. Passages performed with similar samples obtained using the POLFS construct failed to result in CPE. Individual viral isolates were isolated by limiting dilution, and the sequence of the NS7-coding region was determined. In all cases we observed additional changes in either Sla5045 or sequences elsewhere in the viral genome (Fig. 2). In total, we identified 12 different independently derived viable viruses that could be divided into two groups: phenotypic revertant mutants (Rev 1 to 9) that contained mutations that phenotypically restored the base pairing of Sla5045 and suppressor mutants (SupA to -C) that retained the m53 mutations but had additional changes elsewhere. All Rev isolates contained mutations that appeared to restore the stability of Sla5045 by either reintroducing the WT sequence (Rev 2 to 6 and 8) or introducing a stabilizing mutation on the opposite side of the stem (Fig. 2B). In contrast the suppressor mutants retained the m53 mutations but had additional changes in the NS7-coding region. Of the three Sup mutations identified, SupA and SupC had synonymous mutations that did not alter the corresponding amino acid (data not shown), while SupB resulted in a threonine-to-alanine substitution at residue 439 of NS7.
FIG 2.

Sequence analysis of m53 phenotypic revertants and second-site suppressor mutants. (A) Positions mutated in the m53 mutant are shown in bold, with any changes observed in the phenotypic revertants highlighted in gray. The nucleotide sequences shown are for the negative-sense RNA genome. An asterisk indicates that the identified sequence change results in a coding change in the viral NS7 protein (T439A). Revertant mutants are defined as those that phenotypically restore the base pairing of Sla5045. Suppressors are those that retain the mutations in Sla5045 but have additional mutations outside the region. (B) Location of reversion mutations isolated after repeated passage of m53 in cell culture. Note that the suppressor mutations are not shown, as they lie outside the region containing Sla5045.
Both coding and noncoding suppressor mutations in the NS7-coding region complement the defect in MNVm53.
To confirm that SupA to -C can suppress the defect in MNVm53 replication, individual mutations were introduced into full-length cDNA clones from WT MNV or m53 and the effect on virus recovery examined in BSRT7 cells. The BHK-derived BSRT7 cells are permissive for viral replication but not viral infection, with the viral yields being indicative of a single cycle of viral replication (7). All three suppressor mutations were viable in the m53 background, confirming their ability to restore the replication defect of MNVm53 (Fig. 3A). SupA, with an A4922G substitution on the negative-sense RNA, was consistently better at restoring the replication defect of MNVm53 than SupB or -C, improving viral yields by more than 10-fold compared to the virus with both the m53 and either the SupB or SupC mutations. All three Sup mutations were also viable in the WT cDNA background, with SupA producing viral yields that were comparable to those for WT MNV by 48 h after transfection (Fig. 3A and B). Sequence analysis indicated that all viruses were stable for at least three passages in cell culture (data not shown). Given that SupA and -C had synonymous mutations, these results strongly suggest that the suppression of the m53 defect acted on the viral RNA.
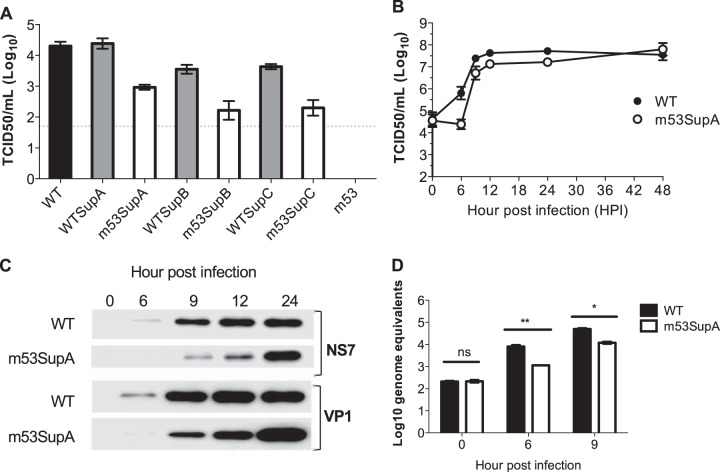
FIG 3.
Characterization of m53 suppressor mutations. (A) Virus yield following reverse genetics rescue of cDNA constructs containing WT or m53 MNV cDNA with or without the additional second-site mutations as detailed in Fig. 2A. Reverse genetics recovery was performed as described in Materials and Methods and then the virus yield at 24 h posttransfection determined by TCID50. The dotted line represents the detection limit of the assay, with error bars representing the SEM. (B to D) Single-step growth curve analysis of m53SupA replication in comparison to that of WT MNV. RAW264.7 cells were infected at a multiplicity of infection of 4 TCID50 per cell and samples harvested at the indicated times postinfection. Infectious virus was released by freeze-thawing and titrated. (B) Infectious virus titers; (C) protein expression levels of NS7 and VP1; (D) viral genomic RNA levels. In panel D, only samples from the exponential growth phase are shown. Error bars represent the SEM.
The replication of MNVm53SupA was further characterized by one-step growth curve analysis (Fig. 3B to D). The production of infectious virus (Fig. 3B), viral protein (Fig. 3C), and viral RNA (Fig. 3D) were all reduced in MNVm53SupA compared to WT MNV. The mutation introduced in SupA results in a silent U4922C mutation on the positive-sense RNA (Fig. 4A). This position corresponded to an alanine at codon 461 in NS7 and enables additional silent mutations to be introduced. We examined whether substitutions to the other three nucleotides at this position would also suppress the m53 defect as well as affect WT MNV. All 4 mutations were well tolerated in a WT MNV background, whereas only the substitutions equivalent to SupA, A4922G on the minus strand or A4922U on the minus-strand RNA, restored replication in the MNVm53 backbone (Fig. 4B). All viable viruses could be repeatedly passaged in cell culture, and the sequences introduced remained stable (data not shown). These data suggest that there is a sequence-specific requirement for the ability of position 4922 to suppress the defect in MNVm53.
FIG 4.
Sequence changes at position 4922 can compensate for a defect in Sla5045. (A) Schematic illustration of the region of the MNV genome mutated in m53 and m53SupA. The sequence of the region shown is represented in the positive polarity to highlight the NS7 reading frame, with the negative-polarity sequence shown below. The amino acids encoded by the region are highlighted, as well as the position of the SupA mutation (in italics) and the mutations introduced in m53. (B) Virus yield following reverse genetics rescue of cDNA constructs containing wild-type (WT) or m53 MNV cDNA with various changes at position 4922. Note that the nucleotide sequences shown represent the sequence of the antisense RNA genome, with G representing the previously isolated SupA suppressor mutation and A representing the nucleotide present in the wild-type cDNA construct. Reverse genetics recovery was performed as described in Materials and Methods and then the virus yield at 24 h posttransfection determined by TCID50. The dotted line represents the detection limit of the assay, with error bars representing the SEM.
Sla5045 function at a different location in the MNV genome.
Due to the location of Sla5045 in the NS7-coding region, mutational analysis was limited to synonymous changes that do not alter the NS7 protein sequence. To facilitate additional mutational analysis, we added a second copy of Sla5045, named Sla2, after the ORF1 termination codon. Sla2 is located within a noncoding intragenic region introduced between the coding sequences of NS7 and VP1 in construct Sla5054Dup (Fig. 5A). The cDNA of Sla5045Dup was engineered to contain either a WT or m53 version of Sla5045 or in the region containing Sla2. In vitro-transcribed and capped RNA was then transfected into cells and the effect on virus yield examined at 24 h posttransfection. RNA-mediated reverse genetics recovery was used to significantly improve the yield of recovered viruses and to enhance our ability to detect viruses that replicate poorly. As expected, RNA generated from the WT MNV cDNA clone resulted in ∼105 infectious units per ml, whereas m53 and a polymerase mutant failed to produce any detectable virus (Fig. 5B). Sla5045Dup containing Sla5045 WT/Sla2 WT (WT/WT) reproducibly yielded viable virus, whereas the WT/m53 construct, containing a mutated Sla2, did not (Fig. 5B). In constructs where Sla5045 contained the m53 mutations, a WT Sla2 restored replication, whereas an m53 Sla2 did not (Fig. 5B). These results demonstrate that only stem-loop sequences positioned at Sla2 function as sgRNA promoters in the context of the Sla5045Dup cDNA constructs. Importantly, however, given that the yield of virus obtained from the Sla5045Dup constructs WT/WT and m53/WT was significantly lower than that of the WT virus (Fig. 5B), it appears that the sgRNA promoter functions less well in the Sla2 position.
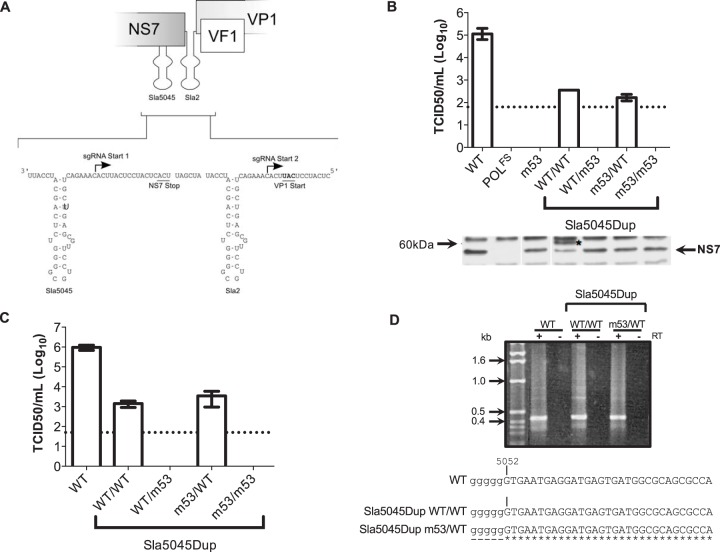
FIG 5.
Duplication of Sla5045 enables mutagenesis in a noncoding context. (A) Schematic illustration of the reconstruction of Sla5045Dup stem-loop duplication constructs. The position of the authentic Sla5045 on the antisense RNA genome is shown, and the introduced additional Sla5045 is shown as Sla2. For simplicity, only wild-type copies of the stem-loop are shown. The positions of the NS7 stop and VP1 start codons on the corresponding positive-sense viral RNA are also shown for reference. (B) Virus yield following reverse genetics rescue of capped in vitro-transcribed RNA of wild type (WT), polymerase frameshift (POLFS), m53, or Sla5045 stem-loop duplication (Sla5045Dup) constructs. The Sla5045Dup constructs are shown using the nomenclature copy1/copy2 as described in the text. Reverse genetics recovery was performed as described in Materials and Methods using in vitro-transcribed, enzymatically capped RNA and the virus yield at 24 h posttransfection determined by TCID50. A Western blot analysis for NS7 expression is shown below the virus yield data. The asterisk indicates a greater-than-full-length NS7 product observed in cells transfected with SLa5045Dup WT/WT. (C) Virus yield assay following a single passage of virus recovered following RNA transfection of wild-type MNV (WT) or various Sla5045 stem-loop duplications. The dotted line represents the detection limit of the assay, with error bars representing the SEM. (D) 5′ RACE analysis of WT MNV or Sla5045 stem-loop duplications. Viral RNA was isolated from infected cells and subjected to 5′ RACE analysis. Samples were prepared with or without reverse transcription (RT) and then subjected to PCR as described in Materials and Methods. The wild-type MNV virus was included as a control. Samples were then sequenced and aligned as shown. The lowercase poly(G) tract is introduced as a result of the 5′ RACE methodology.
Western blot analysis was used as an indirect measure of viral RNA transfection efficiency, as we have previously observed that viral antigen expression after transfection of RNA is due to translation of the capped RNA only and not due to viral replication (9). Similar levels of the NS7 protein were detectable in all cases, although the Sla5045Dup construct that contained Sla5045 WT/Sla2 WT produced a protein detected by the anti-NS7 antisera with higher molecular weight (identified by an asterisk in Fig. 5B). We suspect that this was due to the insertion of an additional stable stem-loop structure in close proximity to the NS7 stop codon resulting in suppression of translational termination, as this has been widely documented in the literature (reviewed in reference 22). This hypothesis is also supported by the absence of the extended product when the Sla5045 was disrupted by the introduction of the m53 mutations. The lack of NS7 expression in the POLFS polymerase mutant was due to the introduction of a frameshift mutation, leading to the production of a truncated NS7 protein that is rapidly degraded (9).
Although the titers of the Sla5045Dup viruses recovered were low, some of the introduced mutations were stable and could be passaged in cell culture, indicating that they are viable (Fig. 5C). Sequence analysis and 5′ RACE confirmed that the introduced mutations were stable for up to 3 passages in cell culture and that the start of the sgRNA produced during authentic norovirus replication in cell culture is position 5052, as has been predicted previously (Fig. 5D) (4).
The sequence, positioning, and stability of Sla5045 are essential for norovirus replication.
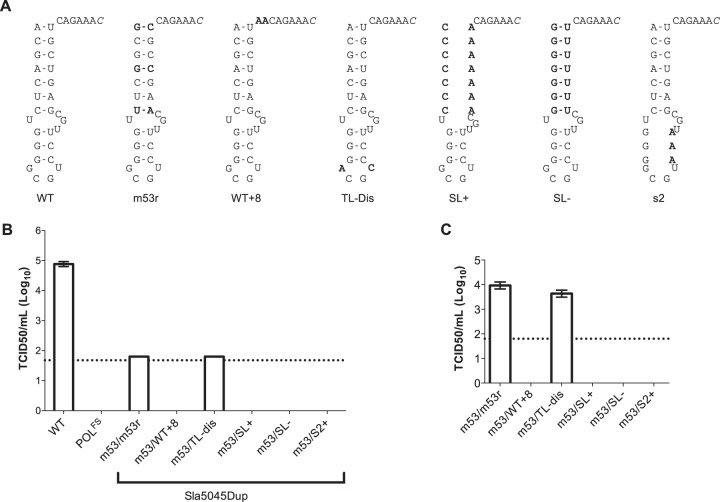
Our data indicate that Sla5045 is essential for norovirus replication and that its function can be complemented by an additional copy in cis in a noncoding region upstream of the capsid-coding sequence. To examine the sequence requirement further, we made constructs that contained a mutated m53 within the NS7-coding region along with various mutant forms in the second copy present in the noncoding region. The construct Sla5045Dup m53/Sla2 m53r contained the m53r derivative of Sla5045, which is predicted to compensate for the disrupted base pairs in m53 (Fig. 6A). As expected, Sla5045Dup m53/Sla2 m53r produced viable virus after reverse genetics recovery (Fig. 6B). The resultant virus could be passaged in cell culture (Fig. 6C) and stably maintain both copies of Sla5045 (data not shown). In contrast, Sla5045Dup m53/Sla2 WT + 8, where the spacing between the stem-loop and the predicted initiation site of the sgRNA was increased from 6 to 8 nt, was not viable. Significant alterations to the base stem sequences (Sla5045Dup m53/Sla2 SL+ or SL−) or disruption of the top stem sequence (Sla5045Dup m53/Sla2 S2) also prevented replication (Fig. 6A to C). In contrast, the introduction of two nucleotide changes in the terminal loop region of Sla5045 in the construct Sla5045Dup m53/Sla2 TL-Dis had no effect on the ability of Sla5045 to complement the defect in the m53 mutant backbone and did lead to the production of viable virus (Fig. 6A to C).
FIG 6.
Mutational analysis of Sla5045 in a noncoding context. (A) Schematic illustration of Sla5045 mutations characterized in the Sla5045Dup cDNA backbone. Sla5045 is shown in the 3′-5′ direction, with the site of subgenomic RNA initiation shown in italics. Mutations introduced are highlighted in bold. (B) Virus yield following reverse genetics rescue of capped in vitro-transcribed RNA of WT, polymerase frameshift (POLFS), m53, or Sla5045 stem-loop duplication (Sla5045Dup) constructs. The Sla5045Dup constructs are shown using the nomenclature copy1/copy2 as described in the text. Reverse genetics recovery was performed as described in Materials and Methods using in vitro-transcribed, enzymatically capped RNA and the virus yield at 24 h posttransfection determined by TCID50. (C) Virus yield assay following a single passage of virus recovered following RNA transfection of the various Sla5045 stem-loop duplications. The dotted line represents the detection limit of the assay, with error bars representing the SEM.
Sla5045 functions as a template for the norovirus RNA polymerase NS7 in vitro.
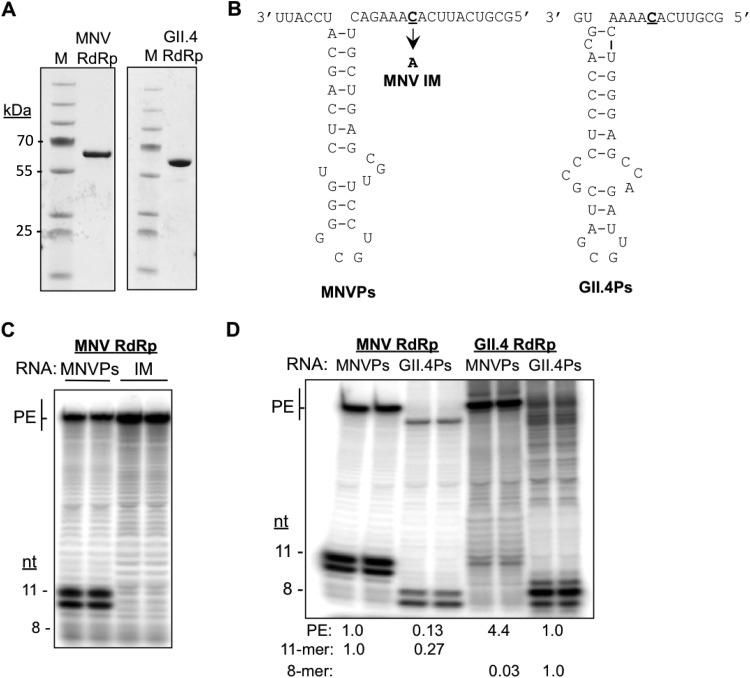
Results from characterization of Sla5045 are all consistent with the hypothesis that Sla5045 forms the core of the promoter for MNV sgRNA synthesis and is used to direct sgRNA synthesis. However, it is unclear whether it functions directly in RNA synthesis. To determine whether this is the case, we examined whether RNAs that contain Sla5054 can direct RNA-dependent RNA synthesis by the recombinant MNV NS7 protein in vitro (Fig. 7A). An RNA containing Sla5045 with a 5′ overhang that contains the initiation nucleotide 5052 was chemically synthesized in an RNA that we termed the MNV proscript (MNVPs). The term proscript is used to denote that the RNA contains both a putative promoter and the template sequence (23) (Fig. 7B). If the MNV proscript directs accurate initiation of RNA synthesis from nt 5052, an 11-nt product should result. Based on comparison with RNAs of known lengths, the products of MNVPs were 11 nt in length. We also observed a 10-nt RNA that is presumed to be a premature termination product that has been previously observed with other recombinant RdRps (Fig. 7C) (24). To ascertain that the RNA was initiated from the expected cytidylate at nt 5052, we tested an RNA, named NMV IM, that is identical to MNVPs except that nt 5052 was replaced with an adenylate. IM failed to direct the synthesis of either the 10- or 11-nt RNA product (Fig. 7C). These data confirm that Sla5045 can accurately direct de novo initiation of the norovirus sgRNA synthesis.
FIG 7.
Sla5045 can direct the initiation of RNA synthesis by the MNV RdRp in vitro. (A) Recombinant RdRps from MNV and the human GII.4 norovirus used in this study. The RdRps was expressed and purified as described in Materials and Methods. The gel images were from SDS-PAGE and Coomassie blue staining. (B) Schematics of the synthetic MNV proscript, MNVPs, and the GII.4 RNA proscript, GII.4Ps, used for the in vitro RNA synthesis assays. The initiation nucleotide used to initiate MNV subgenomic RNA synthesis is in bold and denoted with an arrow. A proscript containing a mutated initiation site named IM is used to control for specificity. (C) Denaturing PAGE analysis of in vitro RNA synthesis assays using the MNV NS7 with either MNVPs or the initiation mutant (IM). The positions of primer extension (PE) and de novo initiation products are highlighted. The lengths of the RNAs were assigned by comparison to RNAs of 8 to 51 nt that were labeled at the 5′ terminus with 32P. (D) Virus genotype-specific RdRp-proscript interaction results in a higher level of RNA-dependent RNA synthesis. The results are representative of six independent experiments.
In the in vitro RNA synthesis reactions, we also detected an RdRp product of ca. 60 nt, longer than the input proscript. This RNA is likely formed by viral RdRp using the 3′-terminal nucleotide of the template RNA to form a primer-extended product (25). This activity is common to numerous viral RdRps (26, 27). Notably, IM generated a higher abundance of the primer-extended RNA products than did MNVPs. Since de novo-initiated RNA product generated from MNVPs was more abundant that those from primer extension, we conclude that the recombinant MNV RdRp has a propensity for de novo-initiated RNA synthesis from the MNV sgRNA.
To examine further whether the MNV Sla5045 can direct genotype-specific RNA synthesis by the MNV RdRp, we performed in vitro RNA synthesis reactions with proscript containing the comparable sequence from a human GII.4 norovirus, named GII.4Ps (Fig. 7B). Accurate de novo-initiated RNA synthesis from GII.4Ps should give rise to an 8-nt product (Fig. 7D). The recombinant MNV RdRp did produce RNAs of 8 nt and 7 nt from GII.4Ps (Fig. 7D). However, in more than six independent assays, the amount of the RNA product was 27% of the amount produced from the MNV RdRp. In contrast, the recombinant GII.4 RdRp produced more than 30-fold the amount of products from the GII.4Ps than from the MNVPs (Fig. 7D). The MNV RdRp reproducibly produced a reduced amount of the primer extension product from the GII.4Ps (13%) (Fig. 7D). Furthermore, the GII.4 RdRp also produced a reduced level of primer extension products from the MNVPs (Fig. 7D). These results suggest that MNV RdRp can specifically recognize the MNVPs.
Negative-sense subgenomic RNA is produced during MNV replication.
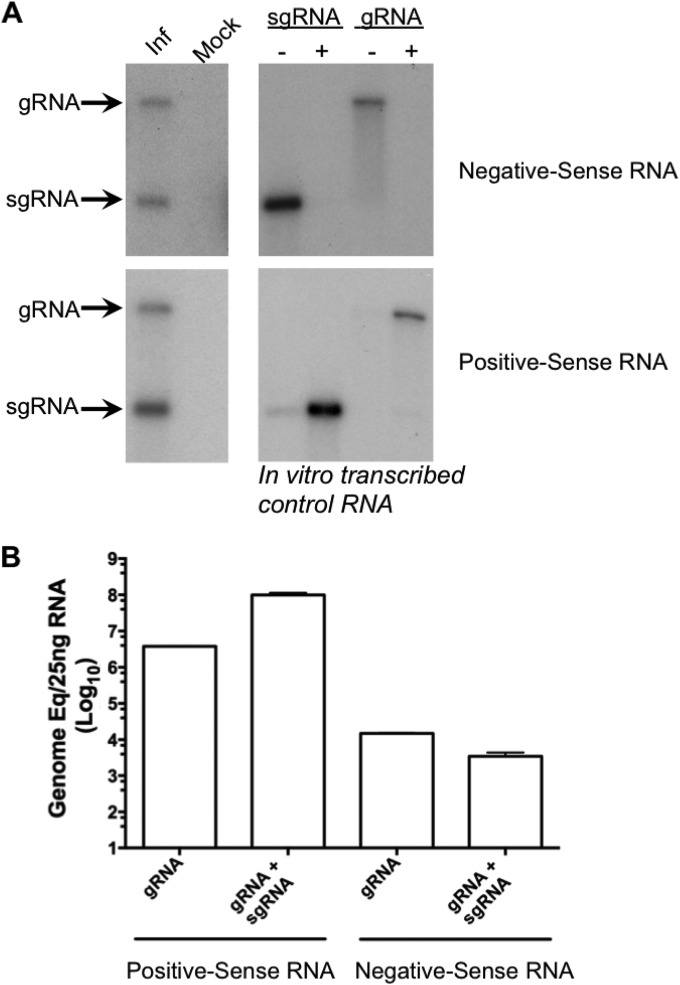
The presence of Sla5045 on the negative-sense genomic RNA and the ability of Sla5045 to function in vitro as a template for priming by the RdRp would fit with the hypothesis that MNV uses a process of internal initiation for the generation of sgRNA. However, negative-sense sgRNA is readily detected in cells infected with feline calicivirus (FCV) (28) and in Norwalk virus replicon-containing cells (29). Northern blot and strand-specific RT-qPCR analysis of RNA isolated from MNV-infected cells also confirmed the presence of a negative-sense sgRNA intermediate (Fig. 8A). While we observed similar levels of negative-sense genomic RNA and sgRNA produced in infected cells, the levels of sgRNA were typically >26-fold higher than the genomic RNA levels (Fig. 8B), fitting with previous observation on FCV and Norwalk virus (28, 29). These data are consistent with the hypothesis that newly synthesized MNV sgRNA may function as a template for further rounds of replication via a negative-sense sgRNA intermediate.
FIG 8.
Negative-sense subgenomic RNA is produced during MNV replication. (A) Northern blot analysis of RNA isolated from either mock-infected (mock) or MNV-infected (Inf) cells. A 1.5-μg portion of RNA harvested 10 h postinfection of cells with a multiplicity of infection of 5 TCID50 per cell was denatured by glyoxylation prior to agarose gel electrophoresis. Positive- and negative-sense RNAs were detected using strand-specific RNA probes as described in Materials and Methods. Control in vitro-transcribed RNAs representing the positive-sense (+) or negative-sense (−) viral RNAs were used as a control. (B). Quantitative real-time PCR analysis of the RNA sample shown in panel A. The RNA was analyzed using a strand-specific RT-qPCR assay designed to detect only the genomic RNA or both genomic and subgenomic RNAs simultaneously. The genome copy number is shown as genome equivalents per 25 ng of total RNA and was determined by comparison to in vitro-transcribed control RNAs. The data shown represent the mean and standard deviation from 4 independent repeats.
DISCUSSION
In this study, we used a combination of genetic and biochemical approaches to characterize a stem-loop RNA sequence that we predicted to be the MNV sgRNA promoter. A mutant MNV that has substitutions that disrupted the predicted secondary structure of the stem-loop was poorly infectious, while nucleotide substitutions that restored the stem-loop structure allowed the virus to regain infectivity. Suppressor mutations found after repeated passages in cell culture also had restored Sla5045 structure, although additional suppressor mutations were also isolated outside the Sla5045. Examination of a second copy of Sla5045 in a noncoding region of the MNV genome revealed that the stem-loop structure functions in cis. Using purified recombinant viral RdRp, we demonstrated that an RNA containing Sla5054 can direct the genotype-specific initiation of RNA synthesis from Sla5045.
A change in the codon of viral RdRp NS7 was found in SupB, which suppressed the changes in m53. Further analysis of this nucleotide position indicated that the effect was mediated in a noncoding context, as changing this codon (ACU) to GUU to code for valine, or to GAU to code for aspartic acid, also restored replication to m53 (data not shown). There are several explanations for our ability to isolate second-site suppressor mutations outside Sla5045. The first is that Sla5045 may form higher-order RNA structures with other regions of the viral genome, including the positions identified as second-site suppressors, and that the second-site suppressors stabilize these interactions. Our bioinformatic analysis has failed to identify any obvious interactions between the regions (data not shown). However, accurate predictions of long- or medium-range RNA-RNA interactions are not currently available, and we cannot at this point rule out that long-range RNA-RNA interactions contribute to Sla5045 function. In addition, the introduction of the m53 mutations could result in promiscuous base pairing of Sla5045 with other sequences flanking the start of the sgRNA. The suppressor mutations then function to disrupt the “off-target” RNA-RNA interactions, favoring the restoration of the Sla5045 structure. Distinguishing between these two possibilities will be challenging; however, our observation that the addition of Sla5045 to the 3′ end of minigenome RNA with a reporter gene in the antisense orientation is not sufficient to drive RNA synthesis and reporter gene expression in infected cells (data not shown) suggests that additional RNA sequences may contribute to RNA synthesis directed by a core promoter that consists of Sla5045.
The production of an sgRNA is conserved in all members of the Caliciviridae, yet the mechanism of its synthesis is poorly understood. In contrast, the synthesis of sgRNA is well characterized in numerous other RNA viruses (reviewed in reference 14). For viruses that possess only a single genomic RNA and generate a single sgRNA, one commonly used mechanism for sgRNA synthesis is the premature termination of negative-strand RNA synthesis that is used by the viral replicase as a template to produce the positive-sense sgRNA. A hallmark of the premature termination mechanism is the production of a truncated negative-sense sgRNA during virus replication. Low levels of negative-sense sgRNA have been identified in cells stably replicating the Norwalk virus replicon (29), in purified replication complex from feline calicivirus-infected cells (28), and as now in MNV-infected cells (Fig. 8). However, our data and those from previous publications (15) suggest an alternative model for MNV sgRNA synthesis. This mechanism is used by plant viruses (23, 30, 31) and alphaviruses (32, 33) and involves the binding of the viral replicase complex to a specific sequence and/or structure present in negative-sense genomic RNA. Our data are consistent with Sla5045 being required as an RNA structure needed for MNV infectivity and to function as a template for RNA synthesis by the RdRp. It is also important to note that in the plant viruses, it is not clear which subunit(s) of the viral replicase can specifically recognized the sgRNA promoter, and our results demonstrate that the norovirus RdRp is sufficient for recognition of the sgRNA core promoter. In addition we observed that a genotype-specific interaction between the RdRp and the subgenomic proscript affected the amount of RNA synthesis (Fig. 7D). Our data would, however, indicate that as negative-sense sgRNA is present in MNV-infected cells, the newly synthesized MNV sgRNA may function as a template for additional rounds of RNA replication by the viral RdRp via a double-stranded RNA (dsRNA) intermediate.
The precise initiation site of the calicivirus sgRNA has been identified previously in a number of studies. The human norovirus sgRNA initiation site has been previously confirmed using a helper virus system to drive the production of viral genomic RNA in cells (34). In the related vesivirus FCV, 5′ RACE was also used to identify the specific sgRNA initiation site (35). To our knowledge, our work provides the first confirmation of the start site of norovirus sgRNA during an authentic infectious norovirus replication cycle.
Our data indicate that the positioning of the core sgRNA promoter is critical for its function, as in the Sla5045Dup constructs, only the sgRNA promoter sequences in Sla2 region functioned (Fig. 5). In addition, increasing the distance between the stem-loop structure and the sgRNA initiation site from 6 to 8 nt also debilitated virus replication (Fig. 6). These data are in agreement with our previous observations using bioinformatics analysis of all calicivirus genomes that indicate an absolute conservation of the spacing, i.e., invariably 6 nt (16).
Overall our data demonstrate that norovirus sgRNA synthesis relies on a sequence- and genotype-specific interaction of the viral RdRp with a stem-loop sequence on the minus-strand RNA. These observations add to our growing understanding of the norovirus life cycle and the molecular mechanisms used by caliciviruses to control viral genome translation and replication.
ACKNOWLEDGMENTS
We thank Stanislav Sosnovtsev for critical comments on this work.
This work was supported by grants from the Wellcome Trust (WT097997MA) and BBSRC (BB/I012303/1) to I.G., from the Indiana Economic Development Corporation to C.C.K., and from the Research University Grant, USM, to M.A.Y. (1001/CIPPT/811233). I.G. is a Wellcome Senior Fellow.
REFERENCES
- 1.Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N Engl J Med 361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall AJ, Lopman BA, Payne DC, Patel MM, Gastañaduy PA, Vinjé J, Parashar UD. 2013. Norovirus disease in the United States. Emerg Infect Dis 19:1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MPG, Estes MK. 2004. Laboratory efforts to cultivate noroviruses. J Gen Virol 85:79–87. doi: 10.1099/vir.0.19478-0. [DOI] [PubMed] [Google Scholar]
- 4.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 5.Wobus CE, Karst SM, Thackray LB, Chang K-O, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward VK, McCormick CJ, Clarke IN, Salim O, Wobus CE, Thackray LB, Virgin HW, Lambden PR. 2007. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc Natl Acad Sci U S A 104:11050–11055. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhry Y, Skinner MA, Goodfellow IG. 2007. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J Gen Virol 88:2091–2100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias A, Ureña L, Thorne L, Yunus MA, Goodfellow I. 2012. Reverse genetics mediated recovery of infectious murine norovirus. J Vis Exp doi: 10.3791/4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yunus MA, Chung LMW, Chaudhry Y, Bailey D, Goodfellow I. 2010. Development of an optimized RNA-based murine norovirus reverse genetics system. J Virol Methods 169:112–118. doi: 10.1016/j.jviromet.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorne LG, Goodfellow IG. 2014. Norovirus gene expression and replication. J Gen Virol 95:278–291. doi: 10.1099/vir.0.059634-0. [DOI] [PubMed] [Google Scholar]
- 11.Clarke IN, Lambden PR. 2000. Organization and expression of calicivirus genes. J Infect Dis 181(Suppl 2):S309–S316. doi: 10.1086/315575. [DOI] [PubMed] [Google Scholar]
- 12.McFadden N, Bailey D, Carrara G, Benson A, Chaudhry Y, Shortland A, Heeney J, Yarovinsky F, Simmonds P, Macdonald A, Goodfellow I. 2011. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog 7:e1002413. doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thackray LB, Wobus CE, Chachu KA, Liu B, Alegre ER, Henderson KS, Kelley ST, Virgin HW. 2007. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J Virol 81:10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sztuba-Solińska J, Stollar V, Bujarski JJ. 2011. Subgenomic messenger RNAs: mastering regulation of (+)-strand RNA virus life cycle. Virology 412:245–255. doi: 10.1016/j.virol.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morales M, Bárcena J, Ramírez MA, Boga JA, Parra F, Torres JM. 2004. Synthesis in vitro of rabbit hemorrhagic disease virus subgenomic RNA by internal initiation on (−)sense genomic RNA: mapping of a subgenomic promoter. J Biol Chem 279:17013–17018. doi: 10.1074/jbc.M313674200. [DOI] [PubMed] [Google Scholar]
- 16.Simmonds P, Karakasiliotis I, Bailey D, Chaudhry Y, Evans DJ, Goodfellow IG. 2008. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res 36:2530–2546. doi: 10.1093/nar/gkn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subba-Reddy CV, Yunus MA, Goodfellow IG, Kao CC. 2012. Norovirus RNA synthesis is modulated by an interaction between the viral RNA-dependent RNA polymerase and the major capsid protein, VP1. J Virol 86:10138–10149. doi: 10.1128/JVI.01208-12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Meyers G. 2007. Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J Virol 81:9623–9632. doi: 10.1128/JVI.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Napthine S, Lever RA, Powell ML, Jackson RJ, Brown TDK, Brierley I. 2009. Expression of the VP2 protein of murine norovirus by a translation termination-reinitiation strategy. PLoS One 4:e8390. doi: 10.1371/journal.pone.0008390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vashist S, Ureña L, Goodfellow I. 2012. Development of a strand specific real-time RT-qPCR assay for the detection and quantitation of murine norovirus RNA. J Virol Methods 184:69–76. doi: 10.1016/j.jviromet.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi G, Deval J, Fan B, Cai H, Soulard C, Ranjith-Kumar CT, Smith DB, Blatt L, Beigelman L, Kao CC. 2012. Biochemical study of the comparative inhibition of hepatitis C virus RNA polymerase by VX-222 and filibuvir. Antimicrob Agents Chemother 56:830–837. doi: 10.1128/AAC.05438-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J Gen Virol 93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siegel RW, Adkins S, Kao CC. 1997. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc Natl Acad Sci U S A 94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y-C, Russell WK, Ranjith-Kumar CT, Thomson M, Russell DH, Kao CC. 2005. Functional analysis of RNA binding by the hepatitis C virus RNA-dependent RNA polymerase. J Biol Chem 280:38011–38019. doi: 10.1074/jbc.M508145200. [DOI] [PubMed] [Google Scholar]
- 25.Kao CC, Yang X, Kline A, Wang QM, Barket D, Heinz BA. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J Virol 74:11121–11128. doi: 10.1128/JVI.74.23.11121-11128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjith-Kumar CT, Gajewski J, Gutshall L, Maley D, Sarisky RT, Kao CC. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J Virol 75:8615–8623. doi: 10.1128/JVI.75.18.8615-8623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomar S, Hardy RW, Smith JL, Kuhn RJ. 2006. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol 80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green KY, Mory A, Fogg MH, Weisberg A, Belliot G, Wagner M, Mitra T, Ehrenfeld E, Cameron CE, Sosnovtsev SV. 2002. Isolation of enzymatically active replication complexes from feline calicivirus-infected cells. J Virol 76:8582–8595. doi: 10.1128/JVI.76.17.8582-8595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang K-O, Sosnovtsev SV, Belliot G, King AD, Green KY. 2006. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 353:463–473. doi: 10.1016/j.virol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Adkins S, Siegel RW, Sun JH, Kao CC. 1997. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA 3:634–647. [PMC free article] [PubMed] [Google Scholar]
- 31.Sivakumaran K, Chen M-H, Roossinck MJ, Kao CC. 2002. Core promoter for initiation of Cucumber mosaic virus subgenomic RNA4A. Mol Plant Pathol 3:43–52. doi: 10.1046/j.1464-6722.2001.00089.x. [DOI] [PubMed] [Google Scholar]
- 32.Levis R, Schlesinger S, Huang HV. 1990. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol 64:1726–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ou JH, Rice CM, Dalgarno L, Strauss EG, Strauss JH. 1982. Sequence studies of several alphavirus genomic RNAs in the region containing the start of the subgenomic RNA. Proc Natl Acad Sci U S A 79:5235–5239. doi: 10.1073/pnas.79.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asanaka M, Atmar RL, Ruvolo V, Crawford SE, Neill FH, Estes MK. 2005. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc Natl Acad Sci U S A 102:10327–10332. doi: 10.1073/pnas.0408529102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neill JD. 2002. The subgenomic RNA of feline calicivirus is packaged into viral particles during infection. Virus Res 87:89–93. doi: 10.1016/S0168-1702(02)00086-2. [DOI] [PubMed] [Google Scholar]