ABSTRACT
To better characterize the assembly of the HIV-1 core, we have used electron cryotomography (ECT) to image infected cells and the viral particles cryopreserved next to them. We observed progressive stages of virus assembly and egress, including flower-like flat Gag lattice assemblies, hemispherical budding profiles, and virus buds linked to the plasma membrane via a thin membrane neck. The population of budded viral particles contains immature, maturation-intermediate, and mature core morphologies. Structural characteristics of the maturation intermediates suggest that the core assembly pathway involves the formation of a CA sheet that associates with the condensed ribonucleoprotein (RNP) complex. Our analysis also reveals a correlation between RNP localization within the viral particle and the formation of conical cores, suggesting that the RNP helps drive conical core assembly. Our findings support an assembly pathway for the HIV-1 core that begins with a small CA sheet that associates with the RNP to form the core base, followed by polymerization of the CA sheet along one side of the conical core toward the tip, and then closure around the body of the cone.
IMPORTANCE During HIV-1 assembly and release, the Gag polyprotein is organized into a signature hexagonal lattice, termed the immature lattice. To become infectious, the newly budded virus must disassemble the immature lattice by proteolyzing Gag and then reassemble the key proteolytic product, the structural protein p24 (CA), into a distinct, mature hexagonal lattice during a process termed maturation. The mature HIV-1 virus contains a conical capsid that encloses the condensed viral genome at its wide base. Mutations or small molecules that interfere with viral maturation also disrupt viral infectivity. Little is known about the assembly pathway that results in the conical core and genome encapsidation. Here, we have used electron cryotomography to structurally characterize HIV-1 particles that are actively maturing. Based on the morphologies of core assembly intermediates, we propose that CA forms a sheet-like structure that associates with the condensed viral genome to produce the mature infectious conical core.
INTRODUCTION
Late events in HIV-1 replication are profoundly dependent upon the biochemical, structural, and enzymatic properties of the virus's two polyproteins, Gag and Gag-Pol (reviewed in reference 1). The Gag polyprotein (p55gag) is the key structural determinant of the nascent viral particle, and its protein components include (in order from the N to C terminus) p17 (matrix, MA), p24 (CA), spacer peptide 1 (SP1), p7 (nucleocapsid, NC), spacer peptide 2 (SP2), and p6 (see Fig. 2A). The Gag-Pol polyprotein (p160gag-pol) is a product of a frameshift in the ribosomal reading frame that occurs at the junction of SP2 and p6. p160gag-pol is produced at a ratio of approximately 1:20 of p55gag and includes the products of viral pol, including the viral enzymes protease (PR), reverse transcriptase (RT), and integrase (IN) (1).
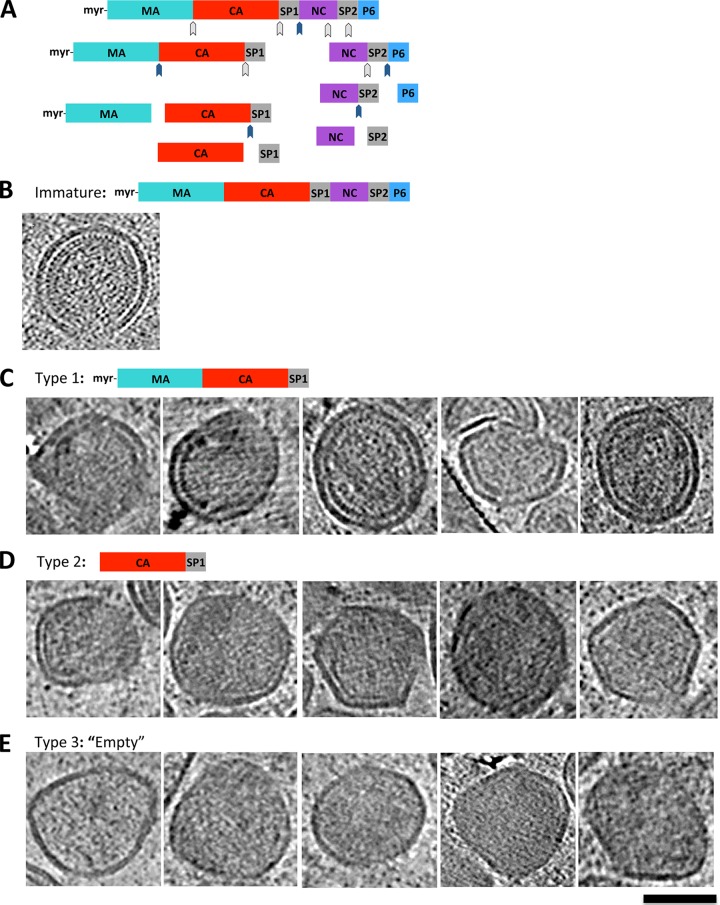
FIG 2.
Proteolytic intermediates formed during HIV-1 maturation. (A) Schematic representation of the proteolysis steps that constitute the HIV-1 maturation process. (B) Immature viral particle from the in cellulo sample. (C to E) Tomographic slices through viral particles that contain structures that represent the indicated maturation intermediate: MA-CA-SP1 polyprotein (C), the last proteolytic intermediate, CA-SP1 (D), and viral particles with fully proteolyzed polyprotein (E). Scale bar, 100 nm.
Both Gag polyproteins are targeted to the plasma membrane for assembly by basic residues in MA that bind to the plasma membrane-specific lipid, phosphatidyl inositol-4,5-bisphosphate. Membrane binding triggers the exposure of a myristoyl group modification at the polyprotein N terminus that functions to stabilize the protein at the plasma membrane (reviewed in reference 1). Targeting of the viral genome to sites of virus assembly is mediated by interactions between the viral RNA psi sequence and the zinc finger motifs within the NC domain of the Gag polyproteins. Assembly of a new viral particle is initiated when membrane-associated Gag multimerizes as a result of both protein-protein interactions among CA domains and RNA-protein interactions between the viral genome and NC. These interactions drive polyprotein self-assembly and their consequent organization as a rigid hexagonal lattice, termed the immature lattice. Progressive assembly of the immature lattice at the cell surface induces membrane curvature and initiates formation of the viral bud. Subsequent recruitment of the cellular endosomal-sorting-complexes-required-for-transport (ESCRT) machinery through the Gag late-domain motif in p6 leads to membrane fission and release of the viral bud as an immature, noninfectious particle (reviewed in reference 1).
Within the newly budded particle, the immature Gag lattice is arranged radially; while the p55gag N-terminal MA is associated with the inner leaflet of the viral membrane, the C-terminal NC bound to the viral RNA extends toward the center of the particle (2, 3). Although the immature lattice is important to particle formation and budding, it does not represent the infectious form of the virus. Formation of the infectious particle requires that the immature lattice be disassembled by PR-mediated proteolysis of Gag at specific cleavage sites (see Fig. 2A) (1). Eventual release of CA from the Gag polyprotein by PR leads to the assembly of CA into a mature conically shaped core within the viral membrane (1).
Both mature and immature particles are structurally defined by distinct hexagonal lattices that are dependent upon the self-assembly properties of CA. However, CA utilizes very different domain contacts to stabilize the two lattices (4, 5), and, as a result, they have profoundly different structural characteristics (2, 6) and distinct functions during viral replication (1). In the immature lattice, polyprotein hexamers are stabilized by CA–C-terminal domain (CTD)/CA-CTD interactions and SP1, while interhexamer contacts are mediated by CA–N-terminal domain (NTD)/CA-NTD interactions. In contrast, mature lattice hexamers are stabilized by both CA-NTD/CA-NTD and CA-NTD/CA-CTD interactions (7). Interhexamer contacts in the mature lattice are the result of the formation of a CTD dimer interface between adjacent hexamers and a hydrophobic trimer interface that mediates hexameric or pentameric assemblies of CA hexamers after final cleavage at the CA-SP1 site (4). Because the CA contacts that stabilize the immature and mature lattices are so different, it is expected that the assembly program for the mature core requires the full disassembly of the immature lattice, followed by de novo reassembly of a mature CA lattice as a conically shaped core (8).
Although the proteolytic steps that lead to disassembly of the immature lattice are biochemically and temporally well defined (9), the process by which CA assembles into the mature core is poorly understood. Previous electron cryotomography (ECT) studies of mature virions led to two distinct models for core assembly. The first model proposes that core assembly is initiated at the wide base of the capsid and proceeds to the narrow tip. This “base-to-tip” assembly model was proposed by Benjamin et al. (6), based on their observations that the capsid bases were often consistent with respect to size, curvature, and distance from the membrane but that the capsid tips were less well ordered and sometimes unclosed. Based on these observations, the authors argued that capsid growth originates at the structurally regular base and proceeds to the tip, sometimes failing to completely close the structure.
In a separate study on the ultrastructure of mature HIV-1 virions (10), the authors noted that, regardless of the virion diameter, the conical capsid extended across the full diameter of the viral particle. The authors therefore proposed a “tip-to-base” capsid growth model in which capsid growth originates at the narrow tip and continues until the opposite membrane is reached, whereupon growth is redirected to close the structure at the base. In this model, the membrane is predicted to be an essential factor for the formation of the conical core. In vitro CA assembly experiments show, however, that purified CA, under conditions of high salt, can assemble cones independent of membranes (11).
Computational simulations suggested a third model in which capsid assembly proceeds as the nonequilibrium growth of an elastic sheet (12). This model suggests that CA forms a sheet that precedes formation of the mature fullerene cone-shaped core and that sheet growth drives curvature of the structure until its edges join in space. The core then incorporates additional CA units until the structure closes, albeit with some defects and gaps, as documented by ECT (13). These simulations could recapitulate not only the conical core structures but also the full spectrum of CA assemblies observed for diverse retroviruses.
Intermediate structures comprising the HIV-1 maturation pathway have been difficult to document due to the rapidity of the process. Previous studies have used mutagenesis to arrest maturation at specific PR cleavage steps (14, 15), while some other studies have used small molecules to inhibit just the final PR cleavage step (8, 16). These studies provided structural information regarding the disassembly of the immature Gag lattice but not the assembly of the viral core. Additionally, accumulation of intermediates may not fully reflect the maturation process due to potential dominant negative effects the intermediates can have on assembly (17).
New work described here avoids the limitations of previous studies by analyzing the maturation process while it is under way. To this end, we have imaged by ECT the released viral particles found assembling on the edge of and after release from infected cells and have characterized the structures found within these particles. Because this sample is asynchronous with respect to the maturation process, we expect that all potential capsid structures representing the assembly pathway are present. As a result, we can characterize capsid assembly without the complication of off-pathway issues that might arise due to the use of drugs or the introduction of mutations to manipulate the maturation process. Morphological characterization and quantitation of the structures present in the maturing particles have allowed us to propose a model for HIV-1 capsid assembly from the initial release of the immature particle to the formation of the mature, infectious, fullerene cone. Our model proposes that viral core assembly proceeds asymmetrically and not as a uniform assembly from one end to another (i.e., base to tip, or tip to base). Instead, our results support an assembly pathway that begins with a small CA sheet that associates with the RNP at the base of the forming viral core. Growth then proceeds rapidly from the base along one side of the capsid structure until it reaches the membrane, which redirects capsid growth back toward the base and leads to the structure's eventual closure.
MATERIALS AND METHODS
Virus construct.
The proviral construct encoding pNLEGFP-BglI was generously provided by Irvin Chen (University of California, Los Angeles [UCLA]). To generate the virus, human embryonic kidney 293T cells (ATCC) were cotransfected with the proviral construct and the plasmid encoding the vesicular stomatitis virus glycoprotein (VSV-G) envelope (pHEF-VSVG obtained from Lung-Ji Chang through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH). Culture supernatants were collected at 36 h posttransfection and subsequently filtered through a 0.45-μm-pore-size filter. Particles were purified and concentrated by ultracentrifugation at 100,000 × g through a 20% sucrose cushion. The virus pellet was resuspended in phosphate-buffered saline (PBS), aliquoted, and stored at −80°C until used for infection.
Infected cell preparation.
Human umbilical vein endothelia cells (HUVECs; Lonza, Walkersville, MD) were seeded in six-well plates containing carbon-coated gold electron microscopy (EM) finder grids (2-μm hole size and 2-μm hole spacing; Quantifoil, Jena, Germany) and cultured for 24 h in a humidified incubator maintained at 37 °C and 5% CO2. Cells were infected with the VSV-G pseudotyped NLEGFPDelta BglIVprX. Infection was carried out by spinoculation at 800 × g for 1 h at 16°C. Spinoculated samples were washed two times with fresh medium prewarmed to 37°C. Two milliliters of fresh medium was then added to each well, and samples were incubated for 36 h to allow for virus expression before cryopreservation.
Purification of endpoint virus sample.
Viral particles were purified and concentrated from the culture supernatants of infected HUVECs. Briefly, HUVECs were infected as described above, and culture supernatants were collected at 36 h postinfection and filtered through a 0.45-μm-pore-size filter. Filtrates were then placed back in the humidified incubator maintained at 37°C and 5% CO2 for an additional 16 h to allow purified particles to fully mature. Filtrates were then treated with 1 unit of Turbo DNase (Ambion) per 1 ml of filtrate supplemented with 10 mM MgCl2 at 37°C for 2 h. Viral particles were concentrated by ultracentrifugation at 100,000 × g over a 20% sucrose gradient, resuspended in Hanks' balanced salt solution (HBSS), and finally filtered through a 0.22-μm-pore-size filter.
Electron cryotomography of HIV-1-infected whole cells.
EM grids on which virus-producing cells were growing were removed from the culture plate using forceps and treated with 3 μl of warm medium containing 10-nm gold fiducial markers. Forceps and grid were transferred to the environment chamber of a Vitrobot Mark III (FEI) maintained at 37°C and 80% relative humidity (rH). Excess liquid was manually blotted from the grids on one side before they were plunged into liquid ethane. Cryopreserved grids were then imaged in a 300-kV FEI G2 Polara or FEI Titan Krios transmission electron microscope, each equipped with a field emission gun and energy filter (slit width set at 20 eV). Data were collected with either a Gatan Ultracam 4,000- by 4,000-pixel lens-coupled charge-coupled device or a K2 Summit direct detector. Tilt series were collected over a series of angles ranging from −60 to +60° using a step size of 1°, magnification of ×22,500 (effective pixel size of raw data is 5Å), a total dose of 150 e/Å2, and a defocus of −6 μm. The UCSF Tomo software program (University of California, San Francisco) was used to collect the tilt series, and three-dimensional (3D) reconstructions were carried out using a weighted back-projection algorithm tracking 10-nm fiducial markers in the IMOD package of software programs.
Gag assemblies, budding particles, and budded virions (immature, maturing, and mature) were identified based on ultrastructural features. Budded particles were classified as either immature, intermediate, or mature based on the core morphology.
Image analysis.
The Amira package was used to segment the electron density maps of the viral particles. Isosurfaces were generated using a thresholding approach to first select the most dense and prominent feature within the particles. Threshold values were then adjusted to minimize noise while maintaining the feature and contiguous densities through multiple tomographic slices. From the resulting segmentations, an isosurface was generated for CA, the RNP, and the viral membrane.
Statistical analysis of RNP localization in viral particles.
In more than 60 tomograms, 1,003 budded particles were identified and evaluated independently by three individuals. Each particle was assessed for the shape of its core and the location of the RNP within the viral particle, and immature particles were excluded from subsequent analysis. Core shape was characterized as one of three categories, cylindrical, conical, or pleomorphic, and the RNP location was identified using a set of X, Y, and Z coordinates to mark its center point. The positional uncertainty of the RNP was calculated from the three coordinates provided by the three individuals, and only particles whose RNP uncertainty was within 28 nm (i.e., half the distance of the reported core diameter of 56 nm) (6) were considered further. We also limited our final analysis of RNP and capsid shape correlation to particles that had a single capsid structure that was either conical or cylindrical. Of the original particles, only 306 met all of these criteria.
For the 306 chosen particles, the average X, Y, and Z coordinates that identified the position of the RNP in the tomogram were used to determine if the RNP was inside or outside the core. The two-tailed P value result reflects the probability of randomly observing a pattern of RNP location and core shape as observed in our data using 306 particles. The two-tailed P value was 2.38 × 10−6. Analyzing the data with Fisher's exact test yielded a significant result (P < 0.001). We conclude that there is a significant correlation between the shape of the core and the location of the RNP.
RESULTS
ECT of HIV-1-infected HUVECs.
We used ECT to image viral particles that were frozen next to infected HUVECs growing on EM grids (Fig. 1A). HUVECs were selected for this study because we found that they are amenable to imaging by ECT; they attach well to the carbon surface that coats the EM grid, grow and divide on the EM grid in a manner indistinguishable from tissue culture plastic ware, and are readily infected by and produce HIV-1 particles. Because the extreme edges of the HUVECs are very thin (<500 nm), these areas of the cells can be imaged by ECT, which is limited by sample thickness due to the high tilt angles that must be included for three-dimensional (3D) volume reconstruction (18).
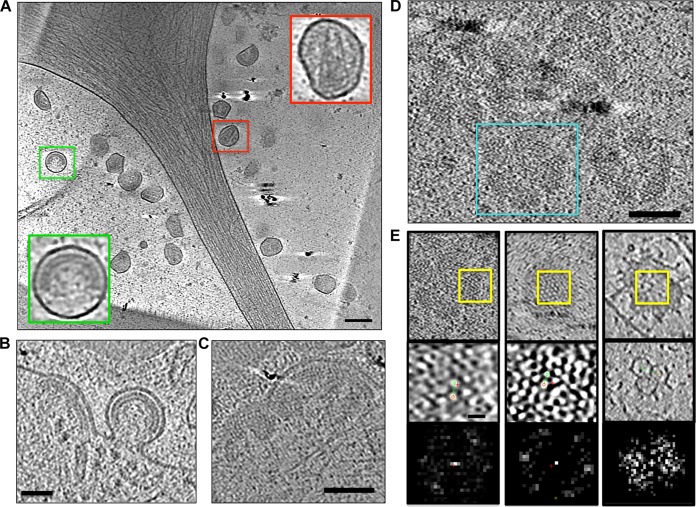
FIG 1.
Ultrastructure of HIV-1 infection in human umbilical vein endothelial cells (HUVECs). (A) Infected cell surrounded by newly released viral particles at various stages of maturation. Insets are enlarged views of an immature viral particle (green boxes) and a mature particle with a conical capsid (red boxes). (B) Late-stage HIV-1 budding profile on the surface of an infected cell. (C) Characteristic half-dome-shaped early assemblies of immature Gag on the cell surface. (D) Flat Gag assemblies on cell surface. (E) Comparison of lattice features from the flat Gag assemblies (left; area from boxed region in panel D), an immature particle (center), and a clathrin-coated vesicle (right). Top panels are tomographic slices through the respective lattices, middle panels are areas of the lattices (yellow boxes) that were compared by Fourier transform, and bottom panels show results. Scale bars: 100 nm (A to D) and 10 nm (E).
Multiple budding profiles demonstrating various stages of virus egress were observed at the plasma membrane (Fig. 1B). The immature Gag lattice underlying the membrane in the budding profiles was readily identified based on the characteristic radial densities (Fig. 1B), as well as the hexagonal arrangement of the lattice in surface views (2, 3) (Fig. 1E). In some cases, the same Gag lattice appeared on the surface of the cells as islands of flat sheets arranged loosely around a center point, similar to the petals of a flower (Fig. 1D). To illustrate that the flat lattice was in fact assembled Gag as opposed to clathrin, which is also known to form flat lattices on the cell surface, we compared the Fourier transforms of the flat Gag lattice (Fig. 1E) with those of a clathrin-coated vesicle (Fig. 1E, compare left and right columns). Indeed, the lattice unit spacings of the two types of lattices were distinct (Fig. 1E). However, clear similarities were apparent when we compared the flat Gag lattice with the Gag lattice of the immature particle, again suggesting that the surface lattice is part of the HIV assembly program. While this petal-like pattern is somewhat unexpected, it may provide an explanation for the observed incompleteness of the Gag lattice in immature virions and impart information regarding the earliest steps of virus assembly.
The budding profiles we observed at the thin edge of the infected HUVECs were primarily half-dome shaped (Fig. 1C), suggesting that this configuration likely represents a kinetically slow assembly intermediate of the budding process. In contrast, more-spherical budding profiles that appeared closer to egress and were linked to the cell surface by a narrow membrane neck (Fig. 1B) were less abundant. These observations are consistent with previously published reports (19, 20) indicating that Gag assembles on the surface of the cells over the course of 9 to 12 min and remains stably associated with the membrane before components of the ESCRT machinery are recruited and more rapidly carry out the membrane scission event.
Collectively, these observations confirmed that the HUVECs were actively infected at the time of cryopreservation and that new viral particles were being continually released into the extracellular space.
Distribution of morphologies among particles in the nonpurified virus sample.
Because the particles observed here were not subjected to any purification process but frozen and imaged directly in the culture environment in which they were produced (Fig. 1A), we refer to this sample as an in cellulo sample. In total, we imaged 2,121 in cellulo particles. For 91 of the particles, we were unable to determine capsid morphology and therefore excluded these particles from subsequent analysis. As summarized in Table 1, the remaining 2,030 particles were characterized as mature (76%), immature (8%), or maturation intermediates (16%).
TABLE 1.
Summary of particle morphologies
| Particle type or shape | No. (%) of unpurified particles (n = 2,030) | No. (%) of purified particles (n = 78) |
|---|---|---|
| Mature | 1,533 (76) | 77 (99) |
| Cone | 1,198 (78)a | 55 (71)a |
| Cylinder | 228 (15)a | 12 (15)a |
| Irregular | 107 (7)a | 10 (13)a |
| Immature | 164 (8) | 1 (1) |
| Intermediate | 333 (16) | NDb |
| Total | 2,030 | 78 |
Values represent the percentages of mature particles.
ND, not detected.
Mature viral particles in our sample demonstrated structural morphologies similar to those reported previously for purified particles (6, 10). Specifically, many of these mature particles contained the characteristic fullerene cone-shaped capsid that extended across the full diameter of the particle membrane (Fig. 1A). Immature particles in the in cellulo sample were also morphologically similar to particles described previously (2, 3) (Fig. 1A). Of particular note was the observation that the immature lattice often did not fully cover the inner membrane surface of the viral particle (Fig. 1A, inset). These findings agree with previous reports (2, 3) suggesting that the immature Gag lattice is incomplete but dispute the findings of Kol and colleagues (21). Collectively, these observations indicate that the mature and immature particles from the in cellulo sample structurally match those found in purified preparations.
An important feature of the in cellulo sample that distinguished it from purified samples was the distribution of mature and immature morphologies within the particle population (Table 1). The in cellulo sample comprised approximately 76% mature particles, compared to approximately 94% reported previously for purified samples (14). Consistent with the observed decrease in the number of mature particles in the in cellulo sample was an increase in the relative number of immature particles. Previous reports indicated that just 3% of the viral particles in a purified population are immature (14), but our sample contained 8% immature particles. Furthermore, 16% of particles in the in cellulo population demonstrated morphologies that were neither mature nor immature, suggesting that these particles contain intermediate capsid structures formed during capsid assembly. The above observations suggest that as much as 24% (i.e., immature and intermediate particles) of the total in cellulo population was in the process of maturing at the time the sample was cryopreserved.
In further support of our conclusion that the in cellulo sample was enriched for particles that were immature or maturing, we purified viral particles from the infected HUVEC cultures and allowed them to fully mature before freezing and imaging them by ECT. This purified sample showed a distribution of morphologies similar to that previously reported for purified particles (Table 1) (14), with the overwhelming majority (99%) of the particles being mature.
Structural characterization of HIV-1 maturation intermediates.
We carefully analyzed the internal structures of the 333 particles in our in cellulo sample that were morphologically neither mature nor immature and were therefore likely maturation intermediates and noticed that they could be categorized into two groups of intermediate forms, proteolytic intermediates and assembly intermediates, with each group containing its own distinct subgroups.
Proteolytic intermediates. (i) MA-CA-SP1 proteolytic intermediate.
Among the intermediate structures observed in the in cellulo sample were particles that had a single, unclosed electron-dense layer that followed closely (within 10 nm) the inner curvature of the membrane (Fig. 2C). These particles constituted just 6% (20/333) of intermediate particles, suggesting that they are very short-lived intermediates. A similar morphology was previously described for viral particles bearing mutations in Gag that arrested p55 processing after cleavage between SP1 and NC (Fig. 2A) (14, 15, 22). Presumably, the structural features of this proteolysis mutant represent the particle following the early SP1-NC cleavage event that separates NC and RNA from the immature lattice. The MA-CA-SP1 protein remains associated with the membrane as a single, thick layer of density, while NC and the RNA condense to form the ribonucleoprotein complex, or RNP.
(ii) CA-SP1 proteolytic intermediate.
Approximately 14% (48/333) of the intermediate particles in the in cellulo sample had a single-density layer that was measurably thinner than that of the MA-CA-SP1 particles (5 nm for CA-SP1 versus 9 nm for MA-CA-SP1) (compare Fig. 2C and D). This thinner-density layer approximated the curvature of the membrane, but it often failed to follow the membrane consistently. These characteristics are similar to the structural features described for the Gag CA5 mutant (15, 22) as well as for particles treated with the maturation inhibitor bevirimat (8, 16). In both cases, the final Gag cleavage event between CA and SP1 is inhibited, and the mature capsid is not formed. The continuous, thin single-layer density is presumably the CA-SP1 protein that is stabilized by a bundle of six SP1 helices that initially form contacts in the immature lattice (2, 3). Failure to fully proteolyze the CA-SP1 prevents maturation and leads to a loss of infectivity (17, 23).
(iii) Fully disassembled proteolytic intermediate.
The final class of particles that we grouped with the proteolytic intermediates included those particles that appeared to have no organized internal structures (Fig. 2E, empty). Overall, these particles were more electron dense than vesicles that were sometimes observed in the sample and were comparable in size to the viral particles but could not be conclusively identified as virions. The existence of such “empty” particles would suggest that the immature lattice is at some point fully disassembled during the maturation process. However, these particles were also rare, comprising just 4% (13/333) of the intermediate particles, which suggests that they are either a very short-lived maturation intermediate or not part of the normal HIV-1 maturation pathway.
Capsid assembly intermediates.
In addition to the proteolytic class of maturation intermediates, we also observed particle morphologies that we classified as assembly intermediates (Fig. 3).
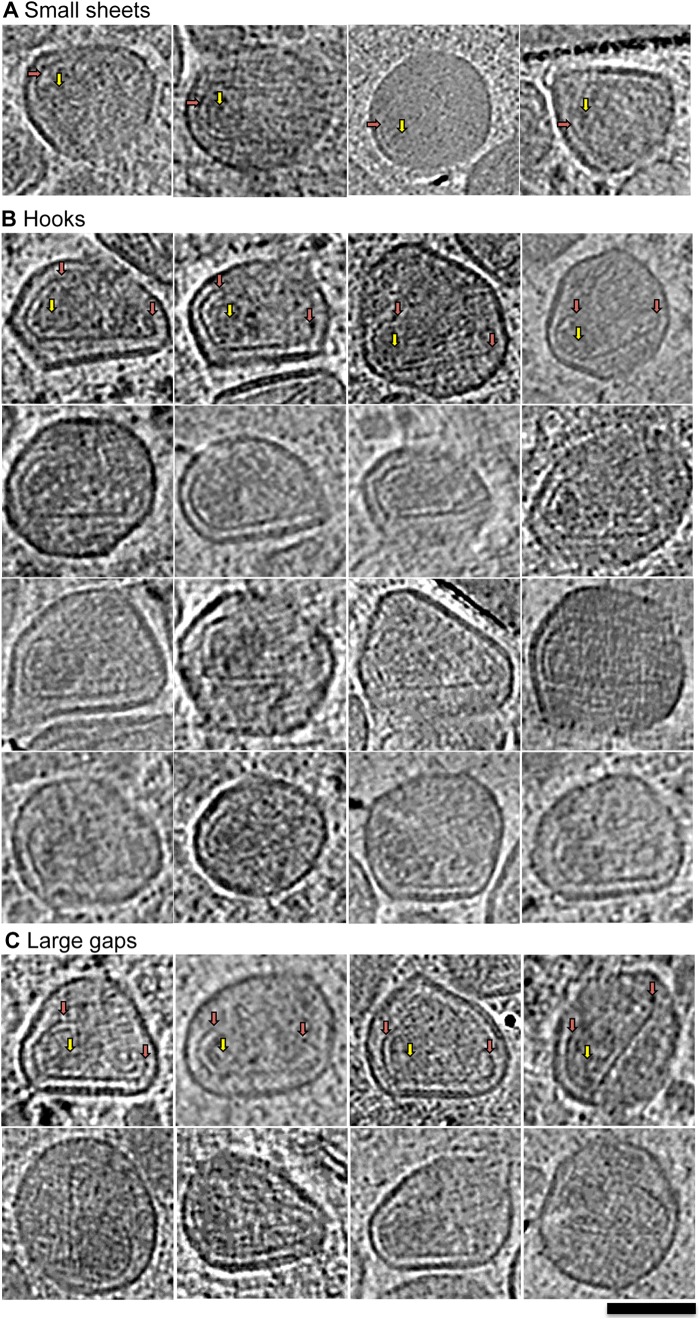
FIG 3.
Assembly intermediates formed during HIV-1 maturation. Shown are tomographic slices through viral particles representing the indicated maturation intermediate: small sheets (A), hooks (B), and large gaps (C). Red arrows identify CA sheets (A) or the ends of the CA sheet forming the hook structure (B); yellow arrows indicate RNP. Scale bar, 100 nm.
(i) Small sheets.
The small-sheet subclass comprised approximately 22% (72/333) of the intermediate forms observed and included particles that contained small sheets of density (Fig. 3A). The particles in this subclass were structurally diverse, with some of the sheets demonstrating high curvature (Fig. 3A, red arrows). Regardless of their shape, however, such densities were typically found near the membrane of the particles and were characteristically thin. We distinguished this class of assembly intermediates from the putative CA-SP1 proteolytic intermediates because they were typically smaller than the CA-SP1 structures that arose as a result of mutation (15) or of treatment with a maturation inhibitor (16). Additionally, CA-SP1 densities typically follow the gradual curvature of the membrane while the small-sheet assembly intermediates demonstrated either a straight edge or a highly curved morphology, both of which are distinct from the morphologies described previously for proteolytic intermediates (15, 16).
In some instances, the small-sheet structures were associated with an electron-dense feature that we interpreted as the RNP (Fig. 3A, yellow arrows). Because RNPs are not clearly defined, the location of putative RNPs was estimated independently by three individuals and only further considered if all three individuals identified the same location (within 25 nm of each other). Of the 43 maturation intermediates exhibiting small sheets, 9 also exhibited putative RNPs (identified in the same position by all three individuals). The average distance from the putative RNP center to the small sheet was 33 nm (±17 nm). When a similar analysis was applied to RNPs and conical capsids in mature particles from the same tomograms, the distance (i.e., from center of RNP to capsid layer at base) was found to be 29 nm (±6 nm). A nonparametric two-tailed t test (P = 0.7567) indicated that the difference in averaged distances was not significant. Together, these observations suggest that the RNP sometimes condenses and associates with capsid sheets soon after assembly begins.
(ii) Hooks.
The hook intermediate subclass included the largest number of intermediate particles (Fig. 3B), with approximately 34% (112/333) of the particles that we classified as intermediate comprising this category. Because the hook subclass represented such a large proportion of our assembly intermediates, we infer that exit from this subclass is a slow step in the assembly pathway. Hooks also had a small curved sheet, but the signature structural feature of this subclass was the presence of a single straight-edge density that extended across the diameter of the particle from the curved end. The curved ends of these structures (Fig. 3B, red arrows) resembled the wide base of a conical mature core and often were associated with the RNP (Fig. 3B, yellow arrows). The similarity of these hook-like structures to the mature conical HIV-1 capsid was striking. Indeed, they appeared to be cones except that they were missing one wall and the narrow tip.
(iii) Large gaps.
The large-gap intermediate subclass accounted for 20% (68/333) of the intermediate particles and was defined by the presence of a clear cone structure with a major gap (Fig. 3C). The gaps observed for these particles were typically localized to one side of the cone or near the cone tip, consistent with earlier reports (6, 13). We consider the large-gap subclass to comprise more advanced assembly intermediates than the hooks subclass and therefore conclude that the final steps of the core assembly pathway involve closing the structure in the areas of high curvature.
Intermediate structures are absent from the fully mature, purified virus sample.
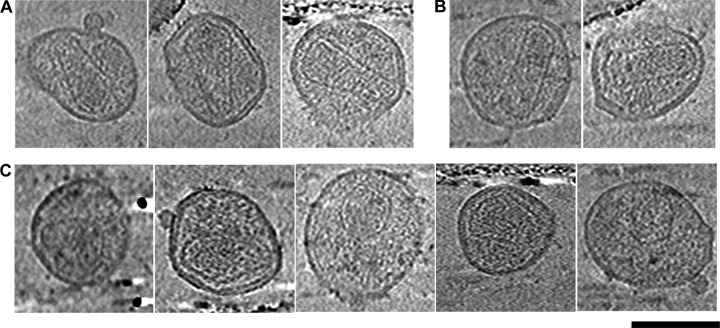
To confirm that the intermediate capsid assemblies observed in nonpurified virus were “on pathway” and not simply aberrant capsids, we purified virus from infected HUVECs and allowed the purified particles time to fully mature before cryopreservation and ECT. The purified virus sample contained primarily mature capsids that were either cones (71%) (Table 1 and Fig. 4A) or cylinders (15%) (Table 1 and Fig. 4B). Approximately 13% of the purified particles showed aberrant morphologies (Table 1). Analysis of the aberrant particles suggested that they are irregular, closed structures (Fig. 4C). Importantly, the purified particles do not appear to contain capsid structures that are similar to the intermediate capsid morphologies (i.e., hooks and large gaps) described above.
FIG 4.
Capsid assembly intermediates are not observed in the fully mature, purified virus sample. Shown are cryotomographic slices through HIV-1 particles purified from cultures of infected HUVECs with a conical capsid (A), a cylindrical capsid (B), or an irregular capsid (C). Scale bar, 100 nm.
Intermediate particles are less spherical than mature, purified particles.
Another characteristic that distinguished the in cellulo intermediate particles from the purified particles was that the in cellulo particles appeared less spherical and in many instances demonstrated flat membrane edges (compare viral particles in Fig. 3 and 4). To quantify the sphericity of individual viral particles, we used a simple ratio of length to width (L/W). We selected the particles shown in Fig. 3B and C for analysis as well as other intermediate and conical capsids found in the same tomograms. For each particle, we measured the length (greatest diameter through center of the particle) and width (diameter through center of the particle orthogonal to the length) and considered the ratio of those values (L/W). For spherical particles the L/W ratio is expected to be 1, and for nonspherical particles, the ratio will be greater than 1.
We carried out the sphericity analysis for intermediate (n = 37) and mature conical (n = 37) particles from the in cellulo sample and for mature conical particles from the purified sample (n = 49). The results of this analysis showed that the intermediate particles from the in cellulo sample were the least spherical (L/W = 1.229), followed by the mature conical in cellulo particles (L/W = 1.150). The purified conical capsids were the most spherical (L/W = 1.118). Direct comparison (unpaired, two-tailed t test) of the average sphericities of the in cellulo intermediates with the in cellulo conical particles gave a P value of 0.0337, suggesting that the difference in sphericities is somewhat significant. The difference in sphericities observed for intermediate and purified conical particles was more significant (P = 0.0003), supporting the idea that intermediate particles are less spherical than their mature counterparts. We also observed a difference in sphericities between the in cellulo conical and purified conical particles (P = 0.0183). This result suggests that the process of purification (i.e., filtration, ultracentrifugation, and resuspension) increases particle sphericity.
RNP localization and mature capsid morphology.
RNP formation precedes the completion of capsid assembly (15) and may help direct the assembly pathway (24). While the overwhelming majority (97%) of conical cores enclosed a density consistent with the features of the RNP (Fig. 5A), cylindrical cores enclosed RNPs just 67% of the time (Fig. 5B and Table 2).
FIG 5.
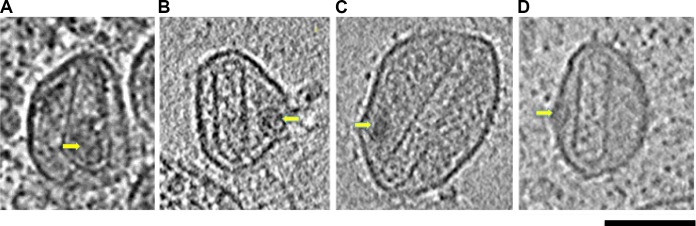
Correlation between cylinder formation and RNP exclusion. Shown are cryotomographic slices through HIV-1 particles with either a conically shaped capsid that encloses the viral RNP (A) or aberrant cylinder-shaped capsids that failed to enclose the RNP (yellow arrows) (B to D). Scale bar, 100 nm.
TABLE 2.
RNP localization in conical versus cylinder capsids
| Capsid morphology (n)a | No. (%) of capsids with:b |
|
|---|---|---|
| RNP inside | RNP outside | |
| Cone (279) | 270 (97) | 9 (3) |
| Cylinder (27) | 18 (67) | 9 (33) |
n, number of capsids.
P = 2.38 × 10−6, probability of randomly observing a pattern of RNP location and core shape.
DISCUSSION
The in cellulo particle population represents a unique experimental system to study the dynamic process of maturation.
We used ECT to structurally characterize HIV particles that were maturing at the edge of infected human cells cultured on EM grids. The particles were cryopreserved and imaged directly in the environment where they were produced and without additional purification. Because there is no purification step that would afford the particles time to fully mature before imaging, our sample contains particles at various stages of assembly, egress, and maturation. Unlike previous studies that characterized viruses with mutations in cleavage sites of Gag (14, 15) or viruses treated with a maturation inhibitor (8, 16), our experimental design avoids the limitations of such strategies that often produce aberrant capsids and noninfectious virus. As a result, we are able to describe maturation and the authentic intermediates that define the process as it occurs unaltered in newly budded viral particles.
While it is not possible to know if every intermediate structure observed is on the assembly pathway to form a conical core, the overwhelming consistency in the hook and large-gap structures and their similarity to the fully mature conical capsid that defines the infectious HIV-1 particle are compelling arguments that these structures are precursors to the mature capsid. Also, the relatively large number (75%) of nonmature/nonimmature particles that display these morphologies (i.e., small sheet, hook, and large gap) strongly suggests that HIV capsids often sample the assembly pathway(s) that lead to these structures and then, often, to a mature conical core. We also note that the final distribution of mature capsid morphologies in our fully mature, purified virus sample is comparable to what has been described previously (10, 14) (Table 1). This suggests that the HIV-1 maturation pathway within the in cellulo sample yields HIV-1 capsids that are morphologically similar to previously described purified particles.
Furthermore, structural analysis of the mature endpoint viruses failed to identify similar intermediate structures among particles that were allowed to mature fully, and to our knowledge the hook and large-gap structures have not been previously reported in the literature. We interpret these points to mean that the intermediate structures observed among the in cellulo particles are not aberrant capsid forms that arise from a dead-end process, but, instead, they are likely structural intermediates that define the HIV-1 capsid assembly pathway. As a result of our unique experimental design, we are able to provide the first description of authentic HIV capsid assembly intermediates.
Maturation-intermediate particles suggest that the HIV-1 capsid initially assembles as a sheet.
The end-to-end models (i.e., tip-to-base or base-to-tip) predict that capsid assembly intermediate forms will be dominated by structures that have either a tip but no base or a fully formed base but no tip. In our sample, such open structures were not routinely observed. Importantly, the asymmetric shape of the hook intermediate structures argues against the end-to-end maturation models that describe core assembly as originating from one of the extreme ends of the viral cone and proceeding circumferentially to the opposite end.
Instead, the morphologies of the assembly intermediates we observed support a capsid assembly pathway that is best described as a hybrid of the three previous models. We propose a model of assembly that begins with the polymerization of a small sheet of CA that associates with the RNP to form the base of the capsid structure. Continued growth of this small sheet from the capsid base progresses along one side of the cone in a manner similar to the simulations of Levandovsky and Zandi (12), which described capsid assembly as the nonequilibrium growth of an elastic sheet. Their simulations demonstrated the rapid growth of the conical shell along one side of its long axis in a manner that could lead to an intermediate structure that is very similar to the hook intermediate we observed in cellulo. Our images suggest that the sheet of CA then grows around the body of the cone to different degrees of completion (13).
The number of particles in each assembly-intermediate subclass suggests relative kinetics of capsid assembly.
The relative number of particles belonging to each of the assembly subclasses likely reflects the kinetics of assembly for the different intermediate structures. Our observation that the small-sheet subclass contains fewer particles than the hook subclass suggests that the initial growth of the capsid along one side of the cone down its long axis proceeds rapidly. The hook subclass is the most abundant assembly intermediate. Because these structures are typically a straight-line density extending from the curved base, we suspect that the slower kinetics may be due to the need for the forming capsid to incorporate pentameric assemblies of CA at the highly curved base or tip in order to continue growing.
The viral membrane and RNP likely guide the core assembly pathway.
In vitro, CA has a strong preference to assemble into tubes, spheres, and sheets but rarely into cones (11). High-salt conditions in vitro help promote the formation of cones (11), but such ionic strength does not reflect conditions found inside the virions in vivo.
In vivo factors that may promote formation of conical cores include the viral membrane and the RNP. The viral particles in our in cellulo sample were often less spherical than those particles in purified samples (compare Fig. 3 and 4). We propose that the apparent membrane deformations are not abnormal but, rather, are a function of the maturation process. Maturation may distort the viral membrane by forcing it to accommodate the growing, rigid capsid structure; for example, growth of a CA sheet along one side of the particle may cause that area of the particle membrane to become flat. Contact of the capsid with the viral membrane may not only affect membrane curvature but also provide feedback to the growing mature lattice (6, 10). In areas where the viral membrane is highly distorted by the capsid, the membrane may prevent continued assembly unless the capsid can adopt a localized curvature such that its growth is no longer impeded. Such curvature could be achieved, presumably, through the incorporation of pentamers at the narrow tip or wide base, as predicted by the fullerene cone model.
Our observation that the assembly-intermediate structures were often associated with a condensed RNP indicates that RNP formation precedes the completion of capsid assembly. Although we are not able to say conclusively when the RNP forms, early formation would suggest that the complex is important to capsid assembly and may bias the assembly pathway toward cones. Additionally, the strong correlation we observed between conical core formation and localization of the RNP to the interior of the capsid also suggests that the RNP influences the capsid assembly pathway. Indeed, such a role for the RNP in determining the structure of the HIV-1 capsid has been proposed (6), and it may be a mechanism of directed capsid assembly across diverse viruses (24). Collectively, these findings suggest that features of the RNP might represent potential therapeutic targets.
Other factors may also nucleate capsid assembly (11). In this case, capsid assembly would begin independent of the RNP, but an assembly pathway that favors formation of a cone would preferentially enclose the RNP in the growing capsid structure. In contrast, a growing capsid that samples an assembly pathway that leads to formation of a cylinder would tend to exclude the RNP from the capsid interior due to the limited space within a uniformly narrow cylinder.
Model for HIV-1 capsid assembly.
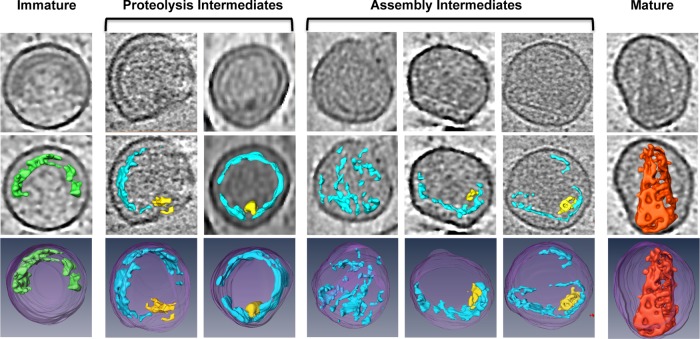
Based on our data, the HIV capsid assembly pathway (Fig. 6) is best described as an asymmetric assembly program mediated by the de novo formation of a CA sheet from dissociated CA subunits. We propose that a small sheet of CA initially forms and associates with the RNP. This CA sheet then grows rapidly along one side toward the narrow tip, followed by slower growth of the structure around the body of the cone. The inherent curvature of the CA sheet together with the RNP ultimately drives cone formation and inclusion of the condensed viral genome within the capsid interior.
FIG 6.
HIV-1 capsid assembly pathway. Tomographic slices through viral particles that contain intermediate capsid structures at various stages of maturation are ordered to suggest progressive assembly of HIV-1 capsid intermediate structures, as follows: top row, single tomographic slices through the particle volume showing the most complete view of the capsid structure; middle row, computational segmentation of the particle's capsid density overlaid on the tomographic slice from the top row; bottom row, full 3D segmentation of the entire particle volume. Purple, membrane; green, immature capsid; blue, capsid assembly intermediate; red, mature capsid; yellow, condensed RNA genome.
ACKNOWLEDGMENTS
We thank Ashley Jensen, Audrey Huang, Taylor Jensen, and Luiza Mendonça for assistance with data analysis and Megan Dobro for useful discussions. Zhiheng Yu and M. Jason de la Cruz of the Howard Hughes Medical Institute CryoEM Shared Resource at Janelia Farm assisted with data collection.
This work was supported by NIH grant 2P50GM082545-06 (to G.J.J.).
REFERENCES
- 1.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. 2009. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A 106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P. 2013. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng X, Zhao G, Yufenyuy E, Ke D, Ning J, Delucia M, Ahn J, Gronenborn AM, Aiken C, Zhang P. 2012. Protease cleavage leads to formation of mature trimer interface in HIV-1 capsid. PLoS Pathog 8:e1002886. doi: 10.1371/journal.ppat.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. 2005. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J Mol Biol 346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yufenyuy EL, Aiken C. 2013. The NTD-CTD intersubunit interface plays a critical role in assembly and stabilization of the HIV-1 capsid. Retrovirology 10:29. doi: 10.1186/1742-4690-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller PW, Huang RK, England MR, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC. 2013. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J Virol 87:13655–13664. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konnyu B, Sadiq SK, Turanyi T, Hirmondo R, Muller B, Krausslich HG, Coveney PV, Muller V. 2013. Gag-Pol processing during HIV-1 virion maturation: a systems biology approach. PLoS Comput Biol 9:e1003103. doi: 10.1371/journal.pcbi.1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs JA, Grunewald K, Glass B, Forster F, Krausslich HG, Fuller SD. 2006. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 12.Levandovsky A, Zandi R. 2009. Nonequilibirum assembly, retroviruses, and conical structures. Phys Rev Lett 102:198102. doi: 10.1103/PhysRevLett.102.198102. [DOI] [PubMed] [Google Scholar]
- 13.Yu Z, Dobro MJ, Woodward CL, Levandovsky A, Danielson CM, Sandrin V, Shi J, Aiken C, Zandi R, Hope TJ, Jensen GJ. 2013. Unclosed HIV-1 capsids suggest a curled sheet model of assembly. J Mol Biol 425:112–123. doi: 10.1016/j.jmb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Marco A, Heuser AM, Glass B, Krausslich HG, Muller B, Briggs JA. 2012. Role of the SP2 domain and its proteolytic cleavage in HIV-1 structural maturation and infectivity. J Virol 86:13708–13716. doi: 10.1128/JVI.01704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Marco A, Muller B, Glass B, Riches JD, Krausslich HG, Briggs JA. 2010. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog 6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. 2011. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol 85:1420–1428. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller B, Anders M, Akiyama H, Welsch S, Glass B, Nikovics K, Clavel F, Tervo HM, Keppler OT, Krausslich HG. 2009. HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J Biol Chem 284:29692–29703. doi: 10.1074/jbc.M109.027144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan L, Jensen GJ. 2012. Electron tomography of cells. Q Rev Biophys 45:27–56. doi: 10.1017/S0033583511000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, Brauchle C, Muller B, Lamb DC. 2011. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol 13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 20.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol 13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kol N, Tsvitov M, Hevroni L, Wolf SG, Pang HB, Kay MS, Rousso I. 2010. The effect of purification method on the completeness of the immature HIV-1 Gag shell. J Virol Methods 169:244–247. doi: 10.1016/j.jviromet.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 22.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Krausslich HG. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol 72:2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Krausslich HG. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J 19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton VL, Dykeman EC, Stonehouse NJ, Ashcroft AE, Twarock R, Stockley PG. 2010. The impact of viral RNA on assembly pathway selection. J Mol Biol 401:298–308. doi: 10.1016/j.jmb.2010.05.059. [DOI] [PubMed] [Google Scholar]