ABSTRACT
Virophages are a unique group of circular double-stranded DNA viruses that are considered parasites of giant DNA viruses, which in turn are known to infect eukaryotic hosts. In this study, the genomes of three novel Yellowstone Lake virophages (YSLVs)—YSLV5, YSLV6, and YSLV7—were identified from Yellowstone Lake through metagenomic analyses. The relative abundance of these three novel virophages and previously identified Yellowstone Lake virophages YSLV1 to -4 were determined in different locations of the lake, revealing that most of the sampled locations in the lake, including both mesophilic and thermophilic habitats, had multiple virophage genotypes. This likely reflects the diverse habitats or diversity of the eukaryotic hosts and their associated giant viruses that serve as putative hosts for these virophages. YSLV5 has a 29,767-bp genome with 32 predicted open reading frames (ORFs), YSLV6 has a 24,837-bp genome with 29 predicted ORFs, and YSLV7 has a 23,193-bp genome with 26 predicted ORFs. Based on multilocus phylogenetic analysis, YSLV6 shows a close evolutionary relationship with YSLV1 to -4, whereas YSLV5 and YSLV7 are distantly related to the others, and YSLV7 represents the fourth novel virophage lineage. In addition, the genome of YSLV5 has a G+C content of 51.1% that is much higher than all other known virophages, indicating a unique host range for YSLV5. These results suggest that virophages are abundant and have diverse genotypes that likely mirror diverse giant viral and eukaryotic hosts within the Yellowstone Lake ecosystem.
IMPORTANCE This study discovered novel virophages present within the Yellowstone Lake ecosystem using a conserved major capsid protein as a phylogenetic anchor for assembly of sequence reads from Yellowstone Lake metagenomic samples. The three novel virophage genomes (YSLV5 to -7) were completed by identifying specific environmental samples containing these respective virophages, and closing gaps by targeted PCR and sequencing. Most of the YSLV genotypes were associated primarily with photic-zone and nonhydrothermal samples; however, YSLV5 had a unique distribution with an occurrence in vent samples similar to that in photic-zone samples and with a higher GC content that suggests a distinct host and habitat compared to other YSLVs. In addition, genome content and phylogenetic analyses indicate that YSLV5 and YSLV7 are distinct from known virophages and that additional as-yet-uncharacterized virophages are likely present within the Yellowstone Lake ecosystem.
INTRODUCTION
Virophages are circular double-stranded DNA viruses that infect giant viruses and their protist hosts and were reported to be distributed widely throughout the world, even including an Antarctic lake (1–3). Sputnik was the first described virophage that was found to inhabit a water-cooling tower in Paris, France, infecting a mamavirus in an Acanthamoeba species (1). Three years later, a virophage designated Mavirus was identified from Cafeteria roenbergensis, a marine phagotrophic flagellate from Texas coastal waters (3). Unlike Sputnik and Mavirus, the genome of Organic Lake virophage (OLV) was identified by de novo assembly of a metagenomic shotgun sequencing database from a hypersaline meromictic lake in Antarctica (2). This was the first example of virophage discovery using culture-independent methods, providing access to novel virophage genomes by exploring metagenomic databases. The fourth reported virophage, almost identical to Sputnik and named Sputnik 2, was associated with contact lens fluid of an individual with keratitis (4, 5). In 2012, we obtained four complete virophage genomes (Yellowstone Lake Lake virophage 1 [YSLV1] to YSLV4) from Yellowstone Lake and one nearly complete genome (ALM) from Ace Lake in Antarctica (6). In 2014, a virophage named Zamilon was reported to be associated with a Mimiviridae host and closely related to Sputnik (7). Virophages, as parasites of the giant viruses, may play a potential role in lateral gene transfer, mediating gene exchange between different giant DNA viruses and enlarging their genome size (1, 8).
Yellowstone Lake occupies a dominant space in Yellowstone National Park, USA, and thus far the largest number of distinct virophages were discovered from this lake ecosystem (6). In a major metagenomic survey of this lake, samples were taken at three general different locations, representing the northern region which is rich in lake floor hydrothermal vent activity (9–11), the West Thumb region where additional lake floor vents occur but that differ in chemistry relative to the northern lake vents (9, 12), and the Southeast Arm region of the lake where as yet there is no known lake floor geothermal activity. These sampling locations represent (i) different hydrothermal vents, (ii) microbial streamers associated with vent openings, (iii) mixing zones, where vent waters mix with lake water, and (iv) photic-zone water column samples, with some taken above sampled vents. Samples were size-fractionated and then extracted DNA subjected to 454 pyrosequencing (∼7.5 Gbp), which is available at http://camera.crbs.ucsd.edu/projects.
In this study, to better understand the distribution, abundance and diversity of YSLVs in Yellowstone Lake, the above-described Yellowstone Lake metagenomic sequences and targeted unique representatives of virophage major capsid proteins (MCPs) were subjected to analysis. Assembly of the distinct virophage genomes present in Yellowstone Lake metagenomes allowed identification of three novel virophages. In addition, a large number of short contigs showing significant similarity with MCPs of known virophages were reconstructed. Our results reveal significantly higher virophage diversity in Yellowstone Lake than previously recognized, implying the important role played by virophages in this ecosystem, as well as the potential possibility to isolate them from this and other freshwater lake ecology.
MATERIALS AND METHODS
Sampling and DNA extraction.
A total of 42 water samples were obtained from different locations of Yellowstone Lake using a remotely operated vehicle in September of 2007 and 2008. Sampling information and methods for biomass collection and DNA extraction have been previously published (13).
Assembly of Yellowstone Lake metagenomic sequences and virophage genomes.
The methods for sequence assembly were similar to the ones described in reference 6, with some modifications. Shotgun metagenomic sequences from Yellowstone Lake samples were assembled de novo using Newbler v2.6 (Roche). Contigs derived from assembly of the entire Yellowstone Lake metagenomic sequence database were constructed as a local database for tBLASTx search for MCPs, which are known to be conserved among all known virophages. Contig sequences with significant similarity (E-value < 10−3) to virophage MCPs were collected as virophage MCP-related sequences. Each contig served as a reference sequence, and then reference assembly was performed using trimmed reads from the Yellowstone Lake metagenomic database with minimum overlap length of 25 bp and minimum overlap identity of 90%. Once an extended sequence with a longer size was obtained, it was used as the next reference sequence for reference assembly of the metagenomic reads. This procedure was repeated until the respective assembled contig sequences stopped extending. All reference assemblies were performed using the bioinformatics platform Geneious Pro (version 7.1.5; Biomatters, Ltd.).
Genome closure using virophage-specific PCR and sequencing.
The existence of the three novel virophages in each water sample was determined by PCR with oligonucleotide primers specific to the MCP from each respective virophage genome (Table 1). The PCRs were conducted using 0.2 nM each respective forward and reverse primer, EconoTaq Plus Green 2× master mix (Lucigen, Middleton, WI) and 10 ng of metagenomic DNA with the following thermal cycling conditions: a touchdown PCR was performed with 10 cycles of 98°C for 20 s, 75 to 65°C for 15 s and 72°C for 5 min, followed by 30 cycles using the same conditions with a 65°C annealing temperature. The PCR amplicons were resolved by agarose gel electrophoresis (SB gel run for 2 h at 165 V) and visualized using ethidium bromide staining on an AlphaImager HP gel documentation system (ProteinSimple, Santa Clara, CA).
TABLE 1.
Primers used in this study
| Virophage | Target region | Primer sequence (5′–3′) |
|
|---|---|---|---|
| Forward | Reverse | ||
| YSLV5 | MCP | ATGAGTGCCGACATTGAGAA | AGCCGAGATAATGTCCTGCT |
| gap | GGCTCGTGGCAGTCGGGATT | CGGCACCGTCGTCCTTCCAT | |
| YSLV6 | MCP | CTAGGCGGTCCTCAACAATC | CAGACATTACACCGCCAGAA |
| gap | ATGAGTTACGCCTGTGCAATTCTTCCA | ACATTTCATAAACACGCTTTAAGGGCT | |
| YSLV7 | MCP | ACGACCAGCGCCGGATTCAA | ACCGTGACGATGGCATTCACT |
| gap | ATGCGACGCCTATGCAATGGC | GCGGCTGAGAATAATGCAGGGC | |
In order to close the gaps of the assembled sequences to form a circular genomic DNA for each of the virophage genomes, primers were designed based on the ends of the assembled sequences (Table 1). Genomic DNA extracted from photic-zone lake samples that resulted in a PCR amplicon using the virophage-specific MCP primer set was used as the template. A KAPA Hifi HotStart ReadyMix (Kapa Biosystems, Inc., Wilmington, MA) was used to perform a touchdown PCR as indicated above. As for sequencing, PCR products were purified using a DNA Clean & Concentrator kit (Zymo Research, Irvine, CA), quantified by using a Qubit dsDNA BR assay kit (Life Technologies, Grand Island, NY) according to manufacturer's protocols, and Sanger sequenced (Lucigen Corp., Middleton, WI). Sequences were trimmed for quality using the CLC Genomics Workbench (CLC Bio, Cambridge, MA) and then used for assembly of virophage genomes using the Geneious Pro bioinformatics package.
Prediction and annotation of virophage encoded genes.
Geneious Pro was used to predict virophage open reading frames (ORFs) with a start codon of ATG, minimum size of 150 bp and standard genetic code. Predicted ORFs were compared to the GenBank database by BLASTp and PSI-BLAST programs (14, 15). The translated ORFs were annotated by using the InterProScan 5 program and NCBI Conserved Domain Search (16, 17). A local virophage database, containing all predicted ORFs in all known virophages, including YSLV1 to -4 and the three new virophages described in the present study, was constructed for further identification and analysis of homologous genes.
Phylogenetic analysis of conserved virophage genetic loci.
Alignments of predicted amino acid sequences of three virophage core genes (ATPase, MCP, and Pro) were performed by MAFFT (version 7) and then concatenated (18). The concatenated alignment was input into RAxML (version 8) for reconstruction of phylogenetic trees using maximum likelihood with 1,000 iterations (19).
Nucleotide sequence accession numbers.
The genomic sequences of the three YSLVs have been deposited in GenBank under the accession numbers KM502589 (YSLV5), KM502590 (YSLV6), and KM502591 (YSLV7).
RESULTS
Abundances and distribution of YSLVs in different Yellowstone Lake samples.
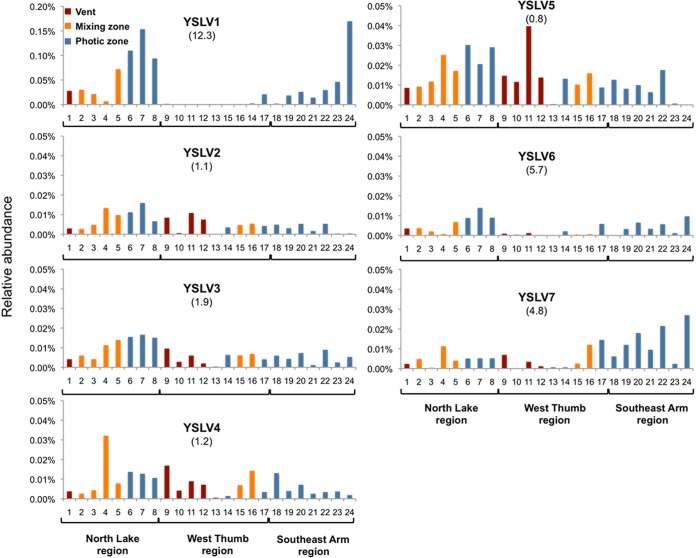
Based on relative abundance data from the metagenomic data sets, YSLV1 was the dominant virophage, contributing up to ca. 0.17% of the total library reads (n = 526,420), and was roughly up to 4- to 10-fold more abundant than the other virophages in the same metagenomes (Fig. 1). YSLV1 distribution was biased toward mixing and photic-zone environments, and potentially interesting, this virophage was either below detection or at relatively very low abundance in all but one of the samples acquired from the West Thumb region of the lake (Fig. 1). In making similar comparisons for the other virophages, there appeared to be no strong evidence of potential lake region provenance, with distributions being of relatively similar abundance across the lake (Fig. 1).
FIG 1.
Distribution and abundance patterns of seven dominant virophages in Yellowstone Lake, Yellowstone National Park as determined by relative representation in 24 different metagenomes. The ratio of relative abundance of each virophage in the photic-zone water to vent samples (i.e., the photic/vent ratio) is provided parenthetically below each virophage identifier. Samples 6, 7, and 8 are size-fractioned samples from a single inflated plain photic-zone water sample, i.e., size classes of 0.1 to 0.8 μm, 0.8 to 3 μm, and 3 to 20 μm, respectively. Similarly, samples 22, 23, and 24 represent a single size-fractioned photic-zone water sample acquired in the Southeast Arm (same respective size classes). Otherwise, all samples are from 0.1- to 0.8-μm size fractions. Note the difference in the y axis scale for YSLV1.
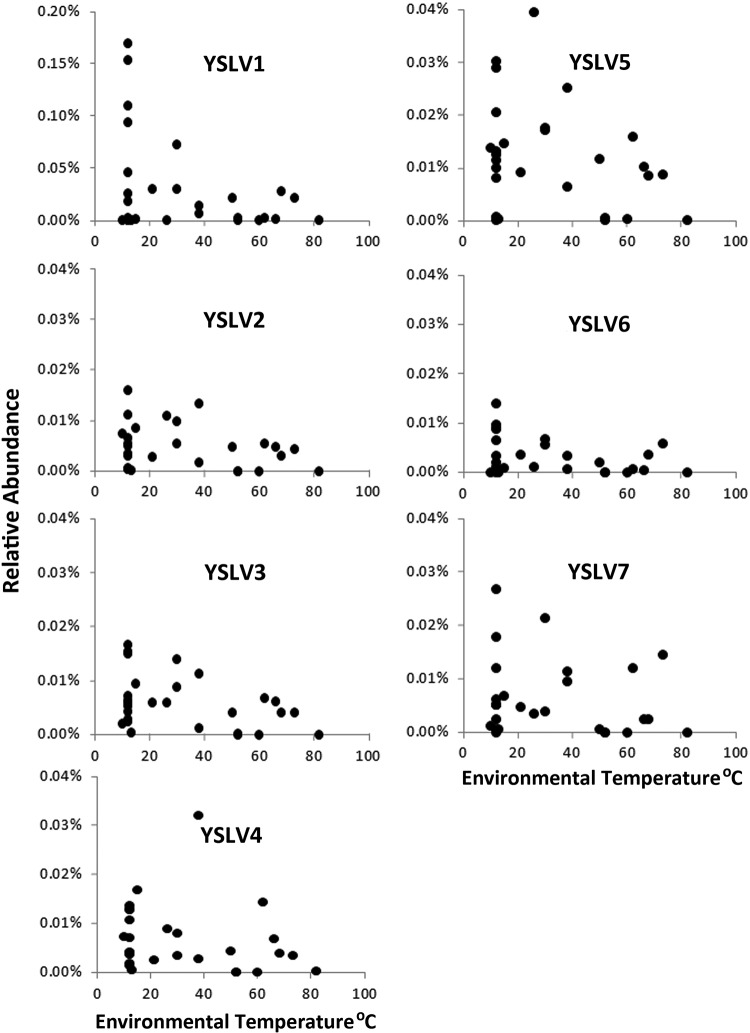
Because of the nature of the lake sampling effort, virophage distribution could also be examined in terms of lake microenvironment and biomass size. To various degrees, all of the virophages were detected in vent water metagenome samples (40 to 68°C), although relative abundances were biased toward the coolest temperatures in the photic-zone samples (Fig. 2). There were no virophages associated with streamer samples (libraries not shown), which are extensive macroscopic community assemblages intimately associated with the orifice of lake floor vents and that were sampled and washed separately from vent waters.
FIG 2.
Relative abundance distributions of each virophage as a function of environmental sample temperature. The y axis scales are the same as those used for Fig. 1. Note again the scale difference for YSLV1.
Separate metagenome libraries from the Northern and the Southeast Arm lake regions were also developed to represent differing biomass size classes of 0.1 to 0.8, 0.8 to 3.0, and 3.0 to 20.0 μm. YSLV1 exhibited an abundance pattern in the Southeast Arm (Fig. 1, samples 22, 23, and 24), suggesting that it and/or its host were more prevalent in the smallest size fraction category (Fig. 1). However, this pattern did not hold in the Northern lake region (Fig. 1, samples 6, 7, and 8). The size distribution of YSLV5 in the Southeast Arm photic zone samples suggested that it predominated in the largest (20.0- to 3.0-μm) category, but again this was also not demonstrated significantly in the northern lake region samples (Fig. 1).
Complete genomes of three novel virophages.
Based on the initial metagenomic assembly, a total of 28 incomplete contigs were identified, revealing a potentially high diversity of virophages that did not have sufficient abundance and genome coverage in order for their respective genomes to be completely assembled. The three largest and nearly complete genomes, ranging from 29 to 22 kb, were selected for more detailed analysis and genome completion by PCR. These virophage genomes were named as YSLV5, YSLV6, and YSLV7, respectively. The existence of the three novel virophages was first identified by PCR targeting unique MCP gene sequences within different Yellowstone Lake samples, and this enabled identification of specific Yellowstone Lake water photic-zone samples that contained these virophage genomes (data not shown) and thus target for targeted PCRs.
We were able to use the metagenomic DNA extracted from the virophage-containing photic-zone samples in order to complete the three novel virophage genomes. The virophage-specific PCR amplicons primed from the ends of the assembled contigs and filled in the predicted gaps. Positive PCR amplicons were first confirmed by agarose gel electrophoresis (data not shown). Sequencing of these amplicons resulted in three DNA sequences that were 1,096 bp for YSLV5, 565 bp for YSLV6, and 403 bp for YSLV7. Each of these sequences successfully assembled with their corresponding virophage genomes, forming three complete and circular virophage DNA sequences.
The genome of YSLV5 was 29,767 bp in length, consisting of 32 predicted ORFs. However, YSLV5 had a G+C content of 51.1%, which is much higher than that (range, 26.7 to 39.1%) of other virophages, including YSLV6 and -7, determined in this study. This might suggest a unique host range for YSLV5. Eleven predicted ORFs of YSLV5 were homologous to that of known virophages. Among these 11 ORFs, the top hits of all were ORFs of YSLVs except ORF06 and ORF11, which were homologous to V21 (Sputnik) or ALM ORF20 (Ace Lake Mavirus), respectively. YSLV6 had a 24,837-bp genome with a 26.8% G+C content and 29 predicted ORFs. Sixteen ORFs were homologous to that of known virophages, 12 of which had the most significant BLAST hits to other YSLVs based on local BLASTp analysis. YSLV7 had a genome size of 23,193 bp, with a 27.3% G+C content and 26 predicted ORFs. Eleven ORFs were homologous to that of known virophages, seven of which had top BLAST hits to other YSLV ORFs. Taken together, these results suggest that YSLVs are rather diverse but more closely related to each other than to virophages identified in other environments instead of Yellowstone Lake.
Conserved virophage genes.
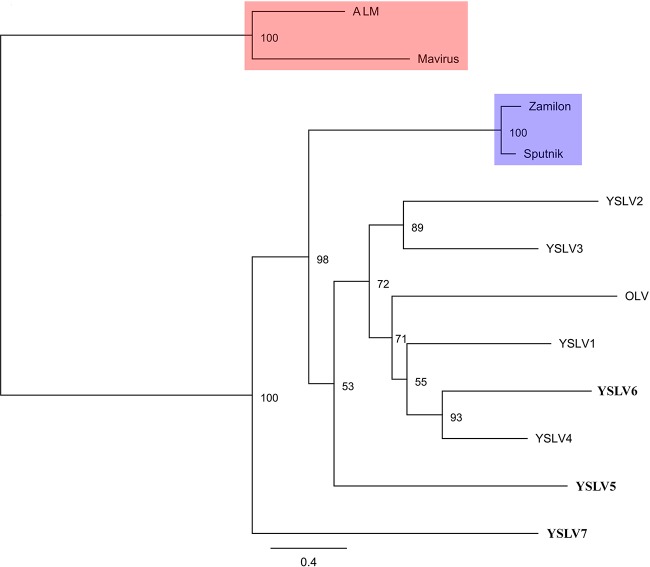
Thus far, five conserved core genes have been detected in all known virophages, including a putative DNA helicase (HEL), packaging ATPase (ATPase), cysteine protease (PRO), major capsid protein (MCP), and minor capsid protein (mCP) (6, 20). They were also identified in YSLVs 5 to 7 through BLASTp and PSI-BLAST analyses (see Data Set S1 in the supplemental material). With the exception of HEL, the other four core genes had high amino acid similarities (42 to 62%) only with their virophage homolog counterparts, respectively, suggesting an early divergent evolution of virophages. Two hypothetical proteins that were only present in OLV and YSLVs 1 to 4 were also shared by YSLVs 5 to 7 and highlighted in red and purple in Fig. 3. In addition, the majority of virophage-homologous genes in YSLVs 5 to 7 had the highest similarity to ORFs of OLV or YSLVs 1 to 4. Accordingly, it suggests a closer evolutionary relationship of YSLVs 5 to 7 with OLV and YSLVs 1 to 4 than with Sputnik, Zamilon, Mavirus, and ALM (Fig. 3).
FIG 3.
Circular maps of the complete genomes of YSLV5, YSLV6, and YSLV7. Homologous genes were labeled by the same color. Gene clusters that have conserved synteny among virophage genomes are highlighted using different kinds of dotted line. The squiggly blue line in the center presents the %G+C skew. Red asterisks indicate the five conserved virophage genes, including HEL, ATPase, PRO, MCP, and mCP.
YSLV6 core MCP, mCP, PRO, and ATPase genes revealed the highest sequence similarities (57 to 67%) to that of YSLV4 (see Data Set S2 in the supplemental material). YSLV5 ATPase and mCP also showed the highest similarities to that of YSLV4 with amino acid identities of 38.7 and 26.9%, respectively (see Data Set S2 in the supplemental material); MCP and PRO displayed high similarities to that of YSLV3 and YSLV7, with 29.4 and 32.0% amino acid identities, respectively (see Data Set S2 in the supplemental material). As for YSLV7, in contrast, ATPase and PRO were the most similar to that of OLV4 (28.9% amino acid identity) and YSLV5 ORF19 (32% amino acid identity) (see Data Set S2 in the supplemental material); MCP and mCP, however, exhibited the greatest similarity to Zamilon ORF06 (22.1% amino acid identity) and ORF05 (28.1% amino acid identity), respectively, rather than to other the YSLVs or OLV (see Data Set S2 in the supplemental material).
YSLV5 ORF31, encoding a putative helicase, showed significant BLASTp hits to YSLV3 ORF11, YSLV6 ORF03, Zamilon ORF09, V13, and MV01 (E-value < 10−5). It contained a fusion domain of primase to SF3 helicase (see Data Set S3 in the supplemental material). The primase-helicase fusion protein is common among viruses (21, 22). The predicted helicase of YSLV6 ORF03 had 30.4 and 24.0% amino acid identities with YSLV3 ORF11 and YSLV5 ORF31, respectively (see Data Set S2 in the supplemental material). The primase-helicase fusion domain was also detected in YSLV6 ORF03 (see Data Set S3 in the supplemental material). In contrast, like YSLV2 ORF10, YSLV7 ORF24 had a superfamily 1/2 helicase domain (see Data Set S3 in the supplemental material). These results suggest that virophage helicases underwent multiple recombination events during the evolution of these viruses.
Interestingly, YSLV6 ORF05, ORF16, and ORF21 contained a GIY-YIG endonuclease domain (see Data Set S4 in the supplemental material). This domain was also identified in Mavirus MV06, Sputnik V14, OLV OLV24, YSLV1 ORF09, and YSLV3 ORF12. YSLV6 ORF16 showed significant similarity to MV06 (34.1% amino acid identity) and YSLV1 ORF09 (35.3% amino acid identity). YSLV6 ORF05 was homologous to OLV01 (35.4% amino acididentity, E-value 3.01−22) and shared 38.0 and 29.9% amino acid identities with MV06 and YSLV1 ORF09, respectively (see Data Set S2 in the supplemental material). Although the GIY-YIG domain was undetectable in OLV01 using InterProScan (data not shown), OLV01 shared significant sequence similarity with YSLV6 ORF05, suggesting either a potentially distantly related GIY-YIG domain or an alternative functional analog in OLV01 (data not shown).
Differences in gene synteny among newly discovered virophage genomes.
All previously described virophage genomes had a cluster of two adjacent genes encoding MCP and mCP with identical synteny. This conserved gene cluster was also present in YSLV5 and YSLV6 (Fig. 3). Another gene cluster consisting of the ATPase gene (ORF29) and a gene of unknown function (ORF01) was also discovered in YSLV6 (Fig. 3). This gene cluster was previously reported in YSLVs 2 to 4 and OLV, suggesting a closer evolutionary affiliation of YSLV6 with these virophages than with the others.
Surprisingly, the gene syntenies observed for the MCP and mCP genes and the third one of the HEL gene and an ORF with unknown function in known virophage genomes was absent within the YSLV7 genome. Although the genome of YSLV7 contained both MCP and mCP, they were separated by 4,843 bp (Fig. 3). This observation supports a distant evolutionary affiliation of YSLV7 with other virophage genomes.
Phylogenetic analysis.
A phylogenetic analysis was first conducted using the predicted MCP amino acid sequences for all available MCP gene sequences recovered from the Yellowstone Lake metagenomic data sets. In total, there were 32 full-length or partial virophage-like MCP sequences identified in distinct contigs. Of these 32 MCPs, 7 correspond to complete virophage genomes (4 previously published and 3 in the present study), leaving 25 MCP sequences that hypothetically correspond to as-yet-uncharacterized virophages. Of these 25 additional MCPs that are not associated with complete virophage genomes, 7 were determined to be complete enough to provide sufficient alignment with other virophage MCP amino acid sequences. Maximum-likelihood analysis revealed that the non-genome-associated MCP sequences affiliated closely with the genome-associated MCP sequences (data not shown).
A phylogenetic analysis was also conducted based on the concatenated amino acid sequences of three core genes—ATPase, PRO, and MCP—in order to shed light on the evolutionary relationships of virophages with complete genome sequences. YSLV6 and YSLV4 appear to form a monophyletic group, which is in agreement with the results of gene content analyses (see above). Accordingly, they are the closest relatives, and YSLV6 is a new member of the virophage lineage comprising YSLVs 1 to 4 and OLV (Fig. 4). YSLV5 was more affiliated with YSLVs, excluding YSLV7, and OLV as well. They seem to have a common ancestor and to diverge from the Sputnik/Zamilon lineage with strong bootstrap support (Fig. 4), which is consistent with previous studies (7). YSLV7 was distantly related to any other virophages and apparently represented a novel virophage lineage.
FIG 4.
Maximum-likelihood-based phylogenetic analysis of the seven Yellowstone Lake virophages based on the concatenated alignment of MCP, PRO, and ATPase amino acid sequences (1,398 amino acids). Bootstrap values (1,000 iterations) are indicated at each node. YSLV5, YSLV6, and YSLV7 obtained in the present study are shown in boldface. The distinct lineages are labeled on the tree.
DISCUSSION
Using the strategy of targeting the conserved major capsid protein as a genomic anchor to assemble shotgun metagenomic sequences derived from different sampling locations in Yellowstone Lake, we detected here the presence of at least 32 distinct virophage genotypes in the unique Yellowstone Lake ecosystem. Three complete novel virophage genomes (YSLVs 5 to 7) are presented here and can now be added to the four complete virophage genomes (YSLVs 1 to 4) previously described (6). This was enabled by de novo PCR efforts using virophage-specific PCR primer sets with lake DNA from specific photic-zone samples that were found to contain these novel virophages. These PCRs extended and closed each of the respective new virophage genome contigs.
The abundance, distribution and diversity of virophages are somehow associated with water chemistry and temperature (Fig. 2). It appears that nonhydrothermal environments, especially with water temperature <30°C, are the primary Yellowstone Lake habitats for most of these viruses and presumably linked to the ecology of their eukaryotic and giant viral hosts, which are unable to tolerate and thrive at high water temperatures. However, the abundance and distribution of YSLV5 appear unusual by comparison. For example, within a deep vent water sample collected in 2007 (sample 14, 52 m deep, 60 to 66°C), YSLV1 was below detection, and YSLVs 2 to 4, 6, and 7 were <0.01% (Fig. 1). However, this sample recorded the highest abundance for YSLV5 (Fig. 1). Indeed, the general abundance pattern for these virophages suggests potentially real differences with respect to their environment preference or tolerance. YSLV5 occurrence in vent samples was nearly equal to photic-zone water (vent/photic ratio of ∼0.8), whereas the average abundance of YSLV1 in photic-zone samples was ∼12-fold greater than in vent water samples. Although it is premature to suggest YSLV5 represents a novel thermophilic virophage, its high G+C content coupled with its divergent phylogenetic position (Fig. 4) and its different lake distribution pattern (Fig. 1 and 2) implies the YSLV5 host ecophysiology somehow differs from the other YSLVs, as well as all other virophages thus far characterized. In this context, it is important to note that all vent samples likely contain at least some non-vent water due to the inability to attain an absolute seal around vent openings with the rim of the sampling cup device of the remote operating vehicle (previously noted by Clingenpeel et al. [13]). The most prominent example would be sample 9, where the vent water emitted from a rock assemblage, within which lake water could easily circulate (see supplemental Movie S3 in Clingenpeel et al. [13]). Accordingly, in terms of virophage ecology, we view the occurrence of the YSLVs in vent fluids with considerable caution. Perhaps an alternative and more reasonable view may be to relate relative abundances of these virophage in vent samples as being an indicator of their prevalence in lake bottom waters surrounding these vents, potentially reflecting their tendency to contribute to lake sediment detritus (sinking) or may be important information with respect to ecological differences of their hosts (e.g., spatial relationships in the photic zone).
Among the three novel virophages described in the present study, YSLV6 appears to be closely related to OLV and YSLVs 1 to 4 and represents a new member in this lineage. This is supported by multiple lines of evidence, including the multilocus phylogenetic analysis (Fig. 4), the presence of three conserved gene clusters with similar gene synteny (Fig. 3), and the number of shared conserved genes. The four core ATPase, Pro, mCP, and MCP genes present within YSLV6 have high percent identities to the corresponding homologous genes of YSLV4, and these two virophages were phylogenetically grouped together with >90% bootstrap support (Fig. 4). This evidence strongly suggests that YSLV6 and YSLV4 are close relatives. In contrast, the phylogenetic analysis indicates that YSLV5 is relatively distinct from the other YSLVs. In addition, the unusual high percent G+C content of YSLV5 also suggests a distinct host range for this virophage compared to other known virophages. Even though the phylogeny of YSLV5 is uncertain, both phylogenetic and gene content analyses indicate it is affiliated with YSLVs rather than with Mavirus or Sputnik lineages. Consequently, like YSLV6, YSLV5 possibly also belongs to the virophage lineage of YSLVs and OLV, albeit with distinct features. Most virophage homologous genes in YSLV7 (7 of 11), including two conserved core genes of ATPase and PRO, reveal high sequence similarity to OLV and other YSLVs, suggesting the affiliation of YSLV7 with this clade. However, the two capsid protein genes that are usually highly conserved in viruses are most similar to that of Zamilon virophage instead of YSLVs, indicating a complicated evolution of YSLV7. In addition, the conserved gene cluster of MCP and mCP present in all other virophages is absent in YSLV7. Taken together, these results indicate that YSLV7 is the first member of a very distinct lineage from known virophages as shown on the phylogenetic tree (Fig. 4). As more virophages are discovered and characterized, the biological significance of these genomic differences and how this impacts the molecular interactions between virophages and their giant virus and eukaryotic hosts may be better understood.
In conclusion, this study has significantly broadened the perspective of virophage diversity and novelty in nature. The Yellowstone Lake metagenome libraries enabled the discovery of new phylogenetic lineages that exhibit distinctly different genome structures as well as apparent distribution patterns that presumably are linked to some degree to host ecological preferences. Their discovery contributes to a long history of other novel finds preserved in the World's first national park.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (no. 41376135), the Doctoral Fund of the Ministry of Education of China (no. 20133104110006), and the Innovation Program of the Shanghai Municipal Education Commission (14ZZ144) to Y.W. and by the Montana Agricultural Experiment Station (project 911310) and the Gordon and Betty Moore Foundation (project 1555) to T.R.M.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03039-14.
REFERENCES
- 1.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, Raoult D. 2008. The virophage as a unique parasite of the giant mimivirus. Nature 455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 2.Yau S, Lauro FM, DeMaere MZ, Brown MV, Thomas T, Raftery MJ, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Gibson JA, Cavicchioli R. 2011. Virophage control of Antarctic algal host-virus dynamics. Proc Natl Acad Sci U S A 108:6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer MG, Suttle CA. 2011. A virophage at the origin of large DNA transposons. Science 332:231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 4.Gaia M, Pagnier I, Campocasso A, Fournous G, Raoult D, La Scola B. 2013. Broad spectrum of mimiviridae virophage allows its isolation using a mimivirus reporter. PLoS One 8:e61912. doi: 10.1371/journal.pone.0061912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desnues C, La Scola B, Yutin N, Fournous G, Robert C, Azza S, Jardot P, Monteil S, Campocasso A, Koonin EV, Raoult D. 2012. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci U S A 109:18078–18083. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou JL, Zhang WJ, Yan SL, Xiao JZ, Zhang YY, Li BL, Pan YJ, Wang YJ. 2013. Diversity of virophages in metagenomic data sets. J Virol 87:4225–4236. doi: 10.1128/JVI.03398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaia M, Benamar S, Boughalmi M, Pagnier I, Croce O, Colson P, Raoult D, La Scola B. 2014. Zamilon, a novel virophage with mimiviridae host specificity. PLoS One 9:e94923. doi: 10.1371/journal.pone.0094923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raoult D, Boyer M. 2010. Amoebae as genitors and reservoirs of giant viruses. Intervirology 53:321–329. doi: 10.1159/000312917. [DOI] [PubMed] [Google Scholar]
- 9.Morgan LA, Shanks WC, Lovalvo DA, Johnson SY, Stephenson WJ, Pierce KL, Harlan SS, Finn CA, Lee G, Webring M, Schulze B, Duhn J, Sweeney R, Balistrieri L. 2003. Exploration and discovery in Yellowstone Lake: results from high-resolution sonar imaging, seismic reflection profiling, and submersible studies. J Volcanol Geoth Res 122:221–242. doi: 10.1016/S0377-0273(02)00503-6. [DOI] [Google Scholar]
- 10.Balistrieri LS, Shanks WCI, Cuhel RL, Aguilar C, Klump JV. 2007. The influence of sub-lacustrine hydrothermal vents on the geochemistry of Yellowstone Lake, p 173–199 InMorgan LA. (ed), Integrated geoscience studies in the greater Yellowstone area: volcanic, tectonic, and hydrothermal processes in the Yellowstone geoecosystem. USGS professional paper 1717. U.S. Geological Survey, Kearneysville, WV. [Google Scholar]
- 11.Morgan LA, Shanks WC III, Pierce KL, Lovalvo DA, Lee GK, Webring MW, Stephenson WJ, Johnson SY, Harlan SS, Schulze B, Finn CA. 2007. The floor of Yellowstone Lake is anything but quiet—new discoveries from high resolution sonar imaging, seismic-reflection profiling, and submersible studies, p 95–126 InMorgan LA. (ed), Integrated geoscience studies in the greater Yellowstone area: volcanic, tectonic, and hydrothermal processes in the Yellowstone geoecosystem. USGS professional paper 1717. U.S. Geological Survey, Kearneysville, WV. [Google Scholar]
- 12.Kan J, Clingenpeel S, Macur RE, Inskeep WP, Lovalvo D, Varley J, Gorby Y, McDermott TR, Nealson K. 2011. Archaea in Yellowstone Lake. ISME J 5:1784–1795. doi: 10.1038/ismej.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clingenpeel S, Macur RE, Kan J, Inskeep WP, Lovalvo D, Varley J, Mathur E, Nealson K, Gorby Y, Jiang H, LaFracois T, McDermott TR. 2011. Yellowstone Lake: high-energy geochemistry and rich bacterial diversity. Environ Microbiol 13:2172–2185. doi: 10.1111/j.1462-2920.2011.02466.x. [DOI] [PubMed] [Google Scholar]
- 14.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yutin N, Raoult D, Koonin EV. 2013. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol J 10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilyina TV, Gorbalenya AE, Koonin EV. 1992. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J Mol Evol 34:351–357. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- 22.Iyer LM, Koonin EV, Leipe DD, Aravind L. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res 33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.