ABSTRACT
Ebola virus (EBOV) transmission is currently poorly characterized and is thought to occur primarily by direct contact with infectious material; however transmission from swine to nonhuman primates via the respiratory tract has been documented. To establish an EBOV transmission model for performing studies with statistical significance, groups of six guinea pigs (gps) were challenged intranasally (i.n.) or intraperitoneally (i.p.) with 10,000 times the 50% lethal dose (LD50) of gp-adapted EBOV, and naive gps were then introduced as cage mates for contact exposure at 1 day postinfection (p.i.). The animals were monitored for survival and clinical signs of disease and quantitated for virus shedding postexposure. Changes in the duration of contact of naive gps with infected animals were evaluated for their impact on transmission efficiency. Transmission was more efficient from i.n.- than from i.p.-challenged gps, with 17% versus 83% of naive gps surviving exposure, respectively. Virus shedding was detected beginning at 3 days p.i. from both i.n.- and i.p.-challenged animals. Contact duration positively correlated with transmission efficiency, and the abrogation of direct contact between infected and naive animals through the erection of a steel mesh was effective at stopping virus spread, provided that infectious animal bedding was absent from the cages. Histopathological and immunohistochemical findings show that i.n.-infected gps display enhanced lung pathology and EBOV antigen in the trachea, which supports increased virus transmission from these animals. The results suggest that i.n.-challenged gps are more infectious to naive animals than their systemically infected counterparts and that transmission occurs through direct contact with infectious materials, including those transported through air movement over short distances.
IMPORTANCE Ebola is generally thought to be spread between humans though infectious bodily fluids. However, a study has shown that Ebola can be spread from pigs to monkeys without direct contact. Further studies have been hampered, because an economical animal model for Ebola transmission is not available. To address this, we established a transmission model in guinea pigs and determined the mechanisms behind virus spread. The survival data, in addition to microscopic examination of lung and trachea sections, show that mucosal infection of guinea pigs is an efficient model for Ebola transmission. Virus spread is increased with longer contact times with an infected animal and is possible without direct contact between an infected and a naive host but can be stopped if infectious materials are absent. These results warrant consideration for the development of future strategies against Ebola transmission and for a better understanding of the parameters involved in virus spread.
INTRODUCTION
Ebola virus (EBOV) is a zoonotic pathogen that causes sporadic outbreaks localized to the humid, remote rainforests of sub-Saharan Africa. Infected individuals develop symptoms similar to those of infection by many other common pathogens, including fever, nausea, diarrhea, and general malaise, before progressing to specific signs characterized by hemorrhagic symptoms, as well as multiorgan failure and a syndrome resembling septic shock, leading to death within 6 to 10 days after the onset of symptoms (1). While EBOV is the most virulent form, with mortality rates of up to 90% in humans, Reston virus is currently the most widespread, with serological evidence of infection among pigs in the Philippines (2) and orangutans in Indonesia (3), as well as bats in China (4) and Bangladesh (5). The erratic nature of these outbreaks, in addition to isolated occasions when the virus can be accidentally introduced into areas of nonendemicity by the import of infected nonhuman primates (NHPs) (6–8), have caused concern among public health authorities. The natural reservoir of EBOV is currently unknown; however, fruit bats have been postulated as a potential culprit due to evidence of asymptomatic infection in wild populations (9). Past experimental inoculation of fruit and insectivorous bats with EBOV resulted in extensive viral replication in bat tissues and high viremia with no apparent clinical symptoms or fatalities associated with disease, but shed live virus was successfully recovered from the feces of only one infected fruit bat (10).
Direct contact with infected materials is generally accepted as the primary mode of transmission, as outbreaks can occasionally be traced back to an index case where the individual had handled infected bush meat (11). Amplification and transmission of virus between humans is subsequently made possible via contact with infected tissue or bodily fluids and by improper use of needles (12), either in a nosocomial setting or during burial rituals. Past reports have also described successful lethal infection of NHPs with aerosolized EBOV (13), where initial infection occurred in the respiratory lymphoid tissues before spread to regional lymph nodes by infected dendritic cells and macrophages (14). Multiple organs, including the liver and spleen, are then infected following extensive viral replication in the lymph nodes. Clinical disease occurs within 7 to 10 days of infection and includes hallmarks of EBOV disease, such as decreasing lymphocyte and platelet levels and elevated liver enzymes, culminating in coagulation disorders and shock (14). In addition, recent studies have demonstrated that pigs can be infected mucosally with EBOV and then transmit the virus to other, naive pigs (15) and to naive NHPs (16). The establishment of interspecies transmission without direct contact, in addition to past epidemiological observations (17), supports the notion that airborne EBOV transmission could potentially contribute, albeit at a low level, to the spread of disease during an outbreak.
Transmission studies in swine and NHPs require the use of precious animal species and necessitate extensive coordination and resources, making them impractical and possibly unsuitable to support scientific progress, such as the early advancement of vaccines and treatments. Previous sequential passaging of EBOV in guinea pigs (gps) has yielded a gp-adapted EBOV variant (GA-EBOV) that is lethal to these animals (18) (19). GA-EBOV has previously been extensively utilized in pathogenesis, vaccine, and treatment studies. The objective of this study was to investigate whether GA-EBOV transmission from direct contact between systemically or mucosally infected gps and naive animals is possible. Factors that may impact transmission, such as the length of contact between the two gps, as well as the prevention of direct physical contact with an infectious animal or material, were also investigated. Survival, weight loss, virus shedding patterns, and evidence of transmission were monitored in this study, providing insight into the factors behind EBOV spread. In addition, the pathology of infected animals was also examined to compare the nature of infection between gps infected systemically, mucosally, or via transmission.
MATERIALS AND METHODS
Ethics statement.
All infection experiments were carried out in the biosafety level 4 (BSL-4) laboratory at the National Microbiology Laboratory (NML) at the Public Health Agency of Canada (PHAC) in Winnipeg, Canada. All procedures involving live animals were approved by the Canadian Science Center for Human and Animal Health Animal Care Committee (CSCHAH-ACC) following the guidelines of the Canadian Council on Animal Care. All protocols were designed to minimize animal discomfort. The approval documentation used for this study was animal use document (AUD) H-11-007.
Study design.
The objective of this study was to establish gps as a useful small-animal model for EBOV shedding and transmission studies. Groups of 6 challenged gps were first infected intranasally (i.n.) or intraperitoneally (i.p.) with 1,000 or 10,000 times the 50% lethal dose (LD50) of GA-EBOV (diluted in a 0.5-ml total volume for i.n. challenge and a 1-ml total volume for i.p. challenge) before the addition of 6 naive contact gps 24 h after infection; one naive animal was pair-housed with one infected animal in an isolated cage and monitored for survival, weight loss, and clinical symptoms. Shedding patterns were characterized via reverse transcriptase (RT)-PCR and by infectivity assays on oral, nasal, and rectal swabs collected at 3, 5, and 7 days p.i. from i.n.- and i.p.-challenged gps, whereas the same samples, in addition to blood, were collected at 13 or 14 and 28 days after initial exposure (p.e.) from i.n. and i.p. contact gps and analyzed in the same manner. The length of contact time with the infectious animal was shortened in a subsequent experiment, as contact animals were added at 3, 5, and 7 days p.i. to the challenged animals in a manner similar to that described above and monitored for survival and weight loss. To determine whether virus transmission could occur without direct physical contact, GA-EBOV-infected gps were separated into groups of three and placed in one half of a single ventilated rabbit/ferret cage separated by two steel meshes spaced 5 cm apart. Groups of three naive gps were then placed into the other half of the cage downwind of the infected animals without animal bedding. The rabbit/ferret cage experiment was then replicated under the same challenge and exposure conditions in the presence of gp bedding (BioFresh) to investigate whether contact with infectious materials is sufficient for virus transmission. Finally, to compare systemic and mucosal infections of gps with GA-EBOV, the lungs, nasal passages, and tracheas of infected animals were harvested at 6 and/or 12 days p.i. and stained to analyze the pathology and the presence of EBOV VP40 antigen.
Ventilated cage system.
A negative-ventilated rabbit/ferret housing cage system (Allentown Inc., PA, USA) was used to conduct the non-direct-contact transmission experiments in gps. The isolators were retrofitted with stainless steel wire mesh to equally divide the compartment of each unit within the isolator. Briefly, guide rails were bolted to both the top and bottom of the metal cage, and two 4-by-4 (1/4-in. by 1/4-in.) square stainless steel inserts were slid into place, creating a 1-in. space between the halves of each of the six units, with 12 compartments in total. The infected animals were housed in the outer half of each unit, while the naive animals occupied the inner half of each unit. Directional airflow of approximately 35 cubic feet per minute was directed over the shedding animals and across the naive animals as it exited the unit.
Animals and challenge.
Outbred 4- to 8-week-old female Hartley strain gps (Charles River) were used for these studies. The animals were infected i.n. or i.p. with 1,000 or 10,000 times the LD50 of GA-EBOV, strain Mayinga (Ebola virus VECTOR/C.porcellus-lab/COD/1976/Mayinga-GPA; GenBank accession number AF272001.1) (18) and monitored every day for 28 or 29 days for survival, weight, and clinical symptoms.
Histopathology and immunohistochemistry.
For histopathology, lung tissue samples were collected from the gps and fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. For immunohistochemistry (IHC), paraffin tissue sections were quenched for 10 min in aqueous 3% H2O2 and then pretreated with proteinase K for 10 min. The primary antibody was a rabbit polyclonal anti-EBOV VP40 antibody that was used at a 1:1,000 dilution for 1 h. The tissue sections were then visualized using a horseradish peroxidase-labeled polymer Envision + system (anti-rabbit) (Dako) and reacted with the chromogen diaminobenzidine (DAB). The sections were then counterstained with Gill's hematoxylin.
Virus titrations.
Oral-, nasal-, and rectal-swab samples were collected in 1 ml of Dulbecco's modified Eagle's medium (DMEM), and the amount of virus present was quantitated by direct titration onto VeroE6 cells or quantitative RT-PCR (RT-qPCR). For titration of live virus, samples were inoculated in 10-fold serial dilutions of DMEM on VeroE6 cells with 3 replicates per sample. Plates were scored 10 to 14 days after infection for cytopathic effect. The 50% tissue culture infective dose (TCID50) titers were calculated using the Reed-Muench method (20) and expressed as TCID50 per ml of sample (TCID50/ml). For detection of GA-EBOV RNA, total RNA was extracted from samples with the QIAamp Viral RNA minikit (Qiagen) and then quantified with the LightCycler 480 RNA Master Hydrolysis Probes kit (Roche) and oligonucleotides targeting the GA-EBOV RNA polymerase gene. The oligonucleotide sequences are as follows: EBOVLF2, CAGCCAGCAATTTCTTCCAT; EBOVLR2, TTTCGGTTGCTGTTTCTGTG; and EBOVLP2 FAM, 6-carboxyfluorescein (FAM)-ATCATTGGCGTACTGGAGGAGCAG-minor groove binder (MGB). RT-qPCR was performed on a Step One Plus RT thermocycler (Applied Biosystems) under the following reaction conditions: 63°C for 3 min and 95°C for 30 s, followed by 40 cycles of 95°C for 15 s and 60°C for 40 s. The results are presented as genome equivalents per ml of sample (GEQ/ml).
Serology.
Sera were collected from naive animals and heat inactivated at 56°C for 45 min. An enzyme-linked immunosorbent assay (ELISA) was used to quantitate glycoprotein (GP)-specific IgG levels. Plates were coated overnight with 30 μg/well of His-tagged EBOV GP (BioTherapeutics Inc.), and the assay was performed after blocking with each sample in triplicate, first diluted to 1:50 and then 2-fold serially in 2% skim milk–phosphate-buffered saline (PBS)–0.05% Tween 20. Washes were performed with Tris-buffered saline (TBS)–0.1% Tween 20, and horseradish peroxidase (HRP)-conjugated goat anti-gp IgG (KPL) was used as a detection antibody. The plates were read using a VMax Kinetic ELISA Microplate Reader (Molecular Devices) at an optical density of 405 nm, and the data were analyzed using CellMaxPro software.
Statistical analysis.
Statistical significance for survival curves was calculated using the log rank Mantel-Cox test. P values of <0.05 were considered significant, P values of<0.01 were considered highly significant, and P values of <0.001 were considered extremely significant. The error bars on graphs, where applicable, indicate 1 standard deviation from the mean. All analyses were performed with GraphPad Prism v.5.01 software.
RESULTS
Survival and clinical symptoms of GA-EBOV-infected and naive contact animals.
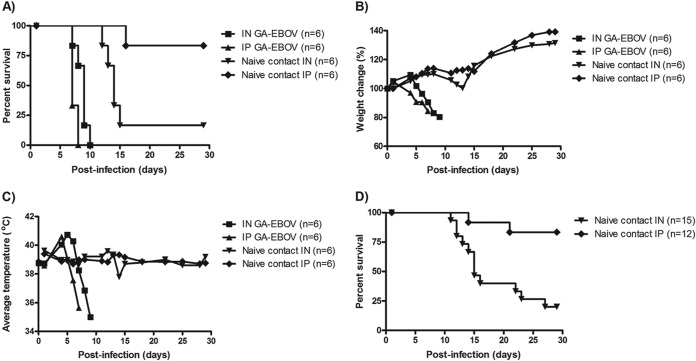
To establish whether EBOV transmission can occur from infected to naive gps through close contact, a naive gp was pair-housed with an experimentally infected animal in a single cage 24 h after challenge. Initially, the challenged animals were infected via either the i.n. or i.p. route with 1,000 or 10,000 times the LD50 of GA-EBOV. Animals challenged i.p. (n = 6) or i.n. (n = 6) with 10,000 times the LD50 succumbed to GA-EBOV infection with an average time to death of 7.3 ± 0.5 or 8.7 ± 1.0 days postinfection, respectively (Fig. 1A). Rapid weight loss was observed by 5 days p.i. (Fig. 1B), in addition to a fever spike of above 40°C preceding hypothermia 2 days before death in both groups (Fig. 1C). One of six naive gps introduced to i.p.-challenged animals lost weight starting 11 days p.e. and succumbed to disease at 14 days p.e., whereas five of six naive gps introduced to i.n.-challenged animals lost weight starting at 7 days p.e. and succumbed to disease with an average time to death of 12.6 ± 1.1 days p.e. (P = 0.01). Surviving naive contact gps did not lose weight or exhibit significant changes in body temperature despite being in close contact with a terminally ill animal (Fig. 1B and C). To determine if these transmission results are reproducible and whether the survival rate of naive contact animals mimics that of human infections, survival data for naive contact gps from repeat experiments (n = 27) were pooled under identical conditions, with the exception that the animals were challenged with 1,000 times the LD50 of GA-EBOV. It was found that exposure of naive animals to i.p.- and i.n.-challenged gps resulted in fatality rates of 17% (2 of 12) and 80% (12 of 15), respectively, with average times to death of 16.5 ± 4.9 days and 15.2 ± 4.9 days p.e. (P = 0.001), respectively (Fig. 1D). Similar survival rates were observed for naive gps pair-housed with animals challenged at 1,000 times the LD50 or 10,000 times the LD50; however, the time to death of naive gps was less variable at the higher challenge dose (Fig. 1A and D), indicating that more uniform transmission occurred at 10,000 times the LD50. Therefore, 10,000 times the LD50 was selected for subsequent experiments, with naive gps introduced 24 h following infection.
FIG 1.
Survival, weight loss, and clinical symptoms of infected and naive contact gps. (A to C) Challenged animals were i.n. or i.p. infected with 10,000 times the LD50 of GA-EBOV, and naive contact gps were introduced at 1 day p.i. as cage mates. All challenged and contact animals were monitored for changes in survival (A), weight loss (B), and body temperature (C). (D) Transmission experiments were repeated for reproducibility and summarized in a survival curve of all exposed contact gps introduced to animals challenged i.n. or i.p. with 1,000 or 10,000 times the LD50 of GA-EBOV.
Shedding patterns of i.n.- versus i.p.-challenged animals and exposed naive animals.
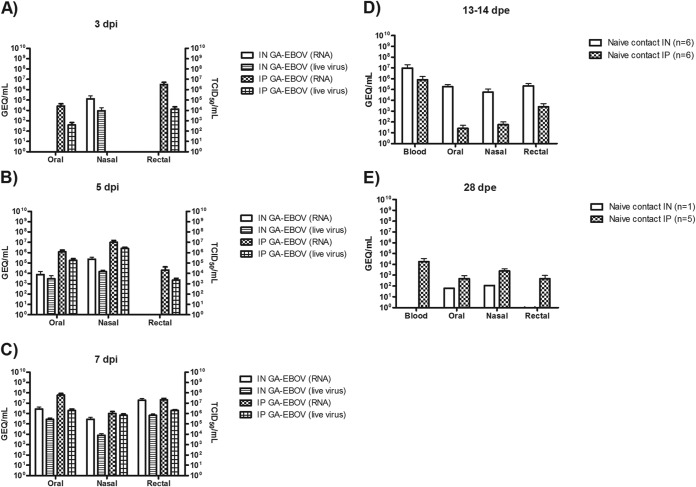
Levels of GA-EBOV shedding from challenged gps (n = 6 per group) were measured from oral, nasal, and rectal swabs sampled 1, 3, 5, and 7 days p.i. and then analyzed for the presence of virus using RT-qPCR specific for the GA-EBOV polymerase gene. Live-virus titrations were performed as confirmation of the PCR results. GA-EBOV could not be detected in any swab samples at 1 day p.i. In i.p.-challenged animals, GA-EBOV could be detected at 3 days p.i. in oral swabs at ∼104 GEQ/ml and in rectal swabs at ∼106 GEQ/ml. Among i.n.-challenged gps, GA-EBOV could be detected at 3 days p.i. in nasal swabs at ∼105 GEQ/ml (Fig. 2A). At 5 days p.i., GA-EBOV could be detected in the oral, nasal, and rectal swabs of i.p.-challenged animals at levels of ∼106, ∼107, and ∼104 GEQ/ml, respectively. Among i.n.-challenged gps, GA-EBOV was detected in the oral and nasal swabs at ∼104 and ∼105 GEQ/ml, respectively (Fig. 2B). At 7 days p.i., GA-EBOV could be detected in the oral, nasal, and rectal swabs of i.p.-challenged animals at ∼108, ∼106, and ∼107 GEQ/ml, respectively. Among i.n.-challenged gps, GA-EBOV was detected in the oral, nasal, and rectal swabs at ∼106, ∼105, and ∼107 GEQ/ml, respectively (Fig. 2C). Live GA-EBOV levels from swabs, expressed in TCID50/ml, were overall lower but were found to correlate with observed RT-qPCR data (Fig. 2A, B, and C).
FIG 2.
GA-EBOV shedding in infected and naive contact gps. (A to C) Oral, nasal, and rectal swabs were sampled from challenged gps infected with GA-EBOV i.n. or i.p. at 3 days p.i. (A), 5 days p.i. (B), and 7 days p.i. (C) and then quantitated for levels of virus by RT-qPCR (left y axes) and virus titration (right y axes). (D and E) Oral, nasal, and rectal swabs were sampled from contact gps at 13 or 14 days p.e. (D) and 28 days p.e. (E) and quantitated for levels of virus by RT-qPCR. The error bars represent standard deviations.
Among contact animals, high levels of GA-EBOV were detected only in nonsurviving naive contact gps exposed to i.p.-challenged (n = 1) and i.n.-challenged (n = 5) gps at the time of death (between 13 and 14 days p.e.). Nonsurviving animals exposed to i.p.-challenged gps had detectable levels of GA-EBOV in the blood, as well as in oral, nasal, and rectal swabs, at ∼106, ∼101, ∼101, and ∼103 GEQ/ml, respectively. Nonsurviving animals exposed to i.n.-challenged gps had comparatively high levels of virus in the blood, as well as oral, nasal, and rectal swabs, at ∼107, ∼105, ∼105, and ∼105 GEQ/ml, respectively (Fig. 2D). At the termination of the experiment at 28 days p.e., low levels of GA-EBOV were detected in the oral and nasal swabs of the lone surviving naive contact (with an i.n.-challenged gp) animal at ∼101 and ∼102 GEQ/ml, respectively; however, live virus was not recovered. Despite the lack of clinical symptoms, GA-EBOV was, surprisingly, detected in the blood, as well as oral, nasal, and rectal swabs, from 3 of 5 naive contact (with i.p.-challenged gps) gps at ∼104, ∼102, ∼103, and ∼102 GEQ/ml, respectively (Fig. 2E).
Efficiency of GA-EBOV transmission is dependent on the length of contact exposure.
The effect of the duration of contact between naive and infected gps on GA-EBOV transmission rates was then evaluated. Animals (n = 18) were challenged i.n. with 10,000 times the LD50 of GA-EBOV, with naive gps (n = 6 per group) introduced as cage mates at 3, 5, or 7 days p.i. I.n.-challenged gps succumbed to infection between 7 and 10 days p.i., with an average time to death of 8.6 ± 0.9 days p.i., during which weight loss and clinical symptoms corresponding to GA-EBOV disease were observed (Fig. 3A and B). Naive contact gps introduced at 3, 5, and 7 days p.i. had survival rates of 33%, 50%, and 83%, respectively, with nonsurvivors succumbing to disease with mean times to death of 14.3 ± 2.0, 13.0 ± 2.0, and 12 ± 0.0 days p.e., respectively (P = 0.24, P = 0.20, and P = 0.04 in comparison to the group of naive gps introduced at 1 day p.i.) (Fig. 3A). Surviving gps did not lose significant weight (Fig. 3B) or exhibit clinical signs of disease.
FIG 3.
Survival and weight loss of infected and naive contact gps introduced at different times after challenge. The challenged animals were infected with GA-EBOV i.n., and naive contact gps were introduced at 3, 5, or 7 days p.i. as cage mates. All challenged and contact animals were monitored for changes in survival (A) and weight loss (B).
Direct contact with infectious materials is needed for GA-EBOV transmission.
A study was then conducted in order to explore the mechanism of GA-EBOV transmission and whether the presence of infectious materials could have played a role in enhanced virus spread. Using a ventilated caging system, gps (n = 6) were separated equally into two different cages and challenged i.n. with 10,000 times the LD50 of GA-EBOV. At 1 day p.i., naive gps (n = 6) were separated equally into two cages downwind of the cages housing the infected animals. Also present in both sides of the cages was animal bedding material that was small enough to be able to pass through the holes in the steel mesh barrier. While all infected gps succumbed to GA-EBOV with an average time to death of 8.0 ± 0.6 days p.i., one out of two naive gp cage mates also lost weight starting at 11 days p.i. and succumbed to disease with an average time to death of 16.7 ± 3.5 days p.e. (Fig. 4D and E). The three surviving naive animals in the remaining cage tested seronegative for GA-EBOV at the termination of the experiment (data not shown). The experiment was then repeated without animal bedding inside the cages of the infected and naive animals. Animals (n = 9) were first separated equally into three different cages, and each was challenged i.n. with 10,000 times the LD50 of GA-EBOV. At 1 day p.i., naive gps (n = 9) were separated equally into three different cages downwind of the cages housing the infected animals, with two steel meshes spaced 1 in. apart physically separating the infected gps from the naive animals (Fig. 4A). While i.n.-challenged gps succumbed to infection between 7 and 9 days p.i. with an average time to death of 8.1 ± 0.8 days p.i., all the naive animals survived the exposure without significant weight loss (Fig. 4B and C). The surviving animals were shown to be seronegative for GA-EBOV by IgG ELISA (data not shown), indicating that transmission did not occur.
FIG 4.
Survival and weight loss of infected and naive gps within a ventilated cage system. (A) A representative ventilated cage with three gps on either side of the steel mesh barrier in the middle. Infected animals were placed on the left side, and naive animals were on the right side. Airflow was from the left to the right. The challenged animals were given GA-EBOV i.n., and naive gps were introduced into the cages downwind of the infected cages, with infected animals prevented from direct contact with the naive animals by the presence of two steel meshes placed 5 cm apart between the two groups. (B and C) All challenged and naive animals were monitored for changes in survival (B) and weight loss (C). (D and E) The same experiment was then repeated with the same cage system but in the presence of gp bedding, which was able to pass through the steel meshes. All challenged and naive animals were monitored for changes in survival (D) and weight loss (E).
Histopathology and immunohistochemistry of infected gps show evidence of respiratory infection.
The lungs, nasal passages, and tracheas of i.p.- and i.n.-challenged gps at 6 days p.i., as well as i.p.- and i.n.-transmission animals at 6 and 12 days p.i., were harvested to establish whether any pathological differences exist between a systemic and a mucosal GA-EBOV infection. One infected animal was cohoused with one naive gp in a single cage for these studies. In the i.p.-challenged group, the observed lung lesions were consistent with a pulmonary interstitial reaction and were characterized by expansion of alveolar septa by edema fluid, inflammatory cells (primarily macrophages and granulocytes), and scattered degenerating cells with pyknotic nuclei (Fig. 5A and B). In the i.n.-infected group, patchy areas of alveolar inflammation were similar to those observed for the i.p.-infected group; however, there were also multifocal, locally extensive areas of severe bronchointerstitial pneumonia (Fig. 5C). Bronchioles contained neutrophils, and there was evidence of a loss of bronchiolar epithelial cells (Fig. 5D). Alveolar walls were expanded due to hyperplasia of type II pneumocytes, as well as the presence of edema, macrophages, granulocytes, and degenerating cells with pyknotic nuclei. The alveolar air space was largely replaced by alveolar macrophages, fibrin, necrotic debris, and granulocytes, many of which were degenerating (Fig. 5D). In the i.n.-transmission group at 12 days p.i., several of the lung sections had lesions similar to the severe bronchointerstitial pneumonia described for the i.n.-challenged group (Fig. 5E). However, this type of lesion was observed in only 2 of 3 animals and on 2 of 15 observed lung sections. This was in contrast to the i.n.-challenged group, in which the lesions were observed on all slides. The rest of the lung sections for the i.n.-transmission group showed an interstitial reaction similar to those observed in the i.p.-infected group. Significant lung lesions were not observed with the i.p.-transmission group at 12 days p.i. (Fig. 5F), nor were they observed with the i.p.- and i.n.-transmission groups at 6 days p.i.
FIG 5.
Histopathology findings. Shown are lungs from i.p.-challenged (A and B), i.n.-challenged (C and D), i.n.-transmission (E), and i.p.-transmission (F) groups. (A) Alveolar walls appear mildly thickened with increased cellularity. Bar = 200 μm. (B) Alveolar walls are expanded by inflammatory cells. Note the presence of scattered degenerating cells with pyknotic nuclei (arrows). Bar = 20 μm. (C) Note the large area of severe bronchointerstitial pneumonia (*) in contrast to the less-affected lung tissue with visible alveolar spaces. Bar = 200 μm. (D) Alveoli are filled with neutrophils and necrotic debris, leading to loss of air spaces (arrow). Bronchiolar lumina contain neutrophils, and there is evidence of degeneration of lining epithelial cells (arrow). The alveolar walls contain hyperplastic type II pneumocytes (arrowheads). Bar = 20 μm. (E) Large areas of severe bronchointerstitial pneumonia (*) were observed on a few lung sections. Bar = 200 μm. (F) There are no significant lesions. Bar = 200 μm.
In the lungs of i.p.-challenged gps, moderate amounts of viral antigen were detected within alveolar walls and bronchiole-associated lymphoid tissue (BALT) (Fig. 6A). Positive immunostaining was observed within macrophages, endothelial cells, and occasional pneumocytes, as well as free within capillaries and vessels (Fig. 6B). Viral antigen was not observed within bronchiolar epithelial cells but was detected in occasional cells within the lumen (likely macrophages), as well as within the bronchiolar submucosal tissue. With the i.n.-challenged animals, most sections showed areas of intense and extensive positive immunostaining, which correlated with the presence of lesions and often appeared to be centered on bronchioles and associated arteries (Fig. 6C). Positive immunostaining was occasionally observed within bronchial and bronchiolar epithelial cells (Fig. 6D). Within the interstitium, the staining was so intense that it was difficult to identify individual cells; however, staining was observed to be present in pneumocytes by double immunolabeling (Fig. 6E) and within macrophages, endothelial cells, BALT, and free within capillaries and vessels, as well as in association with alveolar exudate. In a few areas, the staining pattern was similar to that described for the i.p.-challenged group. In the i.n.-transmission group at 12 days p.i., most of the sections showed a staining pattern similar to that described for i.p.-challenged animals. However, in the 2 sections that showed lesions of bronchointerstitial pneumonia, the staining pattern was similar to that described for the i.n.-challenged group with intense focal staining (Fig. 6F).
FIG 6.
Detection of GA-EBOV antigen by IHC in lungs. Lungs were harvested from i.p.-challenged (A and B), i.n.-challenged (C, D, and E), and i.n.-transmission (F) groups. (A) Viral antigen was detected within alveolar walls throughout most of the section (arrows), as well as within the BALT (arrowhead). Bar = 100 μm. (B) Viral antigen was detected within endothelial cells (arrow) and within cells that have the morphological appearance of macrophages (arrowheads). Bar = 20 μm. (C) Large areas of the section show intense, widespread immunostaining. Bar = 100 μm. (D) Positive immunostaining in bronchiolar epithelial cells (arrows). Bar = 20 μm. (E) Double immunolabeling shows cytokeratin-positive hyperplastic type II pneumocytes (brown; arrows), some of which contain viral antigen (pink; arrowhead). Bar = 10 μm. (F) Viral antigen was observed within occasional bronchiolar epithelial cells (arrow). There is intense staining in the adjacent pulmonary parenchyma and alveolar exudate. Bar = 20 μm.
In samples taken from nasal passages of the i.p.- and i.n.-challenged (at 6 days p.i.) and i.n.-transmission (at 12 days p.i.) groups, viral antigen was detected within the endothelial cells of submucosal vessels; however, no antigen was detected within epithelial cells (Fig. 7A). In the tracheas of the i.n.-challenged group, viral antigen could be detected within the epithelium, as well as the submucosa (Fig. 7B). Within the tracheas of the i.p.-challenged and i.n.-transmission groups, antigen was detected only within the submucosa (Fig. 7C). No viral antigen was detected in any tissues from the i.p.-transmission group at 12 days p.i.
FIG 7.
Detection of GA-EBOV antigen by IHC in the nasal passage and trachea. (A) In sections from nasal passages, viral antigen was detected within endothelial cells of submucosal vessels (arrows), but not in epithelia (arrowhead). (B) In the trachea of the i.n.-challenged group, viral antigen could be detected within the epithelium (arrows) and submucosa (arrowhead). (C) Trachea from the i.p.-challenged group showing viral antigen in the submucosa (arrow) but not within the epithelium (arrowhead). Bars = 50 μm.
DISCUSSION
The direct and indirect parameters contributing to transmission of EBOV between animals, fom animals to humans, and from humans to humans are currently largely undefined. It is therefore important to elucidate and characterize the mechanisms behind EBOV transmission and, in cases where exposure to virus has occurred, to evaluate the effectiveness of specific countermeasures. A better understating of EBOV transmission is also important to maximize rationally based decisions when managing an outbreak.
Since both mucosal and systemic inoculation of NHPs with EBOV causes fatal disease highlighted by similar pathogenesis hallmarks typical of filoviral disease (13, 21), gps were infected i.n. or i.p. to determine whether they were susceptible to both routes of infection. While both methods of inoculation resulted in fatal disease for all challenged animals within comparable time frames, characterized by similar levels of virus isolated from oral, nasal, and rectal swabs at the time of death, there was a marked difference between the transmission properties of mucosal and systemic infections in gps. Naive contact gps caged with i.n.-challenged gps fared significantly worse than those housed with i.p.-challenged animals, as seen by lower rates of survival. Interestingly, the overall mortality rate of 80% observed among the contact gps of i.n.-challenged animals in our studies (Fig. 1D) mimics the case fatality rate (CFR) for all documented EBOV outbreaks among humans, which stands at 1,084 deaths out of 1,381 total cases (CFR = 78.5%) (22), excluding the ongoing 2014 outbreak in West Africa, for which final numbers are not available due to the evolving situation.
The observed difference in transmission efficiency between the i.n.- and i.p.-challenged gp groups could be explained by the higher levels of GA-EBOV shedding from the nasal route of the i.n.-challenged animals early after contact, as seen at 3 days p.i. Another parameter possibly playing a role is the fact that i.p.-challenged gps succumb to disease more quickly than i.n.-challenged animals, perhaps due to a route of infection that more directly promotes systemic virus circulation. Since animals that succumbed to disease were removed from their cage mates immediately after death, it is possible that a comparatively long exposure time between i.n.-infected and naive animals resulted in increased transmission. Naive gps introduced to infected cage mates at progressively later times to decrease the contact time spent with a terminally ill animal showed increased survival when the time of contact with an infected animal was decreased, and this difference became statistically significant between naive gps exposed at 1 versus 7 days p.i. These results suggest that the length of contact time between a naive and an infected animal plays an important role. It is possible that the exposure to a higher virus load early after contact, together with a longer time of exposure, leads to more transmission from the i.n.-challenged group. A third possibility that is currently being evaluated in the laboratory is whether immune components from the infected animals (e.g., antibodies) are present at higher levels in the i.p.-challenged animals and interfere with the transfer of infectious particles to the naive animals. This would explain the sublethal infections documented in the surviving animals (Fig. 2E), which has never been described in gps by limiting dilutions of the challenge virus.
The distribution of EBOV antigens in i.p.-challenged gps suggests that the virus enters the lungs systemically via the bloodstream, whereas in i.n.-challenged gps, the data from the lungs and trachea are consistent with an infection from the airway. The lungs of i.p.-challenged gps did not display severe pathology, suggesting that the animals died mainly from systemic disease. In contrast, i.n.-challenged animals displayed a severe inflammatory response in the lungs, in which the alveolar airspace had been replaced by degenerating immune cells, resulting in pneumonia and progression to the systemic disease that is also observed with gps challenged with aerosolized GA-EBOV (23). The earlier release of the viral load, combined with the presence of viral antigen in the tracheal epithelium in i.n.-challenged gps, supports increased transmissibility from these animals. This is reflected in the survival rates of the naive contact animals in this study. Naive gps cohoused with i.n.-challenged gps died, and evidence of infection through the airway was again observed in the lungs of these animals, albeit at an apparently lower frequency.
It was observed in these studies that GA-EBOV transmission could be abrogated by preventing direct contact between an infected and a naive gp, provided that no infectious materials could travel to expose the naive animals. Therefore, the experimental results do not support airborne spread of EBOV but demonstrate instead that virus transmission can occur through direct contact with infected materials traveling over short distances. Interestingly, the pathology observed in animals presumably infected through contaminated bedding are similar to previous findings with gps infected by aerosolized GA-EBOV, such as severe pneumonia characterized by thickened alveolar septa filled with infected immune cells, as well as infrequent necrosis of bronchiolar epithelial cells (23). In-depth studies will be necessary to further characterize and distinguish the differences between systemic, mucosal, and aerosol GA-EBOV infections and their relation to virus transmissibility.
Although gps do not succumb to infection with wild-type EBOV and an adapted variant has been utilized in its place, the hallmarks of GA-EBOV infection in gps are quite comparable to those of wild-type EBOV infection in NHPs. The availability of a small-animal model capable of supporting transmission studies with statistical significance will serve to accelerate future transmission studies. A better understanding of the parameters involved in EBOV transmission should help researchers and medical professionals to develop better medical countermeasures and to protect against all aspects of EBOV outbreaks whether from natural causes or not.
ACKNOWLEDGMENTS
This work was supported by the Public Health Agency of Canada (PHAC) and a grant from the Canadian Safety and Security Program (CSSP) to G.P.K. and X.Q. G.W. is the recipient of a doctoral research award from the Canadian Institute of Health Research (CIHR).
We thank Shane Jones, Geoff Soule, Jim Strong, Allen Grolla, Anders Leung, and Estella Moffat for their excellent technical assistance, in addition to animal husbandry.
REFERENCES
- 1.Feldmann H, Geisbert TW. 2011. Ebola haemorrhagic fever. Lancet 377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrette RW, Metwally SA, Rowland JM, Xu L, Zaki SR, Nichol ST, Rollin PE, Towner JS, Shieh WJ, Batten B, Sealy TK, Carrillo C, Moran KE, Bracht AJ, Mayr GA, Sirios-Cruz M, Catbagan DP, Lautner EA, Ksiazek TG, White WR, McIntosh MT. 2009. Discovery of swine as a host for the Reston ebolavirus. Science 325:204–206. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- 3.Nidom CA, Nakayama E, Nidom RV, Alamudi MY, Daulay S, Dharmayanti IN, Dachlan YP, Amin M, Igarashi M, Miyamoto H, Yoshida R, Takada A. 2012. Serological evidence of Ebola virus infection in Indonesian orangutans. PLoS One 7:e40740. doi: 10.1371/journal.pone.0040740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan J, Zhang Y, Li J, Wang LF, Shi Z. 2012. Serological evidence of ebolavirus infection in bats, China. Virol J 9:236. doi: 10.1186/1743-422X-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olival KJ, Islam A, Yu M, Anthony SJ, Epstein JH, Khan SA, Khan SU, Crameri G, Wang LF, Lipkin WI, Luby SP, Daszak P. 2013. Ebola virus antibodies in fruit bats, Bangladesh. Emerg Infect Dis 19:270–273. doi: 10.3201/eid1902.120524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anonymous. 1990. Update: filovirus infection in animal handlers. MMWR Morb Mortal Wkly Rep 39:221. [PubMed] [Google Scholar]
- 7.Anonymous. 1992. Viral haemorrhagic fever in imported monkeys. Wkly Epidemiol Rec. 67:142–143. [PubMed] [Google Scholar]
- 8.Jahrling PB, Geisbert TW, Dalgard DW, Johnson ED, Ksiazek TG, Hall WC, Peters CJ. 1990. Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet 335:502–505. doi: 10.1016/0140-6736(90)90737-P. [DOI] [PubMed] [Google Scholar]
- 9.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Delicat A, Paweska JT, Gonzalez JP, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 10.Swanepoel R, Leman PA, Burt FJ, Zachariades NA, Braack LE, Ksiazek TG, Rollin PE, Zaki SR, Peters CJ. 1996. Experimental inoculation of plants and animals with Ebola virus. Emerg Infect Dis 2:321–325. doi: 10.3201/eid0204.960407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson AT, Bauer JT, Mills JN. 2004. Ecologic and geographic distribution of filovirus disease. Emerg Infect Dis 10:40–47. doi: 10.3201/eid1001.030125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen J. 2004. Containing the threat—don't forget Ebola. PLoS Med 1:e59. doi: 10.1371/journal.pmed.0010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson E, Jaax N, White J, Jahrling P. 1995. Lethal experimental infections of rhesus monkeys by aerosolized Ebola virus. Int J Exp Pathol 76:227–236. [PMC free article] [PubMed] [Google Scholar]
- 14.Twenhafel NA, Mattix ME, Johnson JC, Robinson CG, Pratt WD, Cashman KA, Wahl-Jensen V, Terry C, Olinger GG, Hensley LE, Honko AN. 2013. Pathology of experimental aerosol Zaire ebolavirus infection in rhesus macaques. Vet Pathol 50:514–529. doi: 10.1177/0300985812469636. [DOI] [PubMed] [Google Scholar]
- 15.Kobinger GP, Leung A, Neufeld J, Richardson JS, Falzarano D, Smith G, Tierney K, Patel A, Weingartl HM. 2011. Replication, pathogenicity, shedding, and transmission of Zaire ebolavirus in pigs. J Infect Dis 204:200–208. doi: 10.1093/infdis/jir077. [DOI] [PubMed] [Google Scholar]
- 16.Weingartl HM, Embury-Hyatt C, Nfon C, Leung A, Smith G, Kobinger G. 2012. Transmission of Ebola virus from pigs to non-human primates. Sci Rep 2:811. doi: 10.1038/srep00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roels TH, Bloom AS, Buffington J, Muhungu GL, Mac Kenzie WR, Khan AS, Ndambi R, Noah DL, Rolka HR, Peters CJ, Ksiazek TG. 1999. Ebola hemorrhagic fever, Kikwit, Democratic Republic of the Congo, 1995: risk factors for patients without a reported exposure. J Infect Dis 179(Suppl 1):S92–S97. doi: 10.1086/514286. [DOI] [PubMed] [Google Scholar]
- 18.Connolly BM, Steele KE, Davis KJ, Geisbert TW, Kell WM, Jaax NK, Jahrling PB. 1999. Pathogenesis of experimental Ebola virus infection in guinea pigs. J Infect Dis 179(Suppl 1):S203–S217. doi: 10.1086/514305. [DOI] [PubMed] [Google Scholar]
- 19.Volchkov VE, Chepurnov AA, Volchkova VA, Ternovoj VA, Klenk HD. 2000. Molecular characterization of guinea pig-adapted variants of Ebola virus. Virology 277:147–155. doi: 10.1006/viro.2000.0572. [DOI] [PubMed] [Google Scholar]
- 20.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 21.Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert J B, Scott DP, Kagan E, Jahrling PB, Davis KJ. 2003. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol 163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CDC. 2013. Known cases and outbreaks of ebola hemorrhagic fever, in chronological order. CDC, Atlanta, GA. [Google Scholar]
- 23.Twenhafel NA, Shaia CI, Bunton TE, Shamblin JD, Wollen SE, Pitt LM, Sizemore DR, Ogg MM, Johnston SC. 2014. Experimental aerosolized guinea pig-adapted Zaire ebolavirus (variant: Mayinga) causes lethal pneumonia in guinea pigs. Vet Pathol 14:0300985814535612. doi: 10.1177/0300985814535612. [DOI] [PubMed] [Google Scholar]