ABSTRACT
Coronaviruses from both the Alphacoronavirus and Betacoronavirus genera interfere with the type I interferon (IFN) response in various ways, ensuring the limited activation of the IFN response in most cell types. Of the gammacoronaviruses that mainly infect birds, little is known about the activation of the host immune response. We show that the prototypical Gammacoronavirus, infectious bronchitis virus (IBV), induces a delayed activation of the IFN response in primary renal cells, tracheal epithelial cells, and a chicken cell line. In fact, Ifnβ expression is delayed with respect to the peak of viral replication and the accompanying accumulation of double-stranded RNA (dsRNA). In addition, we demonstrate that MDA5 is the primary sensor for Gammacoronavirus infections in chicken cells. Furthermore, we provide evidence that accessory proteins 3a and 3b of IBV modulate the response at the transcriptional and translational levels. Finally, we show that, despite the lack of activation of the IFN response during the early phase of IBV infection, the signaling of nonself dsRNA through both MDA5 and TLR3 remains intact in IBV-infected cells. Taken together, this study provides the first comprehensive analysis of host-virus interactions of a Gammacoronavirus with avian innate immune responses.
IMPORTANCE Our results demonstrate that IBV has evolved multiple strategies to avoid the activation of the type I interferon response. Taken together, the present study closes a gap in the understanding of host-IBV interaction and paves the way for further characterization of the mechanisms underlying immune evasion strategies as well as the pathogenesis of gammacoronaviruses.
INTRODUCTION
Coronaviruses constitute a large family of positive-stranded RNA viruses and cause a range of human and veterinary diseases. Infectious bronchitis virus (IBV) is the prototype avian coronavirus from the Gammacoronavirus genus and the causative agent of a highly contagious respiratory disease of major economic importance to the poultry industry (1). IBV enters the avian host through the respiratory tract, where it causes the destruction of the epithelium, leading to respiratory distress and initiation of secondary bacterial infections. Depending on the strain, IBV also can spread to other epithelial surfaces, such as the gastrointestinal tract, the kidneys, and the oviduct, with the latter causing problems in egg production and quality (1–6). Contrary to coronaviruses from the Alphacoronavirus and Betacoronavirus genera, including human coronavirus HCoV-229E, severe acute respiratory syndrome (SARS-CoV), Middle East respiratory syndrome (MERS-CoV), and mouse hepatitis virus (MHV), very little is known about how gammacoronaviruses, including IBV, evade or interfere with the innate immune responses of their host.
Innate immune responses consist of a network of antimicrobial mechanisms, of which the type I interferon (IFN) response is an essential defense mechanism against viruses. Typically, the type I IFN response, here referred to as the IFN response, is initiated upon the activation of host pattern recognition receptors (PRRs), which are present in all animal cells. Two families of PRRs have been shown to be involved in the recognition of RNA viruses, namely the membrane-bound Toll-like receptors (TLRs) and the cytosolic RIG-I-like receptors (RLRs) (7). The primary ligands for the activation of these PRRs are double-stranded RNA (dsRNA) and 5′ triphosphate-containing RNA, normally absent from uninfected host cells. The activation of RLRs leads to the transcription of genes encoding type I interferons (IFN-α and IFN-β). These interferons are secreted from the infected cell, providing a signal for the infected as well as the neighboring cells that induce the transcription of antiviral effector genes collectively called interferon-stimulated genes (ISGs).
The ability of a virus to replicate and produce infectious progeny depends in large part on its ability to avoid induction or to counteract the IFN response of its host. Indeed, a common feature of alpha- and betacoronaviruses, including HCoV-229E, SARS-CoV, and MHV, is their limited activation of the IFN response (8–13). This limited activation can be explained partially by intracellular membrane rearrangements that might shield dsRNA and other viral components from recognition by host PRRs (14, 15). In addition, coronavirus nsp16 displays 2′-O-methylase activity, which results in 2′-O-methylation of a ribose moiety on the 5′ cap of coronavirus mRNAs, making them indistinguishable from host mRNAs (16). Furthermore, many other coronavirus proteins, such as nsp1, nsp3, the nucleocapsid, and many of the accessory proteins, have been shown to interfere with the IFN response in various ways (reviewed in references 17 and 18).
Interaction between gammacoronaviruses and innate immune responses of their avian hosts is poorly understood. Early studies on gammacoronaviruses in chicken suggest that IBV-induced IFN production is variable and dependent on both virus strain and cell type (19–22). Further, two transcriptional studies on tissues collected after in vivo and in ovo IBV infections found only limited upregulation of ISGs at 1 to 3 days postinfection (23–25). Functional studies using IBV Beaudette showed that it induced cell cycle arrest and apoptosis (26, 27), that IBV interacts with eIF3f (28), and that IBV inhibits protein kinase R activation, thereby maintaining protein synthesis (29). Although these studies provided a number of details on the interactions between IBV and the host cell, most experiments were carried out in Vero cells. This nonavian cell line is one of the very few cell lines in which the IBV-Beaudette strain has been adapted to grow, facilitating in vitro experiments. Vero cells, however, lack the Ifnβ gene, preventing them from mounting a type I IFN response (30, 31) and reducing the value of Vero cells for research on innate immune responses to IBV. In addition, the Beaudette strain is nonpathogenic in vivo with limited replication in host tissues (32), reducing the value of these in vitro studies for translation to in vivo situations. For these reasons, we used pathogenic isolates of IBV to infect primary chicken cells and a chicken cell line, as these isolates are known to infect, spread, and cause clinical disease in vivo.
In the current study, we show that IBV infection leads to a significant induction of Ifnβ transcription through an MDA5-dependent activation of the IFN response, although it is delayed with respect to both virus replication and accumulation of dsRNA. This delayed induction of Ifnβ was further confirmed through RNA fluorescence in situ hybridization (FISH) analysis showing that the accumulation of Ifnβ mRNA is restricted to IBV-infected and not neighboring uninfected cells. Although the time lag between the accumulation of dsRNA and the induction of Ifnβ transcription suggests that IBV interferes with the recognition of dsRNA, we observed that sensing of exogenous (nonself) dsRNA remained functional in IBV-infected cells. Using mutant IBV, we demonstrate that both accessory proteins 3a and 3b are involved in limiting Ifnβ expression, as both 3a and 3b null viruses induced increased Ifnβ expression. Nevertheless, 3a and 3b seem to have differential effects on IFN protein production. Infection with 3a null virus induced lower IFN levels, whereas a 3b null virus increased IFN production compared to that of the parental virus. Altogether, our data suggest that IBV delays but does not prevent detection by MDA5, and that accessory proteins 3a and 3b modulate the IFN response in avian cells. This is the first study addressing immune evasion and interference strategies of IBV in chicken and not in mammalian cells, providing information essential to the further understanding of the pathogenesis of gammacoronaviruses.
MATERIALS AND METHODS
Cells.
Chicken embryonic kidneys were aseptically removed from 17- to 19-day-old chicken embryos (Charles River Laboratories Inc.). A cell suspension was obtained by trypsinization for 30 min at 37°C and filtered through a 100-μm mesh. The resulting chicken embryo kidney (CEK) cells were seeded at 4 × 105 cells/cm2 in 199 medium (Invitrogen) supplemented with 0.5% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PenStrep; Gibco, Invitrogen). Chicken tracheal cells were isolated from 8- to 10-week-old chickens (white leghorn). Tracheas were collected in ice-cold phosphate-buffered saline (PBS), washed, and stripped from adipose tissue. Trachea were filled with a solution of 3.5 U/ml protease type XIV (Sigma), 4 U/ml DNase I (Qiagen), and 1% PenStrep in Eagle's minimum essential medium (EMEM), sealed with clamps, and incubated overnight at 4°C. The next day, cells lining the luminal side of the trachea were flushed with cold EMEM, filtered through a cell strainer, and seeded at 4 × 105 cells/cm2 in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS and 1% PenStrep. The RIG-Iwt, RIG-IKO, MDA5wt, and MDA5KO murine embryonic fibroblasts (MEFs) were provided by S. Akira (33). The MAVSwt and MAVSKO MEFs were provided by Z. J. Chen (34). DF-1, CEC-32, and MEF cells were cultured in DMEM (Gibco, Invitrogen) supplemented with 10% FBS and 1% PenStrep. All cells were incubated in a humidified incubator at 37°C and 5% CO2.
DF-1 Ifnβ-luc reporter cell line.
DF-1 cells were transfected using Fugene (Promega) according to the manufacturer's instructions with a construct expressing firefly luciferase under the control of the bp −110 proximal region of the human IFN-β promoter (35). Stably expressing cells were selected over a period of 3 weeks using Geneticin (500 μg/ml). DF-1 Ifnβ-luc stable cells were cultured in DMEM supplemented with 10% FBS and 1% PenStrep and were not further subcloned.
Viruses.
IBV-M41, IBV-QX, IBV-Italy-O2, Rift Valley fever virus clone 13 (RVFV Cl13), and infectious pancreatic necrosis virus (IPNV) were obtained from Merck Animal Health, Boxmeer, The Netherlands. Sindbis-green fluorescent protein (GFP) was a kind gift from J. Fros (Laboratory of Virology, Wageningen University). IBV Beaudette, strain Beau-R, as well as the generation of the ScAUG3a, ScAUG3b, ScAUG3ab, and ScAUG5ab Beau-R null viruses, have been published previously (36–38). In these mutant IBV viruses, the start codons of the indicated accessory genes were mutated to stop codons. All IBV strains were amplified and titrated on CEK cells. Sindbis-GFP was amplified on baby hamster kidney (BHK) cells and titrated on CEK cells. RVFV Cl13 was amplified and titrated on Vero cells, an African green monkey cell line. IPNV was amplified and titrated on the CHSE-214 Chinook-salmon cell line. IPNV was inactivated by 20 min of UV exposure on a 48W BXT-26-M instrument (Uvitec).
Poly(I·C) stimulation and RNase treatment.
Poly(I·C) sodium was purchased from Sigma, dissolved in nuclease-free water, and stored at −80°C. Poly(I·C) was either directly added to the medium or transfected using Lipofectamine 2000 (Invitrogen) per the manufacturer's instructions. DF-1 cells (3 × 105/well) were cultured in 24-well plates and transfected with 500 ng poly(I·C). RNase treatment of CEK cell culture supernatant was performed by addition of 10 μg/ml RNase A (Invitrogen) before IBV infection or before stimulation with 2 μg/ml poly(I·C).
RNA isolation and cDNA synthesis.
Approximately 8 × 105 CEK cells or 3 × 105 DF-1 cells were lysed in RLT buffer (Qiagen) at various time points after treatment or infection. RLT cell lysis buffer was spiked with 1 ng/sample of luciferase mRNA (Promega) immediately prior to RNA isolation. Luciferase expression later will be used as an external reference gene for normalization during gene expression analysis. Total RNA was isolated using the RNeasy minikit (Qiagen) according to the manufacturer's instructions, including an on-column DNase treatment with RNase-free DNase (Qiagen). Before cDNA synthesis of 0.5 to 1 μg total RNA, a second DNase treatment was performed using amplification-grade DNase I (Invitrogen). The synthesis of cDNA was performed using SuperScript III (Invitrogen) with random primers. cDNA samples were further diluted 1:50 in nuclease-free water before real-time quantitative PCR (RT-qPCR) analysis.
Gene expression analysis.
Real-time quantitative PCR was performed on a Rotor-Gene 6000 (Corbett Research) using brilliant SYBR green quantitative PCR (Stratagene) and primers (39–42) listed in Table 1. Cycle thresholds (CT) and amplification efficiencies were calculated by the Rotor-Gene software (version 1.7). The relative expression ratio of the target gene was calculated using the average reaction efficiency for each primer set and the CT deviation of the sample versus the control at time point 0 h, as described in reference 43. For the calculation of the fold change of IBV total RNA, CT deviation was calculated versus a CT of 30, as no IBV was present in the noninfected cells that were used as the control in all experiments. Because the expression of various housekeeping genes was unstable during virus infections at time points later than 24 h (data not shown), gene expression ratios were normalized using an external reference gene (luciferase).
TABLE 1.
siRNAs and primers used in this study
| Gene product | Category | Orientationa | Sequence (5′–3′) | Accession no. | Reference |
|---|---|---|---|---|---|
| TLR3 | siRNA | S | UCGAAUACUUGGCUUUAAA | NM_001011691 | |
| AS | UUUAAAGCCAAGUAUUCGA | ||||
| ctrl | siRNA | S | AGGUAGUGUAAUCGCCUUG | ||
| AS | CAAGGCGAUUACACUACCU | ||||
| MDA5 | siRNA | S | ACACUGGUAUCAAGUUAUU | GU570144 | |
| AS | AAUAACUUGAUACCAGUGU | ||||
| IFNβ | RQ primer | FW | GCTCTCACCACCACCTTCTC | NM_001024836 | |
| RV | GCTTGCTTCTTGTCCTTGCT | ||||
| IFNα | RQ primer | FW | ATCCTGCTGCTCACGCTCCTTCT | XM_004937096 | 40 |
| RV | GGTGTTGCTGGTGTCCAGGATG | ||||
| IRF3 | RQ primer | FW | CAGTGCTTCTCCAGCACAAA | NM_205372 | |
| RV | TGCATGTGGTATTGCTCGAT | ||||
| IRF1 | RQ primer | FW | CAGGAAGTGGAGGTGGAGAA | NM_205415 | |
| RV | TGGTAGATGTCGTTGGTGCT | ||||
| TLR7 | RQ primer | FW | TTCTGGCCACAGATGTGACC | NM_001011688 | 40 |
| RV | CCTTCAACTTGGCAGTGCAG | ||||
| TLR3 | RQ primer | FW | TCAGTACATTTGTAACACCCCGCC | NM_001011691 | 40 |
| RV | GGCGTCATAATCAAACACTCC | ||||
| MDA5 | RQ primer | FW | TGGAGCTGGGCATCTTTCAG | GU570144 | |
| RV | GTTCCCACGACTCTCAATAACAGT | ||||
| Mx | RQ primer | FW | TTGTCTGGTGTTGCTCTTCCT | GQ390353 | |
| RV | GCTGTATTTCTGTGTTGCGGTA | ||||
| OAS | RQ primer | FW | CACGGCCTCTTCTACGACA | NM_205041 | 41 |
| RV | TGGGCCATACGGTGTAGACT | ||||
| IL-8 | RQ primer | FW | TTGGAAGCCACTTCAGTCAGAC | NM_205498 | 41 |
| RV | GGAGCAGGAGGAATTACCAGTT | ||||
| PKR | RQ primer | FW | CCTCTGCTGGCCTTACTGTCA | NM_204487 | 42 |
| RV | AAGAGAGGCAGAAGGAATAATTTGCC | ||||
| ADAR | RQ primer | FW | TGTTTGTGATGGCTGTTGAG | AF403114 | |
| RV | AGATGTGAAGTCCGTGTTG | ||||
| ISG12 | RQ primer | FW | TAAGGGATGGATGGCGAAG | NM_001002856 | |
| RV | GCAGTATCTTTATTGTTCTCAC | ||||
| MHC-I | RQ primer | FW | CTTCATTGCCTTCGACAAAG | NM_001031338 | 41 |
| RV | GCCACTCCACGCAGGT | ||||
| IFNAR2 | RQ primer | FW | TTCTGTGTGCGGCTTGTTAC | AF082665 | 41 |
| RV | TGTTGGCACAGTTGGATCA | ||||
| IBV-N | RQ primer | FW | GAAGAAAACCAGTCCCAGA | AY851295 | |
| RV | TTACCAGCAACCCACAC | ||||
| Luciferase | RQ primer | FW | TGTTGGGCGCGTTATTTATC | X65316 | |
| RV | AGGCTGCGAAATGTTCATACT |
S, sense; AS, antisense; FW, forward; RV, reverse; RQ primer, primer used in real-time quantitative PCR.
Immunohistochemistry.
CEK cells were seeded on fibronectin-coated glass Biocoat coverslips (BD Biosciences) at a density of 1 × 105 cells/cm2. After incubation at 37°C for 48 h, cells were infected with IBV strain M41 at a multiplicity of infection (MOI) of 1, fixed at different time points with 3.7% paraformaldehyde, and permeabilized using 0.1% Triton X-100. Infected cells were probed with anti-dsRNA antibody (English & Scientific Consulting), and polyclonal chicken serum raised against IBV M41 was obtained from Merck AH. Detection was performed using Alexa 488 goat anti-mouse antibody (Invitrogen) and fluorescein isothiocyanate (FITC)-labeled goat anti-chicken antibody (Kirkegaard and Perry Laboratories). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were imaged using a Zeiss Primo Vert microscope and Axiovision software. Image overlays were made in ImageJ.
RNA FISH.
RNA fluorescence in situ hybridization (FISH) was performed according to previously described protocols (44–46). A set of 40 RNA FISH probes (20 bp), each labeled with one CAL Fluor red 610 fluorophore and targeting chicken Ifnβ (NM_001024836), was designed using the Stellaris probe designer (Biosearch Technologies). The coding sequence of chicken Ifnβ is 601 bp; therefore, to accommodate the optimum number of fluorescent probes (48; explained in reference 44), the 3′-untranslated region was included in the probe design tool. CEK cells were grown on fibronectin-coated coverslips (BD Biosciences) at a density of 2 × 105 cells/cm2. After incubation at 37°C for 48 h, cells were infected with IBV M41 and, at the indicated time points, were fixed in 70% ethanol at 4°C. Hybridization of the probes was performed using the manufacturer's protocol for adherent cells. Imaging was performed using a Roper (Evry, France) spinning disc confocal system on a Nikon Eclipse Ti microscope using a 100× Plan Apo oil immersion objective (numeric aperture, 1.4) and a 491-nm laser line. z stacks were collected with 0.25-μm z intervals. For each channel, maximum z-stack projections were made and processed with ImageJ.
chIFN bioassay.
Bioactive chicken type I interferon (chIFN) was measured using a bioassay based on the CEC-32 quail reporter cell line expressing luciferase under the control of the chicken Mx promoter (47) (kindly provided by Peter Staeheli). Briefly, CEC-32 cells were incubated with serial dilutions of chIFN-containing samples for 6 h, after which luciferase activity was quantified and IFN concentrations calculated using a chIFN standard. To avoid the influence of IBV on the assay, samples were heat inactivated at 56°C for 30 min, which did not influence type I chIFN bioactivity.
Gene silencing.
Short interfering RNAs (siRNAs) targeting chicken Tlr3 and Mda5 were designed by and purchased from Microsynth, Switzerland (sequences are in Table 1). Transfections were performed using siLentFect (Bio-Rad) at a final siRNA concentration of 20 nM. For one well, 160 ng siRNA was combined with 1 μl siLentFect in 100 μl Opti-MEM (Gibco) and incubated for 20 min. The siRNA complexes were added to 2 × 105 DF-1 cells grown in 500 μl medium per well in a 24-well plate. siRNA complexes were left on the cells for 48 h before further experiments were performed.
Statistics.
All statistical analyses were performed in GraphPad Prism 5.0. Quantitative reverse transcription-PCR fold changes first were log transformed and then used for statistical analysis. For all tests, equality of variance was assessed using Bartlett's test. Significant differences (P < 0.01) were determined by a one-way or two-way analysis of variance (ANOVA) (indicated in the figure legend), followed by a Bonferroni post hoc test.
RESULTS
IBV delays the onset of an IFN response during infection of primary chicken cells.
To investigate the kinetics of viral replication and IFN induction upon infection with the avian Gammacoronavirus IBV, we infected primary CEK cells (24) with the IBV M41 strain. To monitor the kinetics of the IFN response in relation to IBV replication, we quantified the transcription of Ifnβ, a set of genes involved in innate immunity, extracellular IFN protein production, virus titers, and IBV RNA in M41-infected CEK cells. In line with previous observations (48), progeny virus was produced after 6 h postinfection (hpi), and virus titers reached a maximum around 24 hpi (Fig. 1A). Total intracellular IBV RNA levels reflected the kinetics of infectious IBV in the supernatant (Fig. 1A), reaching maximum levels around 24 hpi. Ifnβ expression was delayed with respect to the peak of viral replication and remained low until 18 hpi, after which it was strongly upregulated, peaking around 36 hpi (Fig. 1B). IFN protein activity levels were quantified using a chicken IFN-specific Mx-luc cell-based bioassay showing accumulation of IFN from 36 hpi onwards (Fig. 1B). Concomitant with Ifnβ, a subset of genes involved in innate immunity, including Mx, Oas, and Il8, was upregulated, whereas others, such as Tlr7, Adar, Isg12, MHC-I, and Ifnar2, appeared not to be affected or were only marginally affected by IBV infection (Fig. 1C). Pattern recognition receptors Mda5 and Tlr3 and the transcription factor Irf3 also were upregulated (Fig. 1C), which is of interest given the role of these PRRs in virus recognition.
FIG 1.
IBV infection delays Ifnβ upregulation. Chicken embryo kidney (CEK) cells were infected with IBV M41 at an MOI of 0.1. (A) Replication of IBV was quantified by titration in cell culture supernatants of infected cells. In a parallel experiment, intracellular IBV RNA was quantified using RT-qPCR. (B) Ifnβ mRNA levels were determined using RT-qPCR, and IFN protein levels were determined using a chicken IFN-specific mx-luc cell-based bioassay. (C) Expression of genes involved in the antiviral response. All gene expressions were calculated as fold changes relative to uninfected control cells and normalized against an external reference gene (luciferase). For IBV total RNA, fold changes were calculated relative to a CT of 30. Depicted are the results from a representative experiment out of three independent experiments.
The delayed IFN response is independent of the cell type or virus strain.
Ifnβ transcription during infection with coronaviruses such as MHV and SARS-CoV generally is low (9, 10, 12, 13, 49) and was shown to be dependent on cell type and virus strain (50). The delayed induction of Ifnβ transcription observed in IBV M41-infected CEK cells prompted us to investigate whether the induction of Ifnβ would be dependent on the cell type or IBV strain. Epithelial cells isolated from trachea of 10-week-old specific-pathogen-free (SPF) chickens and DF-1 chicken fibroblast cells were infected with IBV M41 or IBV Beaudette (Beau-R) (38). At several time points after infection, Ifnβ levels were monitored by RT-qPCR (Fig. 2A and B). In both cell types, Ifnβ transcription followed the same kinetics as those observed in CEK cells (Fig. 1B), indicating that the induction of Ifnβ by IBV is independent of cell type. To study whether the induction of Ifnβ transcription differs between different strains of IBV, we also infected CEK cells with the QX and ItO2 strains of IBV (Fig. 2C). Although we observed some differences in absolute levels of Ifnβ upregulation induced by QX, ItO2, and M41, the kinetics of Ifnβ transcription were similar, suggesting that the delayed induction of Ifnβ transcription could be considered a general feature of IBV infection in chicken cells.
FIG 2.

Delayed induction of Ifnβ transcription is independent of cell type or IBV strain. (A) Epithelial cells from adult chicken trachea were infected with IBV M41 at an MOI of 0.1. (B) Fibroblast DF-1 cells were infected with IBV Beau-R at an MOI of 0.1. (C) CEK cells were infected with IBV strains M41, QX, and It02 at an MOI of 0.1. Intracellular IBV total RNA (open diamonds) and Ifnβ mRNA (bars) are depicted as fold changes as assessed by RT-qPCR. Gene expression of Ifnβ was calculated as fold changes relative to uninfected control cells and normalized against an external reference gene (luciferase). For IBV total RNA, fold changes were calculated relative to a CT of 30. Error bars indicate standard deviations.
To assess whether CEK and DF-1 cells do have the intrinsic ability to express Ifnβ earlier than 18 h, we stimulated these cells with extracellular poly(I·C), transfected poly(I·C), or the dsRNA virus IPNV. We found that the stimulation of CEK cells with poly(I·C) could induce Ifnβ transcription as early as 1 h after stimulation (Fig. 3A). In DF-1 cells, stimulation with IPNV and transfected poly(I·C), but not poly(I·C), induced Ifnβ already at 4 h (Fig. 3B). The observation that DF-1 cells do not respond to stimulation with extracellular dsRNA is in accordance with previous findings and most likely is due to the lack of surface expression of TLR3 (51). In addition, a 12-h infection of CEK cells with Sindbis, IPNV, or Rift Valley fever virus clone 13 (RVFV Cl13) induced a clear transcription of Ifnβ (Fig. 3C). These results suggest that delayed expression of Ifnβ is a specific feature of IBV infection and not an intrinsic characteristic of chicken cells.
FIG 3.
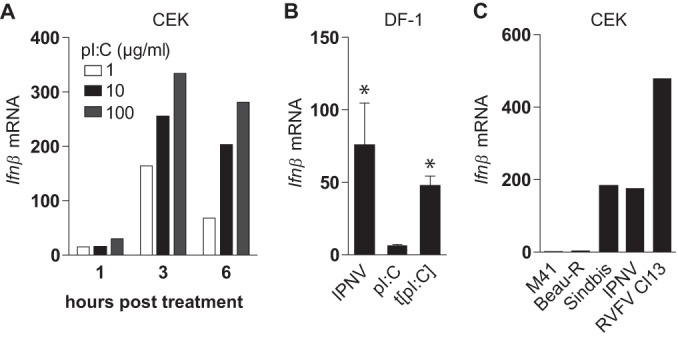
Chicken cells have the intrinsic ability to respond rapidly to dsRNA. (A) CEK cells were seeded in 24-well plates, and 48 h later they were stimulated with extracellular poly(I·C) for the indicated times. (B) DF-1 cells were infected with IPNV, a nonreplicating dsRNA virus, or stimulated with extracellular poly(I·C) (pI:C) or transfected poly(I·C). Four hours later, Ifnβ fold changes were determined by RT-qPCR. Bars represent the means (plus standard deviations) from triplicate wells from a representative experiment. Asterisks indicate significant differences (P < 0.01) with respect to the nonstimulated control as assessed by one-way ANOVA followed by a Bonferroni post hoc test. (C) CEK cells were infected with IBV M41 (MOI, 1), IBV Beau-R (MOI, 1), Sindbis-GFP (MOI, 1), IPNV (MOI, 50), and RVFV Cl13 (MOI, 5). Depicted are Ifnβ fold changes at 12 hpi relative to uninfected control cells as assessed by RT-qPCR.
The intracellular pattern recognition receptor MDA5 is the primary sensor of IBV.
In general, dsRNA has been shown to be the canonical inducer of Ifnβ during infection with alpha- and betacoronaviruses (16, 50). To determine which pattern recognition receptor (PRR) would be involved in sensing (ds)RNA of the Gammacoronavirus IBV, leading to subsequent Ifnβ transcription, we first examined the possibility that IBV-(ds)RNA could be sensed extracellularly by, for example, cell surface receptors. To investigate this, CEK cells were infected with IBV M41 in the presence of RNase A, and Ifnβ expression was analyzed. As a positive control, CEK cells were stimulated with poly(I·C) in the presence or absence of RNase A. The IFN response to poly(I·C) was greatly inhibited by the addition of RNase A, which had no effect on Ifnβ levels induced by infection with IBV M41 (Fig. 4A). These data suggest that Ifnβ upregulation during the late stage (>18 hpi) of IBV infection could be the result of the sensing of IBV-(ds)RNA by an intracellular rather than an extracellular pattern recognition receptor. This is consistent with our observation that IBV infection can be detected by DF-1 cells, which show only a marginal upregulation of Ifnβ transcription in response to extracellular dsRNA (Fig. 3B). In general, dsRNA can be recognized by membrane-bound TLR3 and cytosolic RLRs, such as MDA5 and RIG-I. Genome mining strongly indicates that chickens do not express a RIG-I homologue (52), leaving TLR3 and MDA5 as the two PRRs potentially involved in dsRNA sensing. Silencing of Mda5, but not Tlr3, in DF-1 cells resulted in a 70% decrease in Ifnβ transcription (Fig. 4B). Similar results were obtained with an Ifnβ-luc DF-1 reporter cell line in which silencing of Mda5, but not Tlr3, resulted in a 70% decrease in luciferase activity by the reporter cells (Fig. 4C). Because no antibody against chicken MDA5 currently is available for protein detection, successful knockdown was evaluated using RT-qPCR, demonstrating a silencing efficiency for both Tlr3 and Mda5 of approximately 60% (data not shown). The replication of IBV at the investigated time point was not affected by knockdown of either Tlr3 or Mda5, as measured by both virus titer and intracellular IBV total RNA (Fig. 4D). These results indicate that MDA5 is the primary PRR responsible for sensing Gammacoronavirus IBV-(ds)RNA in chicken cells.
FIG 4.
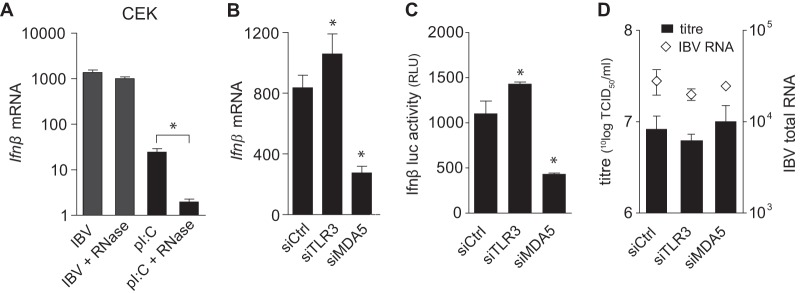
MDA5, not TLR3, is the prime sensor of IBV. (A) CEK cells were infected with IBV M41 for 24 h in the presence or absence of RNase A. Ifnβ expression was analyzed by RT-qPCR. Stimulation with poly(I·C) in the presence or absence of RNase A was included as a positive control. DF-1 cells (B and D) and DF-1 Ifnβ-luc reporter cells (C) were transfected with siRNAs against Tlr3 and Mda5 or a control siRNA, and 48 h later they were infected with IBV M41 (MOI, 0.1). Ifnβ mRNA (B), Ifnβ-luciferase activity (C), and IBV titers and intracellular RNA (D) were analyzed 18 hpi. Bars represent the means (plus standard deviations) from triplicate wells from a representative experiment. Asterisks indicate significant differences (P < 0.01) with respect to the non-RNase A-treated control (A) or to the siRNA control (B and C), as assessed by one-way ANOVA followed by a Bonferroni post hoc test.
Early accumulation of dsRNA in IBV-infected cells does not result in early induction of Ifnβ.
Having assessed that chicken cells can indeed promptly respond to stimulation with dsRNA (Fig. 3) and having identified MDA5 as the primary sensor involved in the detection of IBV (Fig. 4), we investigated whether there would be a temporal difference between IBV-induced accumulation of dsRNA and the upregulation of Ifnβ transcription in CEK cells. Indeed, dsRNA could be detected clearly, even at a low MOI of 0.01, by 12 hpi (Fig. 5A). In contrast, Ifnβ levels at this time point remained low (Fig. 5B) even in cell cultures infected at higher MOIs of 1 or 10 and despite the increased abundance of dsRNA. To further investigate the time lag between early accumulation of dsRNA and late Ifnβ expression, we performed a time course analysis. Foci of dsRNA could be detected as early as 3 h postinfection only in IBV-infected cells (Fig. 5C, inset, 3 hpi), indicating that dsRNA starts accumulating very early in IBV-infected cells but apparently only leads to late (>18 hpi) Ifnβ transcription.
FIG 5.
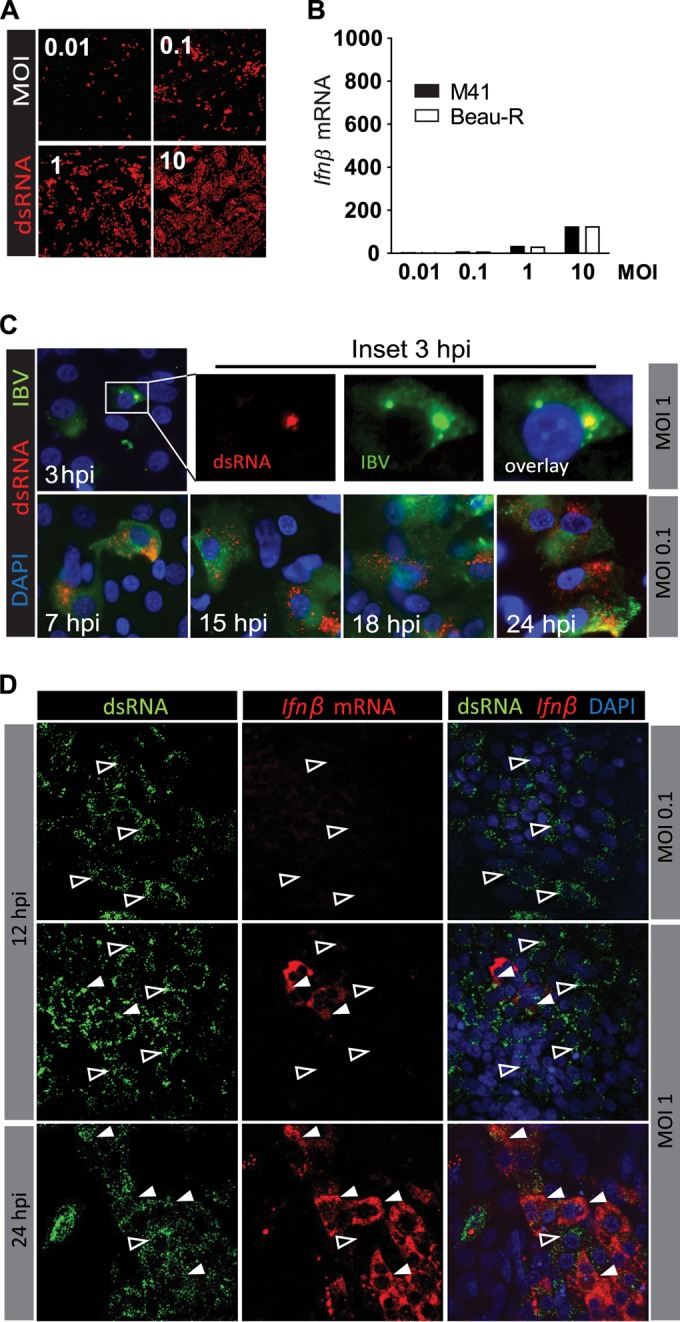
Early accumulation of dsRNA in IBV-infected cells does not result in the early induction of Ifnβ. CEK cells were infected with IBV M41 or IBV Beau-R at the indicated MOIs. At 12 hpi (A), dsRNA was visualized in M41-infected cells using an antibody against dsRNA. (B) Expression of Ifnβ mRNA was analyzed by RT-qPCR. (C) CEK cells were infected with IBV M41, and the accumulation of dsRNA was visualized at the indicated time postinfection. (D) RNA fluorescent in situ hybridization of Ifnβ mRNA in IBV M41-infected CEK cells. Open arrowheads indicate cells that contain dsRNA and no Ifnβ mRNA. Solid white arrowheads indicate cells that contain both dsRNA and Ifnβ mRNA.
Primary CEK cells consist of a heterogeneous mix of cell types. Even at high MOI, IBV M41 infects only ∼70% of the cells, indicating that not all cells are permissive to IBV M41 infection. In order to assess whether the time lag between accumulation of dsRNA and Ifnβ expression could be due to the induction of Ifnβ in bystander rather than IBV-infected cells, we used RNA fluorescent in situ hybridization to visualize Ifnβ mRNA in IBV-infected CEK cell cultures (Fig. 5D). At 12 hpi and low MOI (0.1), with most cells showing clear foci of dsRNA, none of the IBV-infected cells displayed an accumulation of Ifnβ mRNA. At 12 hpi and a higher MOI, a few cells stained positive for Ifnβ mRNA, and only later, at 24 hpi, did most IBV-infected cells also stain positive for Ifnβ mRNA, the kinetics of which closely followed those observed in Fig. 5A. In all cases, the detection of Ifnβ mRNA was restricted to cells that contained dsRNA. Altogether our data show that IBV-infected cells, but not adjacent uninfected cells, upregulate Ifnβ transcription in response to IBV infection. The significant time lag between the accumulation of dsRNA and Ifnβ transcription further suggests the presence of a mechanism adopted by IBV to circumvent the onset of an IFN response.
Accessory proteins 3a and 3b regulate IFN transcription and protein production.
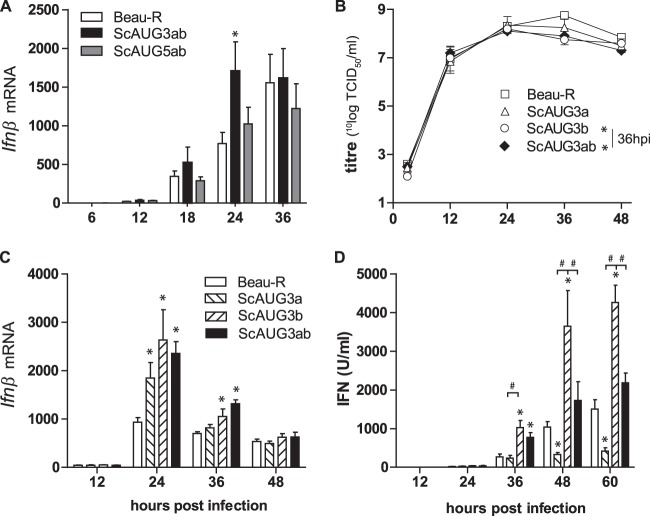
To investigate whether the accessory proteins of IBV play a role in the observed delay in Ifnβ transcription, we infected CEK cells with IBV scAUG3ab and scAUG5ab null viruses and the parental Beau-R virus (scAUG viruses possess a scrambled AUG start codon resulting in transcription but not translation of either open reading frames [ORFs] 3a and 3b or 5a and 5b [36, 37]). Infection for 24 hpi with the scAUG3ab but not the scAUG5ab null virus resulted in increased upregulation of Ifnβ expression (Fig. 6A), indicating that either one or a combination of accessory proteins 3a and 3b play a role in downregulating Ifnβ transcription. The difference in Ifnβ transcription between the scAUG3ab and the parental (Beau-R) virus could not be ascribed to differences in kinetics of virus replication, as all viruses displayed similar growth kinetics until 24 hpi (Fig. 6B). To determine whether 3a, 3b, or both accessory proteins are involved in the observed downregulation of the IFN response, we quantified Ifnβ transcription and IFN protein production in CEK cells infected with scAUG3a, scAUG3b, and scAUG3ab mutant viruses and compared the values to those observed in cells infected with Beau-R (Fig. 6C and D). Infection with all mutant viruses led to increased transcription of Ifnβ compared to that of the Beau-R virus (Fig. 6C), indicating that the presence of either one of the two accessory proteins is sufficient to limit Ifnβ transcription. The kinetics of Ifnβ transcription in response to AUG3a/b differs between Fig. 6A and C. In Fig. 6C there is a significant difference in Ifnβ transcription between AUG3a/b and Beau-R that is absent in Fig. 6A. This difference probably can be attributed to variation in the kinetics of Ifnβ transcription between primary CEK cells isolated from embryos originating from different flocks. Nonetheless, this difference does not affect the conclusion that the knockout of 3a and 3b leads to an increase in the transcription of Ifnβ.
FIG 6.
Accessory proteins 3a and 3b are involved in regulation of IFN transcription and protein production. (A) CEK cells were infected with IBV Beau-R 3a/3b (scAUG3ab) or 5a/5b (scAUG5ab) null virus (MOI, 0.1). Ifnβ levels were determined using RT-qPCR. (B to D) CEK cells were infected with scAUG3a, scAUG3b, or scAUG3ab null IBV virus (MOI, 0.1). In the same cultures, Ifnβ mRNA (B), virus titers (C), and type I IFN protein (D) were quantified. Bars represent the means (plus standard deviations) from triplicate wells from a representative experiment. Significant differences (P < 0.01) relative to the Beau-R virus at the same time point (*) or between the indicated bars (#) as assessed by a two-way ANOVA followed by a Bonferroni post hoc test.
No significant differences in IFN protein production were observed between cells infected with the Beau-R and the scAUG3ab double null virus, except at 36 hpi. However, infection with scAUG3b virus led to an increase in IFN protein levels, whereas infection with the scAUG3a virus led to a decrease in IFN compared to both Beau-R and scAUG3ab double null virus (Fig. 6D). Taken together, these results indicate that accessory proteins 3a and 3b both play a role in the inhibition of Ifnβ transcription but have distinct and opposing effects on protein production. Accessory protein 3b seems to be involved in limiting IFN protein activity, whereas 3a is involved in promoting it.
Signaling of nonself dsRNA remains intact in IBV-infected cells.
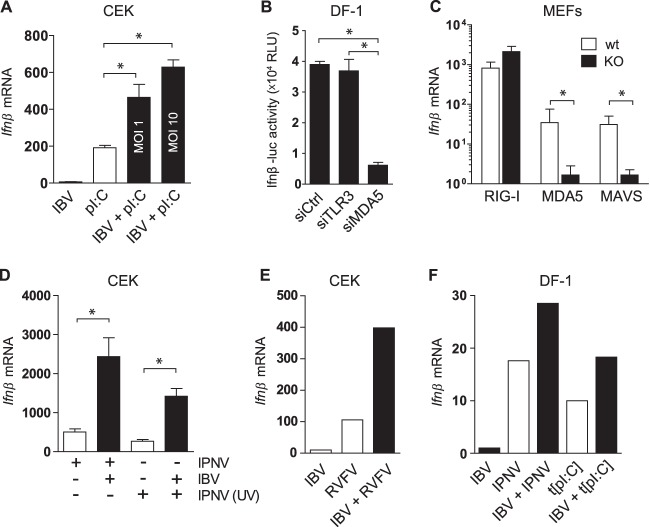
Since IBV showed the intrinsic ability to delay Ifnβ transcription in several cell types (Fig. 1 and 2) even in the presence of high levels of intracellular dsRNA (Fig. 5), we investigated the ability of IBV to interfere with the sensing of nonself dsRNA by TLR3 or MDA5. We infected CEK cells with IBV M41 and subsequently used extracellular poly(I·C) to trigger TLR3 signaling (Fig. 7A). Stimulation with poly(I·C) alone led to a significant increase in Ifnβ transcription, whereas stimulation with poly(I·C) following an infection with IBV led to an enhanced increase in Ifnβ transcription in an MOI-dependent manner. These results indicated that IBV infection does not interfere with TLR3-mediated Ifnβ transcription; on the contrary, IBV infection appears to result in a synergistic activation of the TLR3 pathway triggered by poly(I·C). We next investigated whether IBV infection could interfere with MDA5-mediated transcription of Ifnβ. Although the transfection of poly(I·C) into the intracellular compartment is a commonly used ligand of MDA5, this method induced very little transcription of Ifnβ in primary CEK cells because of its low transfection efficiency (data not shown). As an alternative route to stimulate MDA5 in primary chicken cells, we investigated the use of either RVFV Cl13 or IPNV, which induce Ifnβ transcription in CEK cells (Fig. 3C). RVFV Cl13 is a negative-sense single-stranded RNA virus with a truncated IFN antagonist (53) for which RIG-I, but not MDA5 or TLR3, was previously shown to be the most likely PRR in mammalian cells (54, 55). Since chickens, as opposed to most mammals, do not have a RIG-I homologue, the most likely PRR for RVFV in CEK cells would be MDA5. IPNV is a birnavirus with a dsRNA genome that naturally infects salmonids but has been shown to enter but not replicate in cells of warm-blooded animals (56). To date, the PRR responsible for sensing IPNV dsRNA has not been described. Knockdown experiments in DF-1 Ifnβ-luc reporter cells, using siRNAs against chicken MDA5 or TLR3, revealed that MDA5, but not TLR3, is the prime PRR for IPNV (Fig. 7B). These findings were confirmed using MEFs from knockout mice deficient in the expression of either MDA5, RIG-I, or the downstream adaptor protein MAVS. Here, knockout of either MDA5 or MAVS abrogated sensing of IPNV, as shown by a strong reduction of Ifnβ transcription, whereas the knockout of RIG-I did not (Fig. 7C). Both IPNV and RVFV Cl13 subsequently were used to investigate whether IBV infection could interfere with MDA5-mediated transcription of Ifnβ in CEK cells.
FIG 7.
Signaling of nonself RNA remains intact in IBV-infected cells. (A) CEK cells were infected with IBV M41 for 3 h and stimulated with extracellular poly(I·C) (50 μg/ml) for an additional 3 h, after which Ifnβ transcription was analyzed by RT-qPCR. (B) DF-1 Ifnβ-luc reporter cells were transfected with siRNAs against Tlr3 and Mda5 or a control siRNA, and 48 h later they were infected with IPNV (MOI, 50); at 6 hpi luciferase activity was quantified. (C) Knockout (KO) and wild-type (wt) MEFs were infected with IPNV (MOI, 50) for 8 h. (D) CEK cells were infected with IBV M41 (MOI, 10) for 6 h and superinfected with IPNV or UV-inactivated IPNV (MOI, 50) for an additional 6 h. (E) CEK cells were coinfected with IBV M41 (MOI, 5) and RVFV clone 13 (MOI, 5) and sampled at 6 hpi. (F) DF-1 cells were infected with IBV Beau-R (MOI, 1) for 3 h and superinfected with IPNV (MOI, 50) or transfected with poly(I·C) (t[pI:C]; 500 ng/well) for an additional 4 h. (C to F) Ifnβ levels were quantified by RT-qPCR. Bars represent the means (plus standard deviations) from triplicate wells. Significant differences (P < 0.01) are indicated by an asterisk and were assessed by one-way ANOVA followed by a Bonferroni post hoc test.
Using the quantification of Ifnβ transcription by RT-qPCR as a read out, we showed that IBV infection does not interfere with MDA5-mediated signaling of IPNV (Fig. 7D) or RVFV Cl13 (Fig. 7E); in fact, it had a synergistic effect on Ifnβ transcription, as previously observed for TLR3-mediated signaling (Fig. 7A). Similar results were obtained when stimulating IBV-infected DF-1 cells with IPNV or transfected poly(I·C) (Fig. 7F), indicating that the observed synergistic effect is not specific to CEK cells. Taken together, IBV infection very efficiently prevents sensing of IBV (ds)RNA, but our results indicate that it does not interfere with sensing and downstream signaling of other nonself (ds)RNA ligands.
DISCUSSION
In this study, we performed a comprehensive analysis of the kinetics of IBV infection in avian cells and studied the mechanisms by which IBV interferes with the onset of the type I IFN response. We show that infection with the Gammacoronavirus IBV leads to a considerable activation of the type I IFN response, albeit delayed with respect to the peak of viral replication and accumulation of viral dsRNA. Using an siRNA knockdown approach, we show that MDA5 is the main receptor involved in the induction of Ifnβ expression during IBV infection. We present evidence that IBV accessory proteins 3a and 3b play a role in the modulation of the delayed IFN response by regulating interferon production at both the transcriptional and translational level. In addition, we show that although IBV alone effectively prevents Ifnβ induction in IBV-infected cells, it does not block Ifnβ induction upon stimulation of IBV-infected cells with other RIG-I, MDA5, or TLR3 ligands. To our knowledge, this study provides the most comprehensive analysis of the interplay between a Gammacoronavirus and the avian type I IFN response.
Much of our current knowledge about the interaction of coronaviruses with the innate immune response (reviewed in reference 57) comes from studies in mice and mouse cells using mouse hepatitis virus (MHV). MHV activated IFN production only in specific cell types, and an efficient IFN response was mounted only by plasmacytoid dendritic cells (58), bone marrow-derived macrophages (10, 59), and oligodendrocytes (10). In a recent study on SARS-CoV and MERS-CoV in an epithelial lung cell line, ISGs started to be upregulated at 12 hpi (60), when virus titers already were reaching their maximum. The kinetics of IFN response observed in our study are in line with the aforementioned studies; however, it must be noted that in most cell types, infection with alpha- or betacoronaviruses induced very little, if any, Ifnβ transcription (8–13, 49). This suggests that all of the coronaviruses are able to modulate the activation of the type I IFN response.
We found that IBV infection is detected by various chicken cell types, but until now it was unknown which PRR was involved. MHV has been shown to be detected by MDA5 and not RIG-I or TLR3 in brain macrophages (50), by both MDA5 and RIG-I in an oligodendrocyte-derived cell line (10), and by TLR7 in plasmacytoid dendritic cells (58). The analysis of the chicken genome suggests that chicken lack a RIG-I homologue (52), and basal expression of Tlr7 was found to be very low in CEK cells (data not shown). Therefore, we silenced the remaining candidate RNA sensors MDA5 and TLR3 and were able to show that MDA5, but not TLR3, is involved in the sensing of IBV. The silencing of Mda5 did not lead to an increase in the replication of IBV, suggesting that IBV developed strategies to counteract the activated IFN response.
We recently reported membrane rearrangements in chicken cells infected with IBV (48), similar to those found in cells infected with betacoronaviruses. In theory, the formation of intracellular membrane rearrangements might partly explain the discrepancy observed in the kinetics of dsRNA accumulation and Ifnβ upregulation. Indeed, for SARS-CoV it has been shown that virus-induced double membrane vesicles (DMVs) contain dsRNA (14), suggesting that coronaviruses exploit membrane structures to shield dsRNA from recognition by host PRRs (61). However, the kinetics of Ifnβ transcription were not investigated in these studies. The presence of coronavirus-induced DMVs has been demonstrated as early as 2 hpi in SARS-CoV-infected cells (14). Although we did not demonstrate the presence of DMVs in IBV-infected chicken cells at time points earlier than 7 hpi (48), it is likely that DMVs also are present at earlier time points. As such, the timing of DMV formation in coronavirus-infected cells suggests that membrane rearrangements play a role in the delayed activation of the IFN response by shielding dsRNA from cellular PRRs.
In addition to membrane rearrangements, coronavirus-encoded proteins, including those encoded by numerous accessory genes, have been shown to interfere with the type I IFN response pathway (reviewed in references 17 and 18). To investigate the possible role of IBV accessory proteins in the regulation of the IFN response, we made use of our previously constructed mutant IBV Beau-R viruses that do not express either one or more of the four accessory proteins 3a, 3b, 5a, and 5b. Previously we have demonstrated that the accessory genes of IBV are not essential for replication (36, 37). In the present study, we show that infection of CEK cells with 3a or 3b null virus, as well as a 3a/3b double null virus, led to increased Ifnβ transcription compared to that of Beau-R. Because the kinetics of Ifnβ transcription of 3a, 3b, and 3a/3b null viruses are comparable to those of the parental virus, we conclude that 3a and 3b probably are not responsible for the delay in Ifnβ transcription, suggesting that IBV utilizes additional strategies to delay the transcription of Ifnβ. Apart from their effect on Ifnβ transcription, 3a and 3b seem to have opposing effects on IFN protein production by IBV-infected cells. Infection with the 3b null virus resulted in increased IFN production, whereas infection with the 3a null virus resulted in reduced IFN levels compared to those of the Beau-R virus. Together with the observation that IFN production induced by the 3a/3b double null virus is comparable to that induced by Beau-R virus, our data suggest that accessory proteins 3a and 3b antagonize each other to tightly regulate IFN production (Fig. 6B).
Using the eukaryotic linear motif server (62), we identified a protein phosphatase 1 (PP1)-binding 17KISF20 domain in the IBV 3b protein sequence. The canonical PP1-binding motif is [R/K][V/I/L]X[F/W], in which X can be any amino acid except proline (63). Interestingly, Alphacoronavirus transmissible gastroenteritis coronavirus accessory protein 7 (TGEV-7) has been shown to bind PP1 via a binding motif similar to that found in IBV 3b (64). Similar to IBV scAUG3b, infection with TGEV-Δ7 led to increased mRNA and protein levels of IFN-β (65). The fact that both TGEV-7 and IBV 3b contain a PP1 binding domain indicates that interaction with PP1 is a common strategy of coronaviruses to inhibit the host innate immune response. The mechanism by which interaction of coronavirus accessory proteins with PP1 counteracts the innate immune response still needs to be determined. One clue might come from the PP1-binding domain of measles virus V, which was recently shown to be essential for inhibition of MDA5 signaling (66, 67). Measles V protein binds PP1 and inhibits dephosphorylation of MDA5, which is required for activation and subsequent signaling by MDA5. Motif analysis for IBV 3a protein did not reveal the presence of relevant motives that might explain the observed activity of 3a on IFN regulation. We conclude that both accessory proteins 3a and 3b limit Ifnβ transcription but have distinct and opposing effects on protein production. While 3a seems to promote IFN production, 3b seems to be involved in limiting IFN protein production, possibly through a mechanism similar to that described for protein 7 of TGEV. The fact that IBV 3a and 3b have opposing roles in regulating IFN production indicates that CoVs tightly regulate IFN production to balance their own survival with that of the host. This hypothesis is supported by the observation that field isolates lacking 3a and 3b display reduced virulence in vitro as well as in vivo (68). Elucidation of the exact mechanisms of action of 3a and 3b will be the subject of further investigation.
To investigate whether IBV interferes with a general sensing of (ds)RNA ligands or downstream signaling that leads to Ifnβ transcription, we stimulated IBV-infected cells with TLR3, RIG-I, and MDA5 ligands. Surprisingly, we found that infection with IBV did not reduce Ifnβ transcription but rather increased Ifnβ levels upon stimulation with these PRR ligands. Similar to IBV, MHV has been shown to be unable to inhibit expression of Ifnβ induced by either transfected poly(I·C) or Sendai virus (69, 70), but in these studies no synergistic effect was observed. Currently, we can only speculate about the cause of this synergistic effect. It appears that IBV infection arms the Ifnβ induction pathway without actually triggering it, possibly by enhancing the activity of one or more components of the pathway leading to Ifnβ upregulation. One possibility is that IBV proteins interact with host proteins that regulate this pathway through ubiquitination and phosphorylation (reviewed in reference 71). The fact that stimulation with either TLR3 or MDA5 ligands resulted in exacerbated transcription of Ifnβ indicates that IBV influences a component which is downstream of both MDA5 and TLR3.
Taken together, our study provides the first comprehensive analysis of host-virus interactions of a Gammacoronavirus with the avian innate immune response. We show that the Gammacoronavirus IBV induces activation of the type I IFN response in primary chicken renal cells, tracheal epithelial cells, and a chicken cell line. We show that the activation of the IFN response is dependent on MDA5 but is delayed with respect to the peak of virus replication. We demonstrate that Ifnβ transcription is restricted to IBV-infected, dsRNA-containing cells and provide evidence that accessory proteins 3a and 3b of IBV are involved in regulating transcription as well as protein production of type I IFN.
ACKNOWLEDGMENTS
We thank Peter Staeheli of the University of Freiburg for providing the chicken interferon reporter cell line and chIFN standard and Marleen Scheer and Lieke Golbach from the Cell Biology and Immunology Group of Wageningen University for technical assistance in the RNA silencing experiment and for assistance in microscopy and image processing, respectively. Gorben Pijlman from the Virology Group of Wageningen University and Marjolein Kikkert of the Molecular Virology Laboratory of Leiden University Medical Center are gratefully acknowledged for their critical discussions on the manuscript.
This work was financially supported by MSD Animal Health, Bioprocess Technology & Support, Boxmeer, The Netherlands. H.J.M. and P.B. were supported by The Pirbright Institute and the Biotechnology and Biological Sciences Research Council (BBSRC).
REFERENCES
- 1.Cavanagh D. 2007. Coronavirus avian infectious bronchitis virus. Vet Res 38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 2.Thiel V. 2007. Coronaviruses: molecular and cellular biology. Caister Academic Press, Norfolk, UK. [Google Scholar]
- 3.Cavanagh D. 2005. Coronaviruses in poultry and other birds. Avian Pathol 34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- 4.Cook JK, Jackwood M, Jones RC. 2012. The long view: 40 years of infectious bronchitis research. Avian Pathol 41:239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- 5.Jones RC. 2010. Viral respiratory diseases (ILT, aMPV infections, IB): are they ever under control? Br Poult Sci 51:1–11. doi: 10.1080/00071660903541378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saif YM, Barnes HJ. 2008. Diseases of poultry, 12th ed.Blackwell Publishing Professional, Ames, IA. [Google Scholar]
- 7.Barbalat R, Ewald SE, Mouchess ML, Barton GM. 2011. Nucleic acid recognition by the innate immune system. Annu Rev Immunol 29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 8.Rose KM, Elliott R, Martinez-Sobrido L, Garcia-Sastre A, Weiss SR. 2010. Murine coronavirus delays expression of a subset of interferon-stimulated genes. J Virol 84:5656–5669. doi: 10.1128/JVI.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devaraj SG, Wang N, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Liu Y, Zhang X. 2010. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J Virol 84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kindler E, Jonsdottir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, Geffers R, Fouchier RA, Drosten C, Muller MA, Dijkman R, Thiel V. 2013. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 4:e00611-12. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao L, Rose KM, Elliott R, Van Rooijen N, Weiss SR. 2011. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J Virol 85:10058–10068. doi: 10.1128/JVI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth-Cross JK, Martinez-Sobrido L, Scott EP, Garcia-Sastre A, Weiss SR. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J Virol 81:7189–7199. doi: 10.1128/JVI.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol 76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong Y, Tan YW, Liu DX. 2012. Recent progress in studies of arterivirus- and coronavirus-host interactions. Viruses 4:980–1010. doi: 10.3390/v4060980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu DX, Fung TS, Chong KK-L, Shukla A, Hilgenfeld R. 2014. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res 109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes HC, Darbyshire JH. 1978. Induction of chicken interferon by avian infectious bronchitis virus. Res Vet Sci 25:178–181. [PubMed] [Google Scholar]
- 20.Lomniczi B. 1974. Interferon induction by different strains of infectious bronchitis virus. Acta Vet Acad Sci Hung 24:199–204. [PubMed] [Google Scholar]
- 21.Otsuki K, Nakamura T, Kawaoka Y, Tsubokura M. 1988. Interferon induction by several strains of avian infectious bronchitis virus, a coronavirus, in chickens. Acta Virol 32:55–59. [PubMed] [Google Scholar]
- 22.Otsuki K, Nakamura T, Kubota N, Kawaoka Y, Tsubokura M. 1987. Comparison of two strains of avian infectious bronchitis virus for their interferon induction, viral growth and development of virus-neutralizing antibody in experimentally infected chickens. Vet Microbiol 15:31–40. doi: 10.1016/0378-1135(87)90126-X. [DOI] [PubMed] [Google Scholar]
- 23.Dar A, Munir S, Vishwanathan S, Manuja A, Griebel P, Tikoo S, Townsend H, Potter A, Kapur V, Babiuk LA. 2005. Transcriptional analysis of avian embryonic tissues following infection with avian infectious bronchitis virus. Virus Res 110:41–55. doi: 10.1016/j.virusres.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Rosa AJ, Oliverira HN, Rosa GJ, Guo X, Travnicek M, Girshick T. 2006. Transcriptome of local innate and adaptive immunity during early phase of infectious bronchitis viral infection. Viral Immunol 19:768–774. doi: 10.1089/vim.2006.19.768. [DOI] [PubMed] [Google Scholar]
- 25.Cong F, Liu X, Han Z, Shao Y, Kong X, Liu S. 2013. Transcriptome analysis of chicken kidney tissues following coronavirus avian infectious bronchitis virus infection. BMC Genomics 14:743. doi: 10.1186/1471-2164-14-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li FQ, Tam JP, Liu DX. 2007. Cell cycle arrest and apoptosis induced by the coronavirus infectious bronchitis virus in the absence of p53. Virology 365:435–445. doi: 10.1016/j.virol.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, Xu HY, Liu DX. 2001. Induction of caspase-dependent apoptosis in cultured cells by the avian coronavirus infectious bronchitis virus. J Virol 75:6402–6409. doi: 10.1128/JVI.75.14.6402-6409.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao H, Xu LH, Yamada Y, Liu DX. 2008. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS One 3:e1494. doi: 10.1371/journal.pone.0001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Liao Y, Yap PL, Png KJ, Tam JP, Liu DX. 2009. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J Virol 83:12462–12472. doi: 10.1128/JVI.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emeny JM, Morgan MJ. 1979. Regulation of the interferon system–evidence that vero cells have a genetic defect in interferon-production. J Gen Virol 43:247–252. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- 31.Mosca JD, Pitha PM. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol Cell Biol 6:2279–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodgson T, Casais R, Dove B, Britton P, Cavanagh D. 2004. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J Virol 78:13804–13811. doi: 10.1128/JVI.78.24.13804-13811.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhoj VG, Sun Q, Bhoj EJ, Somers C, Chen X, Torres JP, Mejias A, Gomez AM, Jafri H, Ramilo O, Chen ZJ. 2008. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc Natl Acad Sci U S A 105:14046–14051. doi: 10.1073/pnas.0804717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 36.Hodgson T, Britton P, Cavanagh D. 2006. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J Virol 80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casais R, Davies M, Cavanagh D, Britton P. 2005. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J Virol 79:8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casais R, Thiel V, Siddell SG, Cavanagh D, Britton P. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J Virol 75:12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liniger M, Summerfield A, Zimmer G, McCullough KC, Ruggli N. 2012. Chicken cells sense influenza A virus infection through MDA5 and CARDIF signaling involving LGP2. J Virol 86:705–717. doi: 10.1128/JVI.00742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villanueva AI, Kulkarni RR, Sharif S. 2011. Synthetic double-stranded RNA oligonucleotides are immunostimulatory for chicken spleen cells. Dev Comp Immunol 35:28–34. doi: 10.1016/j.dci.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li YP, Handberg KJ, Juul-Madsen HR, Zhang MF, Jorgensen PH. 2007. Transcriptional profiles of chicken embryo cell cultures following infection with infectious bursal disease virus. Arch Virol 152:463–478. doi: 10.1007/s00705-006-0878-9. [DOI] [PubMed] [Google Scholar]
- 42.Daviet S, Van Borm S, Habyarimana A, Ahanda M-LE, Morin V, Oudin A, Van Den Berg T, Zoorob R. 2009. Induction of Mx and PKR failed to protect chickens from H5N1 infection. Viral Immunol 22:467–472. doi: 10.1089/vim.2009.0053. [DOI] [PubMed] [Google Scholar]
- 43.Forlenza M, Kaiser T, Savelkoul HF, Wiegertjes GF. 2012. The use of real-time quantitative PCR for the analysis of cytokine mRNA levels. Methods Mol Biol 820:7–23. doi: 10.1007/978-1-61779-439-1_2. [DOI] [PubMed] [Google Scholar]
- 44.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raj A, Tyagi S, Nils GW. 2010. Detection of individual Endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol 472:365–386. doi: 10.1016/S0076-6879(10)72004-8. [DOI] [PubMed] [Google Scholar]
- 46.Femino AM, Fay FS, Fogarty K, Singer RH. 1998. Visualization of single RNA transcripts in situ. Science 280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz H, Harlin O, Ohnemus A, Kaspers B, Staeheli P. 2004. Synthesis of IFN-beta by virus-infected chicken embryo cells demonstrated with specific antisera and a new bioassay. J Interferon Cytokine Res 24:179–184. doi: 10.1089/107999004322917025. [DOI] [PubMed] [Google Scholar]
- 48.Maier HJ, Hawes PC, Cottam EM, Mantell J, Verkade P, Monaghan P, Wileman T, Britton P. 2013. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio 4:e00801-13. doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, Peters CJ, Tseng CT. 2010. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One 5:e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karpala AJ, Lowenthal JW, Bean AG. 2008. Activation of the TLR3 pathway regulates IFNbeta production in chickens. Dev Comp Immunol 32:435–444. doi: 10.1016/j.dci.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Barber MRW, Aldridge JR, Webster RG, Magor KE. 2010. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A 107:5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cameron MJ, Kelvin AA, Leon AJ, Cameron CM, Ran L, Xu L, Chu YK, Danesh A, Fang Y, Li Q, Anderson A, Couch RC, Paquette SG, Fomukong NG, Kistner O, Lauchart M, Rowe T, Harrod KS, Jonsson CB, Kelvin DJ. 2012. Lack of innate interferon responses during SARS coronavirus infection in a vaccination and reinfection ferret model. PLoS One 7:e45842. doi: 10.1371/journal.pone.0045842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Habjan M, Andersson I, Klingström J, Schümann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Mühlberger E, Mirazimi A, Weber F. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ermler ME, Yerukhim E, Schriewer J, Schattgen S, Traylor Z, Wespiser AR, Caffrey DR, Chen ZJ, King CH, Gale M, Colonna M, Fitzgerald KA, Buller RML, Hise AG. 2013. RNA helicase signaling is critical for type I interferon production and protection against Rift Valley fever virus during mucosal challenge. J Virol 87:4846–4860. doi: 10.1128/JVI.01997-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orpetveit I, Kuntziger T, Sindre H, Rimstad E, Dannevig BH. 2012. Infectious pancreatic necrosis virus (IPNV) from salmonid fish enters, but does not replicate in, mammalian cells. Virol J 9:228. doi: 10.1186/1743-422X-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kindler E, Thiel V. 2014. To sense or not to sense viral RNA: essentials of coronavirus innate immune evasion. Curr Opin Microbiol 20:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cervantes-Barragan L, Zust R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, Ludewig B. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou H, Zhao J, Perlman S. 2010. Autocrine interferon priming in macrophages but not dendritic cells results in enhanced cytokine and chemokine production after coronavirus infection. mBio 1:e00219-10. doi: 10.1128/mBio.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Menachery VD, Eisfeld AJ, Schafer A, Josset L, Sims AC, Proll S, Fan S, Li C, Neumann G, Tilton SC, Chang J, Gralinski LE, Long C, Green R, Williams CM, Weiss J, Matzke MM, Webb-Robertson BJ, Schepmoes AA, Shukla AK, Metz TO, Smith RD, Waters KM, Katze MG, Kawaoka Y, Baric RS. 2014. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 5:e01174–01114. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hagemeijer MC, Vonk AM, Monastyrska I, Rottier PJ, de Haan CA. 2012. Visualizing coronavirus RNA synthesis in time by using click chemistry. J Virol 86:5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Speck T, Kruger D, Grebnev G, Kuban M, Strumillo M, Uyar B, Budd A, Altenberg B, Seiler M, Chemes LB, Glavina J, Sanchez IE, Diella F, Gibson TJ. 2014. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res 42:D259–D266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M. 2003. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J Biol Chem 278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- 64.Cruz J, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, Zuniga S. 2011. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cruz JL, Becares M, Sola I, Oliveros JC, Enjuanes L, Zuniga S. 2013. Alphacoronavirus protein 7 modulates host innate immune response. J Virol 87:9754–9767. doi: 10.1128/JVI.01032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mesman AW, Zijlstra-Willems EM, Kaptein TM, de Swart RL, Davis ME, Ludlow M, Duprex WP, Gack MU, Gringhuis SI, Geijtenbeek TBH. 2014. Measles virus suppresses RIG-I-like receptor activation in dendritic cells via DC-SIGN-mediated inhibition of PP1 phosphatases. Cell Host Microbe 16:31–42. doi: 10.1016/j.chom.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davis ME, Wang MK, Rennick LJ, Full F, Gableske S, Mesman Annelies W, Gringhuis SI, Geijtenbeek TBH, Duprex WP, Gack MU. 2014. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell Host Microbe 16:19–30. doi: 10.1016/j.chom.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mardani K, Noormohammadi AH, Hooper P, Ignjatovic J, Browning GF. 2008. Infectious bronchitis viruses with a novel genomic organization. J Virol 82:2013–2024. doi: 10.1128/JVI.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou H, Perlman S. 2007. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J Virol 81:568–574. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Versteeg GA, Bredenbeek PJ, van den Worm SH, Spaan WJ. 2007. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gack MU. 2014. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J Virol 88:5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]