ABSTRACT
Opportunistic infection of oligodendrocytes by human JC polyomavirus may result in the development of progressive multifocal encephalopathy in immunocompromised individuals. Neurotropic JC virus generally harbors reorganized noncoding control region (NCCR) DNA interspersed on the viral genome between early and late coding genes. By applying 454 sequencing on NCCR DNA amplified from body fluid samples (urine, plasma, and cerebrospinal fluid [CSF]) from 19 progressive multifocal leukoencephalopathy (PML) patients, we attempted to reveal the composition of the JC polyomavirus population (the quasispecies, i.e., the whole of the consensus population and minor viral variants) contained in different body compartments and to better understand intrapatient viral dissemination. Our data demonstrate that in the CSF of PML patients, the JC viral population is often a complex mixture composed of multiple viral variants that contribute to the quasispecies. In contrast, urinary JC virus highly resembled the archetype virus, and urine most often did not contain minor viral variants. It also appeared that archetype JC virus could sporadically be identified in PML patient brain, although selection of rearranged JC virus DNA was favored. Comparison of the quasispecies from different body compartments within a given patient suggested a strong correlation between the viral population in plasma and CSF, whereas the viral population shed in urine appeared to be unrelated. In conclusion, it is shown that the representation of viral DNA in the CSF following the high-level DNA replication in the brain underlying PML has hitherto been much underestimated. Our data also underscore that the hematogenous route might play a pivotal role in viral dissemination from or toward the brain.
IMPORTANCE For the first time, the JC polyomavirus population contained in different body compartments of patients diagnosed with progressive multifocal encephalopathy has been studied by deep sequencing. Two main findings came out of this work. First, it became apparent that the complexity of the viral population associated with PML has been highly underestimated so far, suggestive of a highly dynamic process of reorganization of the noncoding control region of JC polyomavirus in vivo, mainly in CSF and blood. Second, evidence showing viral dissemination from and/or toward the brain via the hematogenous route was provided, confirming a hypothesis that was recently put forward in the field.
INTRODUCTION
JC polyomavirus (JCPyV) is the causative agent for progressive multifocal leukoencephalopathy (PML), a neurological disorder resulting from lytic infection of oligodendrocytes that may develop in immunosuppressed individuals (e.g., HIV-infected patients) or under therapy-induced immune suppression (1). Like other human polyomaviruses, JC virus is a DNA virus with an ∼5.13-kb double-stranded, circular, supercoiled genome that encompasses early and late genes (2). Small t antigen (stAg) and large T antigen (LTAg) arise from the same primary transcript and are proteins that are expressed early in the viral cycle and that regulate viral replication and transcription (3–5). The viral capsid proteins (VP1, VP2, and VP3) as well as agnoprotein, another regulatory protein, are expressed only during the late stage of the viral cycle. Expression of the viral genes is driven by a bidirectional noncoding control region (NCCR) that lies interspersed between the coding parts of the early and late genes (2). Besides the viral origin of replication, the NCCR harbors multiple DNA sequence motifs upon which host cell-specific transcription factors act and, hence, is key in regulating viral transcription in cells permissive for JCPyV infection (6, 7).
Infection with JC virus is a prerequisite for PML development. The majority of humans have experienced JC virus infection (5, 8), often already in childhood, after which the virus can persist asymptomatically in kidney epithelial cells, urinary tract cells, and lymphoid organs (5). Several reports have also suggested that brain tissue is a site for JCPyV latency (9, 10). About one-quarter of the human population sheds viral DNA in urine, referred to as the DNA of archetype virus, for which the NCCR is well-defined by conserved DNA sequence blocks (domains A to F) (11–13) that can sporadically display modest genomic variations and can exist as a mixture of naturally occurring variants (14).
The onset of PML is caused by lytic infection of oligodendrocytes by JC virus and can often be diagnosed by magnetic resonance imaging (MRI) and detection of viral DNA in cerebrospinal fluid (CSF) during disease progression (1, 15). Whereas in PML cases urinary JCPyV resembles archetype virus, a hallmark of JC virus DNA isolated from PML patient brain or CSF is the presence of multiple, often complex, genomic rearrangements in the NCCR that are hypervariable between patients and are likely to be derived from the archetype by deletion and duplication events (11, 12, 16, 17). In vitro cell studies indicated that such PML-related rearrangements may increase early gene transcription and viral replication and thus mediate cellular tropism and neuropathogenicity (18, 19). Whether rearranged JC virus is required for lytic infection of glial cells or, alternatively, becomes selected as it adapts to its cellular environment in cells previously infected with archetype virus is still a matter of debate.
JC virus has been suggested to disseminate from the initial site of infection via the hematogenous route (5). Besides cell-free JC virus (20), infected B lymphocytes have been proposed to be a major carrier enabling JC virus to transmigrate the blood-brain barrier (BBB) (21–23). The inherent susceptibility of these cells to genomic rearrangements may also contribute to the neurotropic transformation of JC virus (22, 24). Unlike blood-borne virus, urine in which JC virus has been shed likely acts as an important transmission route within and between human populations (25, 26), but whether it also plays a role in viral dissemination between body compartments remains poorly understood.
In this study, we have analyzed the JC virus noncoding control region DNA amplified from different body fluids of PML patients. By applying deep sequencing (454 pyrosequencing), we attempted to identify a representative part of the viral population (the quasispecies) contained in different body reservoirs of PML patients to better understand the nature and genomic diversity of JC virus associated with PML and to gain insight into viral dissemination between body compartments. In addition, we wanted to further investigate the hypothesis, derived from previous work on healthy subjects, that reorganized minor viral variants present in urine might be associated with an increased risk for PML development (14).
MATERIALS AND METHODS
Specimen collection and JC virus load determination.
Body fluid samples (CSF, plasma/serum, and urine) were collected from 19 PML patients at the Laboratory of Molecular Medicine and Neuroscience, NINDS, as part of a Clinical Laboratory Improvement Amendments (CLIA) activity, using validated and certified assays, through the Centers for Medicare and Medicaid Services (CMS), U.S. Department of Health and Human Services (HHS), and were stored at −80°C until further processing. The research conducted by the NINDS was reviewed by the Office of Human Subjects Research Protections (OHSRP), and the research is exempt under 45 CFR 46 101(b) (4) from all 45 CFR part 46 requirements. All PML patients were diagnosed according to their clinical history, MRI evidence of PML lesions, and the presence of viral DNA in their CSF.
For one patient, one CSF and four urine samples were available, while for three other patients, one CSF and one or two plasma/serum samples had been collected. For eight patients, a single CSF sample was available, and for six patients, a single urine sample was available. From one PML patient (Table 1, patient 5) three consecutive CSF samples had been collected over ∼2 months. The JC virus loads from all samples were determined by species-specific quantitative PCR according to a method described earlier (27) and are given in Table 1. Also, the gender of the PML patients as well as their clinical background is indicated in Table 1.
TABLE 1.
Characteristics of PML patients and PML patient samples used in this study
| Patient no. | Gendera | Clinical backgroundb | JC virus load(s) (no. of copies/ml) |
||
|---|---|---|---|---|---|
| Urine | Plasma | CSF | |||
| 1 | M | RRMS | 5,562,426, 179,375,000, 172,000,000, >225,000,000 | 69,718 | |
| 2 | F | Liver transplant | 51,006, 26,912 | 278,000,000 | |
| 3 | F | RRMS | 10,812, 4,063 | 96,438 | |
| 4 | M | RRMS | 5,035 | 2,238,800 | |
| 5 | F | RRMS | 558,967, 756,637, 12,728 | ||
| 6 | M | RRMS | 29,584 | ||
| 7 | F | MS | 15,912 | ||
| 8 | F | Lymphoma | 100,046 | ||
| 9 | M | RRMS | 1,627,025 | ||
| 10 | F | MS | 26,633 | ||
| 11 | F | MS | 181,566 | ||
| 12 | M | MS | 656,020 | ||
| 13 | F | RRMS | 2,501,045 | ||
| 14 | F | CLL | 72,734 | ||
| 15 | M | MS | 43,694,938 | ||
| 16 | F | MS | 94,545,825 | ||
| 17 | M | MS | 189,945,812 | ||
| 18 | M | RRMS | 7,744,225 | ||
| 19 | F | RRMS | 2,804,051 | ||
M, male; F, female.
MS, Multiple sclerosis; RRMS, relapsing-remitting multiple sclerosis; CLL, chronic lymphocytic leukemia.
A DNA template solution containing archetype JC virus NCCR plasmid DNA was spiked before PCR amplification with 1% and 2% of a similar plasmid DNA harboring JC virus with a deleted NCCR sequence, as described earlier (14). These served as control samples.
DNA extraction and amplification of JC virus noncoding control region DNA.
Viral DNA was extracted and concentrated from CSF, plasma, and urine samples using NucliSENS easyMAG system reagents (bioMérieux) and eluted in a 25-μl final volume. Sample volumes varied between 0.3 ml and 1 ml. To minimize PCR errors, JC virus NCCR DNA was amplified in triplicate from the viral DNA using Phusion high-fidelity master mix (2×; New England BioLabs) and sequence-specific Fusion primers, as described earlier (14). For plasma samples (n = 5; viral load range, 4,063 to 51,006 copies/ml; mean viral load, 19,566 copies/ml), 8 μl of extracted DNA was used per PCR replicate, while for urine samples (n = 10; viral load range, 72,734 to >225,000,000 copies/ml; mean viral load, 92,074,501 copies/ml) and CSF samples (n = 15; viral load range, 12,728 to 278,000,000 copies/ml; mean viral load, 19,124,741 copies/ml) less DNA was used as the template. The number of viral copies per sample used as the template for PCR (the input copy number) is given in Table 3. After amplification, triplicate PCR amplicons were pooled and purified, and the DNA integrity and concentration were determined as described earlier (14).
TABLE 3.
Number of NCCR sequences per sample retrieved by 454 sequencing
| Patient no. | No. of reads | JCPyV load (no. of copies/ml) | Input copy no.a | Sampling sizeb |
|---|---|---|---|---|
| 1 | 2,715 | 69,718 | 50,196 | 5.41 |
| 1 | 3,074 | 5,562,426 | 2,669,964 | 0.12 |
| 1 | 3,337 | 179,375,000 | 43,050,000 | 0.01 |
| 1 | 3,500 | 172,000,000 | 41,280,000 | 0.01 |
| 1 | 2,588 | >225,000,000 | >54,000,000 | >0.01 |
| 2 | 2,512 | 278,000,000 | 33,360,000 | 0.01 |
| 2 | 1,511 | 51,006 | 14,690 | 10.29 |
| 2 | 1,390 | 26,912 | 7,750 | 17.94 |
| 3 | 6,195 | 96,438 | 46,290 | 13.38 |
| 3 | 1,932 | 10,812 | 10,380 | 18.61 |
| 3 | 1,860 | 4,063 | 3,900 | 47.69 |
| 4 | 160 | 5,035 | 1,450 | 11.03 |
| 4 | 622 | 2,238,800 | 1,074,624 | 0.06 |
| 5 | 1,677 | 558,967 | 536,608 | 0.31 |
| 5 | 1,567 | 756,637 | 453,982 | 0.35 |
| 5 | 1,453 | 12,728 | 12,219 | 11.89 |
| 6 | 3,265 | 29,584 | 14,200 | 22.99 |
| 7 | 4,839 | 15,912 | 15,276 | 31.68 |
| 8 | 3,515 | 100,046 | 96,044 | 3.66 |
| 9 | 3,982 | 1,627,025 | 488,108 | 0.82 |
| 10 | 2,163 | 26,633 | 25,568 | 8.46 |
| 11 | 1,810 | 181,566 | 174,303 | 1.04 |
| 12 | 809 | 656,020 | 472,334 | 0.17 |
| 13 | 2,987 | 2,501,045 | 1,500,627 | 0.20 |
| 14 | 5,931 | 72,734 | 34,912 | 16.99 |
| 15 | 4,771 | 43,694,938 | 10,486,785 | 0.05 |
| 16 | 5,196 | 94,545,825 | 22,690,998 | 0.02 |
| 17 | 5,834 | 189,945,812 | 45,586,995 | 0.01 |
| 18 | 5,583 | 7,744,225 | 1,858,614 | 0.30 |
| 19 | 5,391 | 2,804,051 | 1,345,944 | 0.40 |
The number of viral copies used for triplicate amplification of the JCPyV NCCR.
Sampling size is defined as [(number of reads/input copy number) × 100].
Dideoxy DNA sequencing of JC virus noncoding control region DNA.
Purified NCCR amplicons were directly used as the template for dideoxy DNA sequencing. Template-specific primers (5′-GATTCCTCCCTATTCAGCACTTTG-3′ [forward primer] and 5′-TCCACTCCAGGTTTTACTAA-3′ [reverse primer]) were used for cycle sequencing PCR using BigDye Terminator (v3.1) cycle sequencing kit reagents (Applied Biosystems) under the following conditions: 96°C for 1 min, followed by 35 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Samples were purified (DyeEx; Qiagen) and run on a 3730xl DNA analyzer (Applied Biosystems). DNA sequences were analyzed with SeqScape (v2.5) and Sequencher (v5.0) software.
454 amplicon sequencing of JC virus noncoding control region DNA.
Equimolar amounts of purified NCCR amplicons were subject to 454 pyrosequencing, performed on a GS Junior platform (Roche) at the VIB Nucleomics Core (http://www.nucleomics.be/; Leuven, Belgium). Only DNA reads of high quality and containing an intact multiplex identifier (MID) sequence for sample identification were used for further analysis. Raw 454 sequencing data have been deposited in the Sequence Read Archive (SRA) database (accession number SRP043413).
Determination of consensus and minor variant noncoding control region DNA sequences.
The 454 sequencing reads were first segregated per sample on the basis of the MID introduced in the PCR amplicon. The sequences were trimmed to remove the low-quality ends as well as 454 sequencing adaptors. Sequences assigned to the same sample were initially clustered using CD-hit software (28) under default settings, i.e., a sequence identity of >0.98. Subsequently, NCCR consensus sequences representing all generated CD-hit clusters per sample were assembled into larger clusters using the CLC Main Workbench program (CLC Bio, Denmark) to correct for technical errors at homopolymeric DNA stretches. After clustering and alignment analysis, all reads per sample were remapped against their sample consensus sequences. In all cases, >99% of all sequences, including those attributed to minor viral variants, were successfully mapped, demonstrating that all minor variant species representing ≥1% of the viral population were detected by this approach. The following criteria were used to evaluate the presence of NCCR viral variants: DNA clusters representing <1% of all reads within a given sample were not further taken into account, while clusters representing ≥1% of all reads were considered genuine viral variants. For patients for whom multiple sample types could be analyzed (i.e., patients 1, 2, and 3), 454 sequences were grouped in forward and reverse reads on the basis of the direction of their sequenced amplification primer. The forward reads were used for K-mer-based tree construction (neighbor joining) with the CLC Genomics Workbench program using a k value of 15 and the Mahalanobis distance (29).
Nucleotide sequence accession numbers.
JC virus noncoding control region DNA sequences deviating from the archetype JC virus sequence (12) were submitted to EMBL/GenBank under accession numbers LK391951 to LK391961.
RESULTS
JC virus load and dideoxy DNA sequencing of the noncoding control region DNA.
Thirty JC virus DNA-positive samples (10 urine, 5 plasma, and 15 CSF samples) originating from 19 PML patients were included in this study. The JC viral load from all PML patient samples was determined by quantitative PCR as described before (27), and the viral loads are given in Table 1. For four PML patients, different body fluid specimens (i.e., either urine and CSF or plasma and CSF) were analyzed, while for one other patient, three consecutive CSF samples were available (Table 1). For all remaining PML patients, a single urine or CSF sample was analyzed.
Following amplification of the JC virus noncoding control region DNA, its genomic organization was analyzed by dideoxy sequencing. The organization of JC virus NCCR DNA amplified from the urine of PML patients always highly resembled the organization of the archetype (12) (NCBI accession number AB038249), with only a single nucleotide variation (i.e., G217A, according to the numbering described previously [2]) occurring in 9 out of 10 samples. This polymorphism was previously attributed to European and Asian JC virus strains (30) and is regularly seen in urinary JC virus (14). One NCCR consensus sequence combined a single nucleotide variation (G219A) and a 2-bp deletion (del221-222) (Table 2) previously reported to be occasionally present in the urine of immunocompetent viral shedders (14). Analysis of JC virus NCCR DNA amplified from PML patient CSF and plasma samples revealed a different and more complex picture. Whereas in 14 out of 15 CSF samples the consensus NCCR sequence was highly rearranged and unique for each patient, 1 CSF sample (from patient 2; Table 2) displayed the archetype NCCR sequence. Also, in both plasma/serum samples from the latter patient, the presence of the archetype NCCR sequence was confirmed (Table 2). In two additional PML cases (patients 3 and 4), both plasma and CSF contained highly rearranged but identical consensus NCCR sequences (Table 2). Three consecutive CSF samples collected within a time frame of ∼2 months were analyzed for PML patient 5, and it was shown that the consensus NCCR sequence remained identical over this period of time. For one CSF sample (from patient 7), no reliable sequence could be obtained, indicative of the existence of multiple viral NCCR variants (quasispecies) present at similar quantities, which was further confirmed by 454 sequencing.
TABLE 2.
Consensus and minor variant JC virus NCCR organization in PML patients
| Patient no. | Matrix | Consensus NCCR sequencea | Presence of minor variantb | Minor variant species (% of isolates)c |
|---|---|---|---|---|
| 1 | Urine (n = 4) | AR (G217A) | n | None |
| 1 | CSF | RR | n | None |
| 2 | Plasma (n = 2) | AR (G217A) | y | del115-183 (7) |
| 2 | CSF | AR (G217A) | y | del115-183 (12) |
| 3 | Plasma (n = 2) | RR | y | del63-136 (2) |
| 3 | CSF | RR | y | del63-136 (3) |
| 4 | Plasma | RR | ND | ND |
| 4 | CSF | RR | ND | ND |
| 5 | CSF | RR | y | del64-168 (23) |
| 5 | CSF | RR | y | del64-168 (23) |
| 5 | CSF | RR | n | None |
| 6 | CSF | RR | y | del61-126 (3) |
| 7 | CSF | ND | y | Highly diverse |
| 8 | CSF | RR | y | del192-274 (2) + del149-250 (11) |
| 9 | CSF | RR | y | del63-139 (7) |
| 10 | CSF | RR | y | Various (n = 6) deletions and rearrangements (2–8) |
| 11 | CSF | RR | y | Highly diverse |
| 12 | CSF | RR | ND | ND |
| 13 | CSF | RR | y | del61-133 (8) |
| 14 | Urine | AR (G217A) | n | None |
| 15 | Urine | AR (G217A) | y | del165-192 (2) |
| 16 | Urine | AR (G217A) | n | None |
| 17 | Urine | AR (G219A + del221-222) | y | ins222 (21 ntd) (2) |
| 18 | Urine | AR (G217A) | n | None |
| 19 | Urine | AR (G217A) | n | None |
AR, archetype; RR, rearranged; G217A, polymorphism at position 217; G219A, polymorphism at position 219; del, deletion.
y, yes [a minor variant(s) was present]; n, no (a minor variant was not present); ND, not determined.
The numbering of the deletion (del) or insertion (ins) is relative to that of the consensus sequence of the corresponding sample. The percentage of samples in which the minor variants were detected is given in parentheses. For simplicity, patients 7 and 11 are defined as highly diverse since up to 10 viral variants contributed to the quasispecies.
nt, nucleotides.
Detection of minor JC viral variants by 454 sequencing of the noncoding control region DNA.
To identify possible minor viral variants and, thus, to be able to characterize a representative part of the viral population (the JC virus quasispecies) contained in the different body compartments of PML patients, 454 sequencing was applied to the above-mentioned samples as well as plasmid control samples. Amplification of JC virus NCCR DNA was performed in triplicate to enhance the likelihood of amplifying viral variants present at low levels as well as to minimize errors possibly introduced by PCR. The highly varying JC viral load assigned to the samples (mean, 40,257,132 copies/ml; minimum, 4,063 copies/ml; maximum, 278,000,000 copies/ml; Table 1) was also taken into account. Hence, for samples with relatively low viral copy numbers, a maximal input of viral DNA was used for PCR, while for samples with high viral copy numbers, less DNA was required to introduce a representative amount of viral copies for deep sequencing. The number of viral copies per sample used in the experimental setup is given in Table 3. Plasmid DNA samples harboring archetype JC virus NCCR with or without a predefined deletion of domain D were processed as described elsewhere (14), and the accuracy of detecting DNA deletions in the JCPyV NCCR by our 454 sequencing approach was confirmed (data not shown).
454 amplicon sequencing resulted in 92,169 high-quality JC virus NCCR sequences (reads) divided over 30 PML samples. For each sample, an average of 3,072 reads (maximum, 6,195 reads; minimum, 160 reads; median, 2,851 reads; Table 3) was used for further analysis. On the basis of the ratio of the number of reads and the input viral copy number per sample, a sampling size was calculated. Since this sampling size was <100 for all samples (minimum, 0.01; maximum, 47.69), no redundancy (i.e., sequencing of the same viral copy multiple times) was introduced in our data set (Table 3), and thus, all sequences obtained were considered to originate from a unique viral copy. To allow reliable determination of JC virus NCCR minor variants representing ≥1% of the viral population, only those samples for which >1,000 reads were generated were used. This means that minorities representing ≥1% of the viral population would be detected by the use of at least 10 DNA sequences. As a consequence, one plasma sample (from patient 4) and two CSF samples (from patients 4 and 12) were withdrawn from further analysis (Tables 2 and 3).
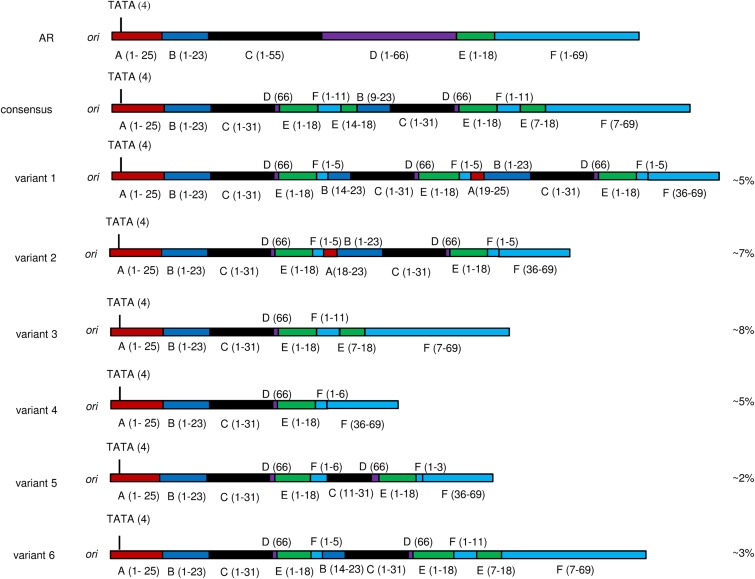
For all samples, the NCCR consensus sequence, as well as the NCCR sequences of the minor viral variants contributing to the quasispecies, were determined from the 454 sequencing data. To do so, sequences attributed to each sample were initially clustered utilizing CD-hit software under default settings (28), and NCCR sequences representing all clusters were further analyzed as described above. In all cases, the consensus NCCR sequence obtained by 454 sequencing agreed with the sequence obtained by dideoxy sequencing. A discrepancy between urinary and CSF/plasma NCCR sequences became immediately apparent. Whereas the sequences of the vast majority of reads obtained for urine samples very much resembled the archetype NCCR sequence, sequences originating from CSF or plasma often appeared to be a mixture of highly diverse sequences whose organization did not share the organization of the archetype sequence. JC virus NCCR minor variants were detected in 11 out of 13 CSF specimens and all plasma/serum samples (n = 4) but in only 2 out of 10 urine samples. In addition, whereas in urine only one minor viral variant contributed to the quasispecies, the JC viruses in CSF appeared to be a highly complex mixture of natural variants. In those samples in which more than one rearranged NCCR sequence was identified, it appeared that longer forms of the NCCR coexisted with shorter forms that were derived therefrom by a single deletion, as exemplified by the NCCR variants detected in patient 10 (Fig. 1). An overview of the NCCR quasispecies identified in PML patient body fluid samples is presented in Table 2.
FIG 1.
JC virus NCCR quasispecies identified in CSF from PML patient 10. Both the consensus NCCR sequence and the sequence of minor variants are schematically shown and compared to the archetype (AR) sequence. The amount of each variant present as a percentage of the total quasispecies is indicated on the right. Numbers in parentheses indicate the original nucleotide sequence position in the archetype sequence.
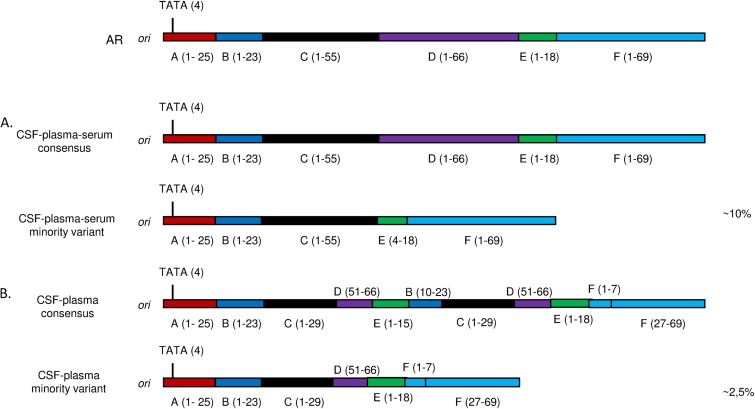
A remarkable feature of JC virus quasispecies in PML cases was the correlation between the viral population identified in CSF and the one identified in plasma/serum, since both the consensus NCCR sequence and the NCCR sequences of the minority variants from both body compartments from the same patients were identical (Table 2; Fig. 2). In contrast, for one PML patient (patient 1) for which urine and CSF were examined, both body compartments contained clearly distinct viral populations (Table 2).
FIG 2.
Comparison of JC virus NCCR organization in CSF and plasma of two PML patients. (A) For patient 2, the consensus NCCR sequence resembled the archetype (AR) sequence, while the minority viral variant shared by CSF and plasma/serum harbored a deletion of domain D and part of domain E. (B) For patient 3, the consensus NCCR sequence was rearranged but was identical in CSF and plasma and contained a duplicated DNA sequence that was absent in the minority viral variant. Numbers in parentheses indicate the original nucleotide sequence position in the archetype sequence.
DISCUSSION
Detailed analysis of the JC virus DNA population contained in different body compartments of PML patients might shed light on viral dissemination within the patient and further characterizes the viral strains associated with PML. In this study, 454 sequencing was used to analyze the noncoding control region DNA in 30 JC virus-positive body fluid samples (CSF, plasma/serum, and urine) from PML patients. Unlike earlier studies in which the consensus NCCR sequence in different PML patient sample types was determined by dideoxy DNA sequencing (11), we aimed at additionally identifying the minor viral variants contributing to the JC virus quasispecies. Moreover, our sample set allowed, at least in a subgroup of patients, comparison of the quasispecies between body fluids within a patient and, thus, to learn about viral dissemination.
In a current understanding, acquisition of neurotropic JC virus occurs at some point during PML development and often leads to the presence of a highly rearranged JC virus NCCR in CSF at the time of or following diagnosis. In vitro cell studies indicated that such rearrangements enhance early viral gene expression and support higher replication rates in glial cells; as a consequence, they increase the cytopathology (18). A notable feature of PML-associated rearranged JCPyV is the total or partial deletion of domain D of the noncoding control region (11, 17). In the majority of the PML patient CSF and plasma samples described here, domain D was also highly affected. However, archetype NCCR was identified in both the CSF and plasma of PML patient 2, a patient that was sampled 6 months after a liver transplant due to hepatitis C cirrhosis. The CSF sample of this patient had an extremely high viral load of 278,000,000 copies/ml, and in addition, ∼12% of this viral population contained a DNA deletion of domain D and part of domain E. Although it is not trivial to interpret to what extent both variants contributed to the high JC viral load in CSF, the fact that the archetype represented the vast majority of the virus detected in CSF indicates that archetype JC virus can be found in vivo in a predominantly glial cell environment. Recently, an unusual archetype organization of the JC virus NCCR has also been identified in the CSF of two PML patients with long-term HIV-1 infection and one HIV-1-positive patient diagnosed with classical PML (16, 31). In addition, in vitro studies showed that NCCR rearrangement is not required per se for activity in glial cells (32, 33), although it might still enhance neurovirulence (18). Taken together, our data support the novel notion that PML can occasionally be associated with archetype JC virus, although in the majority of cases, rearrangement of the NCCR is still favored.
The majority of the human population becomes infected with JC virus, after which the virus can persist lifelong without apparent symptoms. Multiple sites of JC virus latency exist, including kidney epithelial cells, the urinary tract, lymphoid tissue, and the brain (1, 9, 10, 34–36), whereas bone marrow and B lymphocytes originating therefrom have been suggested to be potential sites for JC virus neurotropic transformation (21, 24, 35) due to their inherent susceptibility to DNA rearrangement that might favor the emergence of rearranged JCPyV. Both virus-infected lymphocytes and cell-free virus have been suggested to participate in viral dissemination by crossing the blood-brain barrier (20, 23, 35, 37), whereas cell-mediated viral carriage might have clinical relevance in patients subject to increased peripheral mobilization of hematopoietic progenitor (CD34+) cells due to continued natalizumab treatment (34). Two of the PML patients that we examined were of particular interest since both plasma and CSF samples could be analyzed. Our data support a route of JC virus dissemination between blood and the central nervous system since both the consensus NCCR DNA sequence and the sequence of the quasispecies of virus from those body compartments correlated well. In contrast, the urinary JC virus population appeared to exist as an independent viral population. This phenomenon has been appreciated for some time, but this study now provides concrete evidence for it in a series of PML samples.
Although matching urine and CSF samples could be examined for only one PML patient, the JC virus NCCR DNA present in urine did not resemble the highly rearranged virus DNA generally found in CSF. Recently, we also showed that JCPyV NCCR quasispecies reminiscent of those from PML patients were occasionally present in the urine of healthy viral shedders (14) and hypothesized that this might be reflective of the situation in other body reservoirs. The current data, however, clearly indicate that there is no correlation between urinary and CSF/plasma JCPyV and thus exclude quasispecies analysis in urine as a factor predictive of PML development. The independence between urinary and nonurinary JC virus has also been observed when examining the correlation between mutations in the major capsid protein VP1 in urine and nonurine samples from PML patients, since VP1 mutations present in virus from blood and/or CSF were absent in virus from urine (11).
A major advantage of deep sequencing is its ability to provide an in-depth analysis of the viral population. Whereas JC virus strains from PML cases are often referred to as homogeneous populations, we showed that the JC viral population in CSF and/or blood of PML patients most often exists as a mixture of naturally occurring viral variants, with in some cases up to 10 JC viral variants harboring diverse noncoding control region DNA sequences. Similar observations were previously made by colony sequencing of virus in the CSF of a few PML cases (17), although the colony sequencing technology is much less sensitive than deep sequencing in detecting minor variants, since many fewer DNA sequences are generated compared to the number generated by deep sequencing, and, hence, colony sequencing is likely to underappreciate the true complexity of the viral population.
Despite the high variability of reorganization of the NCCR between different patients, multiple viral variants within a single sample could often be derived from each other by single deletion or duplication events, reflective of a process throughout which the virus continuously adapts to its cellular environment. In contrast, the JC virus population in urine appeared to be stable, and in the urine viral population, archetype virus was continuously selected and additional minor NCCR variants were detected only sporadically (i.e., in 2 out of 10 samples analyzed). The sequences of these minor variants deviated from the consensus NCCR sequence by means of a single deletion, as has previously been observed in the urine of healthy subjects (14).
In conclusion, by applying deep sequencing to JC virus noncoding control region DNA isolated from different body reservoirs of PML patients, we have shown that JC virus can appear as a quasispecies that is shared between CSF and plasma within a single patient but is unrelated to urinary JC virus. Due to this intimate correlation between virus in CSF and plasma, our findings seem to further support the clinical relevance of JC virus viremia.
ACKNOWLEDGMENTS
This work was supported in part by the Agentschap voor Innovatie door Wetenschap en Technologie (IWT; project number 120480).
We thank Els Rousseau (Janssen Diagnostics) for assistance with sample logistics.
REFERENCES
- 1.Major EO. 2010. Progressive multifocal leukoencephalopathy in patients on immunomodulatory therapies. Annu Rev Med 61:35–47. doi: 10.1146/annurev.med.080708.082655. [DOI] [PubMed] [Google Scholar]
- 2.Frisque RJ, Bream GL, Cannella MT. 1984. Human polyomavirus JC virus genome. J Virol 51:458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frisque RJ. 2001. Structure and function of JC virus T′ proteins. J Neurovirol 7:293–297. doi: 10.1080/13550280152537120. [DOI] [PubMed] [Google Scholar]
- 4.Prins C, Frisque RJ. 2001. JC virus T′ proteins encoded by alternatively spliced early mRNAs enhance T antigen-mediated viral DNA replication in human cells. J Neurovirol 7:250–264. doi: 10.1080/13550280152403290. [DOI] [PubMed] [Google Scholar]
- 5.Ferenczy MW, Marshall LJ, Nelson CD, Atwood WJ, Nath A, Khalili K, Major EO. 2012. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev 25:471–506. doi: 10.1128/CMR.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall LJ, Dunham L, Major EO. 2010. Transcription factor Spi-B binds unique sequences present in the tandem repeat promoter/enhancer of JC virus and supports viral activity. J Gen Virol 91:3042–3052. doi: 10.1099/vir.0.023184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall LJ, Major EO. 2010. Molecular regulation of JC virus tropism: insights into potential therapeutic targets for progressive multifocal leukoencephalopathy. J Neuroimmune Pharmacol 5:404–417. doi: 10.1007/s11481-010-9203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stuyver LJ, Verbeke T, Van Loy T, Van Gulck E, Tritsmans L. 2013. An antibody response to human polyomavirus 15-mer peptides is highly abundant in healthy human subjects. Virol J 10:192. doi: 10.1186/1743-422X-10-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Liz G, Del Valle L, Gentilella A, Croul S, Khalili K. 2008. Detection of JC virus DNA fragments but not proteins in normal brain tissue. Ann Neurol 64:379–387. doi: 10.1002/ana.21443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan CS, Ellis LC, Wuthrich C, Ngo L, Broge TA Jr, Saint-Aubyn J, Miller JS, Koralnik IJ. 2010. JC virus latency in the brain and extraneural organs of patients with and without progressive multifocal leukoencephalopathy. J Virol 84:9200–9209. doi: 10.1128/JVI.00609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reid CE, Li H, Sur G, Carmillo P, Bushnell S, Tizard R, McAuliffe M, Tonkin C, Simon K, Goelz S, Cinque P, Gorelik L, Carulli JP. 2011. Sequencing and analysis of JC virus DNA from natalizumab-treated PML patients. J Infect Dis 204:237–244. doi: 10.1093/infdis/jir256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. 1990. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol 64:3139–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen PN, Major EO. 2001. A classification scheme for human polyomavirus JCV variants based on the nucleotide sequence of the noncoding regulatory region. J Neurovirol 7:280–287. doi: 10.1080/13550280152537102. [DOI] [PubMed] [Google Scholar]
- 14.Van Loy T, Thys K, Tritsmans L, Stuyver LJ. 2013. Quasispecies analysis of JC virus DNA present in urine of healthy subjects. PLoS One 8:e70950. doi: 10.1371/journal.pone.0070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bossolasco S, Calori G, Moretti F, Boschini A, Bertelli D, Mena M, Gerevini S, Bestetti A, Pedale R, Sala S, Sala S, Lazzarin A, Cinque P. 2005. Prognostic significance of JC virus DNA levels in cerebrospinal fluid of patients with HIV-associated progressive multifocal leukoencephalopathy. Clin Infect Dis 40:738–744. doi: 10.1086/427698. [DOI] [PubMed] [Google Scholar]
- 16.Delbue S, Elia F, Carloni C, Tavazzi E, Marchioni E, Carluccio S, Signorini L, Novati S, Maserati R, Ferrante P. 2012. JC virus load in cerebrospinal fluid and transcriptional control region rearrangements may predict the clinical course of progressive multifocal leukoencephalopathy. J Cell Physiol 227:3511–3517. doi: 10.1002/jcp.24051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamichi K, Kishida S, Tanaka K, Suganuma A, Sano Y, Sano H, Kanda T, Maeda N, Kira J, Itoh A, Kato N, Tomimoto H, Kurane I, Lim CK, Mizusawa H, Saijo M. 2013. Sequential changes in the non-coding control region sequences of JC polyomaviruses from the cerebrospinal fluid of patients with progressive multifocal leukoencephalopathy. Arch Virol 158:639–650. doi: 10.1007/s00705-012-1532-3. [DOI] [PubMed] [Google Scholar]
- 18.Gosert R, Kardas P, Major EO, Hirsch HH. 2010. Rearranged JC virus noncoding control regions found in progressive multifocal leukoencephalopathy patient samples increase virus early gene expression and replication rate. J Virol 84:10448–10456. doi: 10.1128/JVI.00614-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall LJ, Moore LD, Mirsky MM, Major EO. 2012. JC virus promoter/enhancers contain TATA box-associated Spi-B-binding sites that support early viral gene expression in primary astrocytes. J Gen Virol 93:651–661. doi: 10.1099/vir.0.035832-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapagain ML, Verma S, Mercier F, Yanagihara R, Nerurkar VR. 2007. Polyomavirus JC infects human brain microvascular endothelial cells independent of serotonin receptor 2A. Virology 364:55–63. doi: 10.1016/j.virol.2007.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houff SA, Major EO, Katz DA, Kufta CV, Sever JL, Pittaluga S, Roberts JR, Gitt J, Saini N, Lux W. 1988. Involvement of JC virus-infected mononuclear cells from the bone marrow and spleen in the pathogenesis of progressive multifocal leukoencephalopathy. N Engl J Med 318:301–305. doi: 10.1056/NEJM198802043180507. [DOI] [PubMed] [Google Scholar]
- 22.Tan CS, Dezube BJ, Bhargava P, Autissier P, Wuthrich C, Miller J, Koralnik IJ. 2009. Detection of JC virus DNA and proteins in the bone marrow of HIV-positive and HIV-negative patients: implications for viral latency and neurotropic transformation. J Infect Dis 199:881–888. doi: 10.1086/597117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapagain ML, Nerurkar VR. 2010. Human polyomavirus JC (JCV) infection of human B lymphocytes: a possible mechanism for JCV transmigration across the blood-brain barrier. J Infect Dis 202:184–191. doi: 10.1086/653823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marzocchetti A, Wuthrich C, Tan CS, Tompkins T, Bernal-Cano F, Bhargava P, Ropper AH, Koralnik IJ. 2008. Rearrangement of the JC virus regulatory region sequence in the bone marrow of a patient with rheumatoid arthritis and progressive multifocal leukoencephalopathy. J Neurovirol 14:455–458. doi: 10.1080/13550280802356837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger JR, Miller CS, Mootoor Y, Avdiushko SA, Kryscio RJ, Zhu H. 2006. JC virus detection in bodily fluids: clues to transmission. Clin Infect Dis 43:e9–e12. doi: 10.1086/504947. [DOI] [PubMed] [Google Scholar]
- 26.Bofill-Mas S, Girones R. 2003. Role of the environment in the transmission of JC virus. J Neurovirol 9(Suppl 1):S54–S58. doi: 10.1080/13550280390195306. [DOI] [PubMed] [Google Scholar]
- 27.Ryschkewitsch C, Jensen P, Hou J, Fahle G, Fischer S, Major EO. 2004. Comparison of PCR-Southern hybridization and quantitative real-time PCR for the detection of JC and BK viral nucleotide sequences in urine and cerebrospinal fluid. J Virol Methods 121:217–221. doi: 10.1016/j.jviromet.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentleman JF, Mullin RC. 1989. The distribution of the frequency of occurrence of nucleotide subsequences, based on their overlap capability. Biometrics 45:35–52. doi: 10.2307/2532033. [DOI] [PubMed] [Google Scholar]
- 30.Yogo Y, Zhong S, Shibuya A, Kitamura T, Homma Y. 2008. Transcriptional control region rearrangements associated with the evolution of JC polyomavirus. Virology 380:118–123. doi: 10.1016/j.virol.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 31.Iannetta M, Bellizzi A, Lo Menzo S, Anzivino E, D'Abramo A, Oliva A, D'Agostino C, d'Ettorre G, Pietropaolo V, Vullo V, Ciardi MR. 2013. HIV-associated progressive multifocal leukoencephalopathy: longitudinal study of JC virus non-coding control region rearrangements and host immunity. J Neurovirol 19:274–279. doi: 10.1007/s13365-013-0167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ault GS. 1997. Activity of JC virus archetype and PML-type regulatory regions in glial cells. J Gen Virol 78(Pt 1):163–169. [DOI] [PubMed] [Google Scholar]
- 33.Sock E, Renner K, Feist D, Leger H, Wegner M. 1996. Functional comparison of PML-type and archetype strains of JC virus. J Virol 70:1512–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frohman EM, Monaco MC, Remington G, Ryschkewitsch C, Jensen PN, Johnson K, Perkins M, Liebner J, Greenberg B, Monson N, Frohman TC, Douek D, Major EO. 2014. JC virus in CD34+ and CD19+ cells in patients with multiple sclerosis treated with natalizumab. JAMA Neurol 71:596–602. doi: 10.1001/jamaneurol.2014.63. [DOI] [PubMed] [Google Scholar]
- 35.Monaco MC, Atwood WJ, Gravell M, Tornatore CS, Major EO. 1996. JC virus infection of hematopoietic progenitor cells, primary B lymphocytes, and tonsillar stromal cells: implications for viral latency. J Virol 70:7004–7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monaco MC, Shin J, Major EO. 1998. JC virus infection in cells from lymphoid tissue. Dev Biol Stand 94:115–122. [PubMed] [Google Scholar]
- 37.Tornatore C, Berger JR, Houff SA, Curfman B, Meyers K, Winfield D, Major EO. 1992. Detection of JC virus DNA in peripheral lymphocytes from patients with and without progressive multifocal leukoencephalopathy. Ann Neurol 31:454–462. doi: 10.1002/ana.410310426. [DOI] [PubMed] [Google Scholar]