ABSTRACT
Chronic infection with hepatitis B virus (HBV) is a risk factor for developing liver diseases such as hepatocellular carcinoma (HCC). HBx is a multifunctional protein encoded by the HBV genome; HBx stimulates HBV replication and is thought to play an important role in the development of HBV-associated HCC. HBx can activate the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway in some cell lines; however, whether HBx regulates PI3K/AKT signaling in normal hepatocytes has not been evaluated. In studies described here, we assessed HBx activation of PI3K/AKT signaling in an ex vivo model of cultured primary hepatocytes and determined how this HBx activity affects HBV replication. We report that HBx activates AKT in primary hepatocytes and that the activation of AKT decreases HBV replication and HBV mRNA and core protein levels. We show that the transcription factor hepatocyte nuclear factor 4α (HNF4α) is a target of HBx-regulated AKT, and we link HNF4α to HBx-regulated AKT modulation of HBV transcription and replication. Although we and others have shown that HBx stimulates and is likely required for HBV replication, we now report that HBx also activates signals that can diminish the overall level of HBV replication. While this may seem counterintuitive, we show that an important effect of HBx activation of AKT is inhibition of apoptosis. Consequently, our studies suggest that HBx balances HBV replication and cell survival by stimulating signaling pathways that enhance hepatocyte survival at the expense of higher levels of HBV replication.
IMPORTANCE Chronic hepatitis B virus (HBV) infection is a common cause of the development of liver cancer. Regulation of cell signaling pathways by the HBV HBx protein is thought to influence the development of HBV-associated liver cancer. HBx stimulates, and may be essential for, HBV replication. We show that HBx activates AKT in hepatocytes to reduce HBV replication. While this seems contradictory to an essential role of HBx during HBV replication, HBx activation of AKT inhibits hepatocyte apoptosis, and this may facilitate persistent, noncytopathic HBV replication. AKT regulates HBV replication by reducing the activity of the transcription factor hepatocyte nuclear factor 4α (HNF4α). HBx activation of AKT may contribute to the development of liver cancer by facilitating persistent HBV replication, augmenting the dedifferentiation of hepatocytes by inhibiting HNF4α functions, and activating AKT-regulated oncogenic pathways. AKT-regulated factors may provide therapeutic targets for inhibiting HBV replication and the development of HBV-associated liver cancer.
INTRODUCTION
Chronic infection with hepatitis B virus (HBV) remains a major global health problem. Despite the availability of effective HBV vaccines, there are ∼350 million people worldwide who are chronically infected with HBV. Chronic HBV infection is the major cause of the development of hepatocellular carcinoma (HCC) (1). The level of HBV replication in a chronically HBV-infected individual can vary throughout the course of the infection, and the rise and fall of HBV replication during a chronic infection may affect disease progression (2, 3). There are limited therapeutic options for treating a chronic HBV infection, and the emergence of HBV mutants that are resistant to available therapies is common. Moreover, available anti-HBV drugs typically do not completely eliminate HBV in chronically infected individuals due to the presence and stability of covalently closed circular DNA (cccDNA), a replicative intermediate of HBV that is localized to the nucleus of HBV-infected hepatocytes (4–7). Consequently, there is continued interest in understanding the mechanisms that regulate HBV replication and could serve as potential therapeutic targets.
HBV is an enveloped, partially double-stranded DNA virus that belongs to the Hepadnaviridae family of viruses (8). Although HBV is a DNA virus, it replicates through reverse transcription of an RNA intermediate. HBV replication occurs within the viral capsid in the cytosol of hepatocytes; the capsid is composed of the viral core protein. After the entry of HBV into hepatocytes, the partially double-stranded genome of the virus is delivered to the nucleus, where the genome is repaired by host cell DNA repair machinery to generate cccDNA. cccDNA forms a minichromosome and is the template for all HBV RNA transcripts. Pregenomic RNA (pgRNA), one of the longest HBV transcripts, is enclosed in capsids; pgRNA is covalently linked to the viral DNA polymerase/reverse transcriptase (RT), which reverse transcribes the first DNA strand of the HBV genome by using pgRNA as the template. For reasons that remain incompletely understood, the second, cDNA strand is synthesized to various lengths, giving rise to the partially double-stranded genome of HBV (9–11). The interaction between numerous host cell factors and HBV proteins governs HBV replication in hepatocytes and likely plays an important role in the development of HBV-associated HCC (3, 8, 11, 12).
The shortest open reading frame (ORF) and mRNA transcript of the HBV genome encodes the HBx protein (11, 13). HBx stimulates HBV replication in multiple experimental systems, including cultured primary rat and human hepatocytes, some liver cell lines, livers of normal mice, and chimeric mice with humanized livers (14–19). HBx is also thought to play an important role in the development of HBV-associated HCC due to the regulatory effects of HBx on cellular signaling pathways, DNA damage repair mechanisms, the cell cycle, and apoptosis (14, 20–24). Some HBx activities, such as its role in regulating apoptosis, appear to be context dependent and have varied depending upon the model system that was used to assess this HBx activity (8, 11, 25). While studies conducted with various cell lines have provided important insights concerning the effect of HBx expression on cellular signal transduction pathways, the influence of HBx expression on cellular signal transduction pathways in normal hepatocytes, and in the context of an HBV infection, remains poorly understood. Recent studies with ex vivo-cultured primary hepatocyte model systems and in vivo models have begun to identify HBx effects on normal hepatocytes and how these HBx activities influence HBV replication and affect hepatocyte physiology (8, 11, 14, 23, 26, 27).
The phosphatidylinositol 3-kinase (PI3K)/AKT pathway is a cell signal transduction pathway that can regulate numerous cellular processes, including cellular apoptotic pathways (28, 29). The PI3K/AKT pathway is regulated by growth factors that activate PI3K, which in turn phosphorylates phosphatidylinositol-4,5-bisphosphate (PIP2) to form phosphatidylinositol-3,4,5-trisphosphate (PIP3). PIP3 acts as a docking site for AKT and facilitates the subsequent phosphorylation of AKT at threonine 308 (T308) by phosphoinositide-dependent kinase 1 (PDK1). The complete activation of AKT also requires phosphorylation at serine 473 (S473) of AKT (28, 29). The PI3K/AKT pathway is upregulated in many cancers, and many viruses encode proteins that can activate this pathway (30–33).
Hepatocyte nuclear factor 4α (HNF4α) belongs to the nuclear receptor superfamily and binds to DNA as a homodimer (34). HNF4α is expressed in hepatocytes and is important for the transcription of many hepatocyte-specific genes that affect liver development and hepatocyte differentiation (35, 36). In hepatocytes, HNF4α is mostly nuclear and can undergo covalent modifications such as acetylation and phosphorylation (37–39). HNF4α is phosphorylated at many serine, threonine, and tyrosine residues; however, the identity and location of all phosphorylated amino acids of HNF4α and the kinases responsible for phosphorylation remain incompletely characterized (40, 41). AKT can phosphorylate HNF4α, which causes HNF4α to exit the nucleus and reduces the transcription of HNF4α-dependent genes (42).
In the present study, we assessed the effect of HBx on the regulation of the PI3K/AKT pathway in cultured primary rat and human hepatocytes and analyzed how the PI3K/AKT pathway affects HBV replication. Although previous studies that were conducted with transformed or immortalized cells demonstrated that HBx can activate the AKT/PI3K pathway, those studies were conducted when HBx was expressed on its own and not in the context of the HBV genome. Moreover, those studies did not assess the effect of AKT activation on HBV replication (31–33). Separate studies that did demonstrate that the AKT/PI3K signaling pathway can influence HBV replication did not, in turn, determine whether HBV replication or expression of HBV proteins affected the PI3K/AKT pathway (43, 44). In addition to the HBx protein, the HBV large surface antigen has also been shown to activate AKT in hepatoma cells (45). In our studies, we now demonstrate that HBx activates AKT in cultured primary human and rat hepatocytes, both when HBx is expressed on its own and in the context of the HBV genome. Because HBV naturally infects hepatocytes, our ex vivo hepatocyte systems likely represent more biologically relevant models for studying HBV and HBx effects on hepatocyte signal transduction pathways than similar types of studies with established cell lines. We also show that the activation of either AKT1 or AKT2 diminishes HBV replication in cultured primary hepatocytes; previous studies tested AKT1 regulation of HBV replication in transformed cells only (43, 44). Moreover, we demonstrate that the transcription factor HNF4α is a target of HBx-activated, AKT-mediated regulation of HBV replication. We also show that HBx activation of AKT, both when HBx is expressed on its own and in the context of the HBV genome, prevents apoptosis in cultured primary rat hepatocytes. Finally, we demonstrate that the activation of AKT signaling by HBx is important for inhibiting apoptosis in HBV-expressing hepatocytes. We and others have previously shown that HBx stimulates, and is likely required for, HBV replication (14–19). Therefore, our studies described here in conjunction with previous work suggest that HBx activates multiple cellular signaling pathways to balance HBV replication. Since HBV replicates well in primary hepatocytes, the stimulatory effects of HBx on HBV replication are dominant over the inhibitory effects, but HBx-induced signals that inhibit HBV replication, such as the activation of AKT, may create a cellular environment that prolongs HBV replication by enhancing the survival of HBV-infected cells. We propose that HBx activation of AKT is a critical controller of the noncytopathic characteristics of an HBV infection and that by activating AKT to inhibit apoptosis, HBV sacrifices higher levels of replication to allow hepatocyte survival and, possibly, viral persistence. Our study provides novel insights into the interplay of the PI3K/AKT pathway, HBx expression, and HBV replication in a biologically relevant, ex vivo hepatocyte system.
MATERIALS AND METHODS
Animal studies.
Surgery and isolation of hepatocytes from rats were approved by the Institutional Animal Care and Use Committee of the Drexel University College of Medicine and complied with the Animal Welfare Act, the Public Health Service Policy on Humane Care and Use of Laboratory Animals, and the NIH Guide for the Care and Use of Laboratory Animals (46, 47).
Isolation and maintenance of primary rat hepatocytes.
Primary rat hepatocytes were isolated by a two-step perfusion method as previously described (48). The hepatocytes were plated onto 6-well (34.8-mm) collagen-coated tissue culture plates at ∼1.5 × 106 cells/well. The cells were maintained in Williams E medium supplemented with 1 mM sodium pyruvate, 2 mM l-glutamine, 1 μg/ml insulin-transferrin-selenium (ITS), 5 ng/ml epidermal growth factor (EGF), and 1 μg/ml hydrocortisone at 37°C in 5% CO2. Hepatocyte morphology was monitored, and reverse transcriptase PCR (RT-PCR) was performed, as previously described, to assess the expression of known hepatocyte differentiation markers and to demonstrate that hepatocytes remain differentiated throughout the time course of our experiments (14, 23, 26).
Maintenance of primary human hepatocytes.
Normal primary human hepatocytes in suspension were obtained through the Liver Tissue Cell Distribution System, Pittsburgh, PA, which is funded by NIH contract number HHSN276201200017C. Cultured primary human hepatocytes were maintained under conditions similar to those used for cultured primary rat hepatocytes. The morphology of cultured primary human hepatocytes was monitored, and RT-PCR was performed, as previously described, to assess the expression of known hepatocyte differentiation markers and to demonstrate that cultured primary human hepatocytes remained differentiated throughout the time course of our experiments (26).
Transfections and reagents.
Cultured primary rat hepatocytes were transfected by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. Primary rat hepatocytes were transfected 24 h after plating; 30% to 40% transfection efficiency was regularly obtained, as monitored by cotransfection with a green fluorescent protein (GFP) expression plasmid. Small interfering RNAs (siRNAs) were transfected by using Oligofectamine reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. Tumor necrosis factor alpha (TNF-α) was purchased from Calbiochem (La Jolla, CA), and LY294002 was purchased from Promega (Madison, WI).
Antibodies.
The anti-AKT, anti-phospho-AKT (pAKT), anti-histone H3, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and anti-cleaved caspase-3 antibodies were purchased from Cell Signaling (Danvers, MA). The anti-HBx antibody was purchased from Affinity BioReagents (Golden, CO); the anti-β-actin antibody was purchased from Sigma-Aldrich (St. Louis, MO); the anti-HBV core antibody was purchased from Dako (Carpentaria, CA); the anti-HBsAg antibody was purchased from Meridian Life Science, Inc. (Memphis, TN); and the anti-HNF4α and anti-cytochrome c (A-8) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmids.
The FL1-154 HBx expression plasmid has full-length HBx cloned into the pcDNA3.1(−) vector and a Flag tag at the N terminus of HBx and was previously described (49). The constitutively active myristoylated AKT1 (mAKT1) and mAKT2 plasmids express AKT1 or -2 that has an N-terminal hemagglutinin (HA) tag and is myristoylated at the N terminus; these plasmids were generated by William Sellers (Addgene plasmid numbers 9008 and 9016, respectively). pGEMHBV (payw1.2) and pGEMHBV (HBx−)/payw*7, which expresses all HBV proteins except HBx, were previously described (50–52).
Recombinant adenovirus.
HBV has a narrow host range and naturally infects only human hepatocytes. Some studies, such as HBV replication analyses with cultured primary rat hepatocytes, require that 100% of hepatocytes express the HBV genome, and therefore, for these studies, we used a recombinant adenovirus that expresses a greater-than-unit-length copy of the HBV genome (AdHBV) to infect hepatocytes. The construction of AdHBV was previously described (14). AdHBV also expresses GFP to monitor infection efficiency and to ensure that 100% of hepatocytes were infected.
Cell collection and Western blot analysis.
After a specified time following transfection or AdHBV infection, cells were washed with cold phosphate-buffered saline (PBS), scraped into PBS, pelleted by centrifugation at 2,000 rpm at 4°C for 5 min, and lysed in 0.8% sodium dodecyl sulfate (SDS) buffer (0.8% SDS, 240 mM Tris [pH 6.8], 10% glycerol). The protein concentration of each sample was determined by using the Bio-Rad protein assay (Bio-Rad, Hercules, CA) according to the manufacturer's directions, and equal amounts of protein were subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were then transferred onto a nitrocellulose membrane and blocked for 1 h in 5% nonfat milk. The nitrocellulose membrane was incubated with primary antibody overnight at 4°C, after which the membrane was washed 3 times with Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBST) and incubated with secondary antibody for 1 h. For analyses of AKT, pAKT, HBV core, histone H3, GAPDH, cleaved caspase-3, β-actin, and HBx expression and levels, Alexa Fluor-conjugated secondary antibodies were used, and the proteins were visualized and quantified by using the Odyssey infrared imaging system (Licor Biosciences, Lincoln, NE). For analyses of cytosolic cytochrome c and HNF4α expression and levels, horseradish peroxidase-conjugated secondary antibodies were used, and the proteins were visualized by enhanced chemiluminescence and quantified with ImageJ software. For detection of cleaved caspase-3 as an apoptotic marker, cultured primary hepatocytes were scraped into the cell growth medium so that both live and dead cells were collected. The cells were then processed for SDS-PAGE and Western blot analysis as described above.
Northern blot analysis.
Cultured primary hepatocytes were washed with cold PBS at the indicated times following AdHBV infection. The cells were then collected, and total RNA was isolated by using TRIzol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). mRNA was then isolated from total RNA by using oligo(dT)-cellulose columns (Molecular Research Center, Inc., Cincinnati, OH) according to the manufacturer's instructions, and Northern blot analysis was performed as previously described (53).
HBV replication assay.
Cultured primary hepatocytes were washed with cold PBS at the indicated times post-AdHBV infection; HBV replication was analyzed by Southern blotting as previously described (54).
Analysis of HNF4α nuclear localization.
Cultured primary hepatocytes were washed with cold PBS, scraped into PBS, and pelleted at 2,000 rpm for 5 min at 4°C. The cells were then suspended in cold buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol [DTT]) and incubated on ice for 15 min with occasional mixing, after which NP-40 (0.1%, vol/vol) was added to the cells. The cells were vortexed for 10 s and pelleted at 14,000 rpm for 10 s, and the supernatant was discarded. The pellet was then resuspended in buffer B (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.4 M NaCl, 0.2 mM EDTA, 25% [vol/vol] glycerol, 1 mM DTT) and incubated on ice for 20 min with occasional mixing. The cells were then pelleted at 14,000 rpm for 2 min, and the nuclear extract supernatant was collected for subsequent SDS-PAGE and Western blot analyses and quantification as described above.
Cytochrome c release assay.
Cultured primary hepatocytes were scraped into the culture medium, and both living and dead cells were collected. The cells were then pelleted by centrifugation at 2,000 rpm for 5 min, and the supernatant was discarded. The cytosolic and mitochondrial fractions were separated by using the Mitochondria Isolation kit for mammalian cells (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. Cytosolic cytochrome c levels were then analyzed by Western blotting and quantified as described above.
HBV secretion assay.
Enveloped HBV particles were captured, and quantitative real-time PCR (qRT-PCR) was performed as previously described (55). Briefly, 96-well plates were coated overnight with 250 ng of HBsAg antibody at 4°C, and the plate was then washed three times with TBST. The plates were blocked with 2% bovine serum albumin (BSA) for 1 h at 37°C and again washed with TBST. The antibody-coated plates were then incubated overnight with HBV-containing sample medium at 4°C. The sample medium was removed, and the plate was washed three times with TBST. qRT-PCR was performed on enveloped HBV that remained in the plates; the PCR mixture contained a forward primer (5′-ACTCGTGGTGGACTTCTCTC-3′), a reverse primer (5′-AAGATGAGGCATAGCAGCAGG-3′), and the Power SYBR green PCR master mix (Applied Biosystems, Warrington, United Kingdom), and PCR was conducted according to the master mix manufacturer's instructions. Known copy numbers of HBV plasmid DNA were used in qRT-PCR to generate a standard curve for the quantification of secreted HBV.
Real-time PCR of HNF4α targets.
Cultured primary rat hepatocytes were transfected with the control vector, the pGEMHBV expression plasmid, or the mAKT2 expression plasmid. The cells were harvested 48 h after transfection, and total RNA was isolated by using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. mRNA was converted to cDNA by using Moloney murine leukemia virus (M-MuLV) reverse transcriptase (New England BioLabs, Inc., Ipswich MA) according to the manufacturer's instructions. qRT-PCR was carried out to amplify cDNA by using Power Sybr green PCR master mix (Applied Biosystems, Warrington, United Kingdom) according to the manufacturer's directions. Three specific HNF4α targets, aldolase B (ALDOB), cytochrome p450 family 1 subfamily A polypeptide 2 (CYP1A2), and glucose-6-phosphatase (G6P), were amplified by using the following primers: ALDOB forward primer 5′-AGGTGCCCCGCTTGCAGGAAC-3′ and reverse primer 5′-GCTGGCGTAGCGAGCCAGAGC-3′, CYP1A2 forward primer 5′-GCGCCCTGTTCAAGCACAGTGAGAA-3′ and reverse primer 5′-GCCAATCACCGTGTCCAGCTCCT-3′, and G6P forward primer 5′-AGTCTTGTCAGGCATTGCTGTGGC-3′ and reverse primer 5′-CCCACTCGGGGCGCTCACAC-3′. The fold difference in the expression levels of HNF4α target mRNAs was calculated by the delta-delta threshold cycle (CT) method (56).
RESULTS
HBx activates AKT in cultured primary hepatocytes.
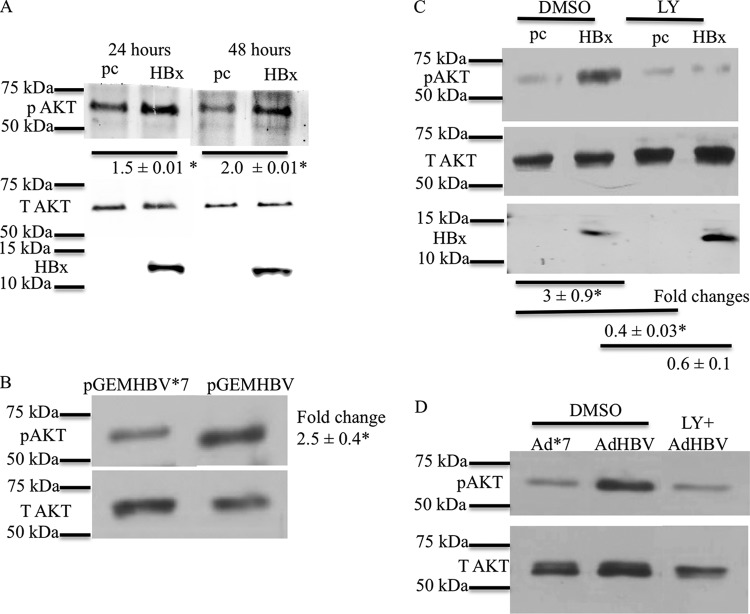
We first tested whether HBx activates AKT in cultured primary rat hepatocytes. Twenty-four hours after isolation and plating, cultured primary rat hepatocytes were transfected with an HBx expression plasmid or the pcDNA control vector. Rat hepatocytes were harvested at 24 and 48 h posttransfection, and Western blot analysis with a phosphospecific AKT antibody was performed to check AKT activation. Phosphorylation of AKT at S473 is a marker for AKT activation (28, 29). HBx-transfected cultured primary rat hepatocytes had higher levels of phosphorylated AKT (S473) than did control-transfected cells, indicating that HBx activates AKT in primary rat hepatocytes. In addition, HBx activation of AKT in primary rat hepatocytes was sustained for at least 48 h after transfection (Fig. 1A). Similar results were obtained with cultured primary rat hepatocytes infected with a recombinant adenovirus that expresses HBx, demonstrating that, when necessary to achieve expression of HBx or HBV in 100% of the cultured hepatocytes, recombinant adenoviruses that express HBV or HBx can be used to study HBV- or HBx-specific effects in cultured primary hepatocytes (J. C. Garner and M. J. Bouchard, unpublished results). These observations are also consistent with our previously reported studies that compared HBx activities in cells transfected with an HBx expression plasmid to those in cells infected with an HBx-expressing recombinant adenovirus (26, 27).
FIG 1.
HBx activates AKT in primary rat hepatocytes. (A) Primary rat hepatocytes were transfected with the HBx expression plasmid or the pcDNA control vector. The cells were collected at different time points, and the levels of pAKT (Ser473) were measured by Western blot analyses. T AKT, total AKT. (B) Primary rat hepatocytes were transfected with the pGEMHBV or pGEMHBV*7 (HBx-deficient HBV) expression plasmid. Western blot analysis was performed to check the levels of pAKT in pGEMHBV- and pGEMHBV*7-transfected cells. (C) Primary rat hepatocytes were transfected with the HBx expression plasmid or the pcDNA control vector. The cells were treated with either the DMSO control or LY294002 (LY) (PI3K inhibitor), and Western blot analysis was performed to check the levels of pAKT in HBx- and pcDNA-transfected cells. (D) Primary human hepatocytes were infected with AdHBV or Ad*7 (without HBx) and then treated with either the vehicle control or LY294002. Western blot analysis was conducted to assess the levels of AKT activation. Each Western blot shows one representative result from at least two independent experiments. The band intensities in each Western blot were measured by using ImageJ software; the band intensities of pAKT were divided by the band intensities of total AKT, and the ratios are represented in the terms of fold differences. The differences indicated are average fold changes from three independent experiments ± standard errors. An asterisk represents a P value of <0.05, determined by Student's t test.
Cultured primary rat hepatocytes were next transfected with HBV expression plasmid pGEMHBV or mutant HBV expression plasmid pGEMHBV*7, which cannot express HBx due to a stop codon inserted in the HBx gene. Expression of HBV from pGEMHBV resulted in the activation of AKT compared to pGEMHBV*7-transfected primary rat hepatocytes (Fig. 1B). Expression of all HBV mRNAs, including HBx, is under the control of their own promoters in the pGEMHBV vector. HBx expression from pGEMHBV in cultured primary rat hepatocytes is below detectable levels by standard Western analysis techniques, similar to observations of HBx expression during an authentic HBV infection; however, we have previously shown that HBx expression can be detected from pGEMHBV when the lysates of a large number of cells are concentrated and analyzed by Western blotting (49, 57). Therefore, the observed activation of AKT in both pGEMHBV- and HBx-transfected cultured primary rat hepatocytes confirmed that that activation of AKT in HBx-transfected primary rat hepatocytes is not an artifact of HBx overexpression and that AKT is activated when HBx is expressed both alone and in the context of the HBV genome. Activation of AKT by HBx in cultured primary rat hepatocytes was blocked by LY294002, a general PI3K inhibitor (Fig. 1C). Although there were variations in the levels of AKT activation in different experiments, they likely reflect slight differences in the transfection efficiencies of different hepatocyte samples and the fact that the hepatocytes were isolated from outbred rats. Importantly, AKT activation was always higher in HBx-expressing hepatocytes than in controls.
We also analyzed the effect of HBx expression on AKT activation in cultured primary human hepatocytes. Cultured primary human hepatocytes were infected with a recombinant adenovirus expressing HBV (AdHBV) or a recombinant adenovirus expressing an HBx-defective mutant HBV (Ad*7). Although cultured primary human hepatocytes can be directly infected with HBV, for these studies and subsequent studies described below, we used AdHBV for infecting cultured primary human hepatocytes because our goal was to directly compare HBx effects in the human and rat primary hepatocyte systems using the recombinant adenovirus system. We also noted in our primary rat hepatocyte studies that in order to detect the consistent but moderate effect of HBx on AKT signaling, a high percentage of cells must express HBx, either alone or in the context of the HBV genome, which can be difficult to achieve by the less-efficient, direct HBV infection of cultured primary human hepatocytes. Finally, due to the extremely low level of replication of HBV*7 (HBx-deficient HBV), isolation and purification of high-titer samples of this HBx mutant HBV are technically difficult, which hampers a comparison of primary human hepatocytes that are directly infected with HBV or HBV*7. AdHBV-infected primary human hepatocytes had higher levels of AKT phosphorylation (S473) than did Ad*7-infected primary human hepatocytes. LY294002 blocked the increase in AKT phosphorylation in AdHBV-infected cultured primary human hepatocytes (Fig. 1D); these observations confirmed that AKT can be activated by HBV, and specifically by HBx, in primary human hepatocytes. In addition to demonstrating HBV activation of AKT in human hepatocytes, the natural site of an HBV infection, these studies also confirmed, as we previously reported, that cultured primary rat hepatocytes can serve as a surrogate system for studying HBV and HBx effects on cellular signal transduction pathways in human hepatocytes (26).
PI3K/AKT signaling reduces HBV replication and HBV mRNA levels.
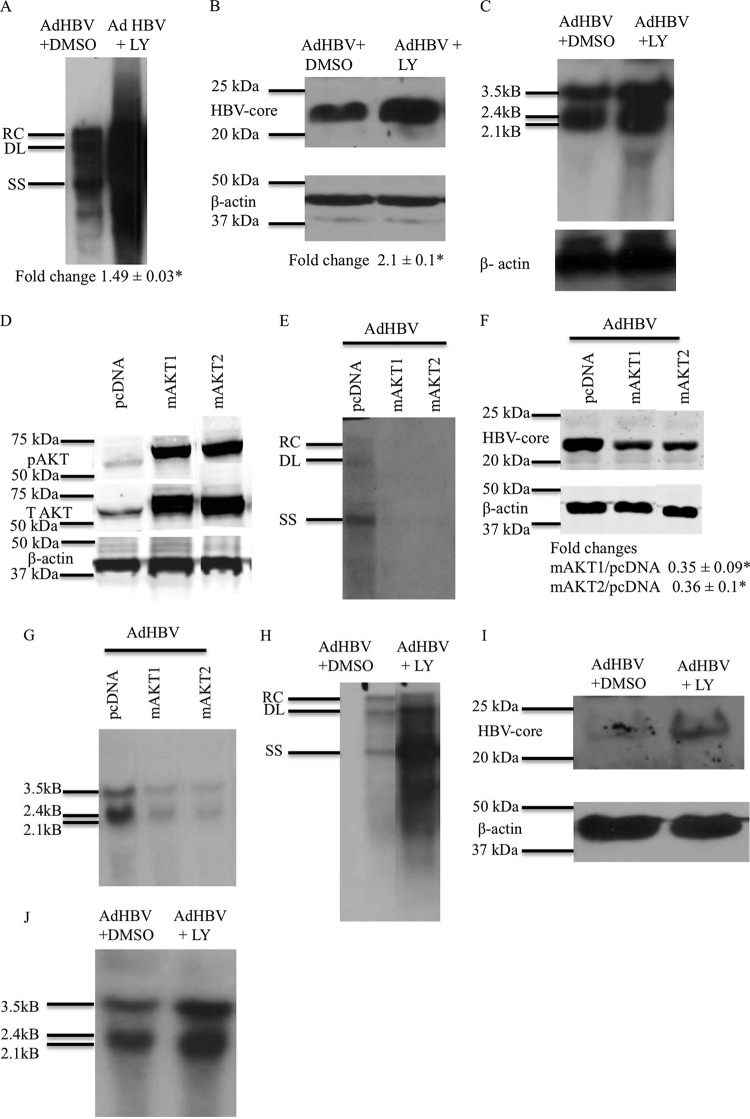
To examine the effect of PI3K/AKT signaling on HBV replication in primary hepatocytes, 24 h after isolation and plating, cultured primary rat hepatocytes were infected with AdHBV; at 12 h post-AdHBV infection, the cells were treated with either LY294002 or the dimethyl sulfoxide (DMSO) vehicle control. Because recombinant adenoviruses can infect 100% of cultured primary rat hepatocytes, the use of AdHBV in our studies facilitates the detection of HBV replication, which can be difficult to detect with the lower level of cultured primary rat hepatocytes that are transfected with the pGEMHBV expression plasmid. AdHBV-infected primary rat hepatocytes were harvested 48 h after AdHBV infection, and HBV replication was analyzed by Southern blotting of cytosolic, encapsidated HBV replicative intermediates, as previously described (54). LY294002 inhibition of PI3K/AKT signaling significantly enhanced HBV replication in cultured primary rat hepatocytes (Fig. 2A). The level of HBV core protein was also elevated when PI3K/AKT signaling was inhibited by LY294002 (Fig. 2B). Overall, these results indicate that inhibition of the PI3K/AKT pathway in cultured primary rat hepatocytes elevates HBV core protein levels and stimulates HBV replication.
FIG 2.
AKT/PI3K signaling reduces HBV replication and HBV mRNA levels. (A) Primary rat hepatocytes were infected with AdHBV, and the cells were treated with either LY294002 or the vehicle control. Southern blot analysis of core particles that were isolated from the infected hepatocytes was performed to assess HBV replication. RC, relaxed circular; DL, double-stranded linear; SS, single stranded. (B) Western blot analysis was performed on the samples from panel A to check the levels of HBV core protein. Southern and Western data shown here are one representative result of three independent experiments, each with duplicate samples. The difference indicated below the Western blot is the average fold change from three independent experiments ± standard error. An asterisk represents a P value of <0.05, determined by using Student's t test. (C) Primary rat hepatocytes were infected with AdHBV, and the cells were treated with either LY294002 or the vehicle control. Northern blot analysis was performed to ascertain the levels of HBV mRNAs. (D) Western blot analysis to check the expression of pAKT in primary rat hepatocytes transfected with mAKT1 or mAKT2 expression plasmids (constitutively active forms of AKTs). T AKT, total AKT. (E) Primary rat hepatocytes were transfected with the pcDNA control vector or the mAKT1 or mAKT2 expression vector and were also infected with AdHBV. Southern blot analysis was performed to check the levels of HBV replication. (F) Western blot analysis was performed on the samples from panel E to check the levels of HBV core protein. (G) Primary rat hepatocytes were treated as described above for panel E, and Northern blot analysis was performed to determine the levels of HBV mRNA. (H) Primary human hepatocytes were treated as described above for panel A, and Southern blot analyses were performed to assess HBV replication. (I) Western blot analyses were performed on the samples from panel H to check the levels of HBV core protein. (J) Primary human hepatocytes were treated as described above for panel A, and Northern blot analyses were performed to check the levels of HBV RNA. The blots shown in panels C to G are representative of at least 2 independent experiments, each with duplicate samples. The differences indicated in panel F are average fold changes from 2 independent experiments ± standard errors. An asterisk represents a P value of <0.05, determined by using Student's t test.
We also compared our observations of primary rat hepatocytes to the effect of AKT signaling on HBV replication in cultured primary human hepatocytes. We observed similar effects of PI3K/AKT signaling on HBV replication in cultured primary human hepatocytes; inhibition of PI3K/AKT signaling enhanced HBV replication and elevated HBV core protein levels (Fig. 2H and I). These results confirm that PI3K/AKT signaling also modulates HBV replication in primary human hepatocytes and again confirm that cultured primary rat hepatocytes can serve as a surrogate model for studying factors that regulate HBV replication in human hepatocytes.
Because PI3K/AKT inhibition increased HBV core protein levels, we speculated that inhibition of the PI3K/AKT pathway might also result in increased transcription of HBV RNAs. Therefore, we examined the effect of the inhibition of PI3K/AKT signaling on HBV mRNA levels. Primary rat hepatocytes were treated as described in the legend of Fig. 2A, and Northern blot analysis was performed to analyze HBV mRNA levels. Inhibition of PI3K/AKT signaling by LY294002 elevated the levels of HBV mRNAs in cultured primary rat hepatocytes (Fig. 2C). We observed similar PI3K/AKT-dependent effects on HBV mRNA levels when PI3K/AKT signaling was inhibited in AdHBV-infected cultured primary human hepatocytes (Fig. 2J).
LY294002 is a general inhibitor of the PI3K/AKT pathway; therefore, to specifically analyze the role of AKT in the regulation of HBV mRNA levels and HBV replication, we utilized constitutively active AKT1 and AKT2 (58). The addition of a myristoylation signal to AKT targets AKT to the plasma membrane, which increases basal AKT phosphorylation and constitutively activates AKT so that AKT initiates downstream signaling in the absence of upstream activators (59). Myristoylated AKT1 (mAKT1) and mAKT2 were used as the constitutively active forms of AKT. We did not assess the effect of the third AKT isoform, AKT3, in our studies because the expression level of AKT3 in liver is very low compared to the expression levels of AKT1 and AKT2 (60). We first confirmed that the mAKT1 and mAKT2 expression plasmids express constitutively active AKT1 and AKT2 (Fig. 2D).
We next examined the effect of mAKT1 or mAKT2 on HBV replication. Cultured primary rat hepatocytes were transfected with mAKT1, mAKT2, or pcDNA, and at 6 h posttransfection, primary rat hepatocytes were infected with AdHBV. Primary rat hepatocytes were collected 48 h after AdHBV infection, and HBV replication was analyzed by Southern blotting. mAKT1 and mAKT2 reduced the levels of HBV DNA compared to the levels in control-transfected cells (Fig. 2E). HBV core protein levels were also reduced by mAKT1 and mAKT2 (Fig. 2F). We also analyzed HBV mRNA levels by Northern blot analyses; mAKT1 and mAKT2 decreased HBV mRNA levels in cultured primary rat hepatocytes (Fig. 2G). We were unable to repeat similar transfection experiments in cultured primary human hepatocytes because transfection efficiencies of primary human hepatocytes are extremely low; however, our previously reported studies and those with cultured primary human hepatocytes described here strongly suggest that HBV and HBx effects are identical in cultured primary human and rat hepatocytes (23, 26, 27). Overall, the results of these AKT studies demonstrate that the activation of either AKT1 or AKT2 decreases HBV mRNA and core protein levels and HBV replication in cultured primary rat hepatocytes.
HNF4α acts downstream of AKT to decrease HBV mRNA levels.
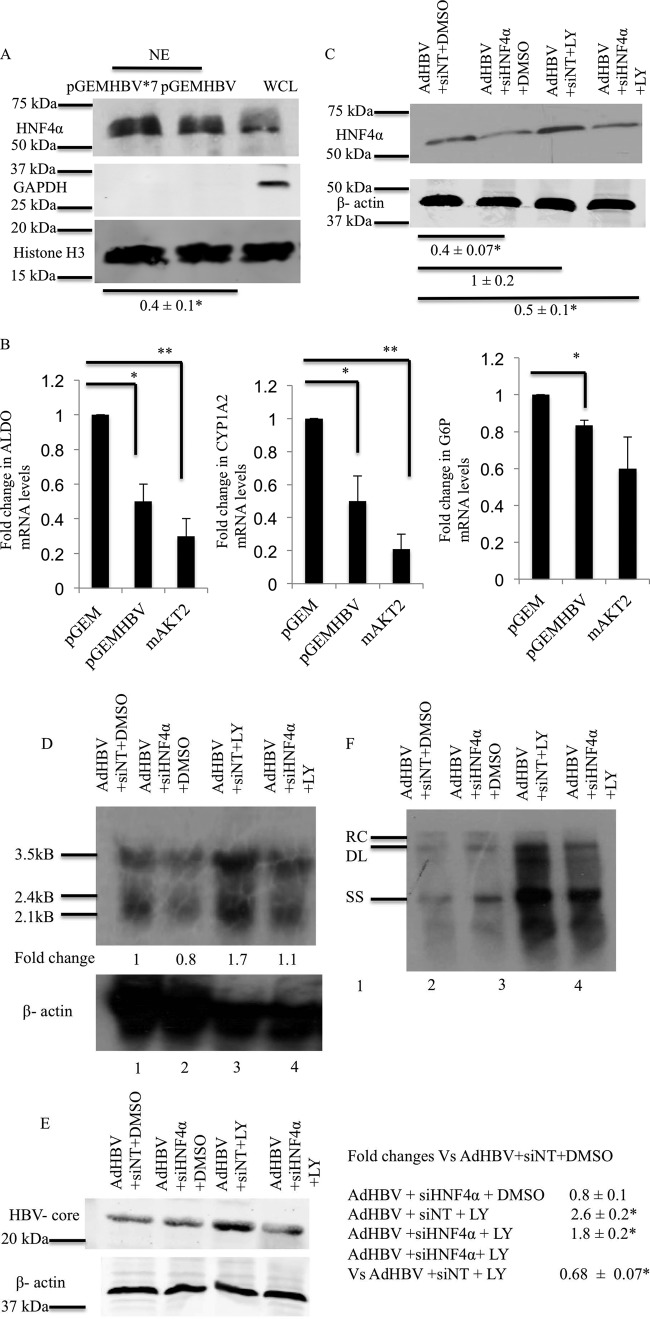
The observed downregulation of HBV mRNA levels suggested that the activation of AKT by HBx might modulate the activity of one or more transcription factors that regulate HBV RNA levels. HNF4α was previously shown to be one transcription factor that can influence the level of HBV replication (61). HNF4α is also a target of AKT; activated AKT can phosphorylate HNF4α, causing translocation of HNF4α out of the nucleus (42). We analyzed the levels of HNF4α in the nucleus of cultured primary rat hepatocytes that were transfected with either pGEMHBV or pGEMHBV*7. Cultured primary rat hepatocytes were harvested at 24 h posttransfection, and the level of HNF4α in nuclear extracts was determined by Western blot analysis. pGEMHBV-transfected primary rat hepatocytes had lower levels of HNF4α in the nucleus than did pGEMHBV*7-transfected primary rat hepatocytes (Fig. 3A). Histone H3 was used as the loading control and nuclear marker, and GAPDH was used as the cytosolic marker and indicator of fraction purity.
FIG 3.
HNF4α acts downstream of AKT to decrease HBV mRNA levels. (A) Primary rat hepatocytes were transfected with the pGEMHBV or pGEMHBV*7 expression plasmid, and nuclear extracts (NE) were prepared after harvesting of the cells. Western blot analysis of the nuclear extracts was performed to determine the levels of HNF4α. Histone H3 was used as the nuclear marker and the loading control, and GAPDH was used as the cytosolic marker. The data are representative of 3 independent experiments, and the difference indicated is the average fold change ± standard error. WCL, whole-cell extracts. (B) Primary rat hepatocytes were transfected with either the pGEM control vector or the pGEMHBV or mAKT2 expression vector. Real-time PCR was conducted by using primers for three HNF4α direct-target genes. ALDO, aldolase; CYP1A2, cytochrome p450 family 1 subfamily A polypeptide 2; G6P, glucose 6 phosphatase. (C) Primary rat hepatocytes were transfected with either siNT (100 nM) or siHNF4α (100 nM), and the cells were infected with AdHBV. The cells were treated with either the vehicle control (DMSO) or LY294002 (LY), and Western blot analysis was performed to check the efficiency of HNF4α knockdown. (D) Primary rat hepatocytes were treated as described above for panel C, and Northern analysis was performed to determine the levels of HBV mRNA. (E) Primary rat hepatocytes were treated as described above for panel C, and Western blot analyses was performed to check the levels of HBV core protein. (F) Primary rat hepatocytes were treated as described above for panel C, and Southern blot analysis was performed to ascertain the levels of HBV replication. The data shown in panels B to F are representative of at least two independent experiments. The differences indicated in panel E are average fold changes from the three independent experiments ± standard errors. Statistical analysis was conducted by using Student's t test, where * and ** represent a P value of <0.05.
To further confirm the regulation of HNF4α by HBV in cultured primary hepatocytes, we analyzed mRNA expression levels of three specific HNF4α target genes, ALDOB, CYP1A2, and G6P, which were previously described to be HNF4α-specific target genes (62). Cultured primary rat hepatocytes were transfected with the control vector pGEM, pGEMHBV, or the mAKT2 expression plasmid, and the cells were harvested 48 h after transfection. Because we observed similar effects on HBV replication in cells transfected with either the mAKT1 or mAKT2 expression plasmids, we used only the mAKT2 expression plasmids in the studies described here. qRT-PCR was performed to determine the mRNA expression levels of the HNF4α target genes; HBV decreased the expression levels of ALDOB, CYP1A2, and G6P, supporting the notion that HBV negatively regulates the activity of HNF4α (Fig. 3B). As expected, mAKT2 also decreased the expression levels of HNF4α targets. This indicates that the activation of AKT in hepatocytes inhibits the transcriptional activity of HNF4α and suggests that HBV could regulate HNFα by activating AKT.
To determine whether HNF4α acts downstream of AKT to regulate HBV replication, we next used siRNAs targeting HNF4α (siHNF4α) to decrease the expression of HNF4α in cultured primary rat hepatocytes. At 100 nM siHNF4α, we saw efficient knockdown of HNF4α compared to that in nontargeting siRNA (siNT)-transfected, cultured primary rat hepatocytes (data not shown). Cultured primary rat hepatocytes were transfected with 100 nM siHNF4α or siNT, and at 24 h posttransfection, primary rat hepatocytes were infected with AdHBV. At 12 h post-AdHBV infection, primary rat hepatocytes were treated with either LY294002 or the vehicle control. The cells were harvested 36 h after LY294002 treatment, and Western blot analyses was performed to assess the efficiency of HNF4α knockdown (Fig. 3C). We also determined whether HNF4α knockdown prevented an increase in the HBV mRNA level when AdHBV-infected primary rat hepatocytes were treated with LY294002. Cultured primary rat hepatocytes were treated as described in the legend of Fig. 3C, and HBV RNA levels were analyzed by Northern blotting. In the presence of siNT and LY294002 treatment, HBV mRNA levels in AdHBV-infected primary rat hepatocytes were higher than those in vehicle control-treated cells (Fig. 3D, lanes 1 and 3). However, when HNF4α was knocked down and cells were treated with LY294002, the level of HBV mRNA was not as high as that in siNT-transfected cells that were treated with LY294002 (Fig. 3D, lanes 3 and 4). In a similar experiment, we also analyzed HBV core protein levels after HNF4α knockdown. LY294002 treatment increased the HBV core protein levels in AdHBV-infected primary rat hepatocytes (Fig. 3E, lanes 1 and 3); however, when HNF4α was knocked down, we did not see an increase in HBV core protein levels after LY294002 treatment (Fig. 3E, lanes 3 and 4). We also analyzed HBV replication by Southern blot analysis of cytosolic, encapsidated HBV replicative intermediates in AdHBV-infected cultured primary rat hepatocytes. When HNF4α was knocked down and primary rat hepatocytes were treated with LY294002, the knockdown of HNF4α expression blocked the LY294002-mediated increase in HBV replication (Fig. 3F). Although the knockdown of HNF4α in AdHBV-infected cells that were treated with LY294002 did not reduce HBV replication to control levels (AdHBV, siNT, and DMSO), this is most likely due to an incomplete knockdown of HNF4α. It is also possible that there is more than one factor downstream of AKT that inhibits HBV replication. Overall, the results of these studies suggest that HBx activation of AKT decreases HNF4α nuclear localization, causing decreases in HBV mRNA levels, core protein levels, and HBV replication.
Inhibition of the PI3K/AKT pathway does not inhibit HBV secretion.
Our results described above indicated that the inhibition of the AKT pathway increased the levels of HBV replicative intermediates contained within cytosolic core particles, which is the focus of our standard HBV replication assay. We next determined if the increase in the level of cytosolic HBV replicative intermediates reflected a true measure of the overall production of enveloped HBV or an accumulation of cytosolic HBV DNA-containing particles resulting from a defect in HBV secretion caused by the inhibition of AKT. Cultured primary rat hepatocytes were infected with AdHBV and then treated with LY294002 or the vehicle control. Primary rat hepatocyte cell culture medium supernatants were collected 48 h after treatment, and enveloped HBV particles were captured by using an anti-HBs antibody as previously described (55). The captured HBV particles were then subjected to real-time PCR with HBV-specific primers to quantify the levels of enveloped HBV particles in the supernatant. A standard curve was made by plotting the change in the CT values with increasing numbers of HBV genomic copies (Fig. 4A). AdHBV-infected primary rat hepatocytes that were treated with LY294002 had higher levels of enveloped HBV particles in the supernatant than did control primary rat hepatocyte medium supernatants (Fig. 4B). The HBV copy number in the supernatant was calculated on the basis of the standard curve shown in Fig. 4A. This result demonstrates that the increased cytosolic core particle HBV DNA level in primary rat hepatocytes after inhibition of the PI3K/AKT pathway is not due to a defect in the secretion of HBV and that the standard cytosolic core particle HBV replication assay, in this case, accurately reflects overall levels of HBV replication.
FIG 4.

Inhibition of the AKT pathway does not inhibit HBV secretion. (A) Standard curve showing the change in CT values with increasing numbers of HBV genomic copies. (B) The supernatant from AdHBV-infected primary rat hepatocytes that were treated with either the vehicle control or LY294002 was used to detect secreted, enveloped HBV particles by qRT-PCR. The copy number of the HBV genome in the supernatant was calculated based upon the standard curve shown in panel A and is represented as the fold difference. The graph represents averages of data from 4 independent experiments, each with triplicate samples. Statistical analysis was conducted by using Student's t test, where the asterisk represents a P value of <0.05.
HBx activates AKT to inhibit apoptosis in cultured primary rat hepatocytes.
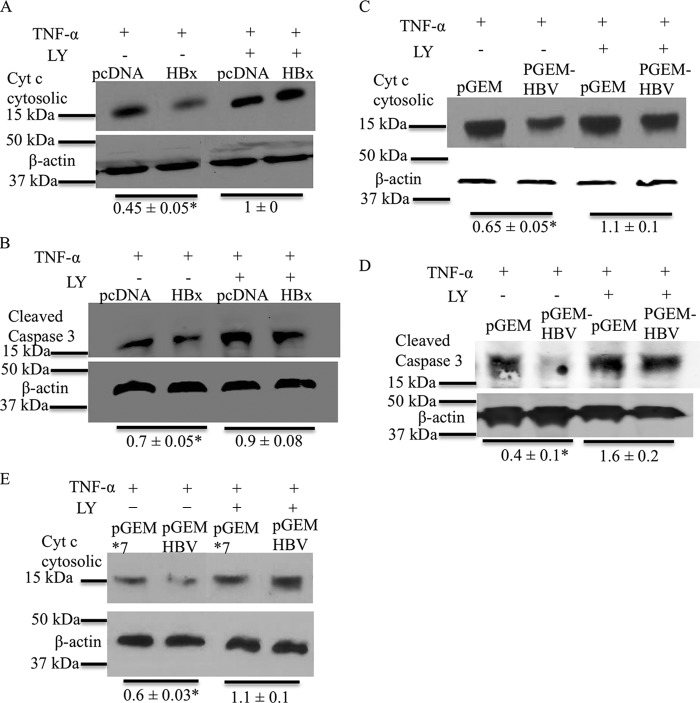
HBx activates AKT in cultured primary rat hepatocytes, even though activation of PI3K/AKT signaling inhibits HBV replication. This led us to question why HBV would activate a pathway that decreases its own replication. The PI3K/AKT pathway is a cell survival pathway (63). We previously demonstrated that HBx inhibits spontaneous and TNF-α-mediated apoptosis in cultured primary rat hepatocytes (14). Therefore, we next determined whether HBx activation of AKT in cultured primary rat hepatocytes is linked to HBx inhibition of apoptosis. Cultured primary rat hepatocytes were transfected with the HBx expression plasmid or the pcDNA control, and at 6 h posttransfection, primary rat hepatocytes were treated with TNF-α and LY294002 or the vehicle control. Primary rat hepatocytes were harvested after 36 h of treatment, and mitochondrial and cytosolic fractions were separated. The cytosolic fractions were then analyzed by Western blotting to determine the levels of cytosolic cytochrome c. Cytochrome c is a mitochondrial protein that is released from mitochondria into the cytosol upon activation of the apoptotic pathways; the level of cytosolic cytochrome c can be used as an indicator of the level of activation of apoptosis (14, 64). In agreement with our previously reported observations, HBx-expressing cultured primary rat hepatocytes had lower levels of cytosolic cytochrome c than did control cells, indicating that HBx inhibited TNF-α-induced apoptosis (Fig. 5A) (14). However, when the activation of AKT was blocked by LY294002, HBx was no longer able to inhibit TNF-α-mediated apoptosis in cultured primary rat hepatocytes, and both HBx- and vector control-transfected cells had similar levels of cytosolic cytochrome c (Fig. 5A). To further confirm our apoptosis results, we analyzed levels of cleaved caspase-3, another apoptosis marker. The levels of cleaved caspase-3 were analyzed by Western blot analysis of cultured primary rat hepatocytes that were treated the same way as described in the legend of Fig. 5A. As shown in Fig. 5B, HBx-expressing primary rat hepatocytes had lower levels of cleaved caspase-3 in response to TNF-α treatment, indicating inhibition of apoptosis, but this inhibition of apoptosis by HBx was lost upon LY294002 treatment. To determine whether similar effects on apoptosis were observed in cultured primary rat hepatocytes transfected with the entire HBV genome, we also analyzed the consequence of apoptosis after blocking the PI3K/AKT pathway in pGEMHBV-transfected cultured primary rat hepatocytes; we previously demonstrated that inhibition of apoptosis in pGEMHBV-transfected hepatocytes was directly linked to HBx expression (14). Cultured primary rat hepatocytes were transfected with either pGEMHBV or the pGEM control, and 6 h after transfection, the cells were treated with TNF-α and LY294002 or the vehicle control. Cultured primary rat hepatocytes were harvested after 16 h of treatment, and either the cells were processed to separate the cytosolic fraction from mitochondria to determine the levels of cytosolic cytochrome c or the samples were directly analyzed to determine the levels of cleaved caspase-3. HBV-expressing cells had lower levels of cleaved caspase-3 and cytosolic cytochrome c, indicating that HBV inhibits TNF-α-mediated apoptosis; however, HBV was not able to inhibit TNF-α-mediated apoptosis when AKT activation was blocked by LY294002 (Fig. 5C and D). We also compared the regulation of apoptosis in pGEM*7- and pGEMHBV-transfected primary rat hepatocytes to more specifically assess HBx regulation of apoptosis in pGEMHBV-expressing cells; the results were similar to those shown in Fig. 5C (Fig. 5E). Overall, these studies demonstrate that HBV, and specifically HBx, activation of the PI3K/AKT pathway in hepatocytes inhibits apoptosis.
FIG 5.
HBx activates the AKT pathway to inhibit apoptosis in primary hepatocytes. (A) Primary rat hepatocytes were transfected with either the pcDNA control vector or the HBx expression plasmid. The cells were then treated with TNF-α and LY294002 or the DMSO vehicle control. The cells were harvested, and Western blot analysis was performed to check the levels of cytosolic cytochrome c. (B) Primary rat hepatocytes were transfected and treated as described above for panel A, and Western blot analysis was performed to check the levels of cleaved caspase-3. (C and D) Primary rat hepatocytes were transfected with either pGEMHBV or the pGEM control vector and treated as described above for panel A. Western analysis was performed to check the levels of cytosolic cytochrome c (C) and cleaved caspase-3 (D). (E) Primary rat hepatocytes were transfected with either the pGEM*7 control vector or the pGEMHBV expression plasmid, and the cells were treated as described above for panel A. Western blot analysis was performed to check the levels of cytosolic cytochrome c. The blots shown are representative of data from 2 independent experiments. The differences indicated are the average fold changes from the 2 independent experiments ± standard errors. An asterisk represents a P value of <0.05, determined by using Student's t test.
DISCUSSION
The HBV HBx protein can modulate numerous cell signal transduction pathways and stimulate HBV replication (27, 54, 65, 66). While some HBx-regulated cell signaling pathways such as the Src–mitogen-activated protein kinase (MAPK) pathway and calcium signaling stimulate HBV replication, others, such as the NF-κB signaling pathway, can inhibit HBV replication in certain cellular contexts (27, 65–67). HBx elevation of cytosolic calcium levels activates Src kinases, which in turn activate the HBV polymerase/reverse transcriptase to stimulate HBV replication (65, 67, 68). HBx also activates the transcription factor NF-κB; however, activation of NF-κB inhibits HBV replication in HepG2 cells by decreasing the stability or formation of HBV core particles (66). The regulation of HBV replication by various cell signal transduction pathways has been studied mostly with immortalized or transformed cell lines, and the results of many of these studies suggest that some HBx activities may be cell context dependent. Therefore, the results of studies conducted with immortalized or transformed cell lines may or may not be relevant to HBx activities in normal hepatocytes and the associated effects on HBV replication (11, 44). Cultured primary rat and human hepatocytes are model systems for studying HBx and HBV effects in normal, differentiated hepatocytes (11, 14, 26, 27, 49).
In the present study, we demonstrated that HBx activates AKT in ex vivo, cultured primary human and rat hepatocyte model systems and that the activation of AKT decreased the transcription of HBV RNAs and HBV replication in these cells. We observed similar HBx activation of AKT when HBx was expressed alone or in the context of the HBV genome. It was previously reported that HBx expression levels might differentially affect transcription pathways and apoptosis in various experimental systems (69–71). In a chronically HBV-infected individual, the HBx expression level is low (72); therefore, expressing HBx in the context of the HBV genome was important to rule out the possibility of an artifact due to HBx overexpression. The mechanism(s) that underlies HBx activation of AKT in primary hepatocytes is under investigation. Previous studies conducted with transformed cell lines have shown that HBx activation of the AKT pathway could be linked to decreases in phosphatase and tensin homolog (PTEN) expression levels (73). PTEN dephosphorylates PI3K, and a reduction in the level of PTEN would facilitate AKT phosphorylation and activation. However, in our studies of cultured primary rat hepatocytes, we did not see any HBx-induced change in PTEN levels, suggesting that the activation of AKT in primary hepatocytes is not due to a reduction in PTEN expression levels (data not shown). Whether HBx affects PTEN activity in normal hepatocytes will be the subject of future studies.
HBV and HBx expression also decreased nuclear levels of HNF4α, which we linked to the activation of AKT. Blocking of the PI3K/AKT signaling pathway increased HBV replication in primary hepatocytes, and this effect was countered by knocking down HNF4α expression, indicating that AKT targeting of HNF4α can decrease HBV replication. HNF4α is a transcription factor that controls the expression of a number of genes in hepatocytes and is regulated by various posttranslational modifications (37–41). Previous studies have also shown that HNF4α binds to transcription enhancers in the HBV genome that stimulate the transcription of HBV mRNAs (61, 74). It was demonstrated previously that AKT can phosphorylate HNF4α to reduce HNF4α nuclear localization (42). Our studies also demonstrate that HBx decreases the nuclear localization of HNF4α and that transcript levels of HNF4α target genes are decreased by HBV, suggesting that the AKT-mediated decrease in HBV replication is linked to the reduction in HNF4α nuclear levels and the associated decreased levels of HBV mRNAs. In support of this notion, inhibition of the PI3K/AKT pathway increased HBV replication, but knockdown of HNF4α in hepatocytes prevented the increase of HBV replication that was observed when PI3K/AKT signaling was inhibited. Cumulatively, our studies suggest that the activation of AKT by HBx results in decreased HNF4α nuclear levels and decreased HBV replication, whereas the inhibition of the PI3K/AKT pathway allows increased nuclear HNF4α levels and increased HBV replication (Fig. 6). Although HNF4α knockdown in LY294002-treated cells did not bring the levels of HBV mRNA and HBV replication to the levels in control hepatocytes, we believe that this was caused by the incomplete knockdown of HNF4α in cultured primary rat hepatocytes due to the lower transfection efficiency of primary hepatocytes combined with the very high AdHBV infection efficiency of cultured primary hepatocytes. It is also possible that additional factors downstream of AKT regulate HBV replication, which will be the focus of future studies.
FIG 6.
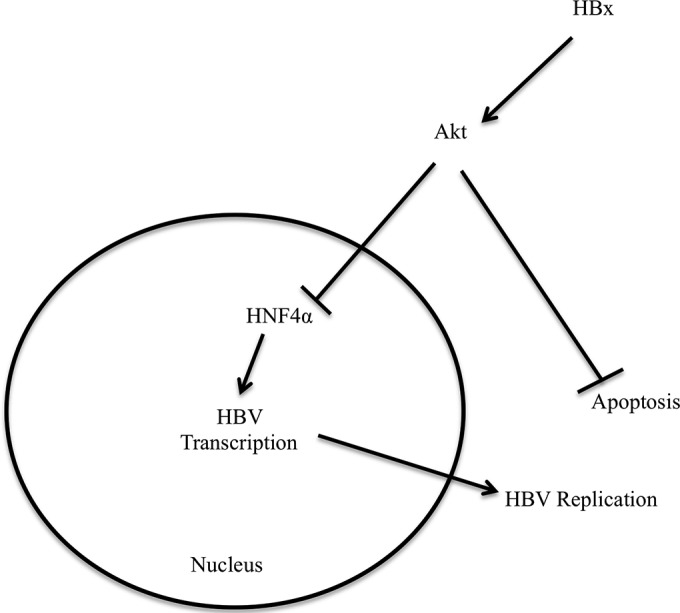
Model. HBx activates AKT in primary hepatocytes, causing a reduction in HNF4α transcription activity. HNF4α is an important transcription factor for the transcription of the HBV genome, and a reduction in HNF4α transcription activity decreases HBV transcription and, consequently, HBV replication. However, activation of AKT by HBx is important for the inhibition of apoptosis in HBV-infected hepatocytes. Overall, activation of the PI3K/AKT pathway decreases HBV replication but inhibits apoptosis, likely providing persistent, noncytopathic replication of HBV in hepatocytes.
Although our observation that the activation of AKT inhibits HBV replication suggests that the replication of an HBx-deficient HBV (HBV*7) might be enhanced by the inhibition of AKT, analysis of the effect of inhibiting AKT on the replication of an HBx-deficient HBV is challenging and perhaps not informative for a number of reasons. The level of HBV replication from HBV*7 is extremely low in cultured primary rat hepatocytes (14), even though, in the absence of HBx, HBV*7 failed to activate AKT (Fig. 1B). In HBV*7-expressing cells, the effect of inhibiting AKT with LY294002 would show the consequence of inhibiting endogenous levels of active AKT on HBV replication rather than the effect of inhibition of HBx-specific activation of AKT. Moreover, this analysis would be conducted with cells where requisite HBx activities that stimulate HBV replication would not be present; HBV*7 does not replicate well in primary hepatocytes because signaling pathways that are activated by HBx to stimulate HBV replication remain inactive (14). We therefore did not explore the effect of the PI3K/AKT pathway on the replication of HBx-deficient HBV.
The observation that HBx activation of AKT decreases HBV replication seems counterintuitive considering that many previous studies, including our own, have shown that HBx stimulates HBV replication (14). The PI3K/AKT pathway is an important cell survival pathway and has been shown to inhibit apoptosis in immortalized hepatocytes in the context of HBx expression (31, 33). We observed that HBx activation of AKT inhibits apoptosis in cultured primary rat hepatocytes. It was previously reported that inhibition of apoptosis in hepatocytes is important for HBV to release infectious progeny (14, 75). HBV is a noncytopathic virus, and inhibition of apoptosis in HBV-infected hepatocytes might be important for persistent replication of HBV in hepatocytes. Our ability to detect elevated levels of HBV replication in hepatocytes where the PI3K/AKT pathway was inhibited likely reflects the short time period of our assays; cells were collected at a time in which apoptotic signals had been activated but the cells had not yet progressed to complete apoptosis. During chronic HBV infection, the short-term HBV replication benefit that might result from not activating AKT would likely be balanced by the long-term benefit of a noncytopathic infection that could facilitate the persistence of HBV infection and the prolonged production of infectious progeny.
Development of HCC in an individual with chronic HBV infection can take decades. Although the underlying causes of HBV-associated HCC remain incompletely understood, three mechanisms that are not mutually exclusive have been proposed as contributors to the development of HBV-associated HCC: recurrent immune-mediated destruction of HBV-infected hepatocytes followed by compensatory liver regeneration; integration of HBV DNA into the host genome, which could alter the expression of cellular factors and, in turn, the control of normal cell functions; and modulation of hepatocyte physiology by HBV proteins such as HBx (8, 76–79). Although HBx is directly oncogenic in some HBx-transgenic mice (80, 81), many studies suggest that HBx has a cofactor role in hepatocyte transformation (11, 82–84). It is likely that subtle and moderate HBx activities, such as modulation of cellular signaling pathways, contribute to processes that link an HBV infection to the development of HCC. The subtlety of HBx effects and the cell-specific consequences of some HBx activities have sometimes made it difficult to precisely define HBx activities in an authentic HBV infection and to directly link these activities to hepatocyte transformation; however, these moderate effects of HBx, such as the ∼2-fold activation of AKT by HBx observed in our studies with cultured primary hepatocytes, are, in fact, consistent with the biology of an HBV infection, including the noncytopathic nature of the infection and the long time frame required for progression from a chronic HBV infection to the development of HCC (11, 85).
Activation of AKT and downregulation of HNF4α transcriptional activity could be independent or interconnected contributors to the development of HBV-associated HCC. HNF4α is an important transcription factor for liver development, and knockout of HNF4α in mice is embryonically lethal (86, 87). HNF4α expression maintains hepatocyte differentiation, and the loss of HNF4α transcriptional activity can lead to the dedifferentiation of hepatocytes and hepatocyte proliferation (36). Inhibition of HNF4α can initiate transformation in immortalized hepatocytes through a microRNA (miRNA) inflammatory feedback loop; perturbation of this feedback loop was also found for human hepatocellular carcinoma (62). The loss of HNF4α has been directly associated with the development of HCC, and HNF4α expression suppressed the development of HCC in a mouse model system (88, 89). AKT is an oncogene and is upregulated in many cancers, providing additional signaling pathways that could link HBx expression to the development of HCC (90–93). Finally, by lowering the level of HBV replication, HBx activation of AKT might influence HBV evasion of immune-mediated destruction of HBV-infected hepatocytes as an additional means to facilitate persistent HBV replication, a chronic HBV infection, and the eventual development of HCC.
Overall, the results of our studies with cultured primary rat and human hepatocytes demonstrate that HBx activation of the PI3K/AKT signaling pathway decreases HBV replication and cellular apoptosis. Although it is now well established that HBx is required for efficient HBV replication, our findings here suggest that HBx activates pathways that both stimulate and diminish HBV replication. While some of these cellular signaling pathways, such as the Src/MAPK and calcium signaling pathways, are important for stimulating HBV replication, others, such as the PI3K/AKT pathway, diminish HBV replication while enhancing cell survival. Therefore, we propose that HBx can function as a rheostat that balances cell survival and HBV replication, possibly to allow for persistent HBV replication. HBx activation of AKT and inhibition of HNF4α might also influence the development of HBV-associated HCC by facilitating the establishment of a chronic HBV infection, enhancing the dedifferentiation of hepatocytes, and activating AKT-regulated oncogenic pathways. Modulation of PI3K/AKT-regulated factors may provide therapeutic targets for inhibiting HBV replication and the development of HBV-associated HCC.
ACKNOWLEDGMENTS
We thank members of the Bouchard lab as well as Jan Hoek, Patrick Loll, Bradford Jameson, and Mauricio Reginato for advice and many helpful discussions.
This project was supported by NIH grant AI064844 to M.J.B.
REFERENCES
- 1.Nguyen VT, Law MG, Dore GJ. 2009. Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat 16:453–463. doi: 10.1111/j.1365-2893.2009.01117.x. [DOI] [PubMed] [Google Scholar]
- 2.Ganem D, Prince AM. 2004. Hepatitis B virus infection—natural history and clinical consequences. N Engl J Med 350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 3.Fattovich G, Bortolotti F, Donato F. 2008. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol 48:335–352. doi: 10.1016/j.jhep.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Cheng PN, Liu WC, Tsai HW, Wu IC, Chang TT, Young KC. 2011. Association of intrahepatic cccDNA reduction with the improvement of liver histology in chronic hepatitis B patients receiving oral antiviral agents. J Med Virol 83:602–607. doi: 10.1002/jmv.22014. [DOI] [PubMed] [Google Scholar]
- 5.Ghany MG, Doo EC. 2009. Antiviral resistance and hepatitis B therapy. Hepatology 49:S174–S184. doi: 10.1002/hep.22900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrillo RP. 2005. Current treatment of chronic hepatitis B: benefits and limitations. Semin Liver Dis 25(Suppl 1):S20–S28. doi: 10.1055/s-2005-915647. [DOI] [PubMed] [Google Scholar]
- 7.Zoulim F. 2004. Mechanism of viral persistence and resistance to nucleoside and nucleotide analogs in chronic hepatitis B virus infection. Antiviral Res 64:1–15. doi: 10.1016/j.antiviral.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard MJ, Navas-Martin S. 2011. Hepatitis B and C virus hepatocarcinogenesis: lessons learned and future challenges. Cancer Lett 305:123–143. doi: 10.1016/j.canlet.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeger C, Mason WS. 2000. Hepatitis B virus biology. Microbiol Mol Biol Rev 64:51–68. doi: 10.1128/MMBR.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locarnini S, Zoulim F. 2010. Molecular genetics of HBV infection. Antivir Ther 15(Suppl 3):S3–S14. doi: 10.3851/IMP1619. [DOI] [PubMed] [Google Scholar]
- 11.Rawat S, Clippinger AJ, Bouchard MJ. 2012. Modulation of apoptotic signaling by the hepatitis B virus X protein. Viruses 4:2945–2972. doi: 10.3390/v4112945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. 2006. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene 25:3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 13.Bouchard MJ, Schneider RJ. 2004. The enigmatic X gene of hepatitis B virus. J Virol 78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clippinger AJ, Gearhart TL, Bouchard MJ. 2009. Hepatitis B virus X protein modulates apoptosis in primary rat hepatocytes by regulating both NF-kappaB and the mitochondrial permeability transition pore. J Virol 83:4718–4731. doi: 10.1128/JVI.02590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leupin O, Bontron S, Schaeffer C, Strubin M. 2005. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J Virol 79:4238–4245. doi: 10.1128/JVI.79.7.4238-4245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang H, Delgermaa L, Huang F, Oishi N, Liu L, He F, Zhao L, Murakami S. 2005. The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J Virol 79:5548–5556. doi: 10.1128/JVI.79.9.5548-5556.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Z, Yen TS, Wu L, Madden CR, Tan W, Slagle BL, Ou JH. 2002. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J Virol 76:2579–2584. doi: 10.1128/jvi.76.5.2579-2584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keasler VV, Hodgson AJ, Madden CR, Slagle BL. 2007. Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J Virol 81:2656–2662. doi: 10.1128/JVI.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuge M, Hiraga N, Akiyama R, Tanaka S, Matsushita M, Mitsui F, Abe H, Kitamura S, Hatakeyama T, Kimura T, Miki D, Mori N, Imamura M, Takahashi S, Hayes CN, Chayama K. 2010. HBx protein is indispensable for development of viraemia in human hepatocyte chimeric mice. J Gen Virol 91:1854–1864. doi: 10.1099/vir.0.019224-0. [DOI] [PubMed] [Google Scholar]
- 20.Yoo YG, Oh SH, Park ES, Cho H, Lee N, Park H, Kim DK, Yu DY, Seong JK, Lee MO. 2003. Hepatitis B virus X protein enhances transcriptional activity of hypoxia-inducible factor-1alpha through activation of mitogen-activated protein kinase pathway. J Biol Chem 278:39076–39084. doi: 10.1074/jbc.M305101200. [DOI] [PubMed] [Google Scholar]
- 21.Becker SA, Lee TH, Butel JS, Slagle BL. 1998. Hepatitis B virus X protein interferes with cellular DNA repair. J Virol 72:266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Lluesma S, Schaeffer C, Robert EI, van Breugel PC, Leupin O, Hantz O, Strubin M. 2008. Hepatitis B virus X protein affects S phase progression leading to chromosome segregation defects by binding to damaged DNA binding protein 1. Hepatology 48:1467–1476. doi: 10.1002/hep.22542. [DOI] [PubMed] [Google Scholar]
- 23.Gearhart TL, Bouchard MJ. 2010. The hepatitis B virus X protein modulates hepatocyte proliferation pathways to stimulate viral replication. J Virol 84:2675–2686. doi: 10.1128/JVI.02196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung JK, Kwun HJ, Lee JO, Arora P, Jang KL. 2007. Hepatitis B virus X protein differentially affects the ubiquitin-mediated proteasomal degradation of beta-catenin depending on the status of cellular p53. J Gen Virol 88:2144–2154. doi: 10.1099/vir.0.82836-0. [DOI] [PubMed] [Google Scholar]
- 25.Benhenda S, Cougot D, Buendia MA, Neuveut C. 2009. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis. Adv Cancer Res 103:75–109. doi: 10.1016/S0065-230X(09)03004-8. [DOI] [PubMed] [Google Scholar]
- 26.Gearhart TL, Bouchard MJ. 2011. The hepatitis B virus HBx protein modulates cell cycle regulatory proteins in cultured primary human hepatocytes. Virus Res 155:363–367. doi: 10.1016/j.virusres.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gearhart TL, Bouchard MJ. 2010. Replication of the hepatitis B virus requires a calcium-dependent HBx-induced G1 phase arrest of hepatocytes. Virology 407:14–25. doi: 10.1016/j.virol.2010.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Courtney KD, Corcoran RB, Engelman JA. 2010. The PI3K pathway as drug target in human cancer. J Clin Oncol 28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engelman JA. 2009. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 30.Yuan TL, Cantley LC. 2008. PI3K pathway alterations in cancer: variations on a theme. Oncogene 27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YI, Kang-Park S, Do SI, Lee YI. 2001. The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J Biol Chem 276:16969–16977. doi: 10.1074/jbc.M011263200. [DOI] [PubMed] [Google Scholar]
- 32.Chung TW, Lee YC, Kim CH. 2004. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: involvement of invasive potential. FASEB J 18:1123–1125. doi: 10.1096/fj.03-1429fje. [DOI] [PubMed] [Google Scholar]
- 33.Shih WL, Kuo ML, Chuang SE, Cheng AL, Doong SL. 2000. Hepatitis B virus X protein inhibits transforming growth factor-beta-induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J Biol Chem 275:25858–25864. doi: 10.1074/jbc.M003578200. [DOI] [PubMed] [Google Scholar]
- 34.Sladek FM, Zhong WM, Lai E, Darnell JE Jr. 1990. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev 4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 35.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. 2001. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. 2012. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4alpha in adult mice. J Biol Chem 287:7345–7356. doi: 10.1074/jbc.M111.334599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soutoglou E, Katrakili N, Talianidis I. 2000. Acetylation regulates transcription factor activity at multiple levels. Mol Cell 5:745–751. doi: 10.1016/S1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 38.Sun K, Montana V, Chellappa K, Brelivet Y, Moras D, Maeda Y, Parpura V, Paschal BM, Sladek FM. 2007. Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol Endocrinol 21:1297–1311. doi: 10.1210/me.2006-0300. [DOI] [PubMed] [Google Scholar]
- 39.Viollet B, Kahn A, Raymondjean M. 1997. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol Cell Biol 17:4208–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang G, Nepomuceno L, Yang Q, Sladek FM. 1997. Serine/threonine phosphorylation of orphan receptor hepatocyte nuclear factor 4. Arch Biochem Biophys 340:1–9. doi: 10.1006/abbi.1997.9914. [DOI] [PubMed] [Google Scholar]
- 41.Ktistaki E, Ktistakis NT, Papadogeorgaki E, Talianidis I. 1995. Recruitment of hepatocyte nuclear factor 4 into specific intranuclear compartments depends on tyrosine phosphorylation that affects its DNA-binding and transactivation potential. Proc Natl Acad Sci U S A 92:9876–9880. doi: 10.1073/pnas.92.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandra V, Holla P, Ghosh D, Chakrabarti D, Padigaru M, Jameel S. 2011. The hepatitis E virus ORF3 protein regulates the expression of liver-specific genes by modulating localization of hepatocyte nuclear factor 4. PLoS One 6:e22412. doi: 10.1371/journal.pone.0022412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo H, Zhou T, Jiang D, Cuconati A, Xiao GH, Block TM, Guo JT. 2007. Regulation of hepatitis B virus replication by the phosphatidylinositol 3-kinase-akt signal transduction pathway. J Virol 81:10072–10080. doi: 10.1128/JVI.00541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ondracek CR, McLachlan A. 2011. Role of peroxisome proliferator-activated receptor gamma coactivator 1alpha in AKT/PKB-mediated inhibition of hepatitis B virus biosynthesis. J Virol 85:11891–11900. doi: 10.1128/JVI.00832-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H, Xu J, Zhou L, Yun X, Chen L, Wang S, Sun L, Wen Y, Gu J. 2011. Hepatitis B virus large surface antigen promotes liver carcinogenesis by activating the Src/PI3K/Akt pathway. Cancer Res 71:7547–7557. doi: 10.1158/0008-5472.CAN-11-2260. [DOI] [PubMed] [Google Scholar]
- 46.National Institutes of Health. 2002. Public Health Service policy on humane care and use of laboratory animals. Office of Laboratory Animal Welfare, National Institutes of Health, Bethesda, MD. [Google Scholar]
- 47.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed.National Academies Press, Washington, DC. [Google Scholar]
- 48.Seglen P. 1993. Isolation of hepatocytes by collagenase perfusion, vol 1 Academic Press, New York, NY. [Google Scholar]
- 49.Clippinger AJ, Bouchard MJ. 2008. Hepatitis B virus HBx protein localizes to mitochondria in primary rat hepatocytes and modulates mitochondrial membrane potential. J Virol 82:6798–6811. doi: 10.1128/JVI.00154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melegari M, Scaglioni PP, Wands JR. 1998. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J Virol 72:1737–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scaglioni PP, Melegari M, Wands JR. 1997. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame. Virology 233:374–381. doi: 10.1006/viro.1997.8594. [DOI] [PubMed] [Google Scholar]
- 52.Scaglioni PP, Melegari M, Wands JR. 1997. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J Virol 71:345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouchard MJ, Puro RJ, Wang L, Schneider RJ. 2003. Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J Virol 77:7713–7719. doi: 10.1128/JVI.77.14.7713-7719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouchard MJ, Wang LH, Schneider RJ. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 55.Lamontagne J, Mills C, Mao R, Goddard C, Cai D, Guo H, Cuconati A, Block T, Lu X. 2013. Screening and identification of compounds with antiviral activity against hepatitis B virus using a safe compound library and novel real-time immune-absorbance PCR-based high throughput system. Antiviral Res 98:19–26. doi: 10.1016/j.antiviral.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Jin YM, Yun C, Park C, Wang HJ, Cho H. 2001. Expression of hepatitis B virus X protein is closely correlated with the high periportal inflammatory activity of liver diseases. J Viral Hepat 8:322–330. doi: 10.1046/j.1365-2893.2001.00308.x. [DOI] [PubMed] [Google Scholar]
- 58.Crabbe T, Welham MJ, Ward SG. 2007. The PI3K inhibitor arsenal: choose your weapon! Trends Biochem Sci 32:450–456. doi: 10.1016/j.tibs.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Kohn AD, Takeuchi F, Roth RA. 1996. Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J Biol Chem 271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 60.Masure S, Haefner B, Wesselink JJ, Hoefnagel E, Mortier E, Verhasselt P, Tuytelaars A, Gordon R, Richardson A. 1999. Molecular cloning, expression and characterization of the human serine/threonine kinase Akt-3. Eur J Biochem 265:353–360. doi: 10.1046/j.1432-1327.1999.00774.x. [DOI] [PubMed] [Google Scholar]
- 61.Tang H, McLachlan A. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci U S A 98:1841–1846. doi: 10.1073/pnas.98.4.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, Iliopoulos D. 2011. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Datta SR, Brunet A, Greenberg ME. 1999. Cellular survival: a play in three Akts. Genes Dev 13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 64.Luetjens CM, Kogel D, Reimertz C, Dussmann H, Renz A, Schulze-Osthoff K, Nieminen AL, Poppe M, Prehn JH. 2001. Multiple kinetics of mitochondrial cytochrome c release in drug-induced apoptosis. Mol Pharmacol 60:1008–1019. [DOI] [PubMed] [Google Scholar]
- 65.Bouchard MJ, Wang L, Schneider RJ. 2006. Activation of focal adhesion kinase by hepatitis B virus HBx protein: multiple functions in viral replication. J Virol 80:4406–4414. doi: 10.1128/JVI.80.9.4406-4414.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biermer M, Puro R, Schneider RJ. 2003. Tumor necrosis factor alpha inhibition of hepatitis B virus replication involves disruption of capsid integrity through activation of NF-kappaB. J Virol 77:4033–4042. doi: 10.1128/JVI.77.7.4033-4042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klein NP, Bouchard MJ, Wang LH, Kobarg C, Schneider RJ. 1999. Src kinases involved in hepatitis B virus replication. EMBO J 18:5019–5027. doi: 10.1093/emboj/18.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klein NP, Schneider RJ. 1997. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol Cell Biol 17:6427–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miao J, Chen GG, Chun SY, Lai PP. 2006. Hepatitis B virus X protein induces apoptosis in hepatoma cells through inhibiting Bcl-xL expression. Cancer Lett 236:115–124. doi: 10.1016/j.canlet.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Terradillos O, Pollicino T, Lecoeur H, Tripodi M, Gougeon ML, Tiollais P, Buendia MA. 1998. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene 17:2115–2123. doi: 10.1038/sj.onc.1202432. [DOI] [PubMed] [Google Scholar]
- 71.Kim HJ, Kim SY, Kim J, Lee H, Choi M, Kim JK, Ahn JK. 2008. Hepatitis B virus X protein induces apoptosis by enhancing translocation of Bax to mitochondria. IUBMB Life 60:473–480. doi: 10.1002/iub.68. [DOI] [PubMed] [Google Scholar]
- 72.Su Q, Schroder CH, Hofmann WJ, Otto G, Pichlmayr R, Bannasch P. 1998. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology 27:1109–1120. doi: 10.1002/hep.510270428. [DOI] [PubMed] [Google Scholar]
- 73.Chung TW, Lee YC, Ko JH, Kim CH. 2003. Hepatitis B virus X protein modulates the expression of PTEN by inhibiting the function of p53, a transcriptional activator in liver cells. Cancer Res 63:3453–3458. [PubMed] [Google Scholar]
- 74.Bar-Yishay I, Shaul Y, Shlomai A. 2011. Hepatocyte metabolic signalling pathways and regulation of hepatitis B virus expression. Liver Int 31:282–290. doi: 10.1111/j.1478-3231.2010.02423.x. [DOI] [PubMed] [Google Scholar]
- 75.Arzberger S, Hosel M, Protzer U. 2010. Apoptosis of hepatitis B virus-infected hepatocytes prevents release of infectious virus. J Virol 84:11994–12001. doi: 10.1128/JVI.00653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feitelson MA, Lee J. 2007. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett 252:157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 77.Bonilla Guerrero R, Roberts LR. 2005. The role of hepatitis B virus integrations in the pathogenesis of human hepatocellular carcinoma. J Hepatol 42:760–777. doi: 10.1016/j.jhep.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 78.Berasain C, Castillo J, Perugorria MJ, Latasa MU, Prieto J, Avila MA. 2009. Inflammation and liver cancer: new molecular links. Ann N Y Acad Sci 1155:206–221. doi: 10.1111/j.1749-6632.2009.03704.x. [DOI] [PubMed] [Google Scholar]
- 79.Bertoletti A, Gehring A. 2007. Immune response and tolerance during chronic hepatitis B virus infection. Hepatol Res 37(Suppl 3):S331–S338. doi: 10.1111/j.1872-034X.2007.00221.x. [DOI] [PubMed] [Google Scholar]
- 80.Kim CM, Koike K, Saito I, Miyamura T, Jay G. 1991. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 81.Koike K, Moriya K, Iino S, Yotsuyanagi H, Endo Y, Miyamura T, Kurokawa K. 1994. High-level expression of hepatitis B virus HBx gene and hepatocarcinogenesis in transgenic mice. Hepatology 19:810–819. doi: 10.1002/hep.1840190403. [DOI] [PubMed] [Google Scholar]
- 82.Dragani TA, Manenti G, Farza H, Della Porta G, Tiollais P, Pourcel C. 1990. Transgenic mice containing hepatitis B virus sequences are more susceptible to carcinogen-induced hepatocarcinogenesis. Carcinogenesis 11:953–956. doi: 10.1093/carcin/11.6.953. [DOI] [PubMed] [Google Scholar]
- 83.Slagle BL, Lee TH, Medina D, Finegold MJ, Butel JS. 1996. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog 15:261–269. doi:. [DOI] [PubMed] [Google Scholar]
- 84.Zheng Y, Chen WL, Louie SG, Yen TS, Ou JH. 2007. Hepatitis B virus promotes hepatocarcinogenesis in transgenic mice. Hepatology 45:16–21. doi: 10.1002/hep.21445. [DOI] [PubMed] [Google Scholar]
- 85.Nguyen DH, Ludgate L, Hu J. 2008. Hepatitis B virus-cell interactions and pathogenesis. J Cell Physiol 216:289–294. doi: 10.1002/jcp.21416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE Jr. 1994. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev 8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 87.Duncan SA, Nagy A, Chan W. 1997. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(−/−) embryos. Development 124:279–287. [DOI] [PubMed] [Google Scholar]
- 88.Lazarevich NL, Cheremnova OA, Varga EV, Ovchinnikov DA, Kudrjavtseva EI, Morozova OV, Fleishman DI, Engelhardt NV, Duncan SA. 2004. Progression of HCC in mice is associated with a downregulation in the expression of hepatocyte nuclear factors. Hepatology 39:1038–1047. doi: 10.1002/hep.20155. [DOI] [PubMed] [Google Scholar]
- 89.Ning BF, Ding J, Yin C, Zhong W, Wu K, Zeng X, Yang W, Chen YX, Zhang JP, Zhang X, Wang HY, Xie WF. 2010. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res 70:7640–7651. doi: 10.1158/0008-5472.CAN-10-0824. [DOI] [PubMed] [Google Scholar]
- 90.Bellacosa A, Kumar CC, Di Cristofano A, Testa JR. 2005. Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv Cancer Res 94:29–86. doi: 10.1016/S0065-230X(05)94002-5. [DOI] [PubMed] [Google Scholar]
- 91.Davies MA. 2011. Regulation, role, and targeting of Akt in cancer. J Clin Oncol 29:4715–4717. doi: 10.1200/JCO.2011.37.4751. [DOI] [PubMed] [Google Scholar]
- 92.LoPiccolo J, Granville CA, Gills JJ, Dennis PA. 2007. Targeting Akt in cancer therapy. Anticancer Drugs 18:861–874. [DOI] [PubMed] [Google Scholar]
- 93.Steelman LS, Stadelman KM, Chappell WH, Horn S, Basecke J, Cervello M, Nicoletti F, Libra M, Stivala F, Martelli AM, McCubrey JA. 2008. Akt as a therapeutic target in cancer. Expert Opin Ther Targets 12:1139–1165. doi: 10.1517/14728222.12.9.1139. [DOI] [PubMed] [Google Scholar]