ABSTRACT
Complement is an innate immune system that most animal viruses must face during natural infections. Given that replication and dissemination of the highly pathogenic Nipah virus (NiV) include exposure to environments rich in complement factors, we tested the in vitro sensitivity of NiV to complement-mediated neutralization. Here we show that NiV was completely resistant to in vitro neutralization by normal human serum (NHS). Treatment of purified NiV with NHS activated complement pathways, but there was very little C3 deposition on virus particles. In in vitro reconstitution experiments, NiV particles provided time- and dose-dependent factor I-like protease activity capable of cleaving C3b into inactive C3b (iC3b). NiV-dependent inactivation of C3b only occurred with the cofactors factor H and soluble CR1 but not with CD46. Purified NiV particles did not support C4b cleavage. Electron microscopy of purified NiV particles showed immunogold labeling with anti-factor I antibodies. Our results suggest a novel mechanism by which NiV evades the human complement system through a unique factor I-like activity.
IMPORTANCE Viruses have evolved mechanisms to limit complement-mediated neutralization, some of which involve hijacking cellular proteins involved in control of inappropriate complement activation. Here we report a previously unknown mechanism whereby NiV provides a novel protease activity capable of in vitro cleavage and inactivation of C3b, a key component of the complement cascade. These data help to explain how an enveloped virus such as NiV can infect and disseminate through body fluids that are rich in complement activity. Disruption of the ability of NiV to recruit complement inhibitors could form the basis for the development of effective therapies and safer vaccines to combat these highly pathogenic emerging viruses.
INTRODUCTION
The complement system constitutes a complex group of soluble and cell-associated proteins that together form an integral part of the innate host defense against pathogens (reviewed in reference 1). Complement serves to link innate and adaptive immunity to viruses through recognition of virions, direct neutralization of infectivity, recruitment and stimulation of leukocytes, opsonization by immune cells, and activation of T and B cell responses (1, 2). Complement activation plays important roles in viral pathogenesis (e.g., see references 3 and 4) and has been the focus of efforts to improve the effectiveness of vaccines and therapeutic vectors. The goal of the work described here was to determine the mechanism by which the paramyxovirus Nipah virus (NiV) is resistant to complement-mediated inactivation by normal human serum (NHS).
The complement cascade can be initiated through three main pathways: the classical pathway, lectin pathway, or alternative pathway (1, 2). These three pathways converge on a central component, C3, which is activated by cleavage into the anaphylatoxin C3a and into C3b, which can bind covalently to viral components to aid in opsonization and phagocytosis. In the case of the alternative pathway, a C3 convertase composed of the complex C3bBb is assembled, which carries out an amplification loop of further C3 cleavage. In the case of the lectin/classical pathway, C4 cleavage into C4a and C4b leads to assembly of a second form of the C3 convertase consisting of C4bC2a. These two convertases can propagate a signal leading to formation of the downstream membrane attack complex (MAC), which is capable of lysing virus particles or infected cells (reviewed in references 1 and 2).
Under normal conditions, inappropriate complement activation is regulated by a complex series of host proteins (5), with one key regulatory step being at the formation and stability of the C3 convertases. For example, CD55 is a membrane-bound host protein that acts to dissociate the C3 convertase to prevent further amplification. An alternative inhibitory mechanism involves the host protease factor I, which blocks the formation of a stable C3 convertase through cleavage of C3b or C4b into inactive forms. Factor I protease activity is highly specific for cleavage of only C3b or C4b and is strictly dependent on a set of soluble or membrane-bound host cofactors, such as factor H, CR1, CD46, or C4 binding protein (C4BP) (6, 7).
The key C3 convertase complex is also a common target for inhibition by many pathogenic microbes (8–10). For example, some enveloped viruses recruit host cell membrane-bound regulators into their envelope (e.g., CD55), which can then act to dissociate C3 convertase complexes that form on the virion surface (e.g., see references 11 and 12). Viruses can also block the C3 convertase by exploiting factor I protease to cleave C3b or C4b into the inactive forms. To date, no viruses have been reported to directly recruit factor I. Instead, viruses can encode analogs or mimics of host cell cofactors that act along with soluble factor I to cleave complement proteins. This is evident in the case of vaccinia virus VCP, a virally encoded mimic of normal cellular cofactors that can function with factor I to cleave C3b (13). Alternatively, cofactors that function with factor I can be supplied through recruitment of host cell proteins, such as in the case of binding of factor H to the West Nile virus NS-1 protein (14) or incorporation of CD46 into a viral envelope (11, 12, 15).
We and others have previously shown that complement is strongly activated by a wide range of negative-strand RNA viruses, including parainfluenza virus 5 (PIV5), mumps virus (MuV), vesicular stomatitis virus (VSV), and Newcastle disease virus (NDV). For each of these viruses, C3 has been shown to play an essential role in effective in vitro neutralization by NHS (11, 15, 16). Conversely, many of these negative-strand RNA viruses exploit normal host cell inhibitors to evade complement pathways, with the association of host cell inhibitors resulting in a delay but not a complete inhibition of in vitro complement-mediated neutralization (e.g., see references 11 and 15). However, the role of complement in neutralization of many emerging highly pathogenic paramyxoviruses is largely unknown.
Nipah virus (NiV) is an emerging zoonotic viral pathogen (genus Henipavirus, family Paramyxoviridae) that can cause severe respiratory and neurological disease. NiV and the closely related Hendra virus (HeV) are considered NIH category C priority pathogens. NiV is of considerable importance, due to its high mortality rate in human infections (>70%), its pandemic potential through human-to-human transmission, and its potential impact on farm economies (17–20). There are currently no approved vaccines or therapeutics against NiV for human use (21). NiV is thought to spread from fruit bats to humans, but person-to-person transmission has become of increasing concern (22, 23). NiV infection of humans can be systemic, where viral antigen can be detected in a number of organs, including the respiratory tract, neurons, lymphoid tissue, and endothelial cells lining blood vessels (reviewed in reference 23). It is proposed that NiV can disseminate through blood in either a free form or through binding to CD3+ lymphocytes (24, 25). Because complement factors can be derived from serum, cells lining the respiratory tract (26), or by recruited lymphocytes, the sites of NiV infection and mechanisms of dissemination suggest that complement is a highly relevant innate immune pathway that NiV encounters during natural infections.
Given the role of complement in neutralization of a range of different paramyxoviruses and that NiV infection involves exposure to body fluids rich in complement activity, we have tested whether NiV is sensitive to neutralization by human complement pathways. We previously showed that VSV-based pseudotypes containing the NiV glycoproteins are resistant to neutralization by NHS in vitro (27). Here we have extended our work to demonstrate that treatment of infectious NiV in vitro with NHS results in activation of complement pathways, but there is neither loss of infectivity nor extensive deposition of C3 on virion particles. Furthermore, our new results using functional assays that reconstituted C3b cleavage in vitro show that purified NiV provides a unique factor I protease activity that is capable of inactivation of C3b. Our data are the first report of a novel mechanism by which a viral pathogen utilizes a virion-associated factor I-like protease to evade human complement pathways.
MATERIALS AND METHODS
Cells, viruses, and neutralization assays.
HeLa and Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (HyClone, Logan, UT), l-glutamine, penicillin, and streptomycin at 37°C in a humidified CO2 incubator (5%). PIV5 was grown in MDBK cells, and its titer was determined on CV-1 cells as described previously (11). NiV (Malaysia strain) and HeV were kindly provided by the Special Pathogens Branch of the Centers for Disease Control and Prevention, Atlanta, GA. For neutralization assays, NiV was grown and its titer was determined on Vero cells. For biochemical analyses of associated cofactors, NiV was grown in HeLa cells, which are known to express CD55 and CD46 (11). The viral titer was determined by plaque assay on Vero cells.
NiV and HeV were purified as described previously (11). Briefly, HeLa cells were infected with virus at a multiplicity of infection (MOI) of 0.1. At 5 days postinfection, the supernatant was collected and clarified by low-speed centrifugation. Clarified medium was then layered over a 20% glycerol cushion in NTE buffer (100 mM NaCl, 10 mM Tris-Cl [pH 7.4], 1 mM EDTA), and virus was concentrated by ultracentrifugation (23,000 rpm for 5 h at 12°C in an SW28 rotor [Beckman Coulter, Fullerton, CA]). Concentrated virus was further purified by centrifugation on a 20 to 60% sucrose gradient (23,000 rpm for 1 h at 12°C in an SW28 rotor), and the banded virus was collected, diluted with NTE buffer, pelleted (23,000 rpm for 5 h at 12°C), and resuspended in 1 ml of DMEM, and aliquots were stored at −80°C. Aliquots were inactivated on dry ice by gamma irradiation (5 megarads). All infectious work was performed in a class II biological safety cabinet (BSC) in a biosafety level 4 (BSL4) laboratory at the Galveston National Laboratory, University of Texas Medical Branch.
For NiV neutralization assays, 2-fold serial dilutions (1:5 to 1:80) of normal human serum (NHS) or heat-inactivated (HI) NHS serum were incubated with 100 PFU of NiV for 1 h at 37°C. Negative-control samples were incubated with phosphate-buffered saline (PBS) alone. NiV-specific mouse immune serum was used as a positive control in this assay. All dilutions of the sera and virus were made in DMEM without fetal bovine serum (FBS). Following the 1-h incubation, each serum-virus mixture was added in triplicate to Vero cells and incubated for another hour at 37°C. Following adsorption, the inoculum was removed, and the cells were overlaid with 0.8% agarose–minimal essential medium (MEM)–10% FBS. Cells were incubated for 3 days, after which cells were fixed and stained with crystal violet, and plaques were counted.
Complement reagents, proteins, and antibodies.
NHS used in this study was previously collected from healthy donors, processed, and divided into small aliquots before being frozen at −80°C as described previously (16). Purified human complement components C3b, C4b, factor I, factor H, and C4bp were from Complement Technologies (Tyler, TX). Purified soluble CD46 (sCD46) was purchased from Sino Biologicals (Beijing, China). Soluble CR1 (sCR1) was a kind gift from Henry Marsh (Celldex Therapeutics, Needham, MA). Goat polyclonal anti-human C3 was from Calbiochem (catalog no. 204869), mouse monoclonal anti-CD46 was from R&D (catalog no. MAB2005), and goat anti-human factor I was from Complement Technology (catalog no. A238).
Factor I protease assays.
In vitro factor I-mediated cleavage activity was assayed as described previously (11). In a standard assay, 5 μg of purified NiV particles derived from infected HeLa cells was incubated for 4 h in PBS (pH 7.4) at 37°C with 3 μg of either C3b or C4b in a total volume of 20 μl. Positive-control samples contained 100 ng of purified factor I. Except where indicated, standard reaction mixtures contained factor H (1 μg), soluble CD46 (1 μg), C4BP (1 μg), or soluble CR1 (1 μg) as cofactors. Reactions were stopped by adding 5 μl of SDS-PAGE sample buffer containing mercaptoethanol and boiling. C3b reaction products were analyzed on 9% SDS-PAGE gels, while C4b reaction products were analyzed on 10% gels. The gels were stained with Gelcode blue stain reagent (Thermo Scientific, IL) to visualize proteins. The level of intact C3b α′ chain remaining after incubation was quantitated using a Kodak Image Station 4000R and compared to C3b α′ chain in standard reaction mixtures lacking cofactor (set at 100%).
Real-time PCR analysis.
Total RNA was purified from 106 HeLa cells using the RNeasy Plus purification system (Qiagen), with spectrophotometric analysis showing very high RNA purity. One microgram of RNA was used to generate cDNA using the Superscript III system (Invitrogen) as described by the manufacturers. One-tenth of the cDNA reaction was then used in real-time PCRs with primers specific for human factor I (Hs00989715_m1; Life Technologies) and the Applied Biosystems TaqMan Universal PCR master mix. In parallel, 10-fold dilutions of a plasmid encoding human factor I (kindly provided by Anna Blom, Lund University, Sweden) were analyzed to generate a standard curve with plasmid copy number, and the HeLa cell copy number was calculated from the corresponding threshold cycle (CT) values. No signals were generated using control samples lacking RNA or which did not have reverse transcriptase added.
N-terminal protein sequencing.
Protein sequence analysis was performed using an Applied Biosystems model 492HT protein/peptide sequencer equipped with an online phenylthiohydantoin (PTH)-amino acid analyzer according to the standard methods for the instrument. Samples were introduced into the sample cartridge as blots on polyvinylidene difluoride (PVDF) membranes previously stained with Coomassie blue to detect the bands of interest. PTH-amino acids released at each cycle of Edman degradation were identified by manual comparison of each chromatogram to the standard PTH-amino acid chromatogram run at the start of the analysis.
ELISA and Western blotting.
Purified NiV (0.01 μg) in PBS was mixed with a 1:10 dilution of NHS (assay volume, 20 μl) and incubated for 2 to 5 min at 37°C. The samples were further diluted 1:500 and analyzed using an OptEIA human C3a kit (BD Biosciences) as described by the manufacturer.
For Western blotting, protein samples were separated on a 0.9% SDS-PAGE gel and analyzed by Western blotting using rabbit polyclonal antisera specific for CD46 (1:500 dilution; Santa Cruz Biotechnology, catalog no. SC-9098). Alternatively, blots were probed with polyclonal goat antisera (Santa Cruz Biotechnology, catalog no. SC-69470) or a mouse monoclonal antibody (R&D Systems, catalog no. MAB3307) specific for the large and small fragments of factor I, respectively. Blots were visualized by horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (Pierce Chemicals).
EM.
C3 deposition on NiV was analyzed by adsorbing 0.5 μg of purified virus on carbon-coated 200-mesh gold grids (CF200-Au; Electron Microscopy Sciences, PA) followed by a 5-min incubation at room temperature in a humidified chamber. After treatment with NHS (1:10 dilution) at 37°C, samples were blocked with 1% bovine serum albumin (BSA) in PBS and then probed with anti-C3 monoclonal antibody (Cell Sciences) at a dilution of 1 μg in 10 μl. Deposition was detected with 6-nm-diameter gold particle-labeled goat anti-mouse antibodies (Jackson ImmunoResearch Laboratories, PA). To detect virion-associated CD46 or factor I, purified NiV was probed with mouse anti-human CD46 or goat anti-human factor I followed by 12-nm-diameter gold particle-labeled anti-mouse or donkey anti-goat (Jackson ImmunoResearch Laboratories, PA), respectively. After labeling, particles were subjected to negative staining with 2% phosphotungstic acid (pH 6.6) and analyzed by electron microscopy (EM) with a Technai transmission electron microscope.
RESULTS
NiV is resistant to complement-mediated neutralization in vitro.
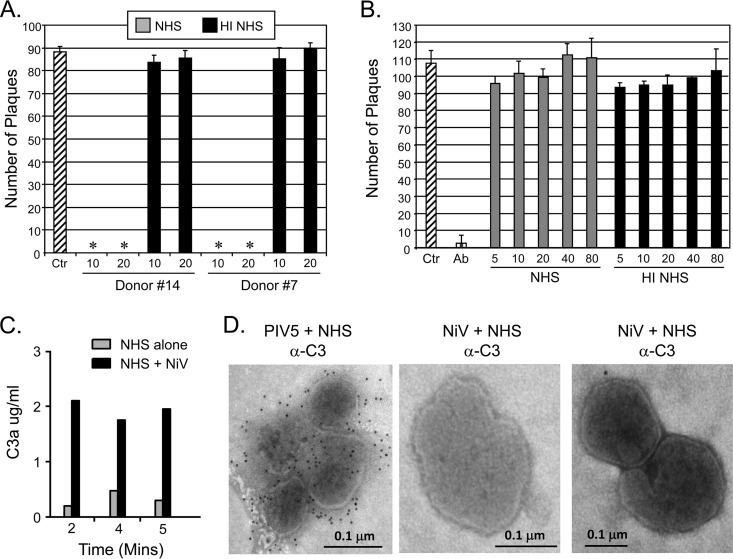
We have previously shown that a wide range of negative-strand RNA viruses are effectively neutralized in vitro by complement pathways. An example of this is shown in Fig. 1A for sera from two individual donors, where incubation of PIV5 with NHS for 1 h at 37°C resulted in complete neutralization (samples with asterisks). In contrast, identical treatment with heat-inactivated (HI) serum had no substantial effect on PIV5 infectivity. To determine if NiV was similarly sensitive to complement-mediated neutralization in vitro, infectious NiV was incubated under BSL4 conditions with PBS as a negative control, a serum containing neutralizing antibodies as a positive control, or dilutions of NHS or HI NHS. After 1 h at 37°C, the remaining infectivity was determined by plaque assay. As shown in Fig. 1B for a representative donor serum, NiV was neutralized by the anti-NiV sera but showed no loss of infectivity even with a 1:5 dilution of NHS. These data indicate that unlike other paramyxoviruses we have tested so far, NiV is refractory to complement-mediated in vitro neutralization. In all subsequent biochemical experiments described below, NiV was purified by sucrose gradient centrifugation, and infectivity was inactivated on dry ice by gamma irradiation.
FIG 1.
Live NiV is not neutralized by NHS in vitro. (A and B) One hundred PFU of PIV5 (A) or NiV (B) was incubated with PBS as a control (Ctr [cross-hatched bar]) or with dilutions of NHS (gray bars) or HI NHS (black bars) for 1 h at 37°C. In addition, one NiV sample contained a neutralizing anti-NiV antibody (Ab) as a positive control. The remaining infectivity was determined by plaque assay. Results represent the average of three assays, with error bars representing standard deviations. Asterisks indicate that no plaques were detected in these samples. (C) NHS (1:20 dilution) was incubated at 37°C alone (gray bars) or with purified NiV (black bars) for 2, 4, or 5 min. Samples were analyzed by ELISA for levels of C3a. (D) Purified PIV5 or NiV was applied to carbon grids and treated with NHS as described in Materials and Methods. Samples were subsequently treated with mouse monoclonal anti-C3 followed by gold-conjugated goat anti-mouse (12-nm-diameter beads) before examination by electron microscopy. The size bar indicates 0.1 μm.
To determine if complement was activated by NiV, NHS was incubated alone or with purified gamma-irradiated virus and complement activation was assayed by an ELISA specific for the C3a cleavage product. As seen in Fig. 1C, incubation of NiV with serum resulted in appearance of C3a cleavage product in as little as 2 min. Immunogold electron microscopy (Fig. 1D) showed extensive deposition of C3 on serum-treated PIV5, consistent with our prior finding that PIV5 activates complement (16) and with the sensitivity of PIV5 to complement-mediated neutralization (Fig. 1A). In contrast, as shown in the two EM examples in Fig. 1D, NHS-treated NiV particles showed very little C3 deposition. While PIV5 and NiV were grown under different cell culture conditions and in different laboratories, these data are consistent with all of our prior results with PIV5 showing deposition of complement. The PIV5 sample served as a control to visualize what C3 deposition looked like for a virus that was sensitive to complement and which contrasted with the lack of C3 deposition on NiV particles. Taken together, these data indicate that treatment of NiV with NHS activates complement pathways in vitro, but this does not result in extensive C3 deposition or loss of infectivity.
NiV contains a factor I-like activity that functions with cofactors to cleave C3b into iC3b in vitro.
The α′ chain of C3b can be cleaved by factor I protease when one of multiple cofactors is present. As shown in the schematic in Fig. 2A, when factor I is coupled with factor H, this cleavage can occur at two possible sites in the α′ chain: R1281 and R1298 (sites 1 and 2 in Fig. 2A). In the presence of CR1, factor I can cleave the α′ chain at sites 1 and 2 but also at an additional site at R932 (site 3).
FIG 2.
Purified NiV promotes factor H- and CR1-dependent cleavage of C3b. (A) The C3b α′ fragment is depicted as a box with vertical arrows denoting three numbered sites of cleavage by factor I in the presence of either of the cofactors factor H (sites 1 and 2) or CR1 (sites 1, 2, and 3). The amino acid sequences surrounding factor I cleavage sites at arginines 932, 1281, and 1298 and the apparent molecular masses (kDa) of cleavage products as seen on SDS gels are shown. (B) Purified NiV (20 μg and 5 μg in lanes 2 and 4, respectively) and noninfected HeLa cell lysate (10 μg, lanes 1 and 3) were analyzed by SDS-PAGE and Gelcode blue staining. The positions of viral P, N, and F1 are indicated. Lane 1 is a marker lane with proteins of the indicated molecular masses (kDa). (C and D) C3b α′ cleavage with cofactor factor H (C) or sCR1 (D) was reconstituted in vitro using the indicated purified proteins. After 4 h at 37°C, samples were analyzed by SDS-PAGE and Gelcode blue staining. The positions of the α′ and β chains of C3b and the cleavage products are indicated. a and b denote the positions of bands excised from the gel for protein sequence analysis. The position of an ∼46-kDa intermediate cleavage product is indicated to the right of panels C and D.
To determine if NiV could supply factors that result in inactivation of C3b as seen for other paramyxoviruses, NiV was isolated from the media of infected HeLa cells by pelleting through a sucrose cushion, followed by conventional sucrose gradient centrifugation as described previously (11). As shown in the Coomassie blue-stained gel in Fig. 2B (lanes 2 and 4), purification of NiV resulted in an enrichment of virus as evidenced by the large amounts of N, P, and F1 proteins and the fact that many of the proteins from noninfected HeLa cells (lanes 1 and 3) were not enriched during the purification process.
In vitro cleavage reactions were reconstituted using purified complement components and purified NiV. After 4 h at 37°C, proteins were analyzed by SDS-PAGE and Coomassie blue staining. As seen for the positive-control samples in Fig. 2C (lane 3), incubation of the C3b substrate with factor H and factor I protease resulted in loss of the C3b α′ chain and appearance of the corresponding 68- and 43-kDa fragments. Incubation of C3b with NiV alone or with NiV plus factor I did not affect levels of the α′ chain (lanes 4 and 6). Most importantly, addition of NiV along with factor H but without any added factor I (lane 5) resulted in loss of the C3b α′ chain to the same extent as the positive-control sample (lane 3). Peptide sequencing (Table 1) revealed that the 43-kDa cleavage product from the positive-control reaction mixture containing factor I plus factor H (Fig. 2C,lane 3, band a) had the same N-terminal amino acid sequence as the 43-kDa cleavage product of NiV plus factor H (Fig. 2C, lane 5, band b). For NiV samples (e.g., Fig. 2C, lanes 4 to 6), an additional protein species of unknown origin was seen running with the dye front. These data indicate that purified NiV has a “factor I-like” activity capable of functioning with factor H to cleave C3b in vitro.
TABLE 1.
Sequences of the peptides examined in this studya
| Sample no. | Gel-lane-band in Fig. 2 | Cofactor + protease | Amino acid sequenceb |
|---|---|---|---|
| 1 | C-3-a | Factor H + factor I (control) | Gly/Ser/Pro-Glu-Glu-Thr/Pro-Lys-Glu-Asn-Glu-Gly-Phe-Thr |
| 2 | C-5-b | Factor H + NiV | Gly/Ser/Pro-Glu-Glu-Thr/Pro-Lys-Glu-Asn-Glu-Gly-Phe-Thr |
| 3 | D-3-a | sCR1 + factor I (control) | Gly/Ser/Pro-Glu-Glu-Thr-Xxx-Glu-Asn-Glu-Gly-Phe-Thr |
| 4 | D-5-b | sCR1+ NiV | Gly/Ser/Pro-Glu-Glu-Thr-Xxx-Glu-Asn-Glu-Gly-Phe-Thr |
C3b cleavage reactions with either factor H (samples 1 and 2) or sCR1 (samples 3 and 4) were reconstituted in vitro using the purified proteins indicated in Fig. 2C and D, respectively. After 4 h at 37°C, samples were analyzed by SDS-PAGE, and the bands labeled a and b in Fig. 2 were processed for N-terminal protein sequencing as described in Materials and Methods.
Xxx indicates the presence of an amino acid that could not be clearly identified, and italic type indicates a possible residue in that position.
C3b can also be cleaved at three sites by factor I in the presence of CR1 (Fig. 2A), resulting in a different cleavage pattern from that seen with factor I plus factor H (Fig. 2D). When C3b was incubated with purified NiV and a soluble version of CR1 (sCR1), a 43-kDa product was generated (Fig. 2D, lane 5). N-terminal peptide sequencing (Table 1) revealed that this cleavage product from the reaction mixture containing NiV plus sCR1 (Fig. 2D, lane 5, band b) had the same amino acid sequence as the cleavage product generated by the positive-control factor I plus CR1 (Fig. 2D, lane 3, band a). Interestingly, the reactions from NiV plus sCR1 generated cleavage of C3b α′ chain at sites 1 and 2 to produce only the 46- and 43-kDa products, respectively. However, the subsequent cleavage at site 3 to generate the 40- and 25-kDa products did not occur (Fig. 2D, compare lanes 3 and 5). Even overnight incubation of NiV with C3b and sCR1 did not promote cleavage of the α′ chain at site 3 (data not shown).
The extent of NiV-mediated cleavage of the C3b α′ chain was dependent on the amount of virus added to the in vitro reactions, but the sensitivity to virus concentration differed depending on whether factor H or sCR1 was included as the cofactor (compare Fig. 3A and B). This is evident in Fig. 3A for NiV plus factor H, where ∼50% of NiV-mediated C3b α′ cleavage was seen with less than 2 μg of virus. In contrast, ∼12 μg of virus was needed to obtain ∼50% of C3b α′ cleavage when NiV was assayed with sCR1 (Fig. 3B). Similarly, time course experiments showed that NiV-mediated C3b α′ cleavage was rapid when factor H was used as a cofactor (one of two representative experiments shown in Fig. 3C), with ∼50% of C3b α′ cleaved by ∼40 min of incubation. In contrast, α′ cleavage was relatively slow and ineffective when sCR1 was used as a cofactor (Fig. 3D), with ∼50% of C3b α′ cleaved by ∼120 min of incubation. In addition, the dependence of NiV-mediated α′ cleavage on factor H concentration differed from that seen with the positive control of purified factor I, since ∼20-fold more factor H was needed to drive 50% α′ cleavage using NiV rather than purified factor I (data not shown).
FIG 3.
Concentration- and time-dependent C3b cleavage by purified NiV. (A and B) C3b cleavage reactions were reconstituted as described in the legend to Fig. 2 using the indicated components, including either factor H (A) or sCR1 (B) as a cofactor. After 4 h at 37°C, samples were analyzed by SDS-PAGE and Gelcode blue staining. Levels of C3b α′ chain remaining in the cleavage reactions were quantitated as a function of NiV concentration and compared to the level of α′ in a control sample containing C3b and factor I without cofactor set to 100%. (C and D) C3b cleavage reactions were reconstituted using 10 μg of NiV along with 2 μg of either factor H (C) or sCR1 (D). After incubation for the indicated times at 37°C, samples were analyzed by SDS-PAGE as described for panels A and B. Data in panels C and D are representative of two independent experiments.
Factor I protease can also function in C3b α′ cleavage by using membrane-bound CD46 as a cofactor (6, 7). Western blotting with anti-CD46 antibodies revealed that CD46 was associated with NiV particles (Fig. 4A) and immunogold EM of NiV particles showed extensive CD46 staining (Fig. 4B). Surprisingly, relative to the positive control of factor I plus soluble CD46 (Fig. 4C, lane 3), in vitro C3b α′ cleavage did not occur to a significant extent when NiV was incubated with C3b alone (Fig. 4C, lane 4) or when reaction mixtures were supplemented with additional soluble CD46 (Fig. 4C, lane 5). This result contrasts with our prior work showing for VSV and mumps virus (MuV) that virion-associated CD46 functions in C3b cleavage in vitro (11). It is noteworthy that addition of exogenous factor I to the reaction (Fig. 4C, lane 6) resulted in a small amount of 43-kDa and 46-kDa cleavage products. This indicates that NiV-associated factor I activity is unusual in that it functions poorly in vitro with virion-associated or soluble cofactor CD46, but in addition, this shows that the activity of the virion-associated CD46 cofactor is only evident when additional factor I is supplied. This may reflect a need for higher levels of factor I when combined with CD46 versus that seen with factor H (see above).
FIG 4.
Soluble CD46 is a poor cofactor for NiV-mediate C3b cleavage. (A) The indicated amounts of purified NiV were analyzed by Western blotting for the presence of CD46. Recombinant CD46 (0.5 μg) was analyzed as a positive control. (B) Purified NiV was applied to carbon grids and treated with mouse monoclonal antibody to CD46 followed by gold-conjugated anti-mouse (12-nm-diameter beads) before examination by electron microscopy. Two representative virions are shown, with the size bar indicating 0.1 μm. (C) C3b cleavage reactions were reconstituted using the indicated components plus sCD46 as a cofactor. After 4 h at 37°C, samples were analyzed as described in the legend to Fig. 2.
NiV-associated protease activity does not function in in vitro cleavage of C4b.
The C3 convertase C4bC2a can be inactivated by factor I cleavage of C4b in the presence of cofactors CR1 or C4BP, with cleavage occurring at two sites within the α′ chain: R937 and R1318 (Fig. 5A). As shown in Fig. 5B (lane 3), in vitro reconstitution of C4b cleavage with purified factor I protease and sCR1 resulted in cleavage of the C4b α′ chain into C4d and a 25-kDa fragment. However, incubation of C4b with NiV alone (lane 4) or with NiV plus CR1 (lane 5) or factor I (lane 6) did not produce significant cleavage of the C4b α′ chain. Similar results were seen when C4b cleavage was reconstituted with C4BP as a cofactor (Fig. 5C), where no detectable loss of the C4b α′ chain was seen following incubation with purified NiV. These data indicate that unlike purified factor I protease, the NiV-associated protease cannot function in in vitro cleavage of the C4b α′ chain.
FIG 5.
NiV does not promote cofactor-dependent cleavage of C4b. (A) The C4b α′ fragment is depicted as a box with vertical lines denoting two numbered sites of cleavage by factor I in the presence of either CR1 or C4BP. The amino acid sequences surrounding the two factor I cleavage sites at arginines 937 and 1318 and the apparent molecular masses of cleavage products as seen on SDS gels are shown. (B and C) C4b cleavage reactions with cofactors sCR1 (B) or C4BP (C) were reconstituted in vitro using the indicated purified proteins. After 4 h at 37°C, samples were analyzed by SDS-PAGE and Gelcode blue staining. The positions of the C4b α′, β, and γ chains, and the C4d and 25-kDa cleavage products are indicated.
Factor I is expressed by HeLa cells and is associated with NiV particles.
The NiV-associated factor I-like activity that cleaves C3b could be derived from the infected HeLa cells. To confirm that HeLa cells express factor I, two independent preparations of HeLa RNA were reverse transcribed and then used in real-time PCRs to quantitate levels of factor I RNA. Results from these samples were compared to those obtained from a standard curve generated from known copy numbers of a plasmid encoding human factor I. As shown in Fig. 6A, approximately 5 × 102 to 1 × 103 copies of factor I RNA per 0.1 μg of cellular RNA were found in two separate preparations. In addition, Western blotting of HeLa cell lysates with two different antibodies showed weak staining for protein bands with mobilities matching those of the factor I heavy and light chains (Fig. 6B). These data indicate that HeLa cells express low levels of factor I as a possible source of protease during NiV infections.
FIG 6.
Factor I is expressed in HeLa cells and is associated with purified NiV particles. (A) One microgram of purified HeLa cell RNA was reverse transcribed, and 1/10 of the reaction mixture was analyzed by real-time PCR using primers specific for human factor I. Known copy numbers of a plasmid expressing factor I were analyzed in parallel and served as a standard curve to determine the number of copies of factor I message in 0.1 μg of total RNA. HeLa RNAs 1 and 2 represent two different preparations of RNA. Error bars show the standard deviations of values from three reactions. (B) HeLa cell proteins (2, 10, and 20 μg in lanes 2 to 4, respectively) were analyzed by Western blotting using antibodies specific for the factor I heavy and light chains indicated by arrows. Lane 1 represents 5 ng of purified factor I as a control. (C) Purified NiV was applied to carbon grids and treated with goat polyclonal antibody specific for human factor I followed by 12-nm-diameter gold particle-labeled donkey anti-goat. Two representative virions are shown, with the size bar indicating 0.1 μm. (D) C3b α′ cleavage with cofactor factor H was reconstituted in vitro using the indicated purified proteins and purified HeV. After overnight incubation at 37°C, samples were analyzed by SDS-PAGE and Gelcode blue staining. The arrows indicate the positions of the 43- and 68-kDa cleavage products.
Attempts to detect NiV-associated factor I by Western blotting with polyclonal goat antiserum gave inconsistent results, due in large part to high background on blotting membranes. As a more sensitive method that can also show localization on virions, NiV particles were analyzed by immunogold EM using polyclonal antibody specific for factor I. As shown in Fig. 6C, NiV particles showed low levels of staining for factor I, much of which was localized to regions consistent with viral membrane structures. These data are consistent with a proposal that NiV-associated factor I is present at low levels on virions but at high enough concentrations to function enzymatically against C3b.
We tested the hypothesis that the closely related henipavirus HeV could also supply factor I-like activity. HeV was grown in HeLa cells and purified by the same conventional sucrose gradient centrifugation approaches used for preparation of NiV before undergoing testing in in vitro cofactor cleavage assays. As shown in Fig. 6D, overnight incubation of HeV with C3b and factor H as a cofactor resulted in a small amount of C3b cleavage. These data indicate that HeV provides a low level of factor I activity compared to that seen with NiV.
DISCUSSION
Many viruses block steps in the complement cascade, with inhibition often focused at the level of formation or stability of the C3 convertase (9, 10). Given our prior findings that most paramyxoviruses are sensitive to complement-mediated neutralization (11, 15) and that NiV replication and dissemination involve exposure to environments rich in complement factors, we have tested whether the emerging pathogen NiV is neutralized by NHS in vitro. Under BSL4 conditions, we demonstrate here that NiV is refractory to neutralization with high concentrations of NHS, a result that contrasts with our results on other enveloped RNA viruses (e.g., PIV5, NDV, MuV, and VSV). Furthermore, C3 is activated in vitro by exposure to NiV, but in contrast to most other paramyxoviruses, there is no evidence of extensive C3 deposition on NiV particles. Our most striking findings are that purified NiV can supply a cofactor-dependent factor I protease activity in functional C3b cleavage reactions in vitro and that NiV particles stain with antibodies to factor I. These results support a novel mechanism of complement evasion by NiV through C3b inactivation by a factor I-like protease. While there is a single prior report of factor I association with the Gram-negative pathogen Prevotella intermedia (28), our work is the first evidence of a functional factor I-like activity with a viral pathogen.
Our previously published work showed that VSV-based pseudotypes containing the NiV glycoproteins were highly resistant to in vitro complement-mediated neutralization and that serum-treated pseudotypes showed no significant C3 deposition (27). As a more rigorous test, we have examined complement interactions with bona fide NiV. Together, these results raise the proposal that factor I is recruited to the NiV surface through interactions with one or both of the viral glycoproteins. NiV encodes two integral transmembrane proteins that are synthesized in the exocytic pathway and are expressed at the infected cell surface: a fusion protein (F) that promotes membrane fusion and the attachment glycoprotein (G) that binds the highly conserved cellular receptors ephrins B2 and B3 (29, 30). Factor I is synthesized as a secreted precursor by a variety of cell types and is transported through the exocytic pathway to accumulate in extracellular fluids at relatively low concentrations (6, 7, 31). This suggested that factor I is produced by HeLa cells, and at some point in its biosynthesis and transport through the exocytic pathway, the protease becomes associated with the NiV glycoproteins. Consistent with this proposal, we show that naive HeLa cells express low levels of factor I RNA and protein. Furthermore, NiV infection has been shown to induce a number of cytokines, such as interleukin-6 (IL-6), which are known to upregulate factor I synthesis (32). This raises the possibility that factor I levels are increased following NiV infection and this leads to enhanced association with NiV particles. Future BSL4 work will test this hypothesis in cells with alterations to factor I biosynthesis and in relevant cell types, such as human respiratory cells, or in animal models.
As an alternative hypothesis, it is formally possible that the particles containing the NiV glycoproteins (NiV particles and VSV pseudotypes) have inherent protease activity capable of the same high specificity for C3b cleavage sites and cofactor dependence as those seen with factor I. Computer-generated sequence alignment has failed to reveal evidence of significant homology between the NiV F or G proteins and human factor I (not shown), although it is conceivable that a three-dimensional folding of one or both of the NiV glycoproteins could create a protease active site with high specificity similar to factor I.
In the presence of factor H or sCR1, purified NiV supplied a protease activity that was capable of cleaving C3b at the same sites known to be processed by bona fide factor I. However, these functional in vitro assays also showed that the cofactor requirement for NiV-associated protease activity was not identical to that seen with purified factor I. This was evident by the finding that while NiV protease activity was very effective in in vitro cleavage of C3b when factor H was used, this cleavage event required higher levels of factor H than were seen with purified factor I. Similarly, the NiV-associated cleavage of C3b differed from that of purified factor I by the relatively low activity when coupled with sCR1 as a cofactor and in that only the first two of three possible C3b cleavage sites were processed. In this regard, this incomplete processing of C3b by NiV factor I protease is still sufficient to inactivate the function of the C3 convertase, as has been shown in previous studies with vaccinia virus complement control protein VCP (13). Finally, there was no detectable NiV-mediated C3b cleavage activity seen with soluble or virion-associated CD46 or when NiV was assayed in vitro against C4b as a substrate. Together, these data indicate that the NiV-associated factor I activity differs functionally from that of soluble factor I.
What is the basis of the unusual cofactor requirements and substrate specificity for this NiV-associated protease? Factor I is an 88-kDa serum protein composed of a series of linked modular domains: a factor I-membrane attack complex (FIMAC) domain, one CD5 domain, two low-density lipoprotein receptor (LDLr) domains, and one serine protease (SP) domain (6, 7, 31). Structural and modeling data suggest that surface-exposed regions of these factor I domains combine to form one part of a tripartite structure along with one of the cofactors and either C3b or C4b as the substrate (6, 33). Formation of this tripartite complex (factor I-substrate-cofactor) is thought to cause a conformational change in factor I such that the catalytic site is then active and able to cleave C3b or C4b. At this time, it is unclear how the NiV-associated factor I protease is bound to the virion particle. However, one possibility is that the tethering of factor I protease to the NiV surface may be through binding to one or more of these key module domains, resulting in restricted access to some but not all substrates and associated cofactors. Thus, the finding that factor H was the most potent at stimulating C3b cleavage by NiV factor I could reflect an easier accessibility of a tethered protease to this particular cofactor or a difference in cofactor-induced conformational changes when binding C3b versus C4b substrates.
As an additional variable, the cofactors that modulate factor I activity can be soluble (e.g., factor H) or membrane bound (CR1, CD46, and C4BP), and the finding that the soluble factor H was the most potent cofactor for the NiV protease suggests that this factor could also contribute to differences between NiV protease and soluble factor I. It is also possible that the preferred use of factor H versus other cofactors (e.g., CD46 or CR-1) could reflect the overall size, sequence, or structure of the different cofactors. In this regard, the factor I-associated cofactors are characterized by having various numbers of tandemly repeated ∼60-amino-acid-long complement control protein (CCP) modules: 30 CCPs for common alleles of CR1, 20 CCPs for factor H, 3 or 8 CCPs for the two chains of C4BP, and 4 CCPs for CD46 (7). Future studies will determine the contributions of virus tethering, soluble versus membrane-bound cofactors, and the cofactor CCP modules in determining the unique cleavage specificity of NiV-associated factor I activity.
When purified HeV was tested in functional assays for in vitro C3b cleavage, only very low levels of factor I-like protease activity were detected. This suggests that the NiV and HeV glycoproteins may differ in their ability to recruit factor I from serum or from within infected cells. The NiV and HeV F and G proteins are very closely related, but the finding of distinct areas of divergence in amino acid sequence may provide a tool to dissect the mechanism of factor I recruitment. The implications of differences in factor I recruitment on NiV and HeV pathogenesis will require further work in animal models.
In conclusion, we have identified a novel mechanism by which NiV evades the human complement system through a factor I-like protease activity. Disruption of the ability of NiV to recruit complement inhibitors could form the basis for the development of safer vaccines or more effective therapies to combat these highly pathogenic emerging viruses.
ACKNOWLEDGMENTS
We thank Ellen Young, Ken Grant, and Olivier Escaffre for excellent technical assistance, Anna Blom for the factor I plasmid, and Henry Marsh (Avant Therapeutics) for kindly providing sCR1.
This work was supported by NIH grants AI083253 (G.D.P.) and AI101675 (G.D.P.) and by Protein Analysis Core Lab services supported by the Comprehensive Cancer Center of Wake Forest University (grant NCI CCSG P30CA012197).
REFERENCES
- 1.Carroll MC. 2004. The complement system in regulation of adaptive immunity. Nat Immunol 5:981–986. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- 2.Blue CE, Spiller OB, Blackbourn DJ. 2004. The relevance of complement to virus biology. Virology 319:176–184. doi: 10.1016/j.virol.2003.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehlhop E, Fuchs A, Engle M, Diamond MS. 2009. Complement modulates pathogenesis and antibody-dependent neutralization of West Nile virus infection through a C5-independent mechanism. Virology 393:11–15. doi: 10.1016/j.virol.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison TE, Fraser RJ, Smith PN, Mahalingam S, Heise MT. 2007. Complement contributes to inflammatory tissue destruction in a mouse model of Ross River virus-induced disease. J Virol 81:5132–5143. doi: 10.1128/JVI.02799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zipfel PF, Sherka C. 2009. Complement regulators and inhibitory proteins. Nat Rev Immunol 9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson SC, Sim RB, Lea SM, Fremeaux-Bacchi V, Blom AM. 2011. Complement factor I in health and disease. Mol Immunol 48:1611–1620. doi: 10.1016/j.molimm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Sim RB, Day AJ, Moffatt BE, Fontaine M. 1993. Complement factor I and cofactors in control of complement system convertase enzymes. Methods Enzymol 223:13–35. doi: 10.1016/0076-6879(93)23035-L. [DOI] [PubMed] [Google Scholar]
- 8.Lambris JD, Ricklin D, Geisbrecht BV. 2008. Complement evasion of human pathogens. Nat Rev Microbiol 6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoermer KA, Morrison TE. 2011. Complement and viral pathogenesis. Virology 411:362–373. doi: 10.1016/j.virol.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zipfel PF, Hallstrom T, Riesbeck K. 2013. Human complement control and complement evasion by pathogenic microbes—tipping the balance. Mol Immunol 56:152–160. doi: 10.1016/j.molimm.2013.05.222. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JB, Lyles DS, Alexander-Miller MA, Parks GD. 2012. Virion-associated CD55 is more potent than CD46 in mediating resistance of mumps virus and VSV to neutralization. J Virol 86:9929–9940. doi: 10.1128/JVI.01154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saifuddin M, Hedayati T, Atkinson JP, Holguin MH, Parker CJ, Spear GT. 1997. Human immunodeficiency virus type 1 incorporates both glycosyl phosphatidylinositol-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement mediated destruction. J Gen Virol 78:1907–1911. [DOI] [PubMed] [Google Scholar]
- 13.Sahu A, Isaacs SN, Soulika AM, Lambris JD. 1998. Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b inactivates the alternative complement pathway. J Immunol 160:5596–5604. [PubMed] [Google Scholar]
- 14.Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, Atkinson JP, Diamond MS. 2006. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory factor H. Proc Natl Acad Sci U S A 103:19111–19116. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biswas M, Kumar S, Johnson J, Parks GD, Subbiah E. 2012. Incorporation of host complement regulatory proteins into Newcastle disease virus enhances complement evasion. J Virol 86:12708–12716. doi: 10.1128/JVI.00886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson JB, Capraro GA, Parks GD. 2008. Differential mechanisms of complement mediated neutralization of the closely related paramyxoviruses simian virus 5 and mumps virus. Virology 376:112–123. doi: 10.1016/j.virol.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton BT, Broder CC, Middleton D, Wang LF. 2006. Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol 4:23–25. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo MK, Rota PA. 2008. The emergence of Nipah virus, a highly pathogenic paramyxovirus. J Clin Virol 43:396–400. doi: 10.1016/j.jcv.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Luby SP, Gurley ES, Hossain MJ. 2009. Transmission of human infection with Nipah virus. Clin Infect Dis 49:1743–1748. doi: 10.1086/647951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luby SP. 2013. The pandemic potential of Nipah virus. Antiviral Res 100:38–43. doi: 10.1016/j.antiviral.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Bossart KN, Broder CC. 2006. Developments towards effective treatments for Nipah and Hendra virus infection. Expert Rev Anti Infect Ther 4:43–55. doi: 10.1586/14787210.4.1.43. [DOI] [PubMed] [Google Scholar]
- 22.Homaira N, Rahman M, Hossain MJ, Epstein JH, Sultana R, Khan MS, Podder G, Nahar K, Ahmed B, Gurley ES, Daszak P, Lipkin WI, Rollin PE, Comer JA, Ksiazek TG, Luby SP. 2010. Nipah virus outbreak with person-to-person transmission in a district of Bangladesh. Epidemiol Infect 138:1630–1636. doi: 10.1017/S0950268810000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rockx B, Winegar R, Freiberg AN. 2012. Recent progress in henipavirus research: molecular biology, genetic diversity, animal models. Antiviral Res 95:135–149. doi: 10.1016/j.antiviral.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Mathieu C, Pohl C, Szecsi J, Trajkovic-Bodennec S, Devergnas S, Raoul H, Cosset FL, Gerlier D, Wild TF, Horvat B. 2011. Nipah virus uses leukocytes for efficient dissemination within a host. J Virol 85:7863–7871. doi: 10.1128/JVI.00549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stachowiak B, Weingartl HM. 2012. Nipah virus infects specific subsets of porcine peripheral blood mononuclear cells. PLoS One 7:e30855. doi: 10.1371/journal.pone.0030855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolger MS, Ross DS, Jiang H, Frank MM, Ghio AJ, Schwartz DA, Wright JR. 2007. Complement levels and activity in the normal and LPS-injured lung. Am J Physiol Lung Cell Mol Physiol 292:L748–L759. doi: 10.1152/ajplung.00127.2006. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JB, Aguilar H, Lee B, Parks GD. 2011. Interactions of human complement with virus particles containing the Nipah virus glycoproteins. J Virol 85:5940–5948. doi: 10.1128/JVI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malm S, Jusko M, Eick S, Potempa J, Riesbeck K, Blom AM. 2012. Acquisition of complement inhibitor serine protease factor I and its cofactors C4b-binding protein and factor H by Prevotella intermedia. PLoS One 7:e34852. doi: 10.1371/journal.pone.0034852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang LF, Eaton BT, Broder CC. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A 102:10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Negrete OE, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian T, Tajyar S, Lee B. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 31.Goldberger G, Arnaout MA, Aden D, Kay R, Rits M, Colten HR. 1984. Biosynthesis and postsynthetic processing of human C3b/C4b inactivator (factor I) in three hepatoma cell lines. J Biol Chem 259:6492–6497. [PubMed] [Google Scholar]
- 32.Escaffre O, Borisevich V, Carmical JR, Prusak D, Prescott J, Feldmann H, Rockx B. 2013. Henipavirus pathogenesis in human respiratory epithelial cells. J Virol 87:3284–3294. doi: 10.1128/JVI.02576-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chamberlain D, Ullman CG, Perkins SJ. 1998. Possible arrangement of the five domains in human complement factor I as determined by a combination of X-ray and neutron scattering and homology modeling. Biochemistry 37:13918–13929. doi: 10.1021/bi9805184. [DOI] [PubMed] [Google Scholar]