ABSTRACT
Chimeric simian immunodeficiency virus (SIV)/human immunodeficiency virus (HIV) (SHIV) infection of macaques is commonly used to model HIV type 1 (HIV-1) transmission and pathogenesis in humans. Despite the fact that SHIVs encode SIV antagonists of the known macaque host restriction factors, these viruses require additional adaptation for replication in macaques to establish a persistent infection. Additional adaptation may be required in part because macaque CD4 (mCD4) is a suboptimal receptor for most HIV-1 envelope glycoprotein (Env) variants. This requirement raises the possibility that adaptation of HIV-1 Env to the macaque host leads to selection of variants that lack important biological and antigenic properties of the viruses responsible for the HIV-1 pandemic in humans. Here, we investigated whether this adaptation process leads to changes in the antigenicity and structure of HIV-1 Env. For this purpose, we examined how two independent mutations that enhance mCD4-mediated entry, A204E and G312V, impact antibody recognition in the context of seven different parental HIV-1 Env proteins from diverse subtypes. We also examined HIV-1 Env variants from three SHIVs that had been adapted for increased replication in macaques. Our results indicate that these different macaque-adapted variants had features in common, including resistance to antibodies directed to quaternary epitopes and sensitivity to antibodies directed to epitopes in the variable domains (V2 and V3) that are buried in the parental, unadapted Env proteins. Collectively, these findings suggest that adaptation to mCD4 results in conformational changes that expose epitopes in the variable domains and disrupt quaternary epitopes in the native Env trimer.
IMPORTANCE These findings indicate the antigenic consequences of adapting HIV-1 Env to mCD4. They also suggest that to best mimic HIV-1 infection in humans when using the SHIV/macaque model, HIV-1 Env proteins should be identified that use mCD4 as a functional receptor and preserve quaternary epitopes characteristic of HIV-1 Env.
INTRODUCTION
Macaque models of human immunodeficiency virus HIV type 1 (HIV-1) infection have been critical to preclinical vaccine and passive-immunization studies and to the understanding of HIV-1 pathogenesis. HIV-1 does not persistently infect macaques because of several species-specific host factors that prevent infection or inhibit viral replication (1). Simian immunodeficiency virus (SIV)/HIV chimeric viruses (SHIVs) encode SIV antagonists of these macaque restriction factors, and such SHIVs serve as surrogates of HIV-1 infection in macaques. Despite the fact that SHIVs incorporate the critical SIV antagonists of known macaque restriction factors, they require additional passage in vivo in order to replicate to high levels and cause persistent infection in macaques (1). Even with the improved understanding of host-virus interactions, there has been variable success in generating SHIVs capable of establishing infection in macaques, and this process remains expensive and labor-intensive.
SHIVs that incorporate the gene for the envelope glycoprotein (Env) of HIV-1 are particularly important for HIV-1 vaccine and passive-immunization studies with macaques because Env is the major target of the host antibody response. Thus, Env proteins from viruses representing those that were transmitted and/or successfully spreading in the population would be ideal; however, all but two SHIVs in current use encode Env sequences derived from chronic infection (2, 3). Moreover, currently available pathogenic SHIVs represent only two of the major circulating HIV-1 subtypes, B and C (2–8). Identifying pathogenic SHIVs based on other subtypes has been hindered by the fact that not all SHIV chimeras replicate in macaque lymphocytes (9). Thus, the current limited collection of SHIVs does not represent the genetic diversity of circulating HIV-1 strains.
All but two of the SHIVs in current use—both carrying a subtype C env (2, 3)—were generated by using virus that was first amplified by replication in culture. Among the SHIVs that have been tested for infection in macaques, all required serial passage to further adapt to cause persistent infection and disease (2–8). Several studies have shown that this process of serial passage resulted in mutations in both the constant and variable regions of Env (8, 10–16). A number of these studies focused on CXCR4 and dual-tropic variants of HIV-1 and showed that the passaged viruses have neutralization profiles that differ from those of the unpassaged viruses from which they were derived, suggesting that adaptation of HIV-1 Env to macaques may alter its antigenicity. In general, the CXCR4- and dual-tropic HIV-1 Env proteins that were passaged in macaques were more resistant to monoclonal antibodies (MAbs). However, there has not been a systematic evaluation of how the process of macaque adaptation impacts the antigenic properties of SHIVs representing transmitted HIV-1 Env proteins, which use the CCR5 coreceptor. Likewise, the role of adaptation of HIV-1 Env to the mCD4 receptor in this process has not been examined.
The requirement for adaptation of SHIVs is not surprising, given that species-specific differences between the human and macaque CD4 (mCD4) receptors restrict the ability of HIV-1 Env variants to infect macaque cells (17, 18). Specifically, a single polymorphism at position 39 in the mCD4 (isoleucine) versus the human CD4 (asparagine) receptor leads to a 1- to 2-log reduction in the entry of circulating HIV-1 variants (18). The SHIVs in common use all have the ability to use the mCD4 receptor more efficiently than circulating HIV-1 variants (18), suggesting that they are able to tolerate this amino acid difference in the mCD4 receptor. Recently, two independent point mutations in Env were identified that increased entry mediated by mCD4 by ∼100-fold for most of the viruses tested (17). Interestingly, these two mutations occur outside the CD4 binding site (CD4bs), in the C2 region (A204E) and the V3 loop (G312V) of the gp120 subunit of Env. A SHIV carrying the G312V change has also been selected in macaques (19).
It is critical that SHIVs that are used in preclinical models for vaccine and passive-immunization studies retain the antigenic features of HIV-1 Env circulating in humans. The recent identification of broadly neutralizing antibodies (bNAbs) against HIV-1 has provided a set of highly sensitive probes to assess changes to the conformation of the Env trimer at distinct epitopes that were not available in earlier studies of SHIV evolution (13-16, 20). These bNAbs include the following: CD4bs antibodies, exemplified by VRC01 and VRC03 (21); a class of antibodies that recognize glycan-dependent epitopes in the V1V2 region, such as PG9, PG16, and PGT145 (20, 22); antibodies that recognize glycan-dependent epitopes in the V3 loop, PGT121 and PGT128 (20); and those that target the membrane-proximal external region (MPER), a recent example of which is 10E8 (23). A number of these bNAbs target quaternary epitopes formed by the interaction of multiple Env protomers and preferentially bind the trimeric form of Env (22, 24, 25). Such bNAbs are useful in detecting conformational changes in the Env trimer.
It is unclear what, if any, effect the changes that result from adaptation of HIV-1 Env proteins circulating in humans for entry using the mCD4 receptor have on the biological and immunological properties of the Env protein. If the process of increasing replication fitness by adapting to mCD4 leads to major biological and/or antigenic changes in Env, then SHIVs based on these Env proteins may not faithfully predict key features of HIV-1 Env proteins spreading in human populations. Thus, understanding how the adaptation impacts Env is a critical consideration in the use of these model systems for screening HIV-1 vaccines and prevention methods. The goals of the present study were to assess the antigenic properties of HIV-1 Env proteins adapted to replication in macaques and to determine if there are conformational changes in the Env trimer that facilitate this adaptation. Our results indicate that Env proteins from SHIVs capable of entry using mCD4 exhibit a pattern of resistance to antibodies that recognize quaternary epitopes that is greater than that of the parental Env while becoming more sensitive to epitopes that are further exposed upon CD4 binding. We propose that adaptation to mCD4 results in conformational changes that allow the Env trimer to more readily adopt the CD4-bound state, disrupting quaternary epitopes in the native Env trimer and increasing sensitivity to CD4-induced antibodies.
MATERIALS AND METHODS
Envelope clones and mutagenesis.
The following envelope clones and their corresponding A204E and G312V mutant proteins were described previously (17, 18): five subtype A clones (Q23ENV.17, MG505.W0M.ENV.H3, BG505.W6M.ENV.B1, QF495.23M.ENV.A3), one subtype C clone (QC406.70M.ENV.F3), and one subtype D clone (QA013.70I.ENV.H1). For the present study, nucleotide changes were introduced by site-directed mutagenesis to generate A204E and G312V Env mutant proteins for envelope clone QA255.662M.C (26) by methods similar to those previously described (17).
The following wild-type envelope clones were also used: seven subtype A clones (QA255.21P.ENV.A15, Q461.d1, QH209.A2, QH359.21M.ENV.C1, QF495.23M.ENV.B2, Q842.d16, QH343.21M.ENV.A10) (27–29), seven subtype B clones (RHPA.4259.7, WITO4160.B33, AD8, TRJO4551.58, CAAN5342.A2, QH0692.42, PVO.4) (30), six subtype C clones (ZM233M.PB6, ZM249.PB1, CAP210.2.00.E8, CAP45.2.00.G3, DU156.12, DU422.1) (30, 31), and three A/D recombinant subtype clones (QG393.60M.ENV.B7, QA790.204I.ENV.A4, QA790.204I.ENV.C1) (28).
The following SHIV envelope clones were used: SHIV AD8-EO (32) and SF162P3 (4). SHIV AD8-EO Env was cloned from a full-length proviral clone into the pCI-neo vector by using EcoRI and SalI restriction enzymes.
Production of virus.
Pseudoviruses were generated by cotransfecting 293T cells with 0.5 μg of each envelope clone of interest with 1.0 μg of an env-deficient subtype A proviral plasmid (Q23Δenv) (27). For these studies, 4 × 105 293T cells were plated into the wells of a six-well dish ∼24 h prior to transfection in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS) and 2 mM l-glutamine (complete DMEM). For each transfection, plasmid DNA was mixed with 6 μl of Fugene 6 transfection reagent (Roche). Pseudoviruses were harvested 48 h posttransfection. Supernatants were spun at 1,200 × g for 5 min at room temperature to remove cell debris, and aliquots of the pseudovirus stocks were stored at −80°C. Green fluorescent protein (GFP) reporter viruses were generated by cotransfecting 5 × 105 293T cells, plated the day prior, with 333 ng of each envelope clone and 667 ng of Q23Δenv-GFP—a derivative of the subtype A Q23Δenv provirus that encodes GFP (27). Transfections were performed as described for pseudoviruses, and supernatants were harvested at 72 h posttransfection. The viral titer of each transfection supernatant was determined by infecting TZM-bl cells and counting the blue cells at 48 h postinfection after staining for β-galactosidase activity (33).
Viral stocks of full-length, replication-competent SHIV 1157ipEL-p (2) were generated by expanding the virus in immortalized pig-tailed macaque lymphocytes as described previously (34).
Neutralization assay.
Approximately 500 infectious pseudoviral particles in 25 μl of complete DMEM were incubated with 5-fold serial dilutions of each MAb or soluble CD4 (sCD4) in duplicate for 60 min at 37°C. A total of 1 × 104 TZM-bl cells in 100 μl of complete DMEM was added to each dilution in the presence of DEAE-dextran at a final concentration of 10 μg/ml. For assays with SHIV 1157ipEL-p, full-length, replication-competent virus was used. Each MAb—VRC01, VRC03 (21), PG9, PG16, PGT145 (22), PGT121, PGT128 (20), 10E8 (23), 447-52D (35), and 697-30D (36)—was tested at a starting concentration of 10 μg/ml. At 48 h postinfection, β-galactosidase activity was measured with the Galacto-Lite system (Applied Biosystems). For assays with MAb 17b (37), pseudovirus was incubated with antibody dilutions for 6 h before infection of TZM-bl cells. Pooled plasma from 30 HIV-1+ individuals from Kenya (38) was tested at 2-fold serial dilutions starting at 1:100. The 50% inhibitory concentration (IC50) was determined as the concentration of antibody or reciprocal plasma dilution at which 50% of the pseudovirus input was neutralized as previously described (33). IC50s represent the average of at least two independent experiments performed in duplicate.
Generation of pseudoviruses incorporating heterotrimeric Env proteins.
Pseudoviruses incorporating heterotrimeric Env proteins were generated by cotransfecting various ratios of Q23.17 A204E mutant Env and Q23.17 parental Env plasmids. Two independent DNA preparations were generated and tested for each mutant and parental plasmid. The concentration of DNA in each preparation was determined with the NanoDrop 2000 spectrophotometer (Thermo Scientific). The probability of any given trimeric protein incorporating a mutant monomer defined as follows: i number of mutant protomers with f fraction of mutant transfected, assuming random assortment of mutant and parental protomers, was determined by using the following formula: (39).
Infection of mCD4 and huCD4 cells.
Cf2Th/syn CCR5 cells that stably express human CCR5 and either mCD4 or hCD4 were plated into the wells of a 24-well plate ∼24 h prior to infection at 4 × 104 cells/well in 500 μl of complete DMEM (18). Cells were infected with HIV-1 Env pseudotyped Q23Δenv-GFP reporter viruses at an estimated multiplicity of infection of 0.1 in 100 μl of DMEM by spinoculation for 90 min at 1,200 × g in the presence of DEAE-dextran at a final concentration of 10 μg/ml. After incubation for 48 h, the cells were washed with 1× phosphate-buffered saline and then incubated with 100 μl of 5 mM EDTA to remove the cells from the plate. The cells were then fixed with 500 μl of 1% paraformaldehyde. Cells were analyzed for GFP expression by flow cytometry on the Canto II (BD Biosciences).
Derivation of the QA255 CD4i variants.
An infectious molecular clone of HIV-1 was generated by cloning the env gene from QA255.662M.C (26) into a full-length plasmid derived from the HIV-1 variant Q23.17 by using the SmaI and XhoI sites flanking the env gene as described previously (40). CD4-independent QA255.662M.C was derived with CD4+ SupT1 T cells and a CD4-negative variant of this line, termed BC7 (41), each of which was transduced with a lentiviral vector to stably express human CCR5 (e.g., SupT1/R5 and BC7/R5). Parental QA255.662M.C was serially passaged in a 1:10 mixture of SupT1/R5 and BC7/R5 cells, respectively, and monitored during each passage for expression of p27gag by immunofluorescence microscopy. When infection of >50% of the cell mixture was documented, virus was passaged cell free for an additional 10 times in BC7/R5 cells alone. Full-length env gene sequences were generated from the culture-adapted virus by standard methods. Briefly, proviral DNA isolated from infected cells was subjected to PCR with primers that amplify the full-length env gene as described previously (26). An infectious molecular clone containing the CD4-independent Env was generated by cloning the env gene into a full-length Q23.17 plasmid as described above. Full-length plasmids containing the CD4-independent env gene or the parental env gene (QA255.662M.C) were transfected into 293T cells to generate an infectious viral stock of each virus. For the viral infection assays, SupT1/R5 and BC7/R5 cells were infected with 50 μg of p24gag for each virus as previously described (42) The resulting PCR products representing CD4-independent env genes were cloned into the pCI-neo vector with the MluI and NotI restriction enzymes. The Env clones were analyzed for fusion function on quail QT6 fibroblast target cells (42, 43).
Data presentation and analysis.
Data were plotted and Spearman correlations were performed with Prism version 6.0c (GraphPad Software). A204 and G312 residues were highlighted on the BG505 SOSIP structure (Protein Data Bank [PDB] code 4NCO) (44) with MacPyMOL.
RESULTS
A204E and G312V mutations that permit entry using mCD4 affect antibody recognition of the V1V2 region.
We assessed whether the A204E and G312V mutations, which lead to increased entry using mCD4 (18), affect the antigenic properties of HIV-1 Env. Seven Env proteins that represent the most common subtypes circulating in sub-Saharan Africa (A, C, and D) were tested, i.e., three that were isolated recently after sexual transmission (23 to 70 days), two isolated in the second year of infection, one from an infant at the first HIV positive time after birth (42 days), and one from the corresponding mother during chronic infection (Fig. 1A). For five of the seven Env variants tested, independent introduction of the A204E and G312V mutations yielded functional Env proteins capable of mediating infection with both the hCD4 and mCD4 receptors (17, 18). For two of the Env proteins (QA013.H1 and QC406.F3), only the A204E mutant proteins encoded an Env protein capable of mediating entry; the G312V mutation yielded an Env protein that could not mediate entry into cells with either human or mCD4, and these two nonfunctional variants were not analyzed in this study. Simultaneous introduction of the A204E and G312V mutations did not produce a functional Env protein for any of the variants tested. The functional Env proteins harboring either the A204E or the G312V mutation, which were selected by passage in vitro, are referred to here as mCD4-adapted Env proteins.
FIG 1.
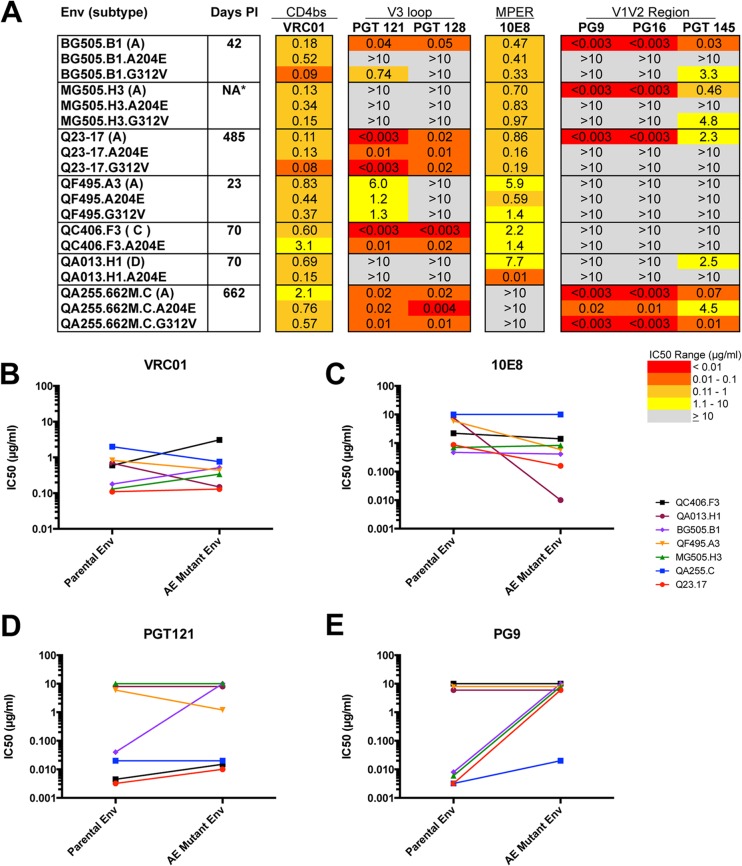
(A) Summary of neutralization profiles of seven parental HIV-1 Env proteins and corresponding A204E/G312V mutant proteins. The envelope clone, the virus subtype (in parentheses), and the estimated number of days postinfection (PI) are indicated in the first two columns. NA indicates that the number of days postinfection is not available. The A204E and G312V mutant proteins are indicated below the corresponding parental Env protein. Darker shading indicates increasing sensitivity to the MAb tested, according to the key at the bottom. Gray shading indicates that 50% neutralization was not reach at the highest concentration tested (10 μg/ml). The IC50s in the chart are average results of at least two independent experiments performed in duplicate. Comparison of IC50s for parental and A204E mutant Env proteins for MAbs VRC01 (B), 10E8 (C), PGT121 (D), and PG9 (E). IC50s (μg/ml) are plotted for the parental (left) and corresponding A204E mutant (right) Env proteins. The key at the right indicates the color coding of each parental and mutant Env pair tested.
We previously reported that the introduction of either the A204E or the G312V mutation resulted in increased sensitivity to sCD4 (17). Despite this change in sensitivity to sCD4, the introduction of these mutations into parental envelope proteins had little (<6-fold) to no effect on neutralization by the CD4-binding site antibody VRC01 (Fig. 1A and B). Similarly, for six of the seven Env proteins tested, the mCD4-adapted mutant proteins exhibited a relatively modest change (<10-fold) in the IC50 of the PGT antibodies recognizing the V3 loop and MPER antibody 10E8 (Fig. 1A, C, and D). The exception to this was BG505, for which the mCD4-adapted Env proteins were resistant to PGT121 and/or PGT128. In contrast, the introduction of A204E or G312V resulted in an increase in resistance to the quaternary V1V2 antibodies PG9, PG16, and PGT145. This was true for all four Env proteins that were originally neutralization sensitive. For three of these four mCD4-adapted Env proteins, the increase in the IC50 of PG9 and PG16 was greater than 300-fold compared to that of the parental virus (Fig. 1A and E). For the other three Env proteins, which were initially resistant to these MAbs, the mCD4-adapted Env proteins showed no detectable change in neutralization sensitivity. In one such case, an Env protein (QF495.A3) that has the N160 residue but does not have the full glycosylation sequon (N-X-S/T) (22, 45) that is required for glycan-dependent recognition by PG9 and PG16, was resistant to neutralization, whether or not the A204E or G312V mutation was present. Two of the envelope proteins (QA013.H1 and QC406.F3) that maintained the N160 glycosylation site and were still resistant to PG9 and PG16 remained resistant when the mutations were introduced.
Thus, while the A204E and G312V mutations had little effect on recognition by the CD4bs antibody VRC01 and V3 and MPER antibodies—MAbs that are directed to epitopes that are largely found within the monomeric protein (20, 23)—these mutations disrupted recognition by MAbs targeting quaternary V1V2 epitopes (PG9, PG16, and PGT145) in Env proteins that were sensitive to these MAbs.
A204E mutant proteins are sensitive to CD4-induced MAb 17b.
Our initial results showing dramatic increases in resistance to PG9, PG16, and PGT145 in the mCD4-adapted Env proteins compared to parental Env and the fact that the mutations are found outside the characterized contact residues of these antibodies prompted us to explore whether these mutations cause structural perturbations by using additional antibodies as probes. The epitopes of PG9 and PG16 depend on interactions involving neighboring gp120 subunits that come into contact in the envelope trimer (22, 25, 45). Of note, the interactions between adjacent gp120 subunits in the Env trimer are also disrupted in the context of CD4 binding (46). We hypothesized that introduction of the A204E and G312V mutations resulted in an open trimer conformation similar to that observed upon CD4 engagement. To explore this hypothesis, we tested the mCD4-adapted Env proteins for neutralization sensitivity to MAb 17b, which recognizes a CD4-induced epitope (37). The neutralization assay was performed in the absence of sCD4 in order to assess the sensitivity of the native trimer to neutralization by MAb 17b. None of the parental envelope proteins were sensitive to MAb 17b at the highest concentration of antibody tested, 10 μg/ml. For five of the seven envelope proteins tested, the A204E mutant protein was more sensitive to neutralization by MAb 17b than the corresponding parental Env protein was (Fig. 2A). None of the G312V mutant proteins exhibited sensitivity to MAb 17b greater than that of the parental envelope protein (Fig. 2A and B). The IC50 for MAb 17b correlated with the reciprocal dilution IC50 for pooled HIV-1+ plasma (Spearman r = −0.81, P < 0.001), suggesting that CD4-induced antibodies similar to MAb 17b may mediate the increased sensitivity of these A204E Env proteins to pooled plasma.
FIG 2.
(A) Summary of the neutralization sensitivities of parental and corresponding A204E/G312V mutant Env proteins to MAbs 17b, VRC03, 697-30D, and 447-52D and to HIV+ pooled plasma. The envelope clone and the virus subtype (in parentheses) are indicated in the first column. The A204E and G312V mutant proteins are indicated below the corresponding parental Env protein. Darker shading indicates increasing sensitivity to the MAb tested or increasing sensitivity to HIV+ pooled plasma, according to the key at the bottom. Gray shading indicates that 50% neutralization was not reach at the highest concentration tested (10 μg/ml) for MAbs or the highest dilution of plasma tested (1:100). Comparison of IC50s for parental and A204E mutant Env proteins for MAbs 17b (B), 697-30D (C), 447-52D (D), and VRC03 (E). IC50s (μg/ml) are plotted for the parental (left) and corresponding A204E mutant (right) Env proteins. The key at the right indicates the color coding of each parental and mutant Env pair tested.
The A204E mutation disrupts quaternary epitopes on the Env trimer.
In order to investigate further the structural changes induced by these adaptive mutations, we first tested a MAb that is predicted to make contacts with the adjacent protomer near the CD4bs (VRC03) (24). VRC03 is a potent and broadly neutralizing antibody, and recently, Lyumkis et al. described the quaternary nature of the epitope recognized by VRC03 (21, 24). Five of the Env proteins were initially sensitive to VRC03, and in all five cases, introduction of the A204E mutation rendered these Env proteins resistant to neutralization by VRC03 at the highest antibody concentration tested (10 μg/ml) (Fig. 2A and E). The subtype A variant Q23.17 exhibited the greatest effect, with a >100-fold increase in the IC50. For four of these Env pairs, the A204E mutation also increased resistance to quaternary V1V2 antibodies (PG9/PG16 and PGT145) (Fig. 1A). The G312V mutation had a more selective effect on VRC03 neutralization, with only two of four initially sensitive variants becoming resistant, both of which also acquired resistance to PG9/PG16 MAbs (Fig. 1A). The Env proteins that were initially resistant to VRC03 remained resistant when mutated to either A204E or G312V.
The above results and the observation that portions of the V2 and V3 domains are occluded in the Env structure (24, 44) led us to hypothesize that the V2 and V3 domains may be more exposed in the mCD4-adapted Env proteins. To address this hypothesis, we tested antibodies that target nonquaternary, conformational epitopes in V2 (697-30D) (36) and V3 (447-52D) (35). All of the parental Env clones tested were resistant to neutralization by 697-30D and 447-52D with IC50s greater than 10 μg/ml (Fig. 2C and D). Three of the Env proteins tested (Q23.17, QA013.H1, and QF495.A3) became sensitive to neutralization by 697-30D and 447-52D upon introduction of the A204E mutation. The A204E mutant forms of these Env proteins were also sensitive to neutralization by MAb 17b (Fig. 2B). The G312V mutant Env proteins did not affect sensitivity to either 697-30D or 447-52D. In the case of 447-52D, this is not unexpected, given that the G312 residue is critical for the binding of 447-52D to the V3 loop of Env (35). Overall, these results indicate that introduction of the A204E mutation disrupts quaternary interactions at the apex of the trimer and at the CD4bs, as evidenced by resistance to PG9/16 and VRC03, and expose regions within the variable loops, as evidenced by sensitivity to 697-30D and/or 447-52D.
Disruption of quaternary epitopes in heterotrimeric Env proteins.
The trimeric conformation of Env is maintained by metastable interactions between protomers. In order to determine whether disruption of the quaternary epitopes in the trimer requires that all protomers encode the determinants for mCD4 entry, we generated pseudoviruses expressing heterotrimeric Env proteins by cotransfecting plasmids encoding mutant Q23.17 A204E and parental Q23.17 Env proteins at mutant Env/total Env fractions of 0.0, 0.2, 0.4, 0.6, 0.8, and 1.0. For each population of heterotrimeric pseudoviruses resulting from the cotransfections, we determined the probability of any given heterotrimeric protein containing >0, >1, and exactly 3 mutant protomers, assuming random assortment of parental and mutant Env proteins (22, 39, 47). For example, at a fraction of 0.2, the probabilities that a given trimer has >0, >1, and exactly 3 mutant protomers are 0.49, 0.10, and 0.008, respectively.
IC50s for VRC03 were determined for each set of heterotrimeric Env proteins. We plotted the VRC03 IC50s of each population of heterotrimeric pseudoviruses independently against the probabilities of producing a protein with >0, >1, and exactly 3 mutant protomers (Fig. 3A to C). As expected, increasing the amount of A204E mutant transfected resulted in an increase in the VRC03 IC50, indicating resistance to neutralization. For the plot of the probability of any given heterotrimeric protein having >1 (2 or 3) mutant protomer, the curve revealed a nearly log-linear relationship between the probability of having >1 mutant protomer and the VRC03 IC50 (Fig. 3B). These results suggest that the heterotrimeric proteins containing two or three mutant protomers are largely responsible for the IC50 increases observed. Thus, the disruption of quaternary interactions between adjacent protomers likely requires the presence of at least two mutant A204E protomers.
FIG 3.
Sensitivity of heterotrimeric pseudoviruses to neutralization by VRC03. IC50s are plotted on the y axis versus the probability of any given trimer containing >0 (A), >1 (B), or 3 (C) mutant protomers. Probabilities were calculated according to the fraction of mutant plasmid transfected (0, 0.2, 0.4, 0.6, 0.8, or 1). Two independent sets of DNA preparations were generated and tested. AE, A204E.
Neutralization profiles of Env proteins from pathogenic SHIV variants.
Because the Env variants carrying changes that conferred increased replication in macaque lymphocytes showed substantial differences in their antigenic profiles compared to parental viruses, we assessed the neutralization profiles of three Env variants from pathogenic SHIVs (SF162P3, 1157ipEL-p, and AD8-EO) that were generated by serial passage in rhesus macaques (2, 4, 32) (Fig. 4A and B). All Env proteins from pathogenic SHIVs that we have tested were able to use mCD4 efficiently, similar to the Env proteins carrying the A204E and G312V mutations (18). Similar to the mCD4-adapted Env proteins, two of the Env proteins from pathogenic SHIVs, both derived from subtype B HIV-1 variants (AD8-EO and SF162P3) were resistant to quaternary V1V2 MAbs (PG9/PG16) (Fig. 4A). The AD8 pair showed a profile with PG9/PG16 similar to that of the parental and A204E/G312V mCD4-adapted pairs—the parental Env protein (AD8) was sensitive to these quaternary MAbs, whereas the macaque-adapted variant (AD8-EO) was not (Fig. 4A). There was also a 17-fold decrease in sensitivity to PGT145 for this pair. In the case of SF162, the parental Env was also resistant to PG9/PG16, reflecting the fact that this variant carries a K rather an N at position 160 (11, 22). SHIV SF162P3 does, however, have an N at position 160; thus, the lack of a potential N-linked glycosylation site does not explain its resistance to PG MAbs (11). The SF162 and AD8 Env proteins were all sensitive to VRC03 and resistant to 697-30D and 447-52D (Fig. 4B).
FIG 4.
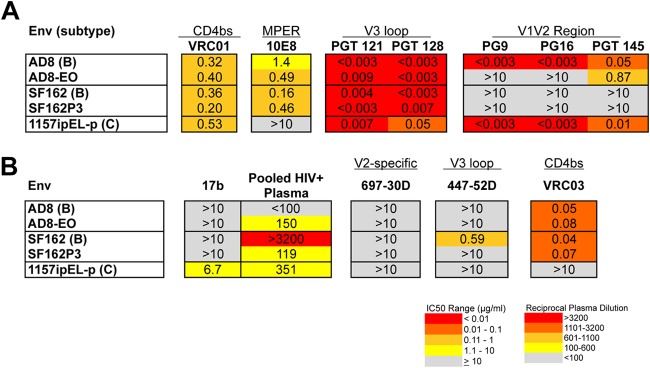
Summary of neutralization profiles of Env proteins from pathogenic SHIVs. The pathogenic SHIVs from which the Env proteins were derived are indicated in the first column along with the subtypes (in parentheses) (A, B). Darker shading indicates increasing sensitivity to the MAb tested or increasing sensitivity to HIV+ pooled plasma, according to the key at the bottom. Gray shading indicates that 50% neutralization was not reach at the highest concentration tested (10 μg/ml) for MAbs or the highest dilution of plasma tested (1:100).
Env from SHIV 1157ipEL-p exhibited an antigenic profile different from that of the subtype B variants (2). Similar to the mCD4-adpated Env proteins, this Env was sensitive to MAb 17b and resistant to VRC03. In contrast to SHIV AD8-EO and SHIV SF162P3, Env from SHIV 1157ipEL-p was sensitive to PG9/PG16 (Fig. 4A). Thus, each of the Env proteins from animal-passaged, pathogenic SHIVs was resistant to some MAbs directed to quaternary epitopes, although they were not uniformly or consistently resistant to particular MAbs of this type.
Changes in the neutralization profile of CD4-independent Env proteins.
The neutralization profiles of the mCD4-adapted Env proteins, particularly the A204E mutant proteins described above, were similar to those previously published for HIV-1 Env proteins that are CD4 independent (CD4i) in that they exhibited increased sensitivity to both sCD4 and MAb 17b (48–50). To test whether CD4i Env proteins show changes in neutralization sensitivity similar to those of mCD4-adpated Env proteins, including disruption of quaternary epitopes, we derived CD4i variants of one of the subtype A HIV-1 Env proteins tested above (QA255.662M.C). For this purpose, a full-length proviral clone encoding the Env of interest was generated and used to produce virus that was passaged in culture containing CD4-negative, CCR5-positive cells. A virus that replicated in these cells was isolated after 10 passages. Full-length Env sequences from the culture-adapted CD4i variants were cloned and tested for the ability to mediate fusion in CD4-negative cells. Two CD4i Env proteins (QA255-CD4iA and QA255-CD4iB) could mediate robust cell-cell fusion on quail QT6 target cell that expressed CCR5 in the absence of CD4 (Fig. 5A). No fusion activity was acquired by these CD4-independent Env proteins on CXCR4 with or without CD4. When cloned into a subtype A provirus, viruses containing the QA255-CD4iB or parental QA255.662M.C Env protein could infect CD4-positive SupT1/CCR5 cells, but only a virus with the QA255-CD4iB Env protein could mediate a spreading infection on a CD4-negative variant of this line (BC7/CCR5) (Fig. 5B).
FIG 5.
Evaluation of CD4-independent Env proteins from QA255.662M.C (A) Cell-cell fusion activity of CD4-independent Env proteins was assessed on QT6 target cells expressing the indicated coreceptors with or without CD4. Fusion was quantified in relative light units (RLU). A vector containing only GFP was used as a negative control. (B) Viral growth in CD4-positive (SupT1/CCR5) and CD4-negative (BC7/CCR5) cell lines is shown for subtype A viruses containing parental QA255.662M.C or CD4-independent QA255-CD4iA and QA255-CD4iB Env proteins. Reverse transcriptase (RT) activity (counts per minute [cpm]) is shown. (C, D) Summary of neutralization profiles of CD4-independent Env proteins. The parental Env protein from which the CD4-independent Env proteins were derived is indicated on the first row with the CD4-independent Env proteins below.
The two CD4-independent Env proteins exhibited neutralization profiles similar to those of the A204E mutant Env proteins (Fig. 5C and D). Compared to the parental Env protein from which they were derived, the CD4-independent Env proteins exhibited an increase in resistance to antibodies that recognize quaternary epitopes (PG9/PG16 and VRC03). This increase in resistance to antibodies that recognize quaternary epitopes corresponded to an increase in sensitivity to the CD4-induced MAb 17b and the MAb 447-52D, which targets a V3-specific epitope that may be buried in the parental Env protein (35). The striking similarity in the antigenic properties of the CD4-independent Env proteins tested and those of A204E mutant Env proteins that evolved to use the mCD4 receptor suggests that similar conformational changes may be required for HIV-1 Env proteins to adapt to mCD4 and to become CD4 independent.
Association between sensitivity to antibodies recognizing quaternary epitopes and mCD4 infectivity.
The ability to use the mCD4 receptor for entry is a prerequisite for establishing persistent infection in macaques. Our neutralization data on the mCD4-adapted Env proteins suggest that adaptation of HIV-1 Env proteins to mCD4 in vitro and in vivo results in disruption of quaternary epitopes that are recognized by some bNAbs within the context of the trimer. These results raise the possibility that HIV-1 Env variants that use mCD4 as a receptor may not be targeted by these neutralizing antibodies, limiting the potential utility of the SHIV model in cases where the immune response targets more complex epitopes. To address this possibility, we investigated whether the ability of HIV-1 Env proteins to use the mCD4 receptor for entry predicts their sensitivity to a MAb that targets a quaternary epitope. We previously tested the ability of 30 HIV-1 Env proteins isolated from patients recently after transmission to enter cells with the mCD4 receptor (18). In the present study, we tested an additional 18 HIV-1 Env variants isolated soon after infection for a total of 48 variants. Forty-three of the HIV-1 Env proteins tested were isolated recently after infection. We focused on recently transmitted variants because SHIVs encoding such Env proteins may be more representative of variants against which an effective vaccine would need to protect than lab-adapted Env proteins or those isolated later in infection. Entry was measured by comparing infection of cells expressing mCD4 with those expressing human CD4 (Fig. 6; see Table S1 in the supplemental material). There was no correlation between sensitivity or resistance to PG9 (Spearman r = 0.12, P = 0.42) or PG16 (Spearman r = 0.16, P = 0.27) and entry using mCD4, suggesting that the ability to use mCD4 for entry is not a determinant of sensitivity to PG9/PG16 for HIV-1 Env proteins. For 83% (40/48) of the HIV-1 Env proteins tested, relative infection of mCD4 cells compared to human CD4 cells was ≤10%. The eight HIV-1 Env proteins that used the mCD4 receptor more efficiently (relative infection of >10%) exhibited a wide range of neutralization sensitivity to PG9/16 (0.003 to 50 μg/ml). Among these variants, two Env proteins were neutralized at a concentration of 0.5 μg/ml, which is comparable to the median IC50 of 0.2 μg/ml for variants tested against PG9 (20). Overall, these results indicate that there is a subset of HIV-1 Env proteins that are able to use the mCD4 receptor for entry and maintain the quaternary epitope targeted by PG9/PG16.
FIG 6.
Relationship between infection of cells expressing mCD4 and sensitivity to MAbs targeting a quaternary epitope in the V1V2 region, PG9 (A) and PG16 (B). The association between infection of cells expressing mCD4 and sensitivity to neutralization by PG9 or PG16 was determined by using infectivity data as described previously (18). Each dot represents a different Env variant tested. The gray dotted line represents the threshold (10% relative infection) used to identify HIV-1 Env proteins that might be suitable for SHIV development.
DISCUSSION
Most HIV-1 Env proteins do not mediate entry using mCD4 or do so poorly, in particular, those representing circulating HIV-1 variants spreading in humans (18). In the context of a SHIV, Env can be readily adapted to increase the replication fitness of the virus in macaque cells, with as little as a single point mutation required to increase infectivity (18). Importantly, SHIVs currently used for vaccine and transmission studies have all undergone adaptation for increased replication fitness in macaques (1). Here we show that the cost of the adaptation of HIV-1 for replication in macaques is a shift in antibody recognition. In particular, our data suggest that the quaternary structure of Env is altered, making adapted Env proteins generally resistant to antibodies that recognize quaternary epitopes or make contacts with the adjacent protomer, such as PG9/PG16 and VRC03. However, there are some examples of circulating HIV-1 variants that can use the mCD4 receptor for entry and retain recognition by quaternary antibodies. We propose that the ability to use mCD4 as a functional receptor with limited disruption of quaternary epitopes should be used as a criterion for the rational design of SIV/HIV chimeric viruses that best recapitulate features of Env proteins of biologically relevant HIV-1 variants.
We previously reported that two independent mutations, A204E and G312V, that were selected as a subtype A-derived SHIV adapted to replicate in macaque cells significantly increase mCD4-mediated entry (18). Although the A204E and G312V mutations were identified by adapting a SHIV encoding a single HIV-1 Env variant (Q23.17), the mutations increased mCD4-mediated entry and replication for HIV-1 Env proteins representing globally circulating HIV-1 variants of diverse subtypes (17, 18). The G312V mutation was also observed in minimally modified HIV-1 passaged in macaques (19). In the present study, we assessed the conformational changes induced during adaptation to mCD4 across seven HIV-1 Env variants, including six cloned directly from the infected individual and several representing transmitted variants. For parental Env proteins that were initially sensitive to the antibodies tested, the mCD4-adapted Env proteins were more resistant to MAbs that recognize quaternary epitopes on the Env trimer. These antibodies included PG9, PG16, and PGT145, which recognize complex epitopes in the V1V2 region (20, 22), and VRC03, which targets the CD4bs (21) and is predicted to have more contact with adjacent protomers than other CD4bs MAbs such as VRC01 (24).
Interestingly, we detected differences in the antibody recognition profiles of the A204E and G312V mutant proteins. The A204E mutation caused a more global change in antibody recognition that included resistance to MAbs targeting quaternary epitopes in V1V2 (PG/PG16 and PGT145) and the CD4bs (VRC03), increased sensitivity to sCD4 and MAb 17b, and in a subset of cases, sensitivity to MAbs that target epitopes in V2 (697-30D) and V3 (447-52D) that may be hidden in the parental Env protein (51, 52). The G312V mutant proteins also showed resistance to MAbs targeting quaternary V1V2 epitopes (PG9/PG16 and PGT145), but only some variants showed resistance to VRC03. In addition, G312V did not affect the sensitivity to MAb 17b or the V2 and V3 MAbs for any of the Env proteins tested, suggesting that there are more limited structural changes because of the G312V mutation than because of the A204E mutation.
Recently, the structure of a soluble, stabilized form of the Env trimer (SOSIP) of a subtype A variant used in this study (BG505) was determined (24, 44). Figure 7 highlights the A204E and G312V mutations mapped onto the SOSIP trimer model (PDB code 4NCO). Previous studies demonstrated that disruption of interactions between V1V2 and V3 within gp120 relaxed the monomer to a state similar to that observed upon CD4 binding, with formation of the bridging sheet and a more ordered CD4bs (53, 54). Thus, there are likely at least two constraints on Env in the context of the trimer: (i) quaternary interactions at the apex of the trimer formed by V1V2 and V3 from adjacent protomers and (ii) tertiary interactions within gp120 subunits including V1V2 and V3 within a given protomer. The observation that the A204E mutation causes both resistance to antibodies that target quaternary epitopes and sensitivity to antibodies that target CD4-induced epitopes suggests that this change is not simply causing local perturbations of a single epitope. Rather, the data are consistent with a model in which this mutation relaxes both constraints on Env in the context of the trimer. Analysis of heterotrimeric Env proteins suggests that at least two mutant protomers are required to disrupt quaternary interactions within a heterotrimeric protein. The G312V mutation caused resistance to quaternary V1V2 antibodies but did not cause increased sensitivity to antibodies targeting CD4-induced epitopes. Thus, the G312V mutation may loosen quaternary contacts between adjacent protomers without disrupting the tertiary interactions within gp120 subunits. The differences in the neutralization profiles of the A204E and G312V mutant proteins suggest that there is a spectrum of conformational changes that can occur as HIV-1 Env proteins adapt to use mCD4 for entry. The A204E mutant proteins may represent mCD4-adapted Env proteins that exhibit more extensive conformational changes similar to those observed for CD4-independent Env proteins, while the G312V mutant proteins represent mCD4-adapted Env proteins with changes limited to the quaternary contacts between adjacent protomers. These findings suggest that it may be possible to identify Env variants that can bind mCD4 and yet retain many of the structural and antigenic features of circulating HIV-1 variants.
FIG 7.
Locations of the A204E and G312V mutations in the context of the structure of the Env trimer. (A) Side view of two gp120 subunits of the BG505 SOSIP structure (44) with the A204 residue highlighted in yellow and the G312 residue in red (obtained with MacPyMOL). (B) Top view of all three protomers of the BG505 SOSIP structure with the A204 and G312 residues highlighted.
The A204E and G312V mutations were selected to permit interaction with mCD4, and they are critical for allowing Env to mediate the entry of a CD4 receptor carrying isoleucine at position 39, as found in the mCD4 molecule (18). Our data indicate that the antigenic changes that accompany adaptation to mCD4 are similar to those that result from adaptation for CD4 independence. The neutralization profiles of the A204E mutant proteins were similar to those of the CD4-independent Env variants derived from one of the Env variants tested in this study (QA255.662M.C), including resistance to quaternary-epitope-directed MAbs and increased sensitivity to MAbs targeting CD4-induced epitopes. Thus, the process of adapting to mCD4 may lead to functional and structural changes similar to those involved in the process of adapting to low human CD4 receptor levels and CD4 independence. Related to this model, prior studies of SIV observed that Env trimers from a CD4-independent SIV by cryo-electron microscopy exhibited an open conformation highly related to CD4-activated Env proteins and markedly different from the more compact CD4-dependent SIV Env protein (46, 55). Of note, the mutations that lead to adaptation to mCD4 also confer the ability to use smaller amounts of the human CD4 receptor for entry (18). Interestingly, previously described variants capable of infecting cells expressing small amounts of CD4 exhibit antigenic properties similar to those identified for A204E and G312V mutant proteins, including resistance to PG9/PG16 and sensitivity to 447-52D (56). Taken together, the results suggest that HIV-1 Env variants may experience similar selective pressures and resulting conformational changes as they adapt to cells expressing either mCD4 or small amounts of human CD4.
Given the changes in conformation in HIV-1 Env that resulted from adaptation to mCD4-mediated entry in vitro, we hypothesized that HIV-1 Env variants encoded by pathogenic SHIVs, which are selected after adaptation for replication in macaques, would show similar disruption of quaternary antigenic determinants. Of note, these Env proteins from pathogenic SHIVs have A at position 204 and G at 312, suggesting that other amino acids are determining the ability of these Env proteins to mediate entry via the mCD4 receptor. These differences allowed us to probe whether general features of the Env protein that confer the ability to use the macaque receptor alter its antigenic properties, irrespective of the specific mutations involved.
Two of the Env proteins tested (AD8-EO and SF162P3) were resistant to PG9/PG16, as reported previously (10), suggesting that they did not form the quaternary epitope required for antibody binding. In the case of AD8-EO, resistance to PG9/PG16 developed after passage of the parental virus (AD8) in animals, an evolution that is similar to that observed for the variants specifically selected for mCD4-mediated entry. Interestingly, Env proteins AD8-EO and SF162P3 exhibited more limited changes in antigenicity that were characteristic of the G312V mutant proteins, including resistance to PG9/PG16 but not impacting VRC03 and 697-30D or 447-52D recognition. Env 1157ipEL-p had a unique profile, exhibiting resistance to VRC03 but none of the other changes associated with either G312V or A204E. Env 1157ipEL-p was neutralized by MAb 17b and HIV-1+ pooled plasma, suggesting that epitopes similar to those exposed upon CD4 binding were formed in the native trimer.
Several prior studies have investigated the effect of serial passage in macaques on the antigenic properties of Env from CXCR4-using or dual-tropic SHIVs and also noted resistance to some MAbs greater than that of the unpassaged viruses (14, 15, 57). In these models, it is unclear what role adaptation to mCD4 plays given that CXCR4-tropic, lab-adapted viruses are able to infect by using the mCD4 receptor (18, 58, 59). Some studies have indicated a role for the host antibody response in driving escape that resulted in resistance to MAbs and plasma from infected macaques (13, 14, 16, 60). Also, this evolution has been attributed in part to changes in coreceptor affinity or specificity (16, 61), and changes in Env that allowed better interaction with the CD4 receptor may have facilitated this coreceptor switch (62). Our findings suggest that, in addition to direct antibody pressure and selection for a change in coreceptor specificity, selection for increased entry via the mCD4 receptor may be an important driver of antigenic variation in SHIVs. It is possible that the SHIVs that advanced through animal testing, which likely represent a small subset of those constructed and tested in cell culture, are the ones that were engineered with the rare HIV-1 Env proteins that utilized the mCD4 receptor with relatively high efficiency. It is also possible that adaption in macaques further enhanced infectivity. In some cases, such as 1157ipEL-p and SF162P3, the virus was passaged during acute infection (2, 63), minimizing competing selection by host neutralizing antibodies. For a virus such as AD8-EO, passage was later infection (32), a time when Env was potentially under competing selective pressures to maximize mCD4-mediated entry and at the same time escape from neutralizing antibodies. In this regard, our findings provide a potentially informative approach to the identification of Env proteins that can use the mCD4 receptor yet have minimal changes in their antigenic profile by adapting Env to mCD4 in the presence of a MAb such as 17b, 697-30D, or 447-52D that would limit global structural effects on antibody recognition that lead to increased sensitivity.
The SHIVs analyzed in this study are important tools for assessing the capacity of antibodies to block infection and provide therapeutic benefits and have led to important findings concerning pathogenesis in macaques. Nonetheless, the disruption of key antigenic determinants in HIV-1 variants adapted for replication in macaques potentially limits the utility of this model for fairly assessing the benefit of antibodies directed to quaternary epitopes. PG9 and PG16 are prototype quaternary, glycan-dependent MAbs that target one of the major epitopes on HIV-1 Env and thus may be an important component of protective antibody responses elicited by a vaccine (64). The potency of these MAbs may be underestimated when using current SHIV models, although our data suggest that the SHIV 1157ipEL-p may be well suited for studies with these MAbs. Similarly, increased neutralization sensitivity to antibodies directed to epitopes that are more concealed on Env trimers could also be misleading. Therefore, there is a need for novel pathogenic SHIVs that maintain quaternary epitopes targeted by antibodies such as PG9/PG16 and are otherwise more representative of the Env structure of viruses circulating in humans. We screened a large panel of HIV-1 Env proteins isolated recently after transmission and identified a subset of unadapted Env proteins that are able to use the mCD4 receptor for entry at levels close to those of human CD4. This subset represents HIV-1 Env proteins of subtypes A, B, and C that exhibit a wide range of sensitivities (IC50s of 0.003 to 50 μg/ml) to the PG9/PG16 MAbs. Identification of HIV-1 Env proteins that are able to use the mCD4 receptor for entry and maintain quaternary epitopes may facilitate the generation of SHIVs that are more representative of transmitted/founder HIV-1 variants.
Supplementary Material
ACKNOWLEDGMENTS
We thank Malcolm Martin for providing the full-length SHIV AD8-EO plasmid; Susan Zolla-Pazner for providing MAbs 447-52D and 697-30D; Xueling Wu and John Mascola for providing MAb VRC01; the IAVI Neutralizing Antibody Consortium for providing MAbs PG9, PG16, PGT121, and PGT128; George J. Leslie for providing the BC7/CCR5 and SupT1/CCR5 cell lines; and the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, for providing the following reagents: MAb 10E8 (Mark Connors), MAb 17b (James E. Robinson), and the viral stock of SHIV 1157ipEL-p (Ruth Ruprecht and the Division of AIDS, NIAID). In addition, we thank Josephine Romano for technical assistance, Kelly Lee and Mike Guttman for helpful comments on the manuscript, and Dara Lehman and Tad Davenport for valuable discussions.
This study was supported by NIH R01 AI103981 (to J.O.) and by R01 AI045378, P30 AI045008 (Viral/Molecular Core, Penn Center for AIDS Research), and a Bill & Melinda Gates Foundation CAVD Award (to J.A.H.). D.F.B. was supported in part by Viral Pathogenesis Training Grant T32AI083203.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02680-14.
REFERENCES
- 1.Hatziioannou T, Evans DT. 2012. Animal models for HIV/AIDS research. Nat Rev Microbiol 10:852–867. doi: 10.1038/nrmicro2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, Li PL, Shai-Kobiler E, Wang T, McCann CM, Zhang H, Wood C, Kankasa C, Secor WE, McClure HM, Strobert E, Else JG, Ruprecht RM. 2006. Molecularly cloned SHIV-1157ipd3N4: a highly replication-competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C env. J Virol 80:8729–8738. doi: 10.1128/JVI.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren W, Mumbauer A, Gettie A, Seaman MS, Russell-Lodrigue K, Blanchard J, Westmoreland S, Cheng-Mayer C. 2013. Generation of lineage-related, mucosally transmissible subtype C R5 simian-human immunodeficiency viruses capable of AIDS development, induction of neurological disease, and coreceptor switching in rhesus macaques. J Virol 87:6137–6149. doi: 10.1128/JVI.00178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harouse JM, Gettie A, Eshetu T, Tan RCH, Bohm R, Blanchard J, Baskin G, Cheng-Mayer C. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIVSF162P3. J Virol 75:1990–1995. doi: 10.1128/JVI.75.4.1990-1995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimura Y, Shingai M, Willey R, Sadjadpour R, Lee WR, Brown CR, Brenchley JM, Buckler-White A, Petros R, Eckhaus M, Hoffman V, Igarashi T, Martin MA. 2010. Generation of the pathogenic R5-tropic simian/human immunodeficiency virus SHIVAD8 by serial passaging in rhesus macaques. J Virol 84:4769–4781. doi: 10.1128/JVI.02279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reimann KA, Li JT, Veazey R, Halloran M, Park IW, Karlsson GB, Sodroski J, Letvin NL. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol 70:6922–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joag SV, Li Z, Foresman L, Stephens EB, Zhao LJ, Adany I, Pinson DM, McClure HM, Narayan O. 1996. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J Virol 70:3189–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li JT, Halloran M, Lord CI, Watson A, Ranchalis J, Fung M, Letvin NL, Sodroski JG. 1995. Persistent infection of macaques with simian-human immunodeficiency viruses. J Virol 69:7061–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luciw PA, Pratt-Lowe E, Shaw KE, Levy JA, Cheng-Mayer C. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc Natl Acad Sci U S A 92:7490–7494. doi: 10.1073/pnas.92.16.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautam R, Nishimura Y, Lee WR, Donau O, Buckler-White A, Shingai M, Sadjadpour R, Schmidt SD, LaBranche CC, Keele BF, Montefiori D, Mascola JR, Martin MA. 2012. Pathogenicity and mucosal transmissibility of the R5-tropic simian/human immunodeficiency virus SHIV(AD8) in rhesus macaques: implications for use in vaccine studies. J Virol 86:8516–8526. doi: 10.1128/JVI.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu M, Harouse JM, Gettie A, Buckner C, Blanchard J, Cheng-Mayer C. 2003. Increased mucosal transmission but not enhanced pathogenicity of the CCR5-tropic, simian AIDS-inducing simian/human immunodeficiency virus SHIVSF162P3 maps to envelope gp120. J Virol 77:989–998. doi: 10.1128/JVI.77.2.989-998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humbert M, Rasmussen RA, Song R, Ong H, Sharma P, Chenine AL, Kramer VG, Siddappa NB, Xu W, Else JG, Novembre FJ, Strobert E, O'Neil SP, Ruprecht RM. 2008. SHIV-1157i and passaged progeny viruses encoding R5 HIV-1 clade C env cause AIDS in rhesus monkeys. Retrovirology 5:94. doi: 10.1186/1742-4690-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etemad-Moghadam B, Karlsson GB, Halloran M, Sun Y, Schenten D, Fernandes M, Letvin NL, Sodroski J. 1998. Characterization of simian-human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected monkeys. J Virol 72:8437–8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cayabyab M, Karlsson GB, Etemad-Moghadam BA, Hofmann W, Steenbeke T, Halloran M, Fanton JW, Axthelm MK, Letvin NL, Sodroski JG. 1999. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2). J Virol 73:976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etemad-Moghadam B, Sun Y, Nicholson EK, Karlsson GB, Schenten D, Sodroski J. 1999. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immunodeficiency virus passaged in vivo. J Virol 73:8873–8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etemad-Moghadam B, Sun Y, Nicholson EK, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. 2000. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J Virol 74:4433–4440. doi: 10.1128/JVI.74.9.4433-4440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humes D, Overbaugh J. 2011. Adaptation of subtype A human immunodeficiency virus type 1 envelope to pig-tailed macaque cells. J Virol 85:4409–4420. doi: 10.1128/JVI.02244-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humes D, Emery S, Laws E, Overbaugh J. 2012. A species-specific amino acid difference in the macaque CD4 receptor restricts replication by global circulating HIV-1 variants representing viruses from recent infection. J Virol 86:12472–12483. doi: 10.1128/JVI.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatziioannou T, Del Prete GQ, Keele BF, Estes JD, McNatt MW, Bitzegeio J, Raymond A, Rodriguez A, Schmidt F, Mac Trubey C, Smedley J, Piatak M, KewalRamani VN, Lifson JD, Bieniasz PD. 2014. HIV-1-induced AIDS in monkeys. Science 344:1401–1405. doi: 10.1126/science.1250761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat S, Wrin T, Simek MD, Investigators PGP, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol G Principal Investigators, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyumkis D, Julien J-P, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Julien J-P, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A 110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch KA, Rainwater S, Jaoko W, Overbaugh J. 2010. Temporal analysis of HIV envelope sequence evolution and antibody escape in a subtype A-infected individual with a broad neutralizing antibody response. Virology 398:115–124. doi: 10.1016/j.virol.2009.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long EM, Rainwater SMJ, Lavreys L, Mandaliya K, Overbaugh J. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res Hum Retroviruses 18:567–576. doi: 10.1089/088922202753747914. [DOI] [PubMed] [Google Scholar]
- 28.Blish CA, Jalalian-Lechak Z, Rainwater S, Nguyen M-A, Dogan OC, Overbaugh J. 2009. Cross-subtype neutralization sensitivity despite monoclonal antibody resistance among early subtype A, C, and D envelope variants of human immunodeficiency virus type 1. J Virol 83:7783–7788. doi: 10.1128/JVI.00673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goo L, Jalalian-Lechak Z, Richardson BA, Overbaugh J. 2012. A combination of broadly neutralizing HIV-1 monoclonal antibodies targeting distinct epitopes effectively neutralizes variants found in early infection. J Virol 86:10857–10861. doi: 10.1128/JVI.01414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BTM, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol 80:11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shingai M, Donau OK, Schmidt SD, Gautam R, Plishka RJ, Buckler-White A, Sadjadpour R, Lee WR, LaBranche CC, Montefiori DC, Mascola JR, Nishimura Y, Martin MA. 2012. Most rhesus macaques infected with the CCR5-tropic SHIV(AD8) generate cross-reactive antibodies that neutralize multiple HIV-1 strains. Proc Natl Acad Sci U S A 109:19769–19774. doi: 10.1073/pnas.1217443109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SMJ, Overbaugh J. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J Virol 80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muñoz NM, Trobridge GD, Kiem H-P. 2009. Ex vivo expansion and lentiviral transduction of Macaca nemestrina CD4+ T cells. J Med Primatol 38:438–443. doi: 10.1111/j.1600-0684.2009.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol 150:635–643. [PubMed] [Google Scholar]
- 36.Gorny MK, Moore JP, Conley AJ, Karwowska S, Sodroski J, Williams C, Burda S, Boots LJ, Zolla-Pazner S. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J Virol 68:8312–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blish CA, Nedellec R, Mandaliya K, Mosier DE, Overbaugh J. 2007. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS 21:693–702. doi: 10.1097/QAD.0b013e32805e8727. [DOI] [PubMed] [Google Scholar]
- 39.Rusert P, Krarup A, Magnus C, Brandenberg OF, Weber J, Ehlert A-K, Regoes RR, Günthard HF, Trkola A. 2011. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. J Exp Med 208:1419–1433. doi: 10.1084/jem.20110196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Provine NM, Puryear WB, Wu X, Overbaugh J, Haigwood NL. 2009. The infectious molecular clone and pseudotyped virus models of human immunodeficiency virus type 1 exhibit significant differences in virion composition with only moderate differences in infectivity and inhibition sensitivity. J Virol 83:9002–9007. doi: 10.1128/JVI.00423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Endres MJ, Clapham PR, Marsh M, Ahuja M, Turner JD, McKnight A, Thomas JF, Stoebenau-Haggarty B, Choe S, Vance PJ, Wells TN, Power CA, Sutterwala SS, Doms RW, Landau NR, Hoxie JA. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745–756. doi: 10.1016/S0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 42.LaBranche CC, Hoffman TL, Romano J, Haggarty BS, Edwards TG, Matthews TJ, Doms RW, Hoxie JA. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J Virol 73:10310–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rucker J, Doranz BJ, Edinger AL, Long D, Berson JF, Doms RW. 1997. Cell-cell fusion assay to study role of chemokine receptors in human immunodeficiency virus type 1 entry. Methods Enzymol 288:118–133. [DOI] [PubMed] [Google Scholar]
- 44.Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang G-Y, Diwanji D, Georgiev I, Do Kwon Y, Lee D, Louder MK, Moquin S, Schmidt SD, Yang Z-Y, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang L-X, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog 6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Kurteva S, Lee S, Sodroski J. 2005. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J Virol 79:3500–3508. doi: 10.1128/JVI.79.6.3500-3508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci U S A 96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edwards TG, Hoffman TL, Baribaud F, Wyss S, LaBranche CC, Romano J, Adkinson J, Sharron M, Hoxie JA, Doms RW. 2001. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J Virol 75:5230–5239. doi: 10.1128/JVI.75.11.5230-5239.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolchinsky P, Kiprilov E, Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol 75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol 78:5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. 2006. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol 80:7127–7135. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A 109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guttman M, Kahn M, Garcia NK, Hu S-L, Lee KK. 2012. Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J Virol 86:8750–8764. doi: 10.1128/JVI.07224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S. 2011. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J Virol 85:12114–12123. doi: 10.1128/JVI.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Connell O, Repik A, Reeves JD, Gonzalez-Perez MP, Quitadamo B, Anton ED, Duenas-Decamp M, Peters P, Lin R, Zolla-Pazner S, Corti D, Wallace A, Wang S, Kong X-P, Lu S, Clapham PR. 2013. Efficiency of bridging-sheet recruitment explains HIV-1 R5 envelope glycoprotein sensitivity to soluble CD4 and macrophage tropism. J Virol 87:187–198. doi: 10.1128/JVI.01834-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Si Z, Cayabyab M, Sodroski J. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J Virol 75:4208–4218. doi: 10.1128/JVI.75.9.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fomsgaard A, Johnson PR, Nielsen C, Novembre FJ, Hansen J, Goldstein S, Hirsch VM. 1995. Receptor function of CD4 structures from African green monkey and pig-tail macaque for simian immunodeficiency virus, SIVsm, SIVagm, and human immunodeficiency virus type-1. Viral Immunol 8:121–133. doi: 10.1089/vim.1995.8.121. [DOI] [PubMed] [Google Scholar]
- 59.Chen Z, Zhou P, Ho DD, Landau NR, Marx PA. 1997. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol 71:2705–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye Y, Si ZH, Moore JP, Sodroski J. 2000. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J Virol 74:11955–11962. doi: 10.1128/JVI.74.24.11955-11962.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karlsson GB, Halloran M, Schenten D, Lee J, Racz P, Tenner-Racz K, Manola J, Gelman R, Etemad-Moghadam B, Desjardins E, Wyatt R, Gerard NP, Marcon L, Margolin D, Fanton J, Axthelm MK, Letvin NL, Sodroski J. 1998. The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques. J Exp Med 188:1159–1171. doi: 10.1084/jem.188.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhuang K, Finzi A, Tasca S, Shakirzyanova M, Knight H, Westmoreland S, Sodroski J, Cheng-Mayer C. 2011. Adoption of an “open” envelope conformation facilitating CD4 binding and structural remodeling precedes coreceptor switch in R5 SHIV-infected macaques. PLoS One 6:e21350. doi: 10.1371/journal.pone.0021350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan RC, Harouse JM, Gettie A, Cheng-Mayer C. 1999. In vivo adaptation of SHIV(SF162): chimeric virus expressing a NSI, CCR5-specific envelope protein. J Med Primatol 28:164–168. doi: 10.1111/j.1600-0684.1999.tb00265.x. [DOI] [PubMed] [Google Scholar]
- 64.West AP, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. 2014. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.