Abstract
Background: The SUMO pathway has been shown to play an important role in tumorigenesis. This report analyzed the involvement of the sole SUMO-Activating Enzyme Subunit 2 (SAE2) in human gastric cancer (GC) progression and prognosis. Methods: Expression of SAE2 was examined by Quantigene Plex, western blotting and immunohistochemistry. The expression of SAE2 and c-MYC were detected in parallel in 276 cases. The molecular mechanisms of SAE2 expression and its effects on cell growth, colony formation, migration and invasion were also explored by CCK8 assay, colony formation experiment, transwell chamber assay with or without matrigel, immunoprecipitation and in vivo tumorigenesis and tumor metastasis. Results: SAE2 was markedly overexpressed in GC cell lines and primary tumor samples of GC, and significantly correlated with deeper tumor depth, distant metastasis, higher pathological stage and stratified survival in human GC. SAE2 positivity was independently associated with a worse outcome in multivariate analysis. Knockdown of SAE2 expression inhibited the proliferation, migration, and invasion of SAE2-overexpressing GC cells. Consistent with the in vitro results, down-regulation of SAE2 in human GC BGC823 cells significantly reduced the tumorigenic and metastatic potential of the cells in vivo. SAE2 protein was significantly associated with the higher expression of c-MYC in primary GC tissues. Moreover, FoxM1 was SUMOylated in GC and that inhibition of SAE2 resulted in a decrease in SUMO1-FoxM1 levels compared with those in the controls. Conclusions: These findings suggest that SAE2 has a pivotal role in the aggressiveness of GC, and highlight its usefulness as a prognostic factor in GC.
Keywords: SAE2, gastric cancer, prognosis, FoxM1, c-MYC
Introduction
Gastric cancer (GC) is the fourth most common malignant disease worldwide and ranks second in terms of global cancer-related mortality [1]. Treatment for GC has improved in recent years, but the majority of patients who are diagnosed with advanced GC are in developing nations, including China. Patients with advanced GC have a poor prognosis and eventually die after surgery as a result of cancer recurrence and metastasis [2]. Therefore, more research is needed to discover the molecular mechanisms that regulate the motility and invasive behavior of GC cells and to develop more effective biomarkers for GC prognosis.
SAE2 is a unique SUMO-activating enzyme subunit that mediates the first step of the SUMO pathway and is conserved from yeast to humans. Patient survival significantly correlates with SAE1/2 levels in MYC-high breast cancer [3]. In hepatocellular carcinoma patients, overexpression of SAE2 has been correlated with poor survival [4]. UBC9, the sole E2 conjugating enzyme for SUMOylation, promotes invasion and metastasis in lung cancer and prostate cancer [5,6]. In Drosophila and other organisms, knockdown of SUMO or SAE1/2 and UBC9 robustly disrupts proliferating cells [7,8], and in adult mice with inducible UBC9 knockout, it has been shown that SUMOylation is essential for the survival of stem cells in the intestinal compartment [9]. This finding is consistent with the notion that the SUMOylation pathway is important for cancer development and progression [10].
SUMO proteins are significantly involved in diverse cellular processes, such as p53 [11], NF-κΒ [12], Hif-1α [13] and Grb2 [14]. They are required to sustain cancer-cell behaviors such as the hypoxia response, cell proliferation, cancer stemness and the epithelial-mesenchymal transition (EMT) [15]. FoxM1 is an oncogenic transcription factor of the Forkhead family and is involved in a wide range of biological processes including embryogenesis, proliferation, migration, invasiveness, angiogenesis and inflammation [16]. FoxM1 has been found to be overexpressed in more than 20 types of human cancer [17], and is an independent prognostic factor in GC [18]. SUMOylation of FoxM1 peaks during G2 and M phase, when FoxM1 transcriptional activity is required [19]. Little is known about the roles of the SUMOylation pathway in gastric carcinogenesis, and it remains unclear whether SAE2 expression is associated with any clinicopathological features of GC.
In this study, we analyzed the expression of SAE2 in GC using Quantigene Plex and immunohistochemistry and determined the relationship between their expression and clinicopathological parameters, including the prognosis of patients. Moreover, in in vitro and in vivo studies, we also examined the relationship between the SAE2 expression and the aggressiveness of GC cells.
Materials and methods
Cell culture and sample collection
AGS, SNU1 and 293FT were obtained from ATCC (Manassas, VA, USA), MKN28 and NUGC3 were obtained from the Health Science Research Resources Bank (Tokyo, Japan) and BGC823, MGC803 and SGC7901 were obtained from the Cell Research Institute (Shanghai, China). The cells were routinely grown in RPMI-1640 medium (GIBCO BRL, Carlsbad, CA), which was supplemented with 10% (v/v) fetal calf serum (FCS, GIBCO) and antibiotics at 37°C in a humidified 5% CO2 atmosphere.
Surgical samples were obtained from 301 patients with GC who underwent surgical resection at the Beijing Cancer Hospital. The patients were diagnosed, and the stage of GC was classified independently by two experienced pathologists according to the American Joint Committee on Cancer stage (AJCC 7th edition). Complete original clinical data were reviewed in the contexts of clinicopathological and follow-up information. Patients receiving chemotherapy or radiotherapy prior to surgery or patients with histories of having other tumors were excluded. The overall survival (OS) was calculated from the date of the surgery to the time of death or the last follow-up. All patients were followed up until 2012. This study was conducted using Clinic Institutional Review Board–approved protocols. Informed consent was obtained from each patient. In the following studies, a portion of the specimen that was removed during surgery was immediately snap-frozen in liquid nitrogen and subsequently stored at -80°C; a portion of this specimen was fixed with 10% buffered formalin for 24 h and embedded in paraffin.
IHC assay for SAE2, c-MYC and FoxM1
Standard laboratory protocols were followed for IHC and quality control measures. Antigen retrieval was conducted on deparaffinized whole specimens by pressure cooking the slides in 10 mmol/L EDTA (pH 8.0) or citrate buffer (pH 6.0) for 3 minutes. Endogenous peroxidase activity was blocked by incubation in 0.3% hydrogen peroxide. Non-specific protein binding was reduced by the addition of normal sleep serum (DAKO, Hamburg, Germany), diluted 1:10 (30 min, room temperature). Consecutive sections were stained with antibodies that were directed against c-MYC (TA150121, Origene, Maryland, USA; diluted 1:250), SAE2 (4A3, Origene, MA, USA; diluted 1:300) and FoxM1 (sc-502, Santa Cruz Biotechnology, Santa Cruz, CA; diluted 1:100). Primary antibodies were then incubated at 4°C overnight. The sections were incubated in a secondary antibody (Dako Envision Plus Dual Link Horseradish Peroxidase Kit; Dako # K4061). The high-sensitivity 3, 3-diaminobenzidine (DAB+) chromogenic substrate system was used for colorimetric visualization followed by counter staining with hematoxylin.
The degree of immunostaining of each tissue section was assessed independently by two experienced pathologists who were blind to the patients’ clinical data. Expression analysis of SAE2 (nucleus), c-MYC (nucleus) and FoxM1 (cytoplasm/nucleus) proteins in malignant cells was performed by comparing staining intensity and the percentage of immunoreactive cells. A semiquantitative approach was used to generate a score for each tissue sample as follows: no nuclear/cytoplasmic staining or nuclear/cytoplasmic staining in < 10% of tumor cells (score 0), faint/barely perceptible staining in > 10% of tumor cells (score 1+), weak-to-moderate staining of the nucleus/cytoplasm in > 10% of tumor cells (score 2+), and strong staining of the nucleus/cytoplasm in > 10% of tumor cells (score 3+). Scores of 0 and 1+ were considered to be negative for SAE2, c-MYC or FoxM1 overexpression, and scores of 2+ and 3+ were considered to be moderate and strong staining, respectively. The case-by-case final consensus result was discussed and determined in a common session.
The QuantiGene 2.0 assay
Tissue homogenates were prepared according to the procedure described in the QuantiGene Sample Processing Kit for FFPE Tissues (Panomics, Inc. Fremont, CA). Briefly, 200 µl of homogenizing solution supplemented with 2 µl of proteinase K (50 µg/µl) was incubated with six deparaffinized 5-µm sections overnight at 65°C. The tissue homogenate was then separated from debris by brief centrifugation and transferred to a new tube.
Standard probe design software was used to design specific oligonucleotide probe sets for target genes for use in the QuantiGene plex 2.0 Reagent System (Panomics, Inc.), which provides 400-fold signal amplification. QuantiGene plex 2.0 Reagent System assays were performed according to manufacturer’s recommended protocols (Panomics, Inc.). Briefly, probe-set oligonucleotides were mixed with the sample, and the mixture was added to an assay well in a 96-well plate. Target RNA was captured during an overnight incubation at 54°C (QuantiGene plex 2.0). Unbound material was removed by three washes with 200 µl of wash buffer followed by sequential hybridization of RNA amplifier molecules, then, pre-amplifier hybridization, amplifier hybridization, and label-probe hybridization were performed. Finally, SAPE working reagent was added to the well to prepare the plate for analysis. Gene expression was quantified in relation to the expression of PGK1 and TBP and the relative quantification method.
Lentiviral production of GC cell lines to stably silence SAE2
Lentivirus was produced by the co-transfection of 293FT cells with a pLenti vector (pGLV3-shControl or pGLV3-shSAE2) and lentiviral packaging mix (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Lentivirus-containing supernatant was harvested 48 h post-transfection, purified by centrifugation and stored at -80°C. For viral transductions, 1 ml of the pGLV3-shControl or pGLV3-shSAE2 lentiviruses was incubated with BGC823, SGC7901 and MKN28 cells overnight at 37°C in a humidified cell culture incubator. Twenty-four hours post-infection, stable GC cells with depleted endogenous SAE2 expression were selected by culturing in puromycin (1 μg/ml).
Cell viability and proliferation assays
Colony formation assays were used to assess the survival capacity of SGC7901, BGC823 and MKN28 cells with and without SAE2. One hundred fifty cells/well were seeded into 6-cm petri dishes. After 12 days of culture, the colonies that formed were fixed with methanol and were then stained with 0.5% crystal violet and manually counted.
Samples of 2 × 103 cells/well for SGC7901 and 3 × 103 cells/well for MKN28 and BGC823 were plated into 96-well plates in triplicate and were allowed to adhere overnight. After 6 hours of starvation (RPMI only), viable cells were quantified using cell–proliferation, ELISA, and a CCK8 assay, and this was recorded as time 0. After 24, 48, 72 and 96 hours, cell viability and proliferation were re-assessed.
Cell migration and invasion assay
Cell migration was measured using the wound-closure assay. Briefly, a confluent cell surface was scratched with a pipette tip, and the migration index was calculated as follows: migration index = [(initial wound width – width of wound at time point tested)/initial wound width] × 100%. The transwell chamber assay with or without a matrigel coating was carried out as described previously [20].
In vivo mouse models of gastric cancer
Animal studies were carried out in strict adherence with institutional guidelines. In vivo tumorigenesis was investigated by tumor xenograft experiments. BGC823 SAE2 shRNA cells or control BGC823 mock shRNA cells (approximately 1 × 106/200 μL per mouse) were injected subcutaneous into the right hind legs of 6-week-old nude mice (10 mice,0 5 per condition). Tumor growth was monitored three times a week by measuring the width and length of the tumors with calipers and 20 days after inoculation, the two groups of mice were killed. The tumor volume was calculated by the formula V = 0.5 × L × W2.
BGC823 cells with or without silencing SAE2 (1 × 106/150 μL per mouse) were injected intravenously via a 30-gauge needle inserted into the tail vein of female BALB/c-nude mice (5-6 weeks old). Six weeks later, the mice were sacrificed, and the lungs were removed and fixed with Bouin’s fixative. The presence of lung metastases was evaluated at autopsy and was confirmed by histopathological examination of the lungs.
Immunoprecipitation and western blotting
Western blotting was carried out as described previously [20] with the following appropriate primary antibodies: anti-GAPDH (Cell signaling Technology, Beverly, MA), anti-SAE2 (Origene, MA, USA), anti-FoxM1, anti-c-MYC (Santa Cruz Biotechnology, Santa Cruz, CA), anti-SUMO1, anti-SUMO2/3 (Abcam, Cambridge, USA). Immunoprecipitation (IP) was performed principally according to the previously described protocol [21]. One milligram of whole-cell extracts was immunoprecipitated with anti-FoxM1 (1:500, sc-500) and anti-FoxM1 (1:500, sc-502) antibody or control IgG (Abcam, Cambridge, USA) at 4°C for 6hrs with rotation and then bound to protein G agarose beads (Roche) and washed eight times with the lysis buffer. The proteins were separated by SDS-PAGE, and the immunoprecipitates were blotted with anti-FoxM1 antibodies. The blots were stripped using a stripping buffer (100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl pH 6.7) and re-probed with anti-Sumo1 antibody.
Statistical analysis
The Chi-squared test for nominal and ordinal variables has been applied to assess the correlations between SAE2 expression and clinicopathological features. Survival curves were estimated by the Kaplan-Meier method and compared with the log rank test. Multivariate analysis was performed using the Cox regression model to assess whether a factor was an independent predictor of OS. Group comparisons were analyzed using Student’s t test, and for correlation analysis of c-MYC and SAE2, we used the Spearman-rank correlation test. The data are shown as the mean ± SD. A two-tailed P-value of < .05 was considered statistically significant. All statistical analyses were performed with SPSS v20.0 software (SPSS Inc. Chicago, IL, USA) or Graphpad Prism 6.0.
Results
Association of SAE2 expression and clinicopathological factors in GC
Compared with paired non-tumor tissues, GC tissues exhibited higher expression levels of SAE2 mRNA (P = .011), protein and SUMO1-conjugated proteins (Figure 1A). Western blotting analyses showed that SAE2 protein expression was markedly up-regulated in all tested GC cell lines (Figure 1B). The Quantigene Plex assay revealed significant expression of SAE2 in GC, which was correlated with SAE2 protein expression (P = .017) (Figure 1I).
Figure 1.
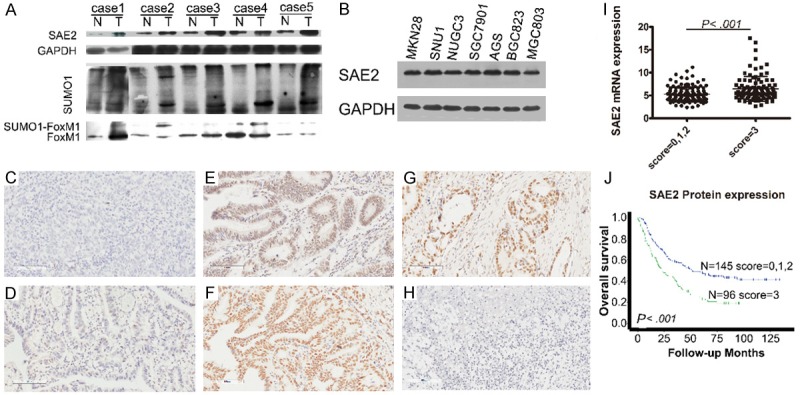
SAE2 expression in GC. (A) Adjacent noncancerous tissues (N) and primary GC tissues (T) were examined by western blotting. (B) Western blotting analysis of SAE2 expression in GC cell lines. (C and D) Negative staining of SAE2 in GC. Cases of no nuclear staining (IHC 0; C), weak staining of the cytoplasm (IHC 1+; D). (E) Moderate staining of the nuclei (IHC 2+). (F) Strong staining of the nuclei (IHC 3+). (G) SAE2 expression at the invasive front in lesions deeper than muscularis propria. (H) Negative control. (I) Quantigene Plex analysis of SAE2 mRNA in 230 GC specimens. (J) Dichotomization of SAE2, Kaplan-Meier survival analysis of OS for radical cure patients.
Additionally, SAE2 protein expression was evaluated by IHC in 301 cases: 21 (7%), 16 (5.3%), 140 (46.5%) and 124 (41.2%) cases were scored as 0, 1+, 2+ and 3+, respectively. Representative examples of SAE2 staining in GC patients are provided in Figure 1C-F. SAE2 protein was mainly localized in the nucleus of GC cells with weak or no cytoplasmic expression, and adjacent non-neoplastic tissues presented no or low levels of SAE2 staining. Figure 1G shows the expression of SAE2 at the invasive front of GC. In patients described as “radical cure”, strong SAE2 (score = 3) expression exhibited significantly poorer overall survival (OS) compared with patients with moderate (score = 2) or negative (score = 0/1) expression (P < .001, Figure 1J). The association between the expression of SAE2 and various clinicopathological parameters is listed in Table 1. High level of SAE2 expression in GC was significantly associated with deeper depth of invasion, distant metastasis and higher pathological stage (all P < .05, Table 1).
Table 1.
Clinicopathological characteristics of GC patients according to SAE2 status
| Variables | Cases | SAE2 expression | P-value | ||
|---|---|---|---|---|---|
|
| |||||
| n (%) | Negative | Positive | |||
|
|
|||||
| 0 or 1+ | 2+ | 3+ | |||
|
|
|||||
| n = 37 (12.3%) | n = 140 (46.5%) | n = 124 (41.2%) | |||
| Gender | .177 | ||||
| Male | 198 (65.8) | 22 (59.5) | 87 (62.1) | 89 (71.8) | |
| Female | 103 (34.2) | 15 (40.5) | 53 (37.9) | 35 (28.2) | |
| Age | .314 | ||||
| (mean ± s.d. years) | 301 (100.0) | 55.0 ± 12.5 | 58.35 ± 12.4 | 57.6 ± 11.9 | |
| T classification | .044 | ||||
| T1 + T2 | 45 (15.0) | 10 (27.0) | 22 (15.7) | 13 (10.5) | |
| T3 + T4 | 256 (85.0) | 27 (73.0) | 118 (84.3) | 111 (89.5) | |
| N classification | .115 | ||||
| No | 67 (22.3) | 13 (35.1) | 31 (22.3) | 23 (18.9) | |
| Yes | 231 (76.7) | 24 (64.9) | 108 (77.7) | 99 (81.1) | |
| Not recorded* | 3 (1) | ||||
| Metastasis | .034 | ||||
| No | 259 (86.0) | 37 (100.0) | 119 (85.0) | 103 (83.7) | |
| Yes | 41 (13.6) | 0 | 21 (15.0) | 20 (16.3) | |
| Not recorded* | 1 (0.4) | ||||
| Vascular invasion | .604 | ||||
| Negative | 120 (39.9) | 17 (45.9) | 57 (40.7) | 46 (37.1) | |
| Positive | 181 (60.1) | 20 (54.1) | 83 (59.3) | 78 (62.9) | |
| Lauren subtype | .220 | ||||
| Intestinal | 48 (15.9) | 9 (24.3) | 18 (12.9) | 21 (16.9) | |
| Diffuse or mixed | 253 (84.1) | 28 (75.7) | 122 (87.1) | 103 (83.1) | |
| TNM Stages | .044 | ||||
| I | 27 (9.0) | 7 (18.9) | 12 (8.6) | 8 (6.5) | |
| II | 61 (20.3) | 11 (29.7) | 26 (18.6) | 24 (19.4) | |
| III | 172 (57.1) | 19 (51.4) | 81 (57.9) | 72 (58.1) | |
| IV | 41 (13.6) | 0 | 21 (15.0) | 20 (16.1) | |
| Resection status | .239 | ||||
| R0 | 248 (82.4) | 34 (91.9) | 115 (82.1) | 99 (79.8) | |
| R1 or R2 | 53 (17.6) | 3 (8.1) | 25 (17.9) | 25 (20.2) | |
Data incomplete.
Increased SAE2 expression predicts worse survival in GC
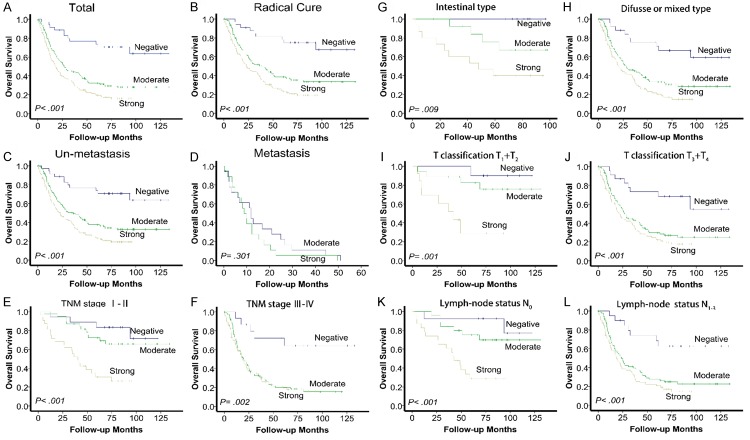
To further analyze the potential of SAE2 to predict prognosis in GC, we found that patients with high levels (score 2/3) of SAE2 expression exhibited significantly poorer 5-year OS compared with patients with low (score 0/1) levels of SAE2 (19.4%, 32.2% and 73.2%, P < .001, Figure 2A). Univariate analysis revealed that SAE2, TNM stage, vascular invasion, Lauren classification and type of surgery were significantly associated with OS (P < .001, Table 2). Further analysis in a multivariate Cox proportional hazards model demonstrated that SAE2, together with TNM stage, vascular invasion, Lauren classification and type of surgery, were strongly associated with OS. After adjustment for the effect of covariates, we found that whether it was moderate or strong, SAE2 expression was an independent prognostic indicator of poor survival (P < .01, Table 2).
Figure 2.
SAE2 expression and survival in patients with GC. Kaplan-Meier survival analysis of overall survival for all patients (A) and radical cure patients (B). Statistical significance of the difference between the curves of SAE2 negative, moderate and strong patients was compared in patient subgroups: M0 (C) and M1 (D) patient subgroups, TNM stage I to II (E) and TNM stage III to IV (F) patient subgroups, Lauren classification intestinal type (G) and diffuse or mixed type (H) patient subgroups, T classification T1 + T2 (I) and T3 + T4 (J) patient subgroups, and lymph node status N0 (K) and lymph node status N1-3 (L) patient subgroups.
Table 2.
Univariate and multivariate Cox Proportional HRs for overall survival
| Parameter | Univariate | P | Multivariate | P |
|---|---|---|---|---|
|
|
|
|||
| 5-year Survival (% ± S.E) | HR (95% CI) | |||
| SAE2 expression | < .001 | |||
| Negative | 73.7 ± 0.075 | 1.00 (reference) | ||
| Moderate | 32.2 ± 0.042 | 2.772 (1.443-5.134) | .002 | |
| Strong | 19.4 ± 0.037 | 2.229 (1.529-3.250) | < .001 | |
| Stage | < .001 | |||
| I | 77.1 ± 0.082 | 1.00 (reference) | ||
| II | 52.1 ± 0.066 | 1.801 (0.786-4.126) | .164 | |
| III | 24.0 ± 0.034 | 2.691 (1.692-4.279) | < .001 | |
| IV | 0 | 2.644 (1.385-5.047) | .003 | |
| Resection status | < .001 | |||
| R0 | 38.1 ± 0.032 | 1.0 (reference) | ||
| R1 or R2 | 2.1 ± 0.021 | 2.126 (1.240-3.646) | .006 | |
| Lauren’s histologic type | < .001 | |||
| Intestinal | 56.6 ± 0.075 | 1.0 (reference) | ||
| Diffuse or mixed | 27.5 ± 0.029 | 1.855 (1.145-3.004) | .012 | |
| Vascular invasion | < .001 | |||
| Negative | 44.0 ± 0.048 | 1.0 (reference) | ||
| Positive | 23.2 ± 0.033 | 1.533 (1.119-2.100) | .008 | |
| Age | .178 | |||
| <60 | 35.3 ± 0.041 | |||
| ≥60 | 28.9 ± 0.039 | |||
| Gender | .746 | |||
| Male | 30.9 ± 0.035 | |||
| Female | 33.3 ± 0.048 |
Kaplan-Meier analysis revealed that the patients with high SAE2 expression had significantly shorter OS compared with patients with low SAE2 expression, whether they were described as “radical cure” or not (P < .001; Figure 2A, 2B). In the subgroup of M0, patients with high SAE2 expression showed a worse outcome compared with those with low SAE2 expression (n = 252, P < .001; Figure 2C), while there was no significant difference in the M1 subgroup, in which all of the patients had high SAE2 expression (n = 37, P = .301; Figure 2D). Similarly, in the “radical cure” group, the OS was significantly shorter in patients with high SAE2 expression in the TNM stage I + II subgroup (n = 88, P < .001; Figure 2E), the TNM stage III + IV subgroup (n = 153, P = .002; Figure 2F), the Lauren classification intestinal-type subgroup (n = 37, P = .009; Figure 2G) and the Lauren classification diffuse or mixed type subgroup (n = 204, P < .001; Figure 2H). Moreover, similar results were shown in the T classification T1 + T2 subgroup (n = 41, P = .001; Figure 2I) and in the T classification T3 + T4 subgroup (n = 200, P < .001; Figure 2J) or in the lymph-node status N0 (n = 62, P < .001; Figure 2K) and lymph-node status N1+3 subgroups (n = 176, P < .001; Figure 2L).
SAE2 regulates cell proliferation in vitro and tumorigenesis in vivo
As shown in Figure 3A, more of the tumor areas with high levels of nuclear SAE2 staining also showed strong c-MYC expression, whereas areas with low SAE2 staining exhibited varied c-MYC degrees of signal (P = .013). Kaplan-Meier survival curves showed that patients with c-MYC-/SAE2- had a significantly better 5-year OS (70%) than those with c-MYC-⁄SAE2+ (26%, log rank X2 = 12.164, P < .001) and those with c-MYC+⁄SAE2+ (14.5%, log rank X2 = 16.232, P < .001) (Figure 3B).
Figure 3.
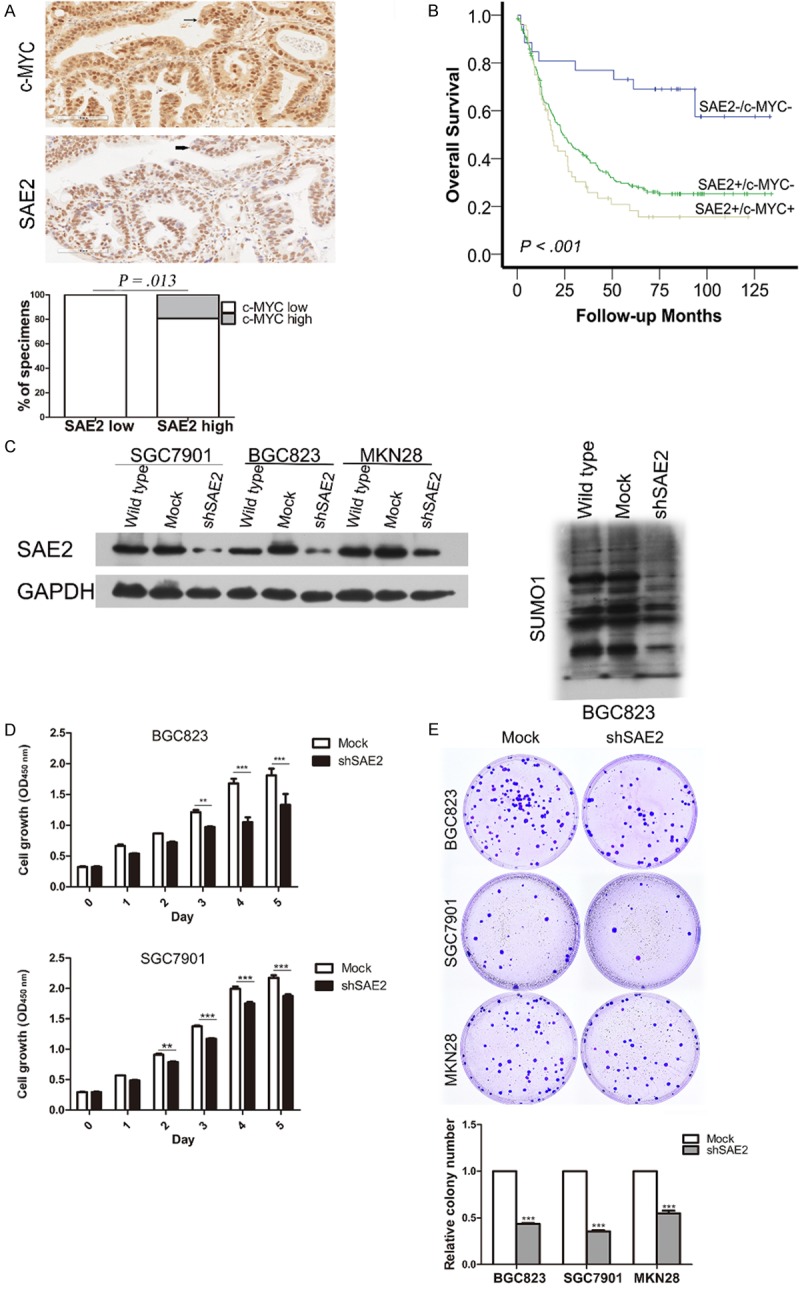
Effects of SAE2 depletion on cell growth in vitro. C-MYC expression levels significantly correlated with SAE2 expression in human GC tissues. One representative case is shown, c-MYC and SAE2 nuclear expression are indicated by arrow, and the percentage of specimens with low or high SAE2 expression, relative to the levels of c-MYC staining (A), Kaplan-Meier survival analysis of overall survival for 276 patients with GC according to the combined expression of SAE2 and c-MYC (B) including SAE2-/MYC-, SAE2+/MYC-, and SAE2+/MYC+. Western blotting analysis of SAE2 expression in SAE2-silenced BGC823, SGC7901 and MKN28 cells and SUMO1 in SAE2-silenced BGC823 cells (C), CCK-8 assay (D) and colony formation assay (E) indicated that the growth rate decreased in the SAE2-silenced cells. The number of colonies was quantified in the colony formation assay. The data shown are the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
We further studied the biological role of SAE2 on GC cell viability and proliferation. We infected BGC823, SGC7901 and MKN28 cells with lentiviruses that encoded small hairpin RNA (shRNA) against SAE2 or for a noncoding shRNA. Immunoblot analysis confirmed that SAE2 protein levels and the amount of SUMO conjugates were reduced but not abrogated after virus infection (Figure 3C). SAE2-depleted GC cells potently inhibited the proliferation and viability over time compared with the cells transfected with the empty vector (Figure 3D), this excluded the possibility of the off-target effect of SAE2 shRNA. Similarly, knockdown of SAE2 limited colony formation to approximately 40%-60% in GC cells compared with the control population (Figure 3E).
We subcutaneously injected shSAE2 and mock BGC823 cells into nude mice to examine whether SAE2 is required for tumor formation. In the group of mice injected with shSAE2 cells, one mouse displayed no tumor nodule. Moreover, xenografts were relatively smaller in the shSAE2 group compared with the control group at every time point (Figure 5A). Tumor weight was markedly reduced in the shSAE2 group compared with the mock group at the endpoint (P = .065, Figure 5B).
Figure 5.
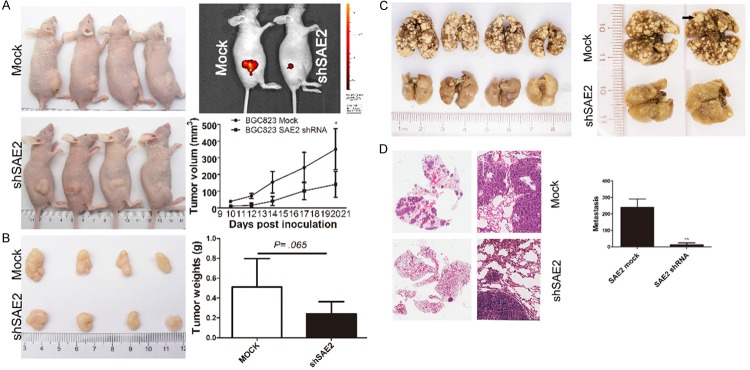
SAE2 depletion inhibits subcutaneous GC xenograft growth and metastasis in vivo. A. Tumor size at different days was measured using a vernier caliper and was expressed as volume (mm3) according to the following formula: tumor volume = (length × width2)/2. B. Twenty days later, the mice were sacrificed and the tumors were collected and weighed. The data are shown as the means ± SD (*p < 0.05, **p < 0.01, ***p < 0.001). C. Representative tumor nodules in the lungs are shown. Arrows indicate the tumor nodules. D. The graph shows representative HE-stained sections of lung metastasis.
Knockdown of SAE2 suppresses GC cell migration, in vitro invasion, and in vivo metastasis
Following the silencing of SAE2 in BGC823, SGC7901 and MKN28 cells, a reduction in wound healing (indicative of decreased migration potential) was noticed in the knockdown-SAE2 groups compared with control cells from the respective groups (Figure 4A). Similarly, down-regulation of SAE2 resulted in a decrease in migration and invasion to the undersurface compared with the control that was transfected with mock cells (Figure 4B).
Figure 4.
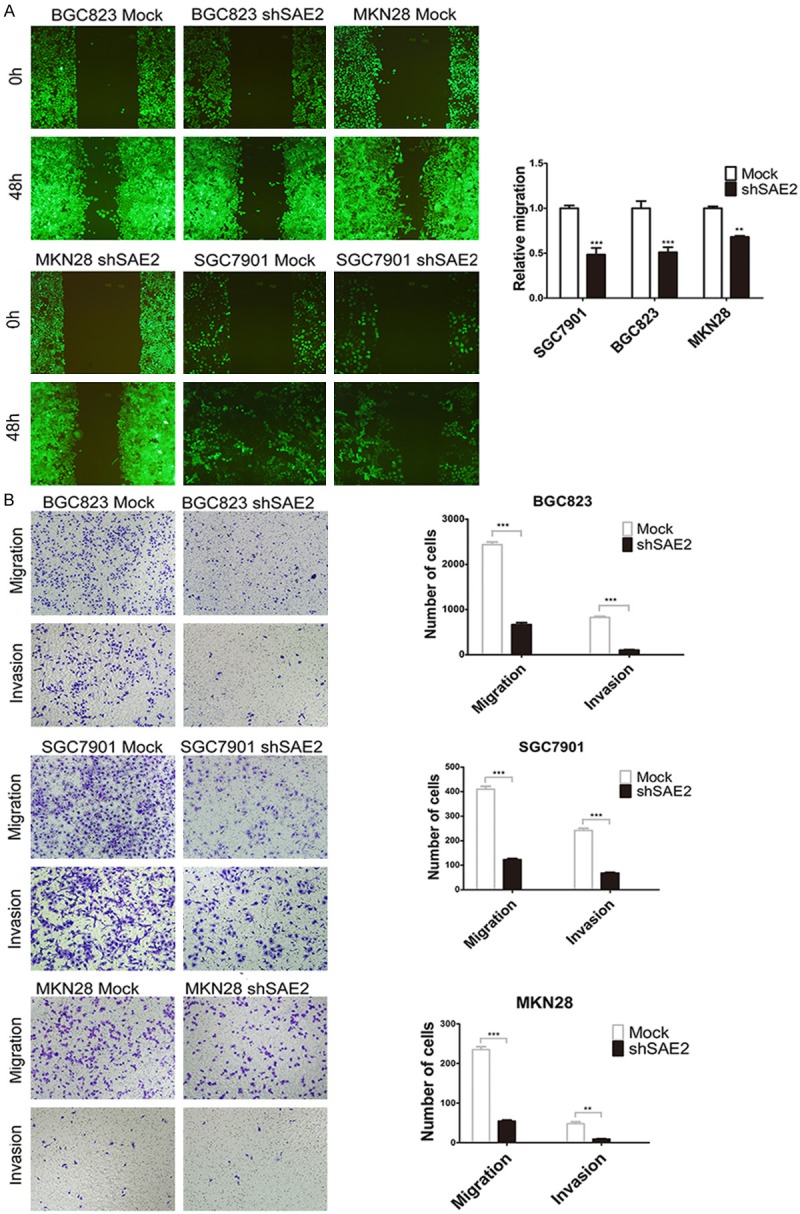
SAE2 depletion attenuated the migration and invasion capabilities of GC cells in vitro. (A and B) The effect of SAE2 silencing on in vitro migration and invasion was assessed via the wound closure assay (A) and matrigel with or without the transwell assay (B), respectively.
To validate the effects of SAE2 on the metastasis of GC cells in vivo, BGC823 cells stably transfected with SAE2-shRNA were intravenously injected into nude mice through the tail vein. Metastatic nodules on the surface of the lungs were counted after 6 weeks. The silencing of SAE2 resulted in a reduction in the number of metastatic nodules compared with those in the control group (Figure 5C). This difference was further confirmed following an examination of the entire lung and through the HE staining of the lung sections (Figure 5D). Significantly lower numbers of metastatic foci were observed in the lungs of mice that had been injected with silenced SAE2 BGC823 cells (P < .01). Our in vivo data complemented the results of functional in vitro studies involving SAE2.
SAE2 was positively correlated with SUMOylated FoxM1 and FoxM1 expression levels in GC
A bioinformatics screening for high-probability SUMOylation sites using a SUMOplotTM (http://www.abgent.com/tools/sumoplot_login) and the SUMOFI (SUMO motif finder, http://csbi.ltdk.helsinki.fi/sumofi/) was performed. As shown in Table 3, seven high-probability SUMOylation sites in human FoxM1 were predicted, and FoxM1 had four NDSM sequences but no consensus PDSM sequences [22,23] (Table 3). IP analysis indicated that FoxM1 was modified by SUMO1 (Figure 6A), and western blot analysis showed the expression levels of FoxM1, SUMOylated FoxM1 were significantly decreased in SAE2-silenced cells (Figure 6B). We also found that SAE2 expression positively correlated with FoxM1 and SUMOylated FoxM1 in the five freshly collected clinical GC samples (Figure 1A). FoxM1 expression in 77 GC patients was analyzed by IHC. FoxM1 was positive in 79.2% of patients, and SAE2 and FoxM1 were co-expressed in the majority (59/77) of the cases (Figure 6C). Only one high-probability SUMOylation sites was predicted in human c-MYC according to the SUMOplotTM prediction database, but we couldn’t detect c-MYC SUMOylation by IP in GC (data not shown).
Table 3.
The high probability SUMO acceptor sites in human FoxM1
| Position | Sequence | Score by SUMOplot | NDSM by SUMOFI | PDSM by SUMOFI |
|---|---|---|---|---|
| 478 | IKVE | 0.94 | YES | NO |
| 460 | IKEE | 0.94 | YES | NO |
| 356 | IKTE | 0.94 | NO | NO |
| 201 | IKQE | 0.94 | YES | NO |
| 218 | VKVE | 0.93 | NO | NO |
| 495 | FKEE | 0.85 | YES | NO |
| 396 | VKVP | 0.82 | NO | NO |
NDSM: the negatively charged amino acid-dependent sumoylation motif. PDSM: phosphorylation-dependent sumoylation motif.
Figure 6.
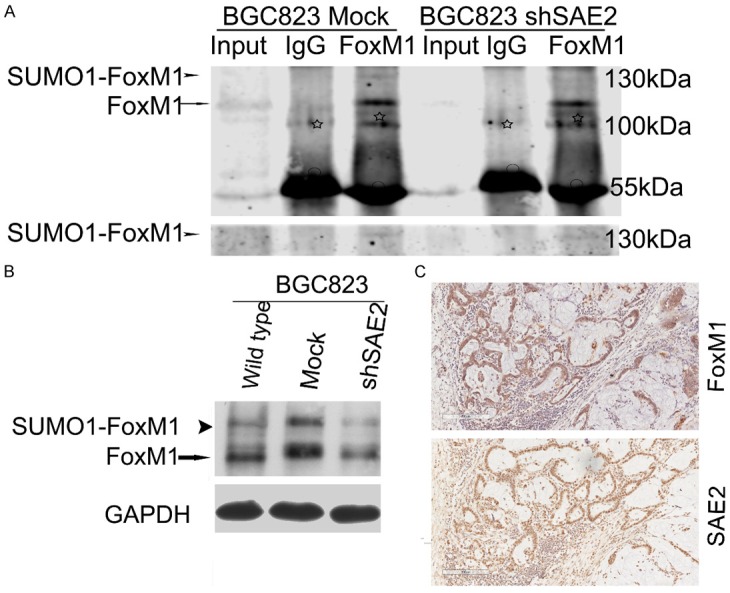
FoxM1 is SUMOylated in vivo, and SAE2 was positively correlated with SUMO1-FoxM1, FoxM1 expression levels in GC. A. Cell lysates were immunoprecipitated and subsequently immunoblotted with the indicated antibodies. SUMOylated FoxM1 and non-SUMOylated FoxM1 are indicated by an arrowhead and an arrow, respectively. White asterisks and circles indicate nonspecific blot and IgG heavy chain, respectively. B. Western blotting analysis of SUMOylated-FoxM1 (SUMO1-FoxM1), total FoxM1 proteins in BGC823. C. One representative case of FoxM1 and SAE2 staining in serial sections of surgically resected GC tissue.
Discussion
SUMOylation has been repeatedly demonstrated as being critically involved in tumorigenesis and cancer metastasis [10,24]. Recently, it has been shown that Sumo1, Sumo2/3, SAE1/2 and UBC9 exhibited similar expression patterns in sperm differentiation [25]. Furthermore, several human tissues and carcinomas have been studied for UBC9 protein expression, but to our knowledge, data concerning GC and SAE2 are lacking.
SAE2 expression was significantly up-regulated in GC tissues when compared with their adjacent tissues. This up-regulation matched our data in cell lines, all of which showed SAE2 overexpression. In GC, high level of SAE2 expression was associated with disease progression and shorter patient survival time. Furthermore, a detailed multivariate Cox analysis demonstrated that SAE2 was an independent prognostic factor for GC patient survival, following surgery. Thus, SAE2 may serve as a novel prognostic marker in patients with GC. Furthermore, when patients were stratified into subgroups according to Lauren classification, T classification, TNM stage, and lymph-node status, high level of SAE2 expression also indicated a shorter OS time. This means that at the time of initial diagnosis of GC, SAE2 expression may be used not only to design optimal, individualized treatment but also to distinguish patients who would benefit from close monitoring after surgery from those who would not.
SAE2 is the only known SUMO E1 enzyme subunit 2 and a key regulator of the SUMOylation pathway. In glioblastoma, CDK6 is modified by SUMO1, and this CDK6 SUMOylation stabilizes the protein and drives the cell cycle for cancer development and progression [26]. In basal breast cancer, SAE2 inhibition leads to the clearing of cells that express CD44?+/hi/CD24-/low markers and the blocking of the outgrowth of cancer xenografts [27]. These data suggest that SAE2 overexpression confers pro-tumorigenic properties to tumor cells. In support of this hypothesis, through IHC assays in 276 cases, we also found a significant correlation between SAE2 and nuclear c-MYC expression in highly proliferative lesions of human GC (P = .013), and GC patients with an SAE2 high/c-MYC high phenotype had significantly shorter survivals than those with the other phenotypes. It has recently been shown that the SAE1 gene is a transcriptional target of c-MYC [28], and SAE2 inhibition switches a transcriptional subprogram of MYC from activated to repressed [3]. Although cellular stresses induced c-MYC SUMOylation, no obvious effects of SUMOylation were detected on MYC stability or activities [29]. Our functional analyses and tumor xenograft model found SAE2 overexpression regulated proliferation, viability and tumorigenesis of GC cells in vitro and in vivo.
In patient tissue samples, the SAE2 expression level was significantly correlated with tumor invasion in GC. Our in vitro and in vivo studies provided that SAE2 plays a crucial role in cancer invasion and metastasis. Since SAE2 is the sole known E1 enzyme subunit for conjugating SUMOs, alterations of SAE2 expression will directly affect SUMOylation [30]. The impact of the SUMOylation pathway on migration and invasion has previously been described, and various molecular mechanisms underlying these capabilities have been proposed [24]. SUMO2/3 modification is greatly up-regulated in metastatic breast cancer cells and promotes 3D cell migration [31]. Additionally, SAE2 overexpression was associated with GC tumorigenesis, this proliferation characteristic could also impact GC cell metastasis in vivo.
Previous reports demonstrated that FoxM1 is extensively SUMOylated and FoxM1 SUMOylation can enhance its transcriptional activity [19,32]. Seven high-probability SUMOylation sites were predicted in human FoxM1 using the SUMOplotTM analysis (Table 3). IP assay in GC cell lines showed that FoxM1 was modified by SUMO1 rather than SUMO2/3, one of the most possible reason may be that the sensitivity of the method we used is lower. Western blotting results showed SUMO1-FoxM1 and other SUMO1 conjugates were strongly up-regulated in GC compared with para-carcinoma tissues. It demonstrated that SUMO1 conjugation was overactive in GC. SUMOylation of FoxM1B plays a functional role in the regulation of its target gene activities in breast cancer [32]. Moreover, FoxM1 directly regulates MMP2 [33], VEGF and Sox2 transcription [17], and plays a central role in cancer initiation and progression. We found that SUMO-FoxM1 was strongly overexpressed in GC tissues compared with paracarcinoma tissues, and FoxM1 expression corresponded to SAE2 expression in 76% of GC patients. Therefore, further research is needed to determine whether the SUMOylation of FoxM1 affects its transcriptional activities in GC.
In summary, we provide clinical and experimental evidence that SAE2 is overexpressed in GC cells and contributes to GC cell progression toward malignancy. SAE2 may be a useful independent prognostic tumor marker to predict metastasis and survival in GC patients.
Acknowledgements
This work was partly funded by National Natural Science Foundation of China (No. 81101879), National Key Technology R&D Program (2012AA02A203-B01, 2012AA02A504-B01, 2012AA020101) and Beijing Municipal Science & Technology Commission (Z121100007512010, D131100005313010).
Disclosure of conflict of interest
The authors disclose no potential conflicts of interest.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, Rao M, Yu P, Dominguez-Vidana R, Liang AC, Solimini NL, Bernardi RJ, Yu B, Hsu T, Golding I, Luo J, Osborne CK, Creighton CJ, Hilsenbeck SG, Schiff R, Shaw CA, Elledge SJ, Westbrook TF. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127:S51–55. doi: 10.1053/j.gastro.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Niu H, Peng Y, Wang J, He P. Ubc9 promotes invasion and metastasis of lung cancer cells. Oncol Rep. 2013;29:1588–1594. doi: 10.3892/or.2013.2268. [DOI] [PubMed] [Google Scholar]
- 6.Moschos SJ, Jukic DM, Athanassiou C, Bhargava R, Dacic S, Wang X, Kuan SF, Fayewicz SL, Galambos C, Acquafondata M, Dhir R, Becker D. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol. 2010;41:1286–1298. doi: 10.1016/j.humpath.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Lan Y, Xu J, Zhang W, Wen Z. SUMO1-activating enzyme subunit 1 is essential for the survival of hematopoietic stem/progenitor cells in zebrafish. Development. 2012;139:4321–4329. doi: 10.1242/dev.081869. [DOI] [PubMed] [Google Scholar]
- 8.Kanakousaki K, Gibson MC. A differential requirement for SUMOylation in proliferating and non-proliferating cells during Drosophila development. Development. 2012;139:2751–2762. doi: 10.1242/dev.082974. [DOI] [PubMed] [Google Scholar]
- 9.Demarque MD, Nacerddine K, Neyret-Kahn H, Andrieux A, Danenberg E, Jouvion G, Bomme P, Hamard G, Romagnolo B, Terris B, Cumano A, Barker N, Clevers H, Dejean A. Sumoylation by Ubc9 regulates the stem cell compartment and structure and function of the intestinal epithelium in mice. Gastroenterology. 2011;140:286–296. doi: 10.1053/j.gastro.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett. 2012;316:113–125. doi: 10.1016/j.canlet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 11.Stindt MH, Carter S, Vigneron AM, Ryan KM, Vousden KH. MDM2 promotes SUMO-2/3 modification of p53 to modulate transcriptional activity. Cell Cycle. 2011;10:3176–3188. doi: 10.4161/cc.10.18.17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mabb AM, Miyamoto S. SUMO and NF-kappaB ties. Cell Mol Life Sci. 2007;64:1979–1996. doi: 10.1007/s00018-007-7005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, Holsboer F, Arzt E. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell. 2007;131:309–323. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 14.Qu YY, Chen Q, Lai XP, Zhu CH, Chen C, Zhao X, Deng R, Xu M, Yuan HH, Wang YL, Yu JX, Huang J. SUMOylation of Grb2 enhances the ERK activity by increasing its binding with Sos1. Mol Cancer. 2014;13:95. doi: 10.1186/1476-4598-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bawa-Khalfe T, Yeh ET. SUMO Losing Balance: SUMO Proteases Disrupt SUMO Homeostasis to Facilitate Cancer Development and Progression. Genes Cancer. 2010;1:748–752. doi: 10.1177/1947601910382555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, Huang S. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–6219. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 17.Halasi M, Gartel AL. FOX(M1) news--it is cancer. Mol Cancer Ther. 2013;12:245–254. doi: 10.1158/1535-7163.MCT-12-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, Huang S, Tan D, Xie K. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schimmel J, Eifler K, Sigurethsson JO, Cuijpers SA, Hendriks IA, Verlaan-de Vries M, Kelstrup CD, Francavilla C, Medema RH, Olsen JV, Vertegaal AC. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell. 2014;53:1053–1066. doi: 10.1016/j.molcel.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Xing X, Lian S, Hu Y, Li Z, Zhang L, Wen X, Du H, Jia Y, Zheng Z, Meng L, Shou C, Ji J. Phosphatase of regenerating liver-3 (PRL-3) is associated with metastasis and poor prognosis in gastric carcinoma. J Transl Med. 2013;11:309. doi: 10.1186/1479-5876-11-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarge KD, Park-Sarge OK. Detection of proteins sumoylated in vivo and in vitro. Methods Mol Biol. 2009;590:265–277. doi: 10.1007/978-1-60327-378-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe M, Itoh K. Characterization of a novel posttranslational modification in neuronal nitric oxide synthase by small ubiquitin-related modifier-1. Biochim Biophys Acta. 2011;1814:900–907. doi: 10.1016/j.bbapap.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Baek SH. A novel link between SUMO modification and cancer metastasis. Cell Cycle. 2006;5:1492–1495. doi: 10.4161/cc.5.14.3008. [DOI] [PubMed] [Google Scholar]
- 25.Vigodner M, Shrivastava V, Gutstein LE, Schneider J, Nieves E, Goldstein M, Feliciano M, Callaway M. Localization and identification of sumoylated proteins in human sperm: excessive sumoylation is a marker of defective spermatozoa. Hum Reprod. 2013;28:210–223. doi: 10.1093/humrep/des317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellail AC, Olson JJ, Hao C. SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat Commun. 2014;5:4234. doi: 10.1038/ncomms5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogachek MV, Chen Y, Kulak MV, Woodfield GW, Cyr AR, Park JM, Spanheimer PM, Li Y, Li T, Weigel RJ. Sumoylation pathway is required to maintain the basal breast cancer subtype. Cancer Cell. 2014;25:748–761. doi: 10.1016/j.ccr.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amente S, Lavadera ML, Palo GD, Majello B. SUMO-activating SAE1 transcription is positively regulated by Myc. Am J Cancer Res. 2012;2:330–334. [PMC free article] [PubMed] [Google Scholar]
- 29.Karvonen U, Jaaskelainen T, Rytinki M, Kaikkonen S, Palvimo JJ. ZNF451 is a novel PML body- and SUMO-associated transcriptional coregulator. J Mol Biol. 2008;382:585–600. doi: 10.1016/j.jmb.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Mo YY, Yu Y, Theodosiou E, Ee PL, Beck WT. A role for Ubc9 in tumorigenesis. Oncogene. 2005;24:2677–2683. doi: 10.1038/sj.onc.1208210. [DOI] [PubMed] [Google Scholar]
- 31.Subramonian D, Raghunayakula S, Olsen JV, Beningo KA, Paschen W, Zhang XD. Analysis of Changes in SUMO-2/3 Modification during Breast Cancer Progression and Metastasis. J Proteome Res. 2014;13:3905–18. doi: 10.1021/pr500119a. [DOI] [PubMed] [Google Scholar]
- 32.Wang CM, Liu R, Wang L, Nascimento L, Brennan VC, Yang WH. SUMOylation of FOXM1B Alters Its Transcriptional Activity on Regulation of MiR-200 Family and JNK1 in MCF7 Human Breast Cancer Cells. Int J Mol Sci. 2014;15:10233–10251. doi: 10.3390/ijms150610233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen PM, Wu TC, Shieh SH, Wu YH, Li MC, Sheu GT, Cheng YW, Chen CY, Lee H. MnSOD promotes tumor invasion via upregulation of FoxM1-MMP2 axis and related with poor survival and relapse in lung adenocarcinomas. Mol Cancer Res. 2013;11:261–271. doi: 10.1158/1541-7786.MCR-12-0527. [DOI] [PubMed] [Google Scholar]