Abstract
The EGFR signaling pathway is important in the control of vital processes in the carcinogenesis of hepatocellular carcinoma (HCC), including cell survival, cell cycle progression, tumor invasion and angiogenesis. In the current study, we aim to assess if genetic variants in the genes of the EGFR signaling pathway are associated with the prognosis of HCC. We genotyped 36 single nucleotide polymorphisms (SNP) in four core genes (EGF, EGFR, VEGF, and VEGFR2) by using DNA from blood samples of 363 HCC patients with surgical resection. The associations between genotypes and overall survival (OS) and disease-free survival (DFS) were estimated using the Kaplan-Meier method. Hazard ratios (HRs) and 95% confident intervals (CIs) were estimated for the multivariate survival analyses by Cox proportional hazards regression models, adjusting for age, gender, family history, HBsAg and AFP. We found that five SNPs in the VEGFR2 gene were significantly associated with clinical outcomes of HCC patients. Among them, four SNPs (rs7692791, rs2305948, rs13109660, rs6838752) were associated with OS (p=0.035, 0.038, 0.029 and 0.028, respectively), and two SNPs (rs7692791 and rs2034965) were associated with DFS (p=0.039 and 0.017, respectively). Particularly, rs7692791 TT genotype was associated with both reduced OS (p=0.037) and DFS (p=0.043). However, only one SNP rs2034965 with the AA genotype was shown to be an independent effect on DFS (p=0.009) in the multivariate analysis. None of the other 31 polymorphisms or 9 haplotypes attained from the four genes was significantly associated with OS or DFS. Our results illustrated the potential use of VEGFR2 polymorphisms as prognostic markers for HCC patients.
Keywords: Hepatocellular carcinoma, survival, EGF, EGFR, VEGF, VEGFR2, genetic polymorphisms
Introduction
Hepatocellular carcinoma (HCC) is diagnosed in more than half a million people worldwide every year, and it is the third leading cause of cancer-related deaths [1]. About half of these cases and deaths are from China, mainly because chronic hepatitis B carriers account for 10% of its population [2]. In 2008, estimated 748,300 new liver cancer cases and 695,900 cancer-related deaths occurred worldwide, making the incidence and mortality rates almost equal [1].
Multiple clinical factors, including large tumor size, positive portal vein thrombosis, increased serum AFP, and advanced TNM stage are involved in poor survival of HCC patients [3]. Although these factors can be used to predict prognosis, recurrence is observed in 77-100% of the patients within 5 years and the 5-year overall survival (OS) rate remains poor, at around 50% [4,5]. Therefore more useful predictive markers are required to identify high-risk patients, thus establishing more appropriate cancer management strategies and improving better clinical outcomes of HCC.
The epidermal growth factor receptor (EGFR) is a tyrosine kinase transmembrane receptor in the ErB family of receptors expressed on the surface of epithelial cells. EGFR regulates important processes in carcinogenesis, including cell survival, cell cycle progression, tumor invasion and angiogenesis [6]. Ligands including epidermal growth factor (EGF) bind to EGFR and they activate signal transduction pathways that upregulate transcription factors leading to proliferation and differentiation of epidermal and epithelial tissue [7]. A few studies have suggested that genetic variants in the EGF and EGFR gene are associated with EGFR amplifications and contribute to cancer risk and prognosis [8-14].
Activation of the EGFR pathway also leads to the up-regulation of the ligand vascular endothelial growth factor (VEGF) and its receptor (VEGFR2) on endothelial cells, thus stimulating angiogenesis and vascular permeability. VEGF is considered to be a key mediator of both physiological and pathological angiogenesis. Angiogenesis, a process involving the growth of new blood vessels, is an important step in the development of cancer and plays a critical role in the primary tumor growth, invasiveness, and metastasis [15]. Several studies have reported the association between VEGF expression and worse prognosis in various cancers [16-19]. As for HCC, results from several reports have indicated that VEGF plays an important role in the development of HCC [20-22], and elevated serum level of VEGF is considered as an independent marker of HCC survival [23,24]. Furthermore, genetic variability of VEGF and VEGFR2 may affect the risk and outcome of various kinds of cancers regulated by angiogenesis [25], and polymorphisms in the VEGF gene have the predictive value on the risk of HCC patients [26,27].
These findings indicate that genetic variations of EGFR and its ligand EGF, VEGF and its ligand VEGFR2 probably affect HCC prognosis, but to our knowledge, the influence of genetic polymorphisms in the EGFR signaling pathway on the clinical outcomes of HCC patients have not been investigated extensively [28,29]. Therefore, the goal of the present study was to determine whether inherited variations in four core genes of the EGFR signaling pathway (EGF, EGFR, VEGF and VEGFR2) modified HCC survival.
Materials and methods
Patients and samples collection
A total of 363 Han Chinese patients newly diagnosed with HCC and receiving surgical resection of HCC tumor were recruited by the Qidong Liver Cancer Institute in Qidong, Jiangsu province, China from April 1996 to September 2009. The clinical outcomes of HCC were recorded until October 2014, with a median follow-up time of 53.0 months (range 2-110 months). The diagnosis of HCC was based on histopathological examination and the National Comprehensive Cancer Network (NCCN) clinical practice guidelines in oncology. All tumors were proven to be HCC by two pathologists. All patients had no other cancers as determined by initial screening examination and were followed up prospectively every 3 months from the time of enrollment by personal or family contacts until death or last time of follow-up.
There were no recruitment restrictions on age, gender and tumor stage. 5 ml whole blood for each subject was extracted. Clinical information was collected at the time the blood specimens were collected from medical records with patients’ consent. The histologic grade of tumor differentiation was assigned by the Edmondson grading system. The clinical typing of tumors was determined according to the TNM classification system of International Union Against Cancer (edition 6). The study endpoints were overall survival (OS), and disease-free survival (DFS). OS was calculated from the date of pathologic diagnosis/recruitment to death regardless of the cause or the end of available follow-up. DFS was defined as the time from pathologic diagnosis/recruitment to disease recurrence, metastasis, disease specific death or last follow-up.
This study was approved by the Department of Scientific Research of Fudan University and the Qidong Liver Cancer Institute, and a written informed consent with a signature was obtained from each patient before enrollment.
SNP selection
To select all the potential functional SNPs of EGF, EGFR, VEGF, and VEGFR2, we utilized the International HapMap Project database (http://hapmap.ncbi.nlm.nih.gov/) [30], and dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) [31] to search for candidate variants in the promoter region, all exons including intron–exon boundaries and the 3’-untranslated region (3’-UTR). We identified 20 potential functional polymorphisms (Table 1). We selected tagging SNPs from 5-kb flanking and within the gene regions of four genes by using the tagger algorithm [32]. 21 tagging SNPs were identified with a cut-off value of r2<0.8 and a minor allele frequency greater than 0.1 in the Chinese population, based on data from the HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) [32]. In addition, functional SNPs and SNPs previously reported to be associated with cancer were also included. Finally, a total of 36 SNPs, including haplotype-tagging SNPs and potential functional SNPs, were selected for genotyping (Table 1).
Table 1.
SNPs selected in 4 EGFR pathway genes for analysis
| Gene | SNP | Allelic change | MAF in Chinese | MAF in this study |
|---|---|---|---|---|
| EGF | rs3756261 | A/G | 0.239 | 0.228 |
| EGF | rs11568835 | A/G | 0.068 | 0.127 |
| EGF | rs4444903 | A/G | 0.367 | 0.450 |
| EGF | rs11568943 | A/T | 0.244 | 0.219 |
| EGF | rs2237051 | A/G | 0.378 | 0.291 |
| EGF | rs11569017 | A/G | 0.256 | 0.229 |
| EGF | rs3733625 | A/G | 0.239 | 0.204 |
| EGFR | rs6965469 | C/T | 0.167 | 0.167 |
| EGFR | rs4947492 | A/G | 0.419 | 0.351 |
| EGFR | rs2227983 | A/G | 0.397 | 0.478 |
| EGFR | rs11977388 | A/C | 0.464 | 0.370 |
| EGFR | rs2293347 | G/A | 0.222 | 0.301 |
| EGFR | rs884225 | G/A | 0.467 | 0.480 |
| VEGF | rs699947 | C/A | 0.278 | 0.266 |
| VEGF | rs833061 | C/T | 0.278 | 0.288 |
| VEGF | rs2010963 | G/C | 0.333 | 0.470 |
| VEGF | rs1413711 | C/T | 0.278 | 0.255 |
| VEGF | rs833070 | A/G | 0.192 | 0.242 |
| VEGF | rs3025000 | C/T | 0.309 | 0.445 |
| VEGF | rs3025033 | A/G | 0.178 | 0.216 |
| VEGF | rs3025035 | C/T | 0.200 | 0.156 |
| VEGF | rs3025039 | C/T | 0.178 | 0.208 |
| VEGF | rs10434 | A/G | 0.189 | 0.212 |
| VEGFR2 | rs2071559 | C/T | 0.300 | 0.335 |
| VEGFR2 | rs7667298 | C/T | 0.284 | 0.337 |
| VEGFR2 | rs1531290 | A/G | 0.233 | 0.273 |
| VEGFR2 | rs6837735 | C/T | 0.362 | 0.411 |
| VEGFR2 | rs7692791 | T/C | 0.411 | 0.398 |
| VEGFR2 | rs2305948 | C/T | 0.156 | 0.141 |
| VEGFR2 | rs2034965 | A/G | 0.178 | 0.221 |
| VEGFR2 | rs1870377 | A/T | 0.467 | 0.459 |
| VEGFR2 | rs13109660 | G/A | 0.278 | 0.297 |
| VEGFR2 | rs6838752 | C/T | 0.438 | 0.467 |
| VEGFR2 | rs17709898 | A/G | 0.389 | 0.303 |
| VEGFR2 | rs1531289 | G/A | 0.200 | 0.213 |
| VEGFR2 | rs7691507 | T/C | 0.156 | 0.182 |
MAF: Minor allele frequency.
DNA extraction, genotyping, and haplotypes reconstruction
Genomic DNA was extracted from blood samples using the QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany). Genotyping was performed with Sequenom MassARRAY iPLEX platform by use of allele-specific MALDI-TOF mass spectrometry assay. Polymerase chain reaction (PCR) and extension primers for these 36 SNPs were designed using the MassARRAY Assay Design 3.0 software (Sequenom). PCR and extension reactions were performed according to the manufacturer’s instructions, and extension product sizes were determined by mass spectrometry using the Sequenom iPLEX system. Duplicate test samples and two water samples (PCR negative controls) that were blinded to the technician were included in each 96-well plate. Genotyping quality was examined by a detailed QC procedure consisting of >95% successful call rate, duplicate calling of genotypes, internal positive control samples.
The linkage disequilibrium (LD) status among SNPs was measured with Lewontin D and r2 by using the Haploview software package (http://www.broad.mit.edu/mpg/haploview). LD blocks were inferred from the definition proposed by Gabriel and colleagues [30]. Probable haplotypes and their frequencies were calculated on the basis of a Bayesian algorithm [33] using PHASE software (ver 2.1.1, Seattle, WA, USA).
Statistical analysis
Associations of genotypes and haplotypes with OS and DFS were estimated using the Kaplan-Meier method, and statistical significance was determined using the log-rank test. The most significant test among the 3 genetic models (general, dominant, and recessive) was used to determine the statistical significance of each SNP. The SNPs or haplotypes with a raw p-value <0.05 in the univariable analysis were included in the multivariate analysis to evaluate their effects on the clinical outcomes. Hazard ratios (HRs) and 95% confident intervals (CIs) were estimated for the multivariate survival analyses by Cox proportional hazards regression models, adjusting for age, gender, family history, HBsAg and AFP.
Data analysis, with the exception of haplotype construction and haplotype frequency estimation, was performed with SPSS software version 19 (SPSS, Chicago, IL). All tests were two-sided and a p<0.05 was considered statistically significant.
Results
Patient characteristics and clinical outcomes
This study included 363 HCC patients with an overall median survival time (MST) of 34.0 months and median follow-up time of 53.0 months. At the time of analysis, 229 (63.1%) of the patients had died. The clinical pathologic characteristics and the association with OS and DFS are summarized in Table 2. By the Kaplan-Meier analysis, tumor capsule and TNM stage were significantly associated with OS and DFS (log-rank p<0.001). AFP was positive in 215 (60.2%) patients and shown to be related with both OS and DFS (p=0.009 and 0.007, respectively). In addition, large tumor size and differentiation were significantly associated with reduced OS and DFS, while venous invasion was a predictor for worse OS (p=0.041) and background cirrhosis for inferior DFS (p=0.044). The HBsAg of 304 (84.0%) cases was positive, but it didn’t demonstrate a relationship with either OS or DFS in the present study.
Table 2.
Correlations between clinicopathologic features and prognosis of HCC patients
| Characteristics | No. of patients | No. of events | Median survival time (95% CI) | Log-rank p | |
|---|---|---|---|---|---|
|
| |||||
| OS | DFS | ||||
| Total | 363 | 229 | 34.0 (27.4-40.6) | ||
| Age (year) | 0.157 | 0.281 | |||
| ≤50 | 187 | 123 | 30.0 (23.7-36.3) | ||
| <50 | 176 | 106 | 42.0 (32.5-51.5) | ||
| Gender | 0.806 | 0.993 | |||
| Female | 63 | 40 | 38.0 (18.6-57.4) | ||
| Male | 300 | 189 | 33.0 (25.9-40.1) | ||
| Family history | 0.834 | 0.866 | |||
| Absent | 263 | 163 | 35.0 (29.6-40.4) | ||
| Present | 81 | 54 | 39.0 (21.1-56.9) | ||
| Unknown | 19 | 12 | |||
| HBsAg | 0.996 | 0.939 | |||
| Negative | 58 | 38 | 20.0 (0.000-41.4) | ||
| Positive | 304 | 190 | 36.0 (29.5-42.5) | ||
| Unknown | 1 | 1 | |||
| AFP | 0.009 | 0.007 | |||
| Negative | 142 | 79 | 51.0 (34.0-68.0) | ||
| Positive | 215 | 144 | 28.0 (21.5-34.5) | ||
| Unknown | 6 | 6 | |||
| Tumor size (cm) | 0.029 | 0.046 | |||
| ≤5 | 145 | 86 | 46.0 (35.6-56.4) | ||
| >5 | 153 | 92 | 24.0 (14.7-33.4) | ||
| Unknown | 65 | 51 | |||
| Differentiation | 0.011 | 0.011 | |||
| I+II | 167 | 89 | 46.0 (33.36-58.64) | ||
| III+IV | 128 | 89 | 30.0 (22.52-37.48) | ||
| Unknown | 68 | 51 | |||
| Tumor capsule | 0.001 | 0.001 | |||
| Absent | 144 | 98 | 26.0 (14.4-37.6) | ||
| Present | 146 | 77 | 47.0 (30.4-63.6) | ||
| Unknown | 73 | 54 | |||
| Cirrhosis | 0.114 | 0.044 | |||
| Absent | 93 | 50 | 40.0 (13.3-66.7) | ||
| Present | 199 | 126 | 33.0 (26.9-39.1) | ||
| Unknown | 71 | 53 | |||
| Venous invasion | 0.041 | 0.078 | |||
| Absent | 199 | 119 | 39.0 (30.0-48.0) | ||
| Present | 77 | 48 | 23.0 (16.0-30.0) | ||
| Unknown | 87 | 62 | |||
| HB history | 0.684 | 0.604 | |||
| Absent | 148 | 92 | 35.0 (27.1-42.9) | ||
| Present | 113 | 65 | 30.0 (20.3-39.7) | ||
| Unknown | 102 | 72 | |||
| Tumor number | 0.491 | 0.442 | |||
| Solitary | 210 | 123 | 37.0 (26.3-42.7) | ||
| Multiple | 65 | 41 | 31.0 (19.2-42.8) | ||
| Unknown | 88 | 65 | |||
| pTNM stage | 0.001 | <0.001 | |||
| I | 70 | 29 | 57.0 (33.2-80.8) | ||
| II | 152 | 97 | 31.0 (23.3-39.9) | ||
| III | 24 | 19 | 19.0 (0.000-44.3) | ||
| IV | 12 | 11 | 6.0 (0.000-17.9) | ||
| Unknown | 105 | 73 | |||
CI: Confidence interval; OS: Overall survival; DFS: Disease-free survival.
As the clinicopathologic information from some patients was not available for several items, such as cirrhosis, venous invasion and TNM stage, and not all clinical factors above could be included in the subsequent multivariate analysis. Therefore we calculated HR and its corresponding p-value using Cox proportional hazard models, adjusted for age, gender, family history, HBsAg and AFP.
Genetic polymorphisms and HCC clinical outcomes
Table 3 shows the data for all the 36 SNPs among 4 genes (EGF, EGFR, VEGF and VEGFR2) analyzed for OS and DFS. In the univariate analysis, of all the 36 SNPs, 5 SNPs (rs7692791, rs2305948, rs13109660, rs6838752 and rs2034965), which are all resided in the VEGFR2 gene, were significantly associated with clinical outcomes. Overall, four SNPs (rs7692791, rs2305948, rs13109660, rs6838752) were associated with OS (Table 3; Figure 1); two SNPs (rs7692791 and rs2034965) were associated with DFS (Table 3; Figure 2). In particular, we observed VEGFR2 rs7692791 CC and CT genotype was significantly associated with improved OS (p=0.037; HR=0.751, 95% CI: 0.574-0.983) and DFS (p=0.043; HR=0.757, 95% CI: 0.579-0.991), compared with the CC/CT genotypes (Table 4), indicating that the rs7692791 variant T allele was significantly protective. On the contrary, we found a significant decreased OS for those carrying the TC genotype of rs2305948 (p=0.04; HR=1.349, 95% CI: 1.013-1.796) or rs6838752 (p=0.066; HR=1.371, 95% CI: 0.980-1.920, respectively). While rs13109660 AA genotype (p=0.033; HR=0.563, 95% CI: 0.333-0.954) was shown to result in a significant improvement in OS (log-rank p=0.029; Figure 1). However, none of the other 31 polymorphisms examined were significantly associated with OS or DFS (Table 3).
Table 3.
Influence of SNPs in 4 EGFR pathway genes on prognosis of HCC patients
| Gene | SNP | Log-rank p for OS | Log-rank p for DFS | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| General | Dominant | Recessive | General | Dominant | Recessive | ||
| EGF | rs3756261 | 0.653 | 0.446 | 0.772 | 0.494 | 0.261 | 0.951 |
| EGF | rs11568835 | 0.966 | 0.865 | 0.87 | 0.852 | 0.914 | 0.571 |
| EGF | rs4444903 | 0.437 | - | - | 0.23 | - | - |
| EGF | rs11568943 | 0.621 | 0.636 | 0.496 | 0.512 | 0.372 | 0.652 |
| EGF | rs2237051 | 0.327 | 0.955 | 0.172 | 0.401 | 0.52 | 0.18 |
| EGF | rs11569017 | 0.713 | 0.716 | 0.537 | 0.699 | 0.493 | 0.759 |
| EGF | rs3733625 | 0.294 | 0.536 | 0.215 | 0.258 | 0.317 | 0.304 |
| EGFR | rs6965469 | 0.969 | 0.831 | 0.935 | 0.883 | 0.817 | 0.631 |
| EGFR | rs4947492 | 0.776 | 0.484 | 0.925 | 0.995 | 0.977 | 0.937 |
| EGFR | rs2227983 | 0.493 | 0.332 | 0.795 | 0.69 | 0.642 | 0.62 |
| EGFR | rs11977388 | 0.602 | 0.586 | 0.573 | 0.737 | 0.976 | 0.464 |
| EGFR | rs2293347 | 0.371 | 0.176 | 0.975 | 0.356 | 0.152 | 0.793 |
| EGFR | rs884225 | 0.435 | 0.631 | 0.198 | 0.345 | 0.611 | 0.793 |
| VEGF | rs699947 | 0.169 | 0.885 | 0.085 | 0.182 | 0.838 | 0.097 |
| VEGF | rs833061 | 0.277 | 0.622 | 0.109 | 0.296 | 0.736 | 0.123 |
| VEGF | rs2010963 | 0.373 | 0.989 | 0.19 | 0.274 | 0.728 | 0.174 |
| VEGF | rs1413711 | 0.231 | 0.716 | 0.09 | 0.251 | 0.794 | 0.103 |
| VEGF | rs833070 | 0.23 | 0.937 | 0.114 | 0.271 | 0.911 | 0.139 |
| VEGF | rs3025000 | 0.436 | 0.795 | 0.274 | 0.362 | 0.6 | 0.286 |
| VEGF | rs3025033 | 0.896 | 0.657 | 0.99 | 0.865 | 0.677 | 0.845 |
| VEGF | rs3025035 | 0.291 | 0.75 | 0.16 | 0.344 | 0.819 | 0.182 |
| VEGF | rs3025039 | 0.891 | 0.923 | 0.673 | 0.815 | 0.844 | 0.602 |
| VEGF | rs10434 | 0.267 | 0.206 | 0.55 | 0.452 | 0.382 | 0.538 |
| VEGFR2 | rs2071559 | 0.437 | 0.352 | 0.584 | 0.282 | 0.331 | 0.369 |
| VEGFR2 | rs7667298 | 0.33 | 0.245 | 0.607 | 0.214 | 0.236 | 0.386 |
| VEGFR2 | rs1531290 | 0.134 | 0.361 | 0.137 | 0.289 | 0.451 | 0.261 |
| VEGFR2 | rs6837735 | 0.788 | 0.51 | 0.994 | 0.679 | 0.406 | 0.972 |
| VEGFR2 | rs7692791 | 0.101 | 0.035 | 0.338 | 0.112 | 0.039 | 0.346 |
| VEGFR2 | rs2305948 | 0.038 | 0.025 | 0.132 | 0.163 | 0.078 | 0.331 |
| VEGFR2 | rs2034965 | 0.103 | 0.091 | 0.082 | 0.017 | 0.027 | 0.019 |
| VEGFR2 | rs1870377 | 0.113 | 0.34 | 0.163 | 0.141 | 0.134 | 0.498 |
| VEGFR2 | rs13109660 | 0.092 | 0.46 | 0.029 | 0.222 | 0.554 | 0.083 |
| VEGFR2 | rs6838752 | 0.028 | 0.263 | 0.066 | 0.063 | 0.123 | 0.272 |
| VEGFR2 | rs17709898 | 0.811 | 0.519 | 0.865 | 0.572 | 0.291 | 0.691 |
| VEGFR2 | rs1531289 | 0.384 | 0.341 | 0.219 | 0.118 | 0.164 | 0.066 |
| VEGFR2 | rs7691507 | 0.751 | 0.452 | 0.794 | 0.259 | 0.24 | 0.171 |
OS: Overall survival; DFS: Disease-free survival.
Figure 1.
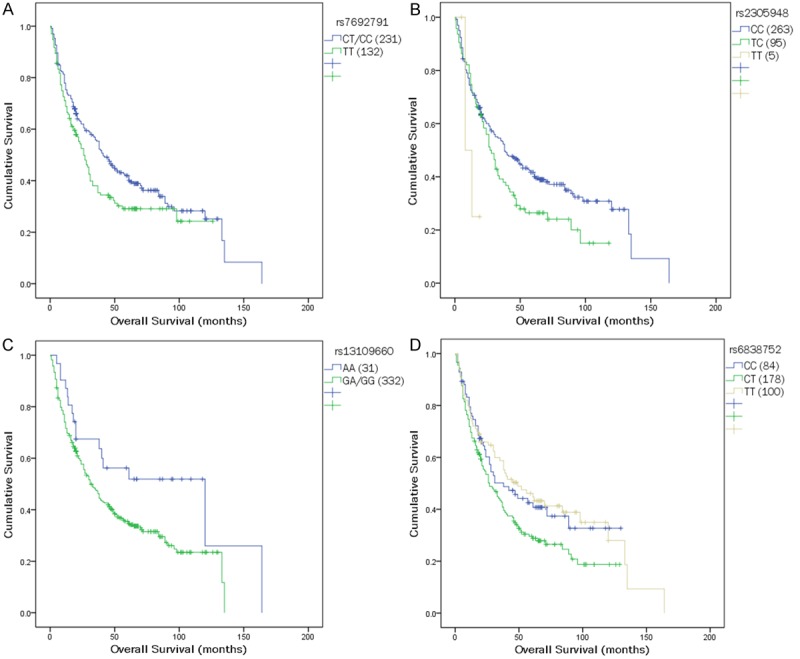
Kaplan-Meier survival curves of overall survival in HCC patients are shown for polymorphisms of (A) rs7692791, (B) rs2305948, (C) rs13109660, and (D) rs6838752.
Figure 2.
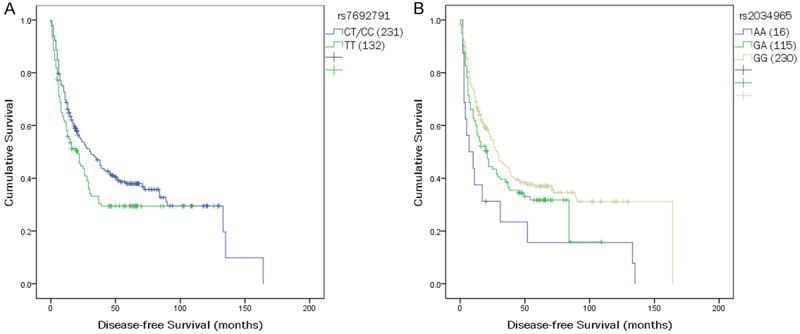
Kaplan-Meier survival curves of disease-free survival in HCC patients are shown for polymorphisms of (A) rs7692791 and (B) rs2034965.
Table 4.
Significant associations of SNPs in 4 EGFR pathway genes with prognosis of HCC patients
| SNP | Outcome | Model | Log-rank p | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| HR (95% CI) | p | HR (95% CI)* | p | ||||
| rs7692791 | OS | Dominant | 0.035 | 0.037 | 0.174 | ||
| TT | 1 | 1 | |||||
| CT/CC | 0.751 (0.574-0.983) | 0.815 (0.608-1.094) | |||||
| rs2305948 | OS | General | 0.038 | 0.027 | 0.185 | ||
| CC | 1 | 1 | |||||
| TC | 1.349 (1.013-1.796) | 0.04 | 1.252 (0.913-1.716) | 0.164 | |||
| TT | 2.543 (0.804-8.040) | 0.112 | 2.281 (0.695-7.484) | 0.174 | |||
| rs13109660 | OS | Recessive | 0.029 | 0.033 | 0.386 | ||
| GA/GG | 1 | 1 | |||||
| AA | 0.563 (0.333-0.954) | 0.753 (0.398-1.428) | |||||
| rs6838752 | OS | General | 0.028 | 0.031 | 0.249 | ||
| CC | 1 | 1 | |||||
| CT | 1.371 (0.980-1.920) | 0.066 | 1.325 (0.933-1.883) | 0.116 | |||
| TT | 0.939 (0.637-1.385) | 0.752 | 1.112 (0.711-1.740) | 0.643 | |||
| rs7692791 | DFS | Dominant | 0.039 | 0.043 | 0.073 | ||
| TT | 1 | 1 | |||||
| CT/CC | 0.757 (0.579-0.991) | 0.769 (0.578-1.025) | |||||
| rs2034965 | DFS | General | 0.017 | 0.021 | 0.024 | ||
| GG | 1 | 1 | |||||
| GA | 1.261 (0.953-1.669) | 0.105 | 0.809 (0.627-1.044) | 0.103 | |||
| AA | 2.029 (1.169-3.522) | 0.012 | 1.672 (1.136-2.460) | 0.009 | |||
OS: overall survival; DFS: disease-free survival; HR: hazard ratio; CI: confidence interval.
Adjusted for age, gender, family history, HBsAg and AFP.
A multivariate analysis of genotype effects on survival was conducted using Cox proportional hazards models adjusted for available clinicopathologic variables. As shown in Table 4, only one SNP rs2034965 remained significant, with the AA genotype presenting an independent negative effect on DFS (p=0.009, HR=1.672, 95% CI: 1.136-2.460), compared to patients who had common homozygous genotype and heterozygous genotype. None of the genetic polymorphisms was identified as an independent prognostic factor for OS.
Furthermore, we examined the associations of the haplotypes with survival outcomes. When examining combinations of SNPs for the EGF, EGFR, VEGF, and VEGFR2, we attained 2 haplotypes of EGF, 2 haplotypes of EGFR, 2 haplotypes of VEGF, and 3 haplotypes of VEGFR2. The inferred haplotypes and their associations with OS and DFS are shown in Table 5. Consistent with the results of individual genotype analyses, none of the haplotypes carrying variant alleles from EGF, EGFR, and VEGF showed a significant association with OS or DFS. The most probable haplotype (CT) was from VEGFR2, which had an estimated frequency of 66.7 percent. However, even though each of the individual SNPs of rs7692791, rs2305948, rs13109660, rs6838752 and rs2034965 in VEGFR2 showed potential prognostic effect on HCC clinical outcomes, the VEGFR2 haplotypes were still significantly related with neither OS nor DFS.
Table 5.
Associations between haplotypes in 4 EGFR pathway genes and prognosis of HCC patients
| Haplotype | No. of alleles | Frequency (%) | No. of events | Log-rank p for OS | Log-rank p for DFS |
|---|---|---|---|---|---|
| EGF-1 | 0.864 | 0.648 | |||
| Haplotype of rs11569017 A/G and rs3733625 A/G | |||||
| AG | 463 | 64.5 | 289 | ||
| GG | 155 | 21.6 | 105 | ||
| AA | 100 | 13.9 | 62 | ||
| EGF-2 | 0.7 | 0.431 | |||
| Haplotype of rs3756261 A/G and rs11568943 A/T | |||||
| AA | 342 | 47.8 | 224 | ||
| GA | 224 | 31.3 | 133 | ||
| AT | 150 | 20.9 | 101 | ||
| EGFR-1 | 0.73 | 0.854 | |||
| Haplotype of rs4947492 A/G and rs6965469 C/T | |||||
| AT | 369 | 51 | 236 | ||
| GC | 279 | 38.5 | 172 | ||
| GT | 72 | 9.9 | 48 | ||
| AC | 4 | 0.6 | 2 | ||
| EGFR-2 | 0.358 | 0.274 | |||
| Haplotype of rs2293347 G/A and rs884225 G/A | |||||
| GG | 350 | 48.9 | 226 | ||
| AA | 212 | 29.6 | 127 | ||
| GA | 153 | 21.4 | 100 | ||
| VEGF-1 | 0.528 | 0.583 | |||
| Haplotype of rs699947 C/A, rs833061 C/T, rs2010963 G/C, rs1413711 C/T, rs833070 A/G, and rs3025000 C/T | |||||
| CTCGGT | 303 | 42 | 200 | ||
| CTGGGC | 211 | 29.2 | 128 | ||
| ACGAAC | 168 | 23.3 | 100 | ||
| Others* | 40 | 5.6 | 28 | ||
| VEGF-2 | 0.159 | 0.236 | |||
| Haplotype of rs3025033 A/G, rs3025035 C/T, rs3025039 C/T, and rs10434 A/G | |||||
| ACCG | 299 | 41.3 | 182 | ||
| ACCA | 163 | 22.5 | 108 | ||
| GCTG | 144 | 19.9 | 90 | ||
| ATCG | 110 | 15.2 | 71 | ||
| Others* | 8 | 1.1 | 7 | ||
| VEGFR2-1 | 0.488 | 0.198 | |||
| Haplotype of rs1531289 G/A and rs2034965 A/G | |||||
| GA | 341 | 47.4 | 216 | ||
| GG | 237 | 32.9 | 144 | ||
| AA | 142 | 19.7 | 96 | ||
| VEGFR2-2 | 0.378 | 0.254 | |||
| Haplotype of rs6838752 C/T, rs17709898 A/G, and rs1870377 A/T | |||||
| CGA | 334 | 46.3 | 209 | ||
| TAT | 210 | 29.1 | 127 | ||
| TGT | 160 | 22.2 | 111 | ||
| Others* | 18 | 2.4 | 11 | ||
| VEGFR2-3 | 0.529 | 0.633 | |||
| Haplotype of rs2071559 C/T and rs7667298 C/T | |||||
| CT | 479 | 66.6 | 304 | ||
| TC | 240 | 33.4 | 153 | ||
Others include rare haplotypes with frequencies less than 5%.
Discussion
Although new treatment modalities changed the global approach to HCC, this disease still represents a therapeutic challenge. While some germline genetic factors have been suspected of playing an important role in prognosis, none have been firmly established [34,35]. The EGFR system regulates important processes within the tumor microenvironment of autocrine and paracrine circuits, including tumor invasion and angiogenesis [6]. Previous studies reported that the intensity of EGF, EGFR, VEGF, VEGFR2 expression correlates with proliferative activity, stage, intrahepatic metastasis and carcinoma differentiation in HCC [20-22,36,37]. In addition, several observations suggested that polymorphisms of these genes might regulate angiogenesis and lymphangiogenesis and thus controlling tumor growth. However, most investigations into SNPs in EGFR pathway (EGF, EGFR, VEGF, and VEGFR2) genes have just focused on their effects on risk rather than prognosis of HCC [38,39], or selected SNPs without a systematic method [27,29,40]. The aim of our study was to evaluate the role of EGFR, EGF, VEGF, VEGFR2 polymorphisms in determining the clinical outcomes of HCC patients. To the best of our knowledge, this is the first evidence showing the relationship between genetic variants of EGFR pathway genes and the prognosis of HCC patients. We found that in the VEGFR2 gene two non-synonymous SNPs, rs7692791 and rs2034965 were significantly associated with DFS and four SNPs (rs7692791, rs2305948, rs13109660, rs6838752) were associated with OS. Once prospectively validated, this finding could be used to predict which patients are at risk for poor clinical outcomes, and the analysis of VEGFR2 SNPs may help to identify HCC patients more likely to benefit from targeted inhibitor therapy [35].
Deregulation of EGF/EGFR signaling pathway is thought to be one of the most important factors in early hepatocarcinogenesis [36,41-44]. Membranous EGFR was observed in 40% HCC patients, and correlated with histological grade. Angiolymphatic invasion was more commonly seen in EGFR-positive cases [45]. A recent study showed that the EGFR signaling system stands at a crossroad between inflammatory signals and intracellular pathways associated with hepatocarcinogenesis. The EGFR ligand amphiregulin AR release and EGFR transactivation by TNF-alpha constitutes a novel link between inflammatory signals and pro-tumorigenic mechanisms in liver cells. Its sheddase ADAM17 increased in pre-neoplastic liver injury further supports its implication in hepatocarcinogenesis [46]. However, the association between the EGFR genotype and prognosis so far has not been described in HCC patients and results from EGF remain controversial [27,29,40]. In our study, none of the seven SNPs of EGF and six SNPs of EGFR investigated was associated with either DFS or OS in HCC patients. This result could be explained partly by the fact that though EGF is only part of a gene expression signature associated with poor OS in HCC patients [47,48], EGF expression alone is not qualified enough to predict HCC prognosis, which is also affected by various clinicopathologic characteristics, such as cirrhosis, venous invasion and TNM stage. As most studies reported that the G allele of EGF rs4444903 was a risk factor for HCC, independently of ethnicity and etiology [39], the variants of EGF/EGFR signaling pathway are more likely to alter the susceptibility rather than prognosis of HCC.
HCC is a highly vascular tumor, which proliferates through angiogenic pathways mediated partly by VEGF and its multiple receptors including VEGFR2. Evidences from preclinical and clinical studies showed that there was a correlation between high VEGFR2 expression, both in tissues and serum, and the metastases or poor prognosis of HCC. VEGFR2 expression was significantly higher not only in the veins and sinusoids of poorly differentiated tumors, but also in the arteries of non-tumorous liver in HCC patients, suggesting that VEGFR2 expression is a feature of poor differentiation and tumor progression [49]. Another study reported that high VEGFR2 expression in HCC was related to large tumor diameter, poor differentiation, high serum alpha-fetoprotein, multifocal gross classification [50], and displayed a trend toward decreased OS [51]. On the other hand, the patients with a low serum level of VEGFR2 had better OS and DFS than those with a high serum level of VEGF [52]. Furthermore, the pre-treatment serum level of VEGFR2 was an independent and significant prognostic factor of survival for HCC patients, and the serum VEGFR2 concentration decrease after transarterial chemoembolization (TACE) may predict favorable OS in patients with HCC [53]. However, limited information is available regarding the role of the VEGF system SNPs, especially its receptor VEGFR2, in HCC. The simultaneous presence of VEGFR2 and VEGF polymorphisms may confer an increased risk of HCC in patients with alcoholics presenting liver disease (ALD) [54]. Certain SNPs of VEGFR2 may affect treatment outcomes and toxicity in patients treated with sunitinib. One study reported that rs7692791 of VEGFR2 was associated with poor OS among patients with gastric or biliary tract cancer who were treated with sunitinib [55]. VEGFR2 alleles C of rs2305948 and VEGF alleles C of rs699947, C of rs2010963 were significant predictors of DFS and OS at univariate analysis, but only rs2010963 resulted to be an independent factor influencing DFS and OS in multivariate analysis [56].
Given curative resection and postoperative treatment, including local radiofrequency ablation (RFA), TACE, radioembolization, and molecular targeted therapy, establishment of more precise prognostic determinants using molecular biology techniques is still warranted to make the best use of these options [57]. Particularly, evidences from studies on EGFR-tyrosine kinase inhibitors for several epithelial cancers are very encouraging. These inhibitors can block the expression of not only EGFR but also VEGF [58]. For example, Vandetanib, an inhibitor of VEGFR2 and EGFR, was showed to suppress tumor development and improve the prognosis of liver cancer [59]. In addition, concomitant inhibition of VEGFR2 and Raf will disrupt oncogenic signaling and efficiently reduce tumor growth and vascularization of HCC in Human HCC cell lines and endothelial cells [60]. Tyrosine kinase inhibitors of VEGFR2, such as sunitinib [61], Sorafenib [62], and foretinib [63], have shown promising preliminary efficacy in patients with HCC. Understanding how these genetic variants work on clinical outcomes of HCC patients may help developing new drugs and achieving personalized therapeutic regimen.
In our study, only one SNP rs2034965 remained significant in the multivariate analysis, with the AA genotype presenting an independent negative effect on DFS. None of additional genetic polymorphisms reached significance and could be served as an independent prognostic factor for OS. Maybe we can find clues through our sample sources. We obtain blood samples from each HCC patient treated with surgery. However, the expression of VEGFR2 in tissues and serum may have different prognostic influence on HCC patients. Another explanation is that even though we selected and investigated these SNPs in a systematical way, due to limited techniques, labor and resources, we missed some key SNPs which play a predominant role in regulating the expression of the EGFR pathway genes. For this reason, we are not capable of concluding that the SNPs of these four genes are not associated with the prognosis of HCC at present. Instead, a more comprehensive analysis of polymorphisms in the EGFR pathway is imperative to illustrate the close correlation between EGFR pathway genes and HCC prognosis.
It is worth mentioning that there were a number of limitations in our study. Firstly, the cohort size was relatively small, and we didn’t recruit enough cases for validation. The significant association found in the univariate analysis should be viewed as generating hypothesis or a clue for related researches afterwards. Therefore, larger well-designed longitudinal follow-up studies and functional evaluation are warranted to confirm these findings. Secondly, though several clinical and pathologic characteristics showed significant associations with OS and/or DFS, including tumor size, differentiation, tumor capsule, cirrhosis, venous invasion and TNM stage, it is regretful that we failed to collect adequate and accurate information of these factors in our study. In order to make the greatest use of the genotype polymorphisms information we got from the 363 HCC patients, we had to operate the multivariate analysis without adjusting all these potential prognostic factors. Future studies are essential to investigate the role of genetic polymorphisms in patients with more complete and comprehensive clinicalpathologic characteristics. Last but not the least, as mentioned above, all of our samples are blood from each HCC patients treated with surgery. This major drawback not only confined our results to the expression of EGFR pathway genes in serum rather than tissues, but also restricted criteria for patients who can be only treated with surgery. However, most patients with HCC are diagnosed at advanced stages when curative treatments, such as hepatic resection and liver transplantation, are not feasible [57]. Accordingly, analyses of tissue samples are urgent to figure out the unknown modulation of these genes in HCC survival.
In summary, our results demonstrated the potential use of VEGFR2 polymorphisms as prognostic markers for HCC patients. However, neither the SNPs in EGF, EGFR, and VEGF genes nor the haplotypes from the EGFR pathway genes were significantly associated with HCC prognosis.
Acknowledgements
We thank all the participants. This study was supported by the National Natural Science Foundation of China (31100895 and 81472618 to D.-K.J.), the National Key Sci-Tech Special Project of China (2008ZX10002-020 and 2013ZX10002010 to L.Y.), as well as supported from an intramural research grant for outstanding young scholar from Fudan University (to D.-K.J.), an intramural research grant for new young teachers from Fudan University (to D.-K.J.), an intramural research grant for promotion of the scientific research ability of young teachers from Fudan University (to D.-K.J.), an intramural research grant from Huashan Hospital, Fudan University (to J.X.), and an intramural research grant from Center for Genetic Epidemiology, Fudan University (to J.X.).
Disclosure of conflict of interest
There are no known conflicts of interest.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao B, Lu J, Yin J, Liu H, Guo X, Yang Y, Ge N, Zhu Y, Zhang H, Xing J. A functional polymorphism in PER3 gene is associated with prognosis in hepatocellular carcinoma. Liver Int. 2012;32:1451–1459. doi: 10.1111/j.1478-3231.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- 4.Torzilli G, Belghiti J, Kokudo N, Takayama T, Capussotti L, Nuzzo G, Vauthey JN, Choti MA, De Santibanes E, Donadon M, Morenghi E, Makuuchi M. A snapshot of the effective indications and results of surgery for hepatocellular carcinoma in tertiary referral centers: is it adherent to the EASL/AASLD recommendations?: an observational study of the HCC East-West study group. Ann Surg. 2013;257:929–937. doi: 10.1097/SLA.0b013e31828329b8. [DOI] [PubMed] [Google Scholar]
- 5.Takayasu K, Arii S, Ikai I, Omata M, Okita K, Ichida T, Matsuyama Y, Nakanuma Y, Kojiro M, Makuuchi M, Yamaoka Y. Prospective cohort study of transarterial chemoembolization for unresectable hepatocellular carcinoma in 8510 patients. Gastroenterology. 2006;131:461–469. doi: 10.1053/j.gastro.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 6.De Luca A, Carotenuto A, Rachiglio A, Gallo M, Maiello MR, Aldinucci D, Pinto A, Normanno N. The role of the EGFR signaling in tumor microenvironment. J Cell Physiol. 2008;214:559–567. doi: 10.1002/jcp.21260. [DOI] [PubMed] [Google Scholar]
- 7.Stoscheck CM, King LE Jr. Role of epidermal growth factor in carcinogenesis. Cancer Res. 1986;46:1030–1037. [PubMed] [Google Scholar]
- 8.Hansen TF, Sorensen FB, Spindler KL, Olsen DA, Andersen RF, Lindebjerg J, Brandslund I, Jakobsen A. Microvessel density and the association with single nucleotide polymorphisms of the vascular endothelial growth factor receptor 2 in patients with colorectal cancer. Virchows Arch. 2010;456:251–260. doi: 10.1007/s00428-009-0878-8. [DOI] [PubMed] [Google Scholar]
- 9.Kang D, Gridley G, Huang WY, Engel LS, Winn DM, Brown LM, Bravo-Otero E, Wu T, Diehl SR, Hayes RB. Microsatellite polymorphisms in the epidermal growth factor receptor (EGFR) gene and the transforming growth factor-alpha (TGFA) gene and risk of oral cancer in Puerto Rico. Pharmacogenet Genomics. 2005;15:343–347. doi: 10.1097/01213011-200505000-00010. [DOI] [PubMed] [Google Scholar]
- 10.Brandt B, Hermann S, Straif K, Tidow N, Buerger H, Chang-Claude J. Modification of breast cancer risk in young women by a polymorphic sequence in the egfr gene. Cancer Res. 2004;64:7–12. doi: 10.1158/0008-5472.can-03-2623. [DOI] [PubMed] [Google Scholar]
- 11.Choi JE, Park SH, Kim KM, Lee WK, Kam S, Cha SI, Kim CH, Kang YM, Kim YC, Han SB, Jung TH, Park JY. Polymorphisms in the epidermal growth factor receptor gene and the risk of primary lung cancer: a case-control study. BMC Cancer. 2007;7:199. doi: 10.1186/1471-2407-7-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Q, Cheung YB, Jada SR, Lim WT, Kuo WL, Gray JW, Lee AS, Chowbay B. EGFR Intron 1 polymorphism in Asian Populations and its correlation with EGFR gene expression and amplification in breast tumor tissues. Cancer Biol Ther. 2006;5:1445–1449. doi: 10.4161/cbt.5.11.3457. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Stoehlmacher J, Park DJ, Yang D, Borchard E, Gil J, Tsao-Wei DD, Yun J, Gordon M, Press OA, Rhodes K, Groshen S, Lenz HJ. Gene polymorphisms of epidermal growth factor receptor and its downstream effector, interleukin-8, predict oxaliplatin efficacy in patients with advanced colorectal cancer. Clin Colorectal Cancer. 2005;5:124–131. doi: 10.3816/ccc.2005.n.025. [DOI] [PubMed] [Google Scholar]
- 14.Forsti A, Jin Q, Altieri A, Johansson R, Wagner K, Enquist K, Grzybowska E, Pamula J, Pekala W, Hallmans G, Lenner P, Hemminki K. Polymorphisms in the KDR and POSTN genes: association with breast cancer susceptibility and prognosis. Breast Cancer Res Treat. 2007;101:83–93. doi: 10.1007/s10549-006-9265-1. [DOI] [PubMed] [Google Scholar]
- 15.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 16.Delongchamps NB, Peyromaure M, Dinh-Xuan AT. Role of vascular endothelial growth factor in prostate cancer. Urology. 2006;68:244–248. doi: 10.1016/j.urology.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Gasparini G. Prognostic value of vascular endothelial growth factor in breast cancer. Oncologist. 2000;5(Suppl 1):37–44. doi: 10.1634/theoncologist.5-suppl_1-37. [DOI] [PubMed] [Google Scholar]
- 18.Giatromanolaki A, Sivridis E, Koukourakis MI. Angiogenesis in colorectal cancer: prognostic and therapeutic implications. Am J Clin Oncol. 2006;29:408–417. doi: 10.1097/01.coc.0000221317.56731.4e. [DOI] [PubMed] [Google Scholar]
- 19.Rudlowski C, Pickart AK, Fuhljahn C, Friepoertner T, Schlehe B, Biesterfeld S, Schroeder W. Prognostic significance of vascular endothelial growth factor expression in ovarian cancer patients: a long-term follow-up. Int J Gynecol Cancer. 2006;16(Suppl 1):183–189. doi: 10.1111/j.1525-1438.2006.00307.x. [DOI] [PubMed] [Google Scholar]
- 20.Iavarone M, Lampertico P, Iannuzzi F, Manenti E, Donato MF, Arosio E, Bertolini F, Primignani M, Sangiovanni A, Colombo M. Increased expression of vascular endothelial growth factor in small hepatocellular carcinoma. J Viral Hepat. 2007;14:133–139. doi: 10.1111/j.1365-2893.2006.00782.x. [DOI] [PubMed] [Google Scholar]
- 21.Park YN, Kim YB, Yang KM, Park C. Increased expression of vascular endothelial growth factor and angiogenesis in the early stage of multistep hepatocarcinogenesis. Arch Pathol Lab Med. 2000;124:1061–1065. doi: 10.5858/2000-124-1061-IEOVEG. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki K, Hayashi N, Miyamoto Y, Yamamoto M, Ohkawa K, Ito Y, Sasaki Y, Yamaguchi Y, Nakase H, Noda K, Enomoto N, Arai K, Yamada Y, Yoshihara H, Tujimura T, Kawano K, Yoshikawa K, Kamada T. Expression of vascular permeability factor/vascular endothelial growth factor in human hepatocellular carcinoma. Cancer Res. 1996;56:3004–3009. [PubMed] [Google Scholar]
- 23.Jinno K, Tanimizu M, Hyodo I, Nishikawa Y, Hosokawa Y, Doi T, Endo H, Yamashita T, Okada Y. Circulating vascular endothelial growth factor (VEGF) is a possible tumor marker for metastasis in human hepatocellular carcinoma. J Gastroenterol. 1998;33:376–382. doi: 10.1007/s005350050099. [DOI] [PubMed] [Google Scholar]
- 24.Poon RT, Lau C, Pang R, Ng KK, Yuen J, Fan ST. High serum vascular endothelial growth factor levels predict poor prognosis after radiofrequency ablation of hepatocellular carcinoma: importance of tumor biomarker in ablative therapies. Ann Surg Oncol. 2007;14:1835–1845. doi: 10.1245/s10434-007-9366-z. [DOI] [PubMed] [Google Scholar]
- 25.Schneider BP, Radovich M, Miller KD. The role of vascular endothelial growth factor genetic variability in cancer. Clin Cancer Res. 2009;15:5297–5302. doi: 10.1158/1078-0432.CCR-08-2576. [DOI] [PubMed] [Google Scholar]
- 26.Giacalone A, Montalto G, Giannitrapani L, Balasus D, Terranova A, Cervello M, Soresi M, Marasa L. Association between single nucleotide polymorphisms in the cyclooxygenase-2, tumor necrosis factor-alpha, and vascular endothelial growth factor-A genes, and susceptibility to hepatocellular carcinoma. OMICS. 2011;15:193–196. doi: 10.1089/omi.2010.0095. [DOI] [PubMed] [Google Scholar]
- 27.Suenaga M, Yamada S, Fujii T, Fuchs BC, Okumura N, Kanda M, Kobayashi D, Tanaka C, Nakayama G, Sugimoto H, Koike M, Nomoto S, Fujiwara M, Takeda S, Hayashi K, Tanabe KK, Goto H, Kodera Y. A functional polymorphism in the epidermal growth factor gene predicts hepatocellular carcinoma risk in Japanese hepatitis C patients. Onco Targets Ther. 2013;6:1805–1812. doi: 10.2147/OTT.S53625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong SY, Park JW, Lee JA, Park JE, Park KW, Hong EK, Kim CM. Association between vascular endothelial growth factor gene polymorphisms and survival in hepatocellular carcinoma patients. Hepatology. 2007;46:446–455. doi: 10.1002/hep.21720. [DOI] [PubMed] [Google Scholar]
- 29.Wu LM, Xie HY, Zhou L, Yang Z, Zhang F, Zheng SS. A single nucleotide polymorphism in the vascular endothelial growth factor gene is associated with recurrence of hepatocellular carcinoma after transplantation. Arch Med Res. 2009;40:565–570. doi: 10.1016/j.arcmed.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 30.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 31.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 32.A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin F, Xiong WJ, Jing JC, Feng Z, Qu LS, Shen XZ. Evaluation of the association studies of single nucleotide polymorphisms and hepatocellular carcinoma: a systematic review. J Cancer Res Clin Oncol. 2011;137:1095–1104. doi: 10.1007/s00432-010-0970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishida N, Kudo M. Recent advancements in comprehensive genetic analyses for human hepatocellular carcinoma. Oncology. 2013;84(Suppl 1):93–97. doi: 10.1159/000345897. [DOI] [PubMed] [Google Scholar]
- 36.Ito Y, Takeda T, Sasaki Y, Sakon M, Yamada T, Ishiguro S, Imaoka S, Tsujimoto M, Higashiyama S, Monden M, Matsuura N. Expression and clinical significance of the erbB family in intrahepatic cholangiocellular carcinoma. Pathol Res Pract. 2001;197:95–100. doi: 10.1078/0344-0338-00016. [DOI] [PubMed] [Google Scholar]
- 37.Zhong JH, You XM, Gong WF, Ma L, Zhang Y, Mo QG, Wu LC, Xiao J, Li LQ. Epidermal growth factor gene polymorphism and risk of hepatocellular carcinoma: a meta-analysis. PLoS One. 2012;7:e32159. doi: 10.1371/journal.pone.0032159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abu DBK, Yang M, Fuchs BC, Karl DL, Yamada S, Sninsky JJ, O’Brien TR, Dienstag JL, Tanabe KK, Chung RT. A functional polymorphism in the epidermal growth factor gene is associated with risk for hepatocellular carcinoma. Gastroenterology. 2011;141:141–149. doi: 10.1053/j.gastro.2011.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z, Wu Q, Shi Y, Nie Y, Wu K, Fan D. Epidermal growth factor 61A>G polymorphism is associated with risk of hepatocellular carcinoma: a meta-analysis. Genet Test Mol Biomarkers. 2012;16:1086–1091. doi: 10.1089/gtmb.2012.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshiya S, Fujimoto Y, Bekki Y, Konishi H, Yamashita Y, Ikegami T, Yoshizumi T, Shirabe K, Oda Y, Maehara Y. Impact of epidermal growth factor single-nucleotide polymorphism on recurrence of hepatocellular carcinoma after hepatectomy in patients with chronic hepatitis C virus infection. Cancer Sci. 2014;105:646–650. doi: 10.1111/cas.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komuves LG, Feren A, Jones AL, Fodor E. Expression of epidermal growth factor and its receptor in cirrhotic liver disease. J Histochem Cytochem. 2000;48:821–830. doi: 10.1177/002215540004800610. [DOI] [PubMed] [Google Scholar]
- 42.Morimitsu Y, Hsia CC, Kojiro M, Tabor E. Nodules of less-differentiated tumor within or adjacent to hepatocellular carcinoma: relative expression of transforming growth factor-alpha and its receptor in the different areas of tumor. Hum Pathol. 1995;26:1126–1132. doi: 10.1016/0046-8177(95)90275-9. [DOI] [PubMed] [Google Scholar]
- 43.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 44.Berasain C, Avila MA. The EGFR signalling system in the liver: from hepatoprotection to hepatocarcinogenesis. J Gastroenterol. 2014;49:9–23. doi: 10.1007/s00535-013-0907-x. [DOI] [PubMed] [Google Scholar]
- 45.Bassullu N, Turkmen I, Dayangac M, Yagiz Korkmaz P, Yasar R, Akyildiz M, Yaprak O, Tokat Y, Yuzer Y, Bulbul Dogusoy G. The Predictive and Prognostic Significance of c-erb-B2, EGFR, PTEN, mTOR, PI3K, p27, and ERCC1 Expression in Hepatocellular Carcinoma. Hepat Mon. 2012;12:e7492. doi: 10.5812/hepatmon.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berasain C, Nicou A, Garcia-Irigoyen O, Latasa MU, Urtasun R, Elizalde M, Salis F, Perugorria MJ, Prieto J, Recio JA, Corrales FJ, Avila MA. Epidermal growth factor receptor signaling in hepatocellular carcinoma: inflammatory activation and a new intracellular regulatory mechanism. Dig Dis. 2012;30:524–531. doi: 10.1159/000341705. [DOI] [PubMed] [Google Scholar]
- 47.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, Reich M, Chan JA, Glickman JN, Ikeda K, Hashimoto M, Watanabe G, Daidone MG, Roayaie S, Schwartz M, Thung S, Salvesen HB, Gabriel S, Mazzaferro V, Bruix J, Friedman SL, Kumada H, Llovet JM, Golub TR. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoshida Y, Villanueva A, Sangiovanni A, Sole M, Hur C, Andersson KL, Chung RT, Gould J, Kojima K, Gupta S, Taylor B, Crenshaw A, Gabriel S, Minguez B, Iavarone M, Friedman SL, Colombo M, Llovet JM, Golub TR. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology. 2013;144:1024–1030. doi: 10.1053/j.gastro.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aucejo F, Kim R, Zein N, Quintini C, Uso TD, Lopez R, Eghtesad B, Fung J, Miller C, Yerian L. Vascular endothelial growth factor receptor 2 expression in non-tumorous cirrhotic liver is higher when hepatoma is beyond Milan criteria. Liver Transpl. 2009;15:169–176. doi: 10.1002/lt.21678. [DOI] [PubMed] [Google Scholar]
- 50.Huang J, Zhang X, Tang Q, Zhang F, Li Y, Feng Z, Zhu J. Prognostic significance and potential therapeutic target of VEGFR2 in hepatocellular carcinoma. J Clin Pathol. 2011;64:343–348. doi: 10.1136/jcp.2010.085142. [DOI] [PubMed] [Google Scholar]
- 51.Patel SH, Kneuertz PJ, Delgado M, Kooby DA, Staley CA 3rd, El-Rayes BF, Kauh JS, Sarmiento JM, Hanish S, Cohen C, Farris AB 3rd, Maithel SK. Clinically relevant biomarkers to select patients for targeted inhibitor therapy after resection of hepatocellular carcinoma. Ann Surg Oncol. 2011;18:3384–3390. doi: 10.1245/s10434-011-1775-3. [DOI] [PubMed] [Google Scholar]
- 52.Zheng YB, Meng QW, Zhao W, Liu B, Huang JW, He X, Li Y, Hu BS, Lu LG. Prognostic value of serum vascular endothelial growth factor receptor 2 response in patients with hepatocellular carcinoma undergoing transarterial chemoembolization. Med Oncol. 2014;31:843. doi: 10.1007/s12032-014-0843-5. [DOI] [PubMed] [Google Scholar]
- 53.Meng QW, Li Y, Hu BS, Shao PJ, Zhan MX, Yu XY, Lu LG. [Prognostic significance of serum level of vascular endothelial growth factor receptor-2 in hepatocellular carcinoma patients after transcatheter arterial chemoembolization] . Zhonghua Yi Xue Za Zhi. 2013;93:341–344. [PubMed] [Google Scholar]
- 54.Machado MV, Janeiro A, Miltenberger-Miltenyi G, Cortez-Pinto H. Genetic polymorphisms of proangiogenic factors seem to favor hepatocellular carcinoma development in alcoholic cirrhosis. Eur J Gastroenterol Hepatol. 2014;26:438–443. doi: 10.1097/MEG.0000000000000044. [DOI] [PubMed] [Google Scholar]
- 55.Maeng CH, Yi JH, Lee J, Hong JY, Choi MK, Jung HA, Park JO, Park SH, Park YS, Kang WK, Lim HY. Effects of single nucleotide polymorphisms on treatment outcomes and toxicity in patients treated with sunitinib. Anticancer Res. 2013;33:4619–4626. [PubMed] [Google Scholar]
- 56.Scartozzi M, Faloppi L, Svegliati Baroni G, Loretelli C, Piscaglia F, Iavarone M, Toniutto P, Fava G, De Minicis S, Mandolesi A, Bianconi M, Giampieri R, Granito A, Facchetti F, Bitetto D, Marinelli S, Venerandi L, Vavassori S, Gemini S, D’Errico A, Colombo M, Bolondi L, Bearzi I, Benedetti A, Cascinu S. VEGF and VEGFR genotyping in the prediction of clinical outcome for HCC patients receiving sorafenib: the ALICE-1 study. Int J Cancer. 2014;135:1247–1256. doi: 10.1002/ijc.28772. [DOI] [PubMed] [Google Scholar]
- 57.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 58.Pore N, Jiang Z, Gupta A, Cerniglia G, Kao GD, Maity A. EGFR tyrosine kinase inhibitors decrease VEGF expression by both hypoxia-inducible factor (HIF)-1-independent and HIF-1-dependent mechanisms. Cancer Res. 2006;66:3197–3204. doi: 10.1158/0008-5472.CAN-05-3090. [DOI] [PubMed] [Google Scholar]
- 59.Inoue K, Torimura T, Nakamura T, Iwamoto H, Masuda H, Abe M, Hashimoto O, Koga H, Ueno T, Yano H, Sata M. Vandetanib, an inhibitor of VEGF receptor-2 and EGF receptor, suppresses tumor development and improves prognosis of liver cancer in mice. Clin Cancer Res. 2012;18:3924–3933. doi: 10.1158/1078-0432.CCR-11-2041. [DOI] [PubMed] [Google Scholar]
- 60.Lang SA, Brecht I, Moser C, Obed A, Batt D, Schlitt HJ, Geissler EK, Stoeltzing O. Dual inhibition of Raf and VEGFR2 reduces growth and vascularization of hepatocellular carcinoma in an experimental model. Langenbecks Arch Surg. 2008;393:333–341. doi: 10.1007/s00423-008-0292-8. [DOI] [PubMed] [Google Scholar]
- 61.Zhu AX, Duda DG, Sahani DV, Jain RK. Development of sunitinib in hepatocellular carcinoma: rationale, early clinical experience, and correlative studies. Cancer J. 2009;15:263–268. doi: 10.1097/PPO.0b013e3181af5e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu W, Gu K, Yu Z, Yuan D, He M, Ma N, Lai S, Zhao J, Ren Z, Zhang X, Shao C, Jiang GL. Sorafenib potentiates irradiation effect in hepatocellular carcinoma in vitro and in vivo. Cancer Lett. 2013;329:109–117. doi: 10.1016/j.canlet.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 63.Huynh H, Ong R, Soo KC. Foretinib demonstrates anti-tumor activity and improves overall survival in preclinical models of hepatocellular carcinoma. Angiogenesis. 2012;15:59–70. doi: 10.1007/s10456-011-9243-z. [DOI] [PubMed] [Google Scholar]


