Abstract
Over-activation of SUMOylation is correlated with poor prognosis in multiple myeloma (MM), with the mechanism unclear. Wnt signaling is one of the aberrantly regulated pathways related to cancer tumorigenesis and progression. Whether SUMOylation is involved in regulating the activity of Wnt/β-catenin pathway, however, has not been reported in MM. Here we found that the TOPflash reporter activity and the expression of Wnt/β-catenin target genes can be down-regulated after interference with SUMOylation through SUMO-1 small interfering RNA (siRNA). SUMOylation inhibition down-regulated β-catenin at protein level via promotion of ubiquitin-proteasomal mediated degradation. Furthermore, over-expression of β-catenin rescued Wnt/β-catenin pathway activity and partially prevented increased apoptosis and growth inhibition induced by SUMOylation inhibition, indicating that β-catenin was responsible for the observed effect on Wnt/β-catenin pathway. To gain a clearer view, we exploited the inter-protein interactions of β-catenin and SUMO-1 in myeloma cell lines. Immunoprecipitation and immunofluorescence assay proved that β-catenin is subjected to SUMOylation in vivo, which may, at least partially explain the impact of SUMOylation inhibition on β-catenin. The association of SUMO-1 and β-catenin was confirmed in myeloma patient samples. Taken together, our data proved that SUMOylation inhibition down-regulates Wnt/β-catenin pathway by promoting the ubiquitin-proteasomal mediated degradation of β-catenin. SUMOylation of β-catenin is part of the mechanisms involved in the dysregulated proliferation of myeloma cells.
Keywords: Multiple myeloma, SUMO-1, SUMOylation, Wnt/β-catenin
Introduction
As the second most common hematological malignancy, multiple myeloma (MM) is characterized with clonal proliferation of abnormal plasma cells and elevated serum and urine monoclonal paraprotein [1]. Despite of various treatment options, MM remains incurable with a median survival of only 6 years [2]. Patient survival is highly heterogeneous, highlighting the necessity to identify critical targets involved in the mechanism of neoplasia and disease progression of MM [3].
SUMOylation is a vital post-translational modification characterized by covalent and reversible binding of SUMO (small ubiquitin-like modifier) proteins to target proteins [4]. Expression of SUMO proteins is markedly increased in processes associated with carcinogenesis, such as cell growth, differentiation, senescence, oxidative stress as well as apoptosis [5]. The SUMOylation pathway is aberrantly activated in MM, associated with adverse clinical results, which could be a specific target for management of MM, with prognostic and therapeutic value [3].
Previous studies indicated that myeloma cells exhibit decreased survival after SUMOylation inhibition [3], but the mechanism remained to be elucidated. Among the aberrantly regulated pathways, Wnt signaling plays important roles in the survival of myeloma cells. Derksen et al.[6] reported that stimulation of Wnt/β-catenin signaling increases cell proliferation, while blocking Wnt/β-catenin signaling would interfere with the growth of MM cells, providing a critical role of Wnt/β-catenin pathway in myeloma survival. β-catenin is the downstream effector of the Wnt/β-catenin signaling pathway, regulating genes including cyclinD1, c-myc, survivin [7-9]. It contributes to enhanced carcinogenetic, proliferative and metastatic properties of MM [10], and considered to be the central component of the Wnt/β-catenin pathway [11]. Here our data showed that Wnt/β-catenin pathway participates in the mechanism of SUMOylation inhibition in myeloma cells via accelerated degradation of β-catenin; SUMOylation of β-catenin is part of the mechanisms involved in the dysregulated proliferation of myeloma cells.
Materials and methods
Cell lines and culture conditions
The human multiple myeloma cell lines RPMI-8226 and NCI-H929 were maintained in RPMI-1640 medium with 10% (v/v) fetal bovine serum (FBS) (Hyclone), 100 units/ml penicillin and 100 μg/ml streptomycin in a humidified atmosphere of 95% air and 5% carbon dioxide (CO2) at 37°C.
Normal and malignant plasma cells
MM bone marrow samples were isolated from newly diagnosed patients by undergoing routine diagnostic aspirations, with informed consent. Normal, human adult bone marrow was from healthy people who underwent physical examination, also with informed consent. Cells were separated by Ficoll density gradient centrifugation and then incubated with anti-CD138 antibodies coupled with magnetic beads and positively selected on a magnetic affinity column (Miltenyi Biotech, Auburn, CA). The amount of CD138-positive plasma cells was determined using light microscopy.
Immunoprecipitation assay
Cells were washed once with ice-cold PBS, lysed in non-denaturing buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2.5 mM sodium pyrophosphate, 1 mM β-sodium glycerophosphate, 1% TritonX-100, 1 mM sodium vanadate, 1 mM PMSF) containing 1 tablet/10 mL of protease inhibitor cocktail (Roche, Basel, Switzerland) and 50 mM NEM (Sigma, St. Louis, MO). The cells were rotated at 4°C for 20 min, centrifuged at 12,000 rpm for 15 minutes. The supernatant was removed, incubating with the appropriate antibody (IgG, anti-β-catenin polyclonal antibody) and protein-A-Sepharose. The mixture was rotated at 4°C for 16 h, then washed with ice cold PBS three times and 30 μL SDS-loading buffer was added to the pellet. The sample was then boiled for 10 minutes, centrifuged for 15 minutes at 12,000 rpm. Then 20 μL was loaded onto Tricine-SDS-PAGE gels, and probed with anti-β-catenin polyclonal antibody (rabbit, CST). The remaining 10 μL was loaded on another gel, probing with anti-SUMO-1 antibody (rabbit, Abcam).
Total RNA isolation and real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen), according to manufacturer’s instructions. RNA concentration and integrity were then checked with a UV/VIS spectrophotometer NanoDrop 1000 (Thermo Scientific, Wilmington, DE, USA). Complementary DNA was generated by M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). Then the complementary DNA was analyzed with quantitative PCR using SYBR® Premix Ex TaqTM (TaKaRa, Dalian, China). The samples analyzed were in duplicate and normalized to actin expression using the 2-ΔΔCT method.
Western blot
Cell lysates were subjected to electrophoresis through Tricine-SDS-PAGE and blotted with antibodies for SUMO-1 (rabbit, Abcam), β-catenin (rabbit antibody for western blot, from BD Biosciences; mouse antibody for immunofluorescence staining, from Cell Signaling Technology) and β-actin (Upstate, Billerica, MA, USA). The immunoblots were visualized with the Enhanced Chemiluminescence (ECL) Western Blotting System (GE Healthcare Life Sciences, Pittsburgh, PA, USA).
RNA interference, plasmids and transfection
Three short interfering RNA (siRNA) duplexes targeting human SUMO-1, 1# (CTGGGAATGGAGGAAGAAG), 2# (CAATGAATTCACTCAGGTT), 3# (GGACAGGATAGCAGTGAGA) were provided by RiboBio Co., Ltd (Guangzhou, China). Negative control siRNA from RiboBio was confirmed not to interact with any mRNA sequence else. Another negative control siRNA (with cy3) was used to determine transfection efficiency. Twenty-four hours after transfection, transfection efficiency was determined by the percentage of cells with cy3 (red) under fluorescence microscope. The mutant β-catenin plasmid (pcDNA3-S33Y, 19286#) and wild-type β-catenin (pcDNA3, 97380#) plasmid were purchased from Addgene. Standard recombinant DNA techniques were used to construct the following plasmids: pXF3H-His/SUMO-1 and pSG5-Myc/SENP1. The transfection of siRNA duplexes and plasmids was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Immunofluorescence staining
Myeloma cells were cultured on coverslips in 6-well plates for 48 hours. Then supernant was removed and cells were washed softly with ice-cold PBS, fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100 for 5 minutes, and then washed with cold PBS. Then fixed cells were blocked with PBS containing 5% BSA for 1 hour at room temperature. The cells were then incubated with the indicated primary antibodies overnight followed by incubation with fluorescence-labeled secondary antibody. The cells were finally stained with Hoechst (1 μg/ml) and then examined under a confocal microscope.
MTS assay
To assess cell viability following different treatment, MTS assay was performed. 10 × 103 myeloma cells were plated in each well of a 96-well plate. Following treatment with various reagents, 20 ml of MTS (Promega) solution was added to each well and the cells were incubated for 2 hours in a humidified incubator at 37°C with 5% CO2. The absorbance at 490/690 nm was measured by FlexStation 3 (Molecular Devices). MTS assays were performed in in triplicate and repeated at least twice.
Flow cytometry and luciferase reporter assay
The Flow cytometry assay was performed using the Annexin V-FITC-PI kit (Kaiji, China). Cells were collected, washed with PBS for twice, and thus stained according to the producer’s manual. Flow cytometer (BD FACS calibur 4, America) was performed immediately. Each groups detected in triplicate experiments and mean were calculated. The Luciferase Reporter Assay was performed as previously described [10]. Briefly, MM cells (4.0 × 105) were transfected with TOPflash or FOPflash Wnt/β-catenin reporter plasmids containing wild-type or mutant TCF DNA binding sites by using Fugene (Roche) according to the manufacturer’s protocol. Cells were also cotransfected with the hRL-Null Renilla plasmid. We have the results normalized to the activity of renilla. Experiments were performed in triplicate and repeated twice.
Statistical analysis
Each experiment was performed for at least two times. The values shown are the mean ± SD. Student’s t-test (two-sided) was used to test for significant differences between groups. * And ** indicates p values of < 0.05 and 0.01, respectively.
Results
SUMOylation inhibition represses the Wnt/β-catenin pathway
First, we investigated the role of SUMOylation in the regulation of Wnt/β-catenin signaling. SUMO-1 is evolutionally conserved from yeast to humans and has been extensively studied in the SUMO family [12]. Besides, the vast majority of SUMO-1 is conjugated to proteins [13]. Then we tested the effect of SUMOylation inhibition by SUMO-1 siRNA. Negative control siRNA (Cy3) was used to evaluate transfection efficiency, which proved to be greater than 90% (Supplementary Figure 1). Three different SUMO-1 siRNAs were employed at a dose of 100 nM. The siRNA#3 effectively decreased the expression of SUMO-1 at mRNA level (Figure 1A, left). Whole cell lysates were probed with anti-SUMO-1 antibody after transfection. SUMO-1 was indicated by arrow and was about 15 kDa in size, while SUMOylation pattern was detected as conjugated proteins with SUMO-1 modification. We found that siRNA#3 effectively decreased the expression of SUMO-1 as well as SUMOyaltion pattern (Figure 1A, right), and it was selected for the following experiments.
Figure 1.
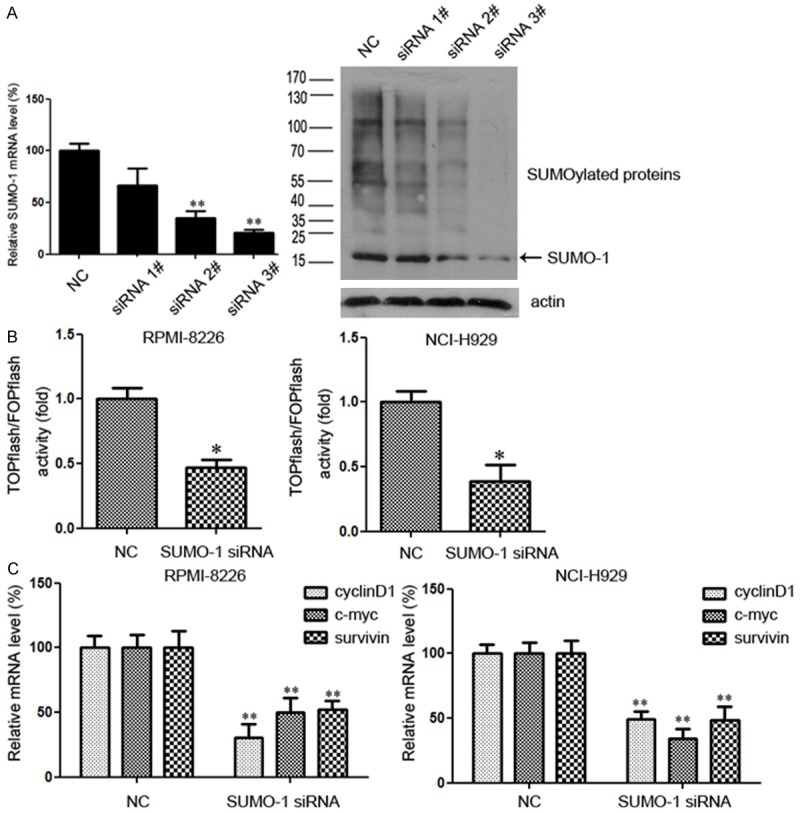
SUMOylation inhibition represses the Wnt/β-catenin pathway. A. For myeloma cells NCI-H929, the silencing efficiency with SUMO-1 siRNA was tested 48 hours after transfection. Total RNA was then extracted, and SUMO-1 mRNA level was determined by real time RT-PCR. Whole cell lysate was collected and lysed and immunoblotted with anti-SUMO-1 antibody. SUMO-1 was indicated by arrow and was about 15 kDa in size, while SUMOylation pattern was detected as conjugated proteins with SUMO-1 modification. B. Effects of SUMO-1 inhibition on TCF/β-catenin reporter activity. Myeloma cells were transfected with control siRNA or siRNA targeting SUMO-1. After 48 h incubation, cells were transfected with TOPflash reporter plasmid or FOPflash-negative control plasmid. Twenty-four hours after re-transfection, cells were harvested and luciferase activity was measured as described in Materials and Methods. All results were normalized for transfection efficiency using the hRL-Null Renilla plasmid. Fold induction corresponds to luciferase activity of positive TOPflash reporter over negative FOPflash reporter. Data represent mean ± S.D.; *p < 0.05 by Student’s t-test; Experiments were performed in triplicate and repeated twice. C. Myeloma cells were treated with SUMO-1 siRNA or negative control for 48 hours. Total RNA was then extracted, and the downstream genes of Wnt/β-catenin pathway, including c-myc, cyclinD1 and survivin, were determined by real time RT-PCR. (*p < 0.05, **p < 0.01 by Student’s t-test). The experiment was performed three times.
The Wnt/β-catenin pathway activity in myeloma cells was detected using the TOPflash reporter and checked with FOPflash activity as a control for background activity of TOPflash [14]. We transiently transfected myeloma cells with either TOPflash or FOPflash (negative control) plasmids after transfection with SUMO-1 siRNA or negative control, and measured the luciferase activity. Results showed that compared with negative control, knockdown of SUMO-1 significantly inhibited TOPflash reporter activity in both RPMI-8226 and NCI-H929 cell lines (Figure 1B). To further confirm the effect of SUMOylation inhibition on the regulation of Wnt/β-catenin pathway, the expression of Wnt/β-catenin target genes, c-myc, cyclinD1 and survivin, was analyzed by quantitative real time PCR. As shown in Figure 1C, all three target genes were significantly down-regulated following SUMOylation inhibition in both cell lines, which was consistent with the changes in reporter activity. Taken together, our results indicated that SUMOylation inhibition could repress the Wnt/β-catenin pathway in myeloma cells.
SUMOylation inhibition promotes the degradation of β-catenin via the ubiquitin-proteasomal pathway
The Wnt/β-catenin signaling could be regulated at multiple steps. As a central effector of Wnt/β-catenin pathway, β-catenin plays crucial role in MM tumor progression. Herein, we assessed the effect of SUMOylation inhibition on β-catenin. As shown in Figure 2A, interference with SUMO-1 had no perceptible effect on the mRNA level of β-catenin. However, β-catenin protein was down-regulated following interference with SUMOylation in RPMI-8226 and NCI-H929 myeloma cell lines (Figure 2B). In addition, the expression of β-catenin was in association with SUMO-1 protein level. When myeloma cells were transfected with mounting doses of SUMO-1 siRNA (30 nM, 50 nM, 100 nM), a lower level of β-catenin was observed (Figure 2C). Taken together, our results suggest that SUMOylation inhibition could down-regulate β-catenin, probably at post-transcriptional level.
Figure 2.
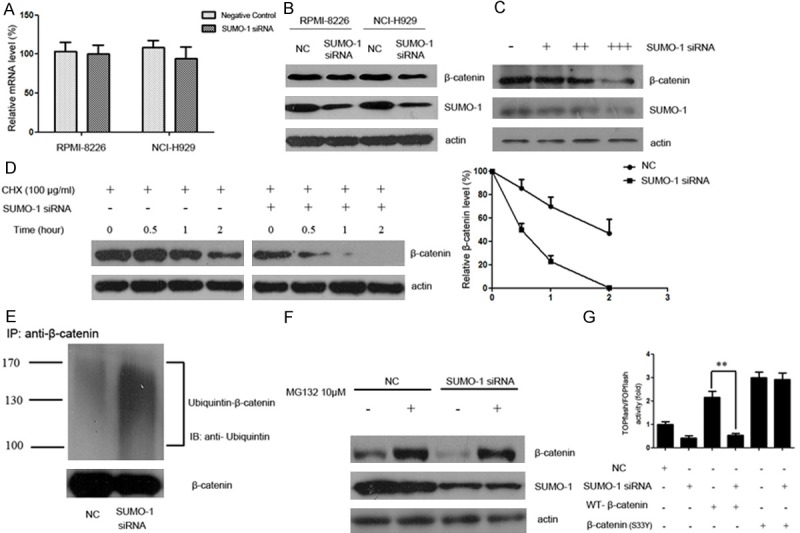
SUMOylation inhibition promotes the degradation of β-catenin via the ubiquitin-proteasomal pathway. A. Myeloma cells were treated with SUMO-1 siRNA or negative control for 48 hours. Total RNA was then extracted, and β-catenin mRNA level was determined by real time RT-PCR. B. Myeloma cells NCI-H929 and RPMI-8226 were transfected with control or siRNA targeting SUMO-1. After 48 h incubation, whole cell lysate was collected and lysed and immunoblotted with anti-SUMO-1 or anti-β-catenin antibody respectively. C. Myeloma cells NCI-H929 were treated with mounting doses of SUMO-1 siRNA (30 nM, 50 nM, 100 nM) for 48 h. Then whole cell lysate was collected and lysed and immunoblotted with anti-SUMO-1 or anti-β-catenin antibody respectively. D. Myeloma cells were transfected with control or siRNA targeting SUMO-1. After 48 hours, cells were treated with CHX (100 μg/ml) for indicated time periods (0, 0.5, 1, 2 hours). Cell lysate was subjected to western blot with anti-β-catenin antibody. The relative intensity of β-catenin proteins at the indicated time points was quantified using SmartViewsoftware. In the negative control group, the intensity was standardized to 100% of the control sample (CHX 0 h); while in the SUMO-1 siRNA group, it was standardized to 100% of the control sample in the SUMO-1 siRNA group (CHX 0 h). This result is a representative of three independent experiments. E. Myeloma cells NCI-H929 were transfected with SUMO-1 siRNA or negative control. After 48 hours, the cells were harvested under denaturing conditions and immunoprecipitated with anti-β-catenin antibody. The immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-ubiquitin and anti-β-catenin antibodies respectively. F. Myeloma cells NCI-H929 were transfected with SUMO-1 siRNA or negative control. 48 hours after transfection, cells were treated with MG132 (10 μM) for 5 hours. The protein extracts were then analyzed by western blot using antibodies against β-catenin, SUMO-1 or actin as loading control. G. Myeloma cells were transfected with SUMO-1 siRNA together with β-catenin (WT) or β-catenin (S33Y) plasmids. After 48 h incubation, cells were transfected with TOPflash reporter plasmid or FOPflash-negative control plasmid. Twenty-four hours after retransfection, cells were harvested and luciferase activity was measured as described in Materials and Methods. All results were normalized for transfection efficiency using the hRL-Null Renilla plasmid. Fold induction corresponds to luciferase activity of positive TOPflash reporter over negative FOPflash reporter. (Data represent mean ± S.D.; **p < 0.01 by Student’s t-test).
To determine whether SUMOylation inhibition affects the degradation of β-catenin, we transfected cells with SUMO-1 siRNA and measured β-catenin protein level in the presence of cycloheximide (CHX), an inhibitor of protein biosynthesis. Immunoblot analysis evidenced an increased degradation of β-catenin in SUMO-1 siRNA transfected cells, confirming that SUMOylation inhibition could promote β-catenin degradation (Figure 2D). Since proteasome plays a major role in the degradation of β-catenin, we set out to examine whether the decrease in β-catenin protein level after SUMO-1 inhibition was mediated by the ubiquitin-proteasomal degradation system. Myeloma cells transfected with SUMO-1 siRNA or negative control were immunoprecipitated with anti-β-catenin antibody. The immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-ubiquitin and anti-β-catenin antibodies respectively. Results showed that SUMO-1 knock-down was associated with enhanced β-catenin ubiquitination, indicating that protein degradation was promoted during the attachment of poly-ubiquitin to the β-catenin protein (Figure 2E).
We further compared the changes of β-catenin in myeloma cell line NCI-H929 transfected with SUMO-1 siRNA, with or without MG132, a specific proteasome inhibitor (Figure 2F). Forty eight hours after SUMO-1 siRNA transfection, myeloma cells were treated in the presence or absence of MG132 (10 μM) for 5 hours, and analyzed by western blot using anti-β-catenin antibody. In the group without MG132 treatment, β-catenin protein reduced after SUMO-1 siRNA transfection (lane 1 and 3). In the negative control group, β-catenin level increased after the addition of MG132 (lane 1 and 2). Similar results were also observed in myeloma cells transfected with SUMO-1 siRNA, in which the addition of MG132 protected β-catenin protein from decreasing (lane 3 and 4). Important to note, the addition of MG132 in the SUMO-1 siRNA transfected group restored the β-catenin level to that in the negative control group (lane 2 and 4), indicating that the ubiquitin-proteasomal mediated degradation plays a major role in the decreased level of β-catenin after SUMOylation inhibition.
As previously reported, Ser33 is an important site for GSK-3β mediated degradation of β-catenin. To determine whether SUMOylation inhibition related β-catenin degradation was dependent on Ser33, we cotransfected SUMO-1 siRNA together with wild-type or mutant β-catenin (S33Y), then measured the TOPflash reporter activity as mentioned above (Figure 2G). We found that SUMO-1 siRNA inhibited the wild-type β-catenin activation of TOPFflash luciferase activity, while had no effect on mutant β-catenin activation, indicating that the effect of SUMOylation on Wnt signaling is dependent on the phosphorylation of the Ser33.
Overexpression of β-catenin rescued Wnt/β-catenin pathway activity and partially prevented increased apoptosis and growth inhibition induced by SUMOylation inhibition
We next set out to test whether the effect of SUMOylation inhibition was mediated by β-catenin down-regulation. Firstly, changes in Wnt/β-catenin downstream effectors (c-myc, cyclinD1, survivin) were compared in cells that were subjected to the knockdown of SUMO-1, overexpression of β-catenin (S33Y), or both. Consistent with previous findings, SUMO-1 siRNA treatment decreased the protein level of β-catenin as well as down-stream effector proteins. Overexpression of β-catenin (S33Y) rescued myeloma cells from the loss of proteins (Figure 3A).
Figure 3.
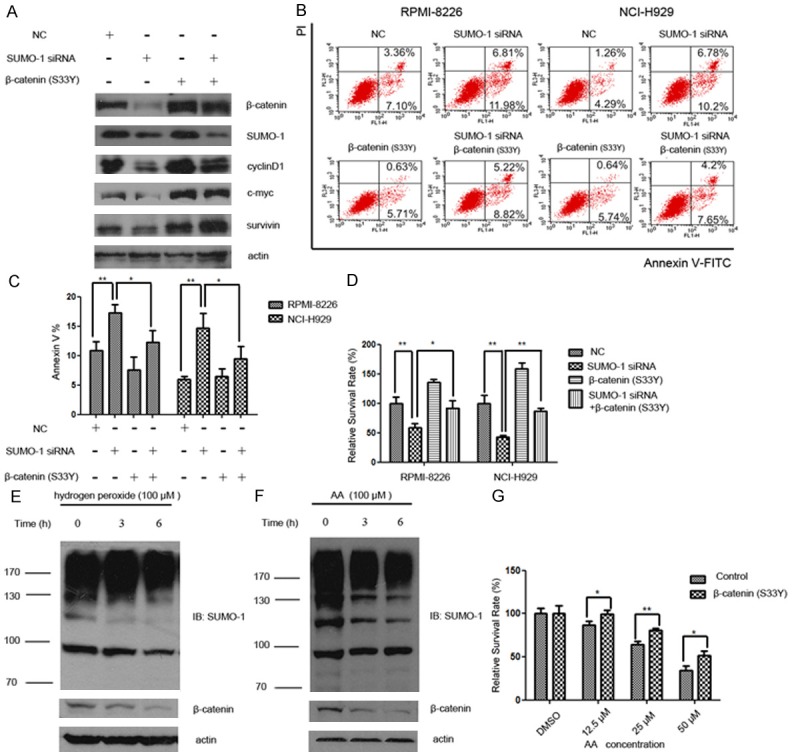
Overexpression of β-catenin rescued Wnt/β-catenin pathway activity and partially prevented increased apoptosis and growth inhibition induced by SUMOylation inhibition. A. Myeloma cells NCI-H929 were treated with SUMO-1 siRNA and/or β-catenin (S33Y) plasmid for 48 hours. Protein samples were subjected to western blot analysis for the Wnt/β-catenin downstream effector proteins (c-myc, cyclinD1, survivin) with indicated antibodies. B. The distribution of apoptotic cells following treatment of myeloma cells with SUMO-1 siRNA, β-catenin (S33Y) expression vector, or both were evaluated using annexin V and propidium iodide (PI) double staining. Early apoptotic cells (Annexin-V+ and PI-) were displayed in the lower right quadrant and late apoptotic cells (Annexin-V+ and PI+) were shown in the upper right quadrant. C. The percentage of all the apoptotic cells (including Annexin-V+ PI- and Annexin-V+ PI+ cells) were indicated by Annexin-V+ cells shown as mean ± SD from three independent experiments. D. The growth inhibition of myeloma cells following different treatments was shown by the percentage of relative survival rate (OD value of sample/OD value of control in each group×100%) (*p < 0.05, **p < 0.01 by Student’s t-test). These experiments were performed three times. E. Myeloma cells (NCI-H929) were exposed to hydrogen peroxide (100 μM) for indicated time intervals (0, 3 h, 6 h). Cell lysate was subjected to western blot with anti-β-catenin and anti-SUMO-1 antibody, respectively. F. Myeloma cells (NCI-H929) were exposed to anacardic acid (AA) (100 μM) for indicated time intervals (0, 3 h, 6 h). Cell lysate was subjected to western blot with anti-β-catenin and anti-SUMO-1 antibody, respectively. G. 48 hours after transfection of β-catenin plasmid, cells were exposed to different concentrations of anacardic acid (AA) (12.5 μM, 25 μM, 50 μM). 48 hours later, cell viability was evaluated using MTS assay as described under “Materials and methods”. These experiments were performed three times. Relative survival rate was calculated as “OD value of sample / OD value of DMSO sample in each transfection group×100%”. MTS assays were performed in in triplicate and repeated at least twice (*p < 0.05, **p < 0.01 by Student’s t-test).
The distribution of apoptotic cells following treatment of myeloma cells with SUMO-1 siRNA, β-catenin expression vector, or both were evaluated using annexin V and propidium iodide (PI) double staining (Figure 3B). The annexin V positive cells were recognized as apoptotic cells, which represented both early and late apoptotic cells. We found that following SUMO-1 silencing with siRNA, the proportion of apoptotic cells increased, though not quite obviously, from 10% to 18% in RPMI-8226, and 5% to 17% in NCI-H929, respectively (Figure 3C). Moreover, β-catenin (S33Y) overexpression partially prevented the cell death induced by SUMO-1 inhibition. Then we measure the cell viability by MTS assay. Consistent with apoptotic results, the cell viability was decreased after SUMO-1 knock-down, which could be partially rescued by overexpression of β-catenin (S33Y) (Figure 3D). It is important to note that overexpression of β-catenin (S33Y) did not fully rescue cells from dying or growth inhibition, suggesting that there might be other pathways also involved in the network regulation of SUMO-1 mediated cell survival.
We further exposed myeloma cells to hydrogen peroxide and anacardic acid (AA), the two known inhibitors of SUMOylation (Figure 3E and 3F). After treatment with hydrogen peroxide or anacardic acid for indicated times, the SUMOylated proteins as well as β-catenin were decreased in a dose dependent manner. To confirm that the cell toxicity of anacardic acid was related to decreased β-catenin protein level, β-catenin (S33Y) was overexpressed in myeloma cells that were then treated with anacardic acid. As shown in Figure 3G, the growth inhibition induced by anacardic acid was reduced from 13% to 1% at 12.5 μM, 35% to 19% at 25 μM, and 66% to 49% at 50 μM after transfection of β-catenin (S33Y) respectively, indicating that β-catenin could protect myeloma cell from the toxicity of SUMOylation inhibitor.
Endogenous β-catenin is modified by SUMO-1 in myeloma cells
Since above results revealed a relationship between SUMOylation and β-catenin, we next assessed SUMO-1 interaction with β-catenin in myeloma cells. Cell lysates from RPMI-8226 and NCI-H929, two myeloma cell lines, were examined by immunoprecipitation (IP) using anti-β-catenin antibody. The IP lysates were further analyzed by western blot with anti-β-catenin or anti-SUMO-1 antibody, which showed the physical combination of SUMO-1 and endogenous β-catenin (Figure 4A).
Figure 4.
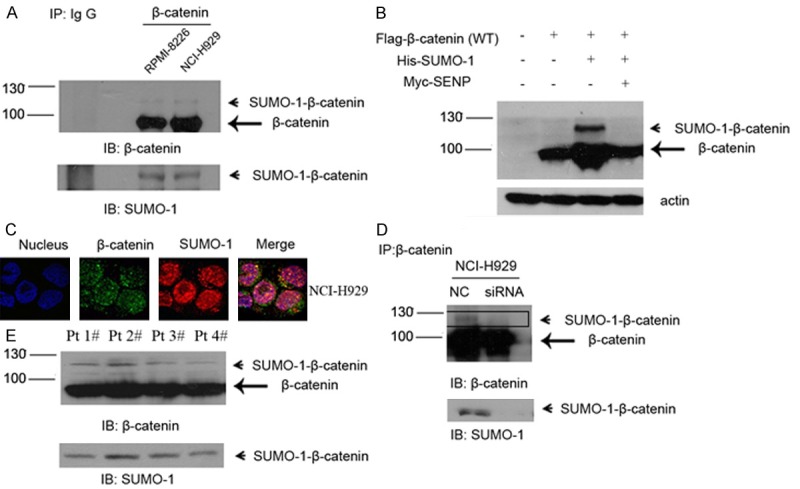
Endogenous β-catenin is modified by SUMO-1 in myeloma cells. A. Myeloma cells RPMI-8226 and NCI-H929 were harvested under denaturing conditions as described under “Experimental Procedures” and immunoprecipitated with anti-β–catenin rabbit antibody or IgG as a control. The immunoprecipitates (IP) were resolved by SDS-PAGE and immunoblotted with anti-β-catenin and anti-SUMO-1 antibodies respectively. SUMOylated β-catenin and non-SUMOylated β-catenin were indicated by arrow head and arrow, respectively. B. NCI-H929 was transfected with Flag-tagged β-catenin or together with His-tagged SUMO-1 or Myc-tagged SENP1, after 48 hours, cells were harvested and then subjected to western blot with anti-Flag antibody. C. Endogenous β-catenin co-localized with SUMO-1. Myeloma cells NCI-H929 were fixed and stained with anti-β-catenin mouse antibody (green) and anti-SUMO-1 rabbit antibody (red). DNA was stained with DAPI. The images were taken by confocal microscopy as described under “Experimental Procedures”. D. After transfection of negative control and/or SUMO-1 siRNA for 48 hours, whole cell lysates were immunoprecipitated with anti-β-catenin antibody. The immunoprecipitates (IP) were resolved by SDS-PAGE and immunoblotted with anti-β-catenin and anti-SUMO-1 respectively. SUMOylated β-catenin was indicated by arrow head. E. Bone marrow samples were isolated from myeloma patients. Total lysate was prepared from 5 × 106 cells in each sample and immunoprecipitated with anti-β–catenin rabbit antibody as mentioned above. The immunoprecipitates (IP) were resolved by SDS-PAGE and immunoblotted with anti-β-catenin and anti-SUMO-1 antibodies respectively. SUMOylated β-catenin and non-SUMOylated β-catenin were indicated by arrow head and arrow, respectively.
To confirm that was the SUMOylated β-catenin instead of coincidence, β-catenin expression vector and SUMO-1 expression vector were also transfected into myeloma cells (Figure 4B). High-molecular-weight β-catenin was detected (lane 3). Besides, the SUMOylated β-catenin disappeared after we co-transfected it with SUMO-specific protease SENP1 (lane 4). Moreover, endogenous combination of β-catenin and SUMO-1 was also confirmed by confocal microscopy analysis (Figure 4C). As shown in Figure 4C, β-catenin was detected both in the cytoplasm and the nucleus of myeloma cells, mainly merged with SUMO-1 in the nucleus. Next we transfected myeloma cells with either SUMO-1 siRNA or negative control (NC). A decreased SUMOylation pattern of β-catenin was observed in the SUMO-1 siRNA treated group by means of IP (Figure 4D). These results suggest that endogenous β-catenin is a SUMOylation substrate and can be modified by SUMO-1 in MM cells.
Finally, we tested the expression of SUMOylated β-catenin in four MM patient samples (Figure 4E). Lysates from CD138-positive plasma cells were immunoprecipitated with anti-β-catenin antibody and probed with a polyclonal antibody specific to either β-catenin or SUMO-1. An in vivo association of β-catenin and SUMO-1 was observed in all four myeloma patients.
Discussion
Both SUMOylation and Wnt/β-catenin pathway are aberrantly activated in MM, and we are interested in the effect of SUMOylation on the Wnt/β-catenin pathway activity. Here our results showed that SUMOylation inhibition through SUMO-1 siRNA down-regulates Wnt/β-catenin pathway by accelerated degradation of β-catenin via the ubiquitin-proteasomal system. Besides, β-catenin is also a target of SUMOylation both in myeloma cell lines and primary myeloma cells, providing a new prospect for the post-translational research for β-catenin.
As one of the most important post-translational modifications, SUMOylation plays an important role in the development of various cancers [6,7]. In mammals, there are four different isoforms of SUMO protein, termed SUMO-1, -2, -3 and -4. They get attached to target proteins via the mediation of three factors, namely E1-activating enzyme (SAE1/SAE2), E2-conjugating enzyme (UBC9), and E3-ligating enzyme (PIAS, Pc2, and RANBP2) [8]. The covalently conjugated SUMO could be cleaved by a family of proteins named SENP (sentrin-specific protease) [9]. Expression of SUMO proteins is markedly increased in processes associated with carcinogenesis, such as cell growth, differentiation, senescence, oxidative stress as well as apoptosis [10]. A number of intracellular signaling molecules have been identified the targets of SUMOylation, such as IκBα, p53, and eIF4F, which are all correlated with carcinogenesis. Driscoll JJ et al. [3] recently demonstrated that the SUMOylation pathway is remarkably activated in myeloma patients compared to normal plasma cells. Activation of SUMOylation pathway conferred multiple properties on tumorigenesis, while destruction of the SUMOylation pathway negatively affected myeloma cell proliferation and adhesion to bone marrow stromal cells. SUMOylation inhibition is emerging as a novel promising therapeutic target in MM treatment. However, the precise mechanisms have not been well defined. In this study, we found that SUMOylation inhibition induced increased apoptosis as well as growth inhibition in myeloma cells, together with down-regulated Wnt/β-catenin pathway, which can be partially reversed by the salvage Wnt/β-catenin signaling activation. The Wnt pathway is probably involved in the mechanism of SUMOylation in MM.
In addition to the previous reported phosphorylation, ubiquitination and acetylation [15,16], we proved that β-catenin is a potential substrate of SUMOyaltion in this study. SUMOylation occurs in different subcellular locations, such as cytoplasmic compartments, plasma membrane, though mainly in the nucleus [17,18]. In most myeloma cell lines, Wnt/β-catenin pathway was activated, with β-catenin distributed in both nucleus and cytoplasm. Besides, β-catenin undergoes free nucleo-cytoplasmic shuttle, making it possible that the SUMOylation of β-catenin occurs in both nucleus and cytoplasm, which is consistent with our immunofluorescence results. As far as we know, this is the first report on the SUMOylation of β-catenin in vivo.
Our data showed that the protein level of β-catenin decreased after SUMO-1 knock-down, accompanied with enhanced cell apoptosis. According to previous reports, apoptosis could induce cleavage of β-catenin by caspase-3 [19,20]. To find out whether the loss of β-catenin following SUMO-1 silencing was due to apoptosis, we further measured the effect of caspase-3 on β-catenin protein level during apoptosis, by using the caspase inhibitor z-VAD-fmk (Supplementary Figure 2). Consistent with previous study, z-VAD-fmk completely abrogated caspase-3 activation. Besides, it partially restored the protein level of β-catenin, indicating that caspase activation is involved in the down-regulation of β-catenin in the SUMO-1 interfering group. However, inhibition of caspase activity did not fully restore β-catenin protein level, suggesting that there are also other mechanisms involved. As previous reported, β-catenin is degraded mainly through the ubiquitin-proteasome system [21], while SUMO competes with ubiquitin for protein stabilization [22]. Our data showed that SUMOylation inhibition promoted ubiquitination and subsequent degradation of β-catenin, while MG-132, the proteasome inhibitor reversed the process. Moreover, in the SUMO-1 siRNA treated group, overexpression of β-catenin with mutation on phosphorylation site which is required for subsequent proteasome degradation restored the protein level as well as transcriptional activity of β-catenin (as showed in Figure 2). We may infer that decreased level of β-catenin SUMOylation promoted β-catenin phosphorylation and subsequent degradation via the ubiquitin-proteasomal pathway, and participates, at least partially in the regulation of the Wnt pathway. Since SUMO-1 plays a global role in living cells, and β-catenin is a target of many important molecules; it is possible that there may be other factors lead to the decrease of β-catenin protein, such as the Axin [23], adenomatous polyposis coli (APC) tumor suppressor protein [24], ubiquitin ligases SCFβ-Trcp [25] and SIAHs [26], which needs further study in the future.
Conjugation of SUMO to substrates generally occurs at a lysine residue within the classical consensus site ψwKxE/D (ψ represents a large hydrophobic residue, and x is any amino acid) [27]. However, SUMO substrates can also be modified on a lysine residue outside consensus motifs as it has been shown in recent studies where approximately 26% of confirmed SUMOylation sites contain a non-consensus motif [28]. In order to find a SUMOylation site within β-catenin, we used SUMOplot™ SUMO acceptor site search program (http://www.abgent.com/tools/sumoplot), a useful tool for predicting potential SUMOylation sites. K258 was identified as the possible SUMOylation site. We replaced K258 lysine with arginine in β-catenin plasmid; however, overexpressed β-catenin K258R could still be SUMOylated in vivo, indicating that K258 is not the key binding site for SUMOylation (data not show). Our further study is still on going for checking other potential SUMOylation sites, especially those outside the consensus motifs. Identification of the potential sites will provide us with a clear view of the biological effect of SUMOylation on β-catenin.
It remains unclear and controversial how canonical Wnt/β-catenin pathway is involved in the dysregulation of MM growth. Derksen et al. [6] reported that stimulation of Wnt/β-catenin signaling with Wnt3α or overexpression of β-catenin could increase cell proliferation, while blocking Wnt/β-catenin signaling would interfere with the growth of MM cells, providing a vital role of Wnt/β-catenin pathway in the survival of myeloma cells. Therapeutic strategy targeting the β-catenin/TCF transcriptional complex has also been used in the treatment of multiple myeloma [10]. On the contrary, some other investigators found that the increased β-catenin in response to Wnt3a or LiCl has no impact on myeloma cells proliferation [29,30]. Different results may due to the use of different cell lines, varied experiment designs and type of cancer cells. Further studies are still needed to make a final conclusion in vivo in MM patients. Our study proved that the activation of Wnt/β-catenin pathway was associated with increased survival in myeloma cell lines RPMI-8226 and NCI-H929, and SUMOylation may regulate MM survival by the involvement of Wnt/β-catenin signaling. Anti-apoptosis effect was also observed, though not dramatically.
Previous studies reported that PIASy (an E3 ligase) and Axam (a deSUMOylation enzyme) participate in the Wnt signaling pathway. PIASy SUMOylated Lef-1, and inhibits its transcriptional activity [29,30]. Tcf-4 is also a target of SUMOylation, while SUMOylation of Tcf-4 enhances its transcriptional activity [29,30]. Axam is recognized as an Axin-binding protein which regulates the Wnt signaling pathway negatively by inducing the degradation of β-catenin [29,30]. The above informations suggest a complicated interaction of SUMOyaltion and the componets of the Wnt pathway. Our results provided direct evidence for SUMOylation of β-catenin in myeloma cells, as well as the effect of SUMOylation inhibition on the activity of the Wnt pathway. More detailed work is needed to clarify the mechanisms of SUMOylation in regulating β-catenin as well as its association with other components in the Wnt pathway in the future.
It is interesting to explore the crosstalk of SUMOylation and Wnt/β-catenin signaling, which may help to better understanding the two important signaling pathways in vivo, reinforcing the clinical potential of therapies targeting SUMOylation pathway in MM.
Acknowledgements
We thank Jia Li of The National Center for Drug Screening for kindly providing the lab resources and critical revisions of the manuscript. We thank Yubo Zhou of The National Center for Drug Screening, Ernesto Diaz-Flores and Lasater Elisabeth of UCSF for the critical revisions of the manuscript. This work was supported in part by grants from the National Natural Science Foundation of China (Grant No. 81172248), Shanghai Committee of Foundation for Outstanding Leaders in Science (11XD1406600), and Shanghai Leading Scientists Foundation.
Disclosure of conflict of interest
The authors have no conflict of interests for this manuscript.
Supporting Information
References
- 1.Kyle RA, Rajkumar SV. Multiple myeloma. N Engl J Med. 2004;351:1860–1873. doi: 10.1056/NEJMra041875. [DOI] [PubMed] [Google Scholar]
- 2.Reece DE. Management of multiple myeloma: the changing landscape. Blood Rev. 2007;21:301–314. doi: 10.1016/j.blre.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR, Barlogie B, Tai YT, Anderson KC, Shaughnessy JD Jr, Annunziata CM, Munshi NC. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood. 2010;115:2827–2834. doi: 10.1182/blood-2009-03-211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 5.Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett. 2012;316:113–125. doi: 10.1016/j.canlet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Derksen PW, Tjin E, Meijer HP, Klok MD, MacGillavry HD, van Oers MH, Lokhorst HM, Bloem AC, Clevers H, Nusse R, van der Neut R, Spaargaren M, Pals ST. Illegitimate WNT signaling promotes proliferation of multiple myeloma cells. Proc Natl Acad Sci U S A. 2004;101:6122–6127. doi: 10.1073/pnas.0305855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 8.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 9.Lepourcelet M, Chen YN, France DS, Wang H, Crews P, Petersen F, Bruseo C, Wood AW, Shivdasani RA. Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell. 2004;5:91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 10.Sukhdeo K, Mani M, Zhang Y, Dutta J, Yasui H, Rooney MD, Carrasco DE, Zheng M, He H, Tai YT, Mitsiades C, Anderson KC, Carrasco DR. Targeting the beta-catenin/TCF transcriptional complex in the treatment of multiple myeloma. Proc Natl Acad Sci U S A. 2007;104:7516–7521. doi: 10.1073/pnas.0610299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willert K, Nusse R. Beta-catenin: a key mediator of Wnt signaling. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 12.Barry J, Lock RB. Small ubiquitin-related modifier-1: Wrestling with protein regulation. Int J Biochem Cell Biol. 2011;43:37–40. doi: 10.1016/j.biocel.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 14.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 15.Sukhdeo K, Mani M, Hideshima T, Takada K, Pena-Cruz V, Mendez G, Ito S, Anderson KC, Carrasco DR. Beta-catenin is dynamically stored and cleared in multiple myeloma by the proteasome-aggresome-autophagosome-lysosome pathway. Leukemia. 2012;26:1116–1119. doi: 10.1038/leu.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T. Acetylation of beta-catenin by CREB-binding protein (CBP) J Biol Chem. 2002;277:25562–25567. doi: 10.1074/jbc.M201196200. [DOI] [PubMed] [Google Scholar]
- 17.Rajan S, Plant LD, Rabin ML, Butler MH, Goldstein SA. Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell. 2005;121:37–47. doi: 10.1016/j.cell.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 18.Wilson VG, Rosas-Acosta G. Wrestling with SUMO in a new arena. Sci STKE. 2005;2005:pe32. doi: 10.1126/stke.2902005pe32. [DOI] [PubMed] [Google Scholar]
- 19.Steinhusen U, Badock V, Bauer A, Behrens J, Wittman-Liebold B, Dorken B, Bommert K. Apoptosis-induced cleavage of beta-catenin by caspase-3 results in proteolytic fragments with reduced transactivation potential. J Biol Chem. 2000;275:16345–16353. doi: 10.1074/jbc.M001458200. [DOI] [PubMed] [Google Scholar]
- 20.Rice PL, Kelloff J, Sullivan H, Driggers LJ, Beard KS, Kuwada S, Piazza G, Ahnen DJ. Sulindac metabolites induce caspase- and proteasome-dependent degradation of beta-catenin protein in human colon cancer cells. Mol Cancer Ther. 2003;2:885–892. [PubMed] [Google Scholar]
- 21.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura T, Hamada F, Ishidate T, Anai K, Kawahara K, Toyoshima K, Akiyama T. Axin, an inhibitor of the Wnt signalling pathway, interacts with beta-catenin, GSK-3beta and APC and reduces the beta-catenin level. Genes Cells. 1998;3:395–403. doi: 10.1046/j.1365-2443.1998.00198.x. [DOI] [PubMed] [Google Scholar]
- 24.Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 25.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Stevens J, Rote CA, Yost HJ, Hu Y, Neufeld KL, White RL, Matsunami N. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol Cell. 2001;7:927–936. doi: 10.1016/s1097-2765(01)00241-6. [DOI] [PubMed] [Google Scholar]
- 27.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, He Y, Qiang B, Yuan J, Peng X, Pan XM. A novel method for high accuracy sumoylation site prediction from protein sequences. BMC Bioinformatics. 2008;9:8. doi: 10.1186/1471-2105-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiang YW, Endo Y, Rubin JS, Rudikoff S. Wnt signaling in B-cell neoplasia. Oncogene. 2003;22:1536–1545. doi: 10.1038/sj.onc.1206239. [DOI] [PubMed] [Google Scholar]
- 30.Qiang YW, Shaughnessy JD Jr, Yaccoby S. Wnt3a signaling within bone inhibits multiple myeloma bone disease and tumor growth. Blood. 2008;112:374–382. doi: 10.1182/blood-2007-10-120253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


