Abstract
Various signal transduction pathways seem to be involved in chemoresistance mechanism of glioblastomas (GBMs). miR-21 is an important oncogenic miRNA which modulates drug resistance of tumor cells. We analyzed the expression of 5 miRNAs, previously found to be dysregulated in high grade gliomas, in 9 pediatric (pGBM) and in 5 adult (aGBM) GBMs. miR-21 was over-expressed, with a significant difference between pGBMs and aGBMs represented by a 4 times lower degree of expression in the pediatric compared to the adult series (p = 0.001). Doxorubicin (Dox) seems to be an effective anti-glioma agent with high antitumor activity also against glioblastoma stem cells. We therefore evaluated the chemosensitivity to Dox in 3 GBM cell lines (A172, U87MG and T98G). Dox had a cytotoxic effect after 48 h of treatment in A172 and U87MG, while T98G cells were resistant. TUNEL assay verified that Dox induced apoptosis in A172 and U87MG but not in T98G. miR-21 showed a low basal expression in treated cells and was over-expressed in untreated cells. To validate the possible association of miR-21 with drug resistance of T98G cells, we transfected anti-miR-21 inhibitor into the cells. The expression level of miR-21 was significantly lower in T98G transfected cells (than in the parental control cells). Transfected cells showed a high apoptotic rate compared to control after Dox treatment by TUNEL assay, suggesting that combined Dox and miR-21 inhibitor therapy can sensitize GBM resistant cells to anthracyclines by enhancing apoptosis.
Keywords: miRNA, glioblastoma multiforme, expression analysis, CNS tumors, pediatric brain tumors
Introduction
Glioblastoma multiforme (GBM) is one of the most lethal forms of brain tumor, with 5-year survival rates ranging from 5% to 10%. Pediatric GBMs are often associated with distinct cytogenetic and molecular alterations, which differ from those observed in the more common adult counterparts. Relatively few molecular studies have been performed on pediatric GBM chemoresistance, with somewhat conflicting results [1-5].
MicroRNAs (miRNAs) are endogenous single-stranded RNA molecules that constitute a novel class of gene regulators, and are involved in the control of cell differentiation, proliferation, apoptosis, anti-viral defense and cancer. Recent studies have shown deregulation of miRNA expression in various tumor types [6-9], demonstrating also a fundamental role in tumor progression and invasion [10]. They also seem to modulate drug sensitivity/resistance of the tumor cells [11].
Alterations in miRNA expression levels are associated with a number of neural diseases [12-15] and brain tumors [16-18]. Recently, specific miRNA expression profiles have been identified in GBMs [17,18], but there are only limited data on the role of miRNA in pediatric GBM [19,20].
miR-21 is an important oncogenic miRNA that promotes cell invasion, by regulating multiple genes, including PTEN, RECK and MARCKS, in several types of cancers, such as glioma, ovarian epithelial carcinoma and prostate cancer [21-23].
miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and poor prognosis [24], and its dysregulation plays a critical role in Doxorubicin (Dox) resistance of breast cancer via targeting PTEN [25].
Therefore, investigation of mir-21 expression profile can be useful to determine whether it is involved in determining the chemotherapy response of brain tumors.
In the present study we investigated the expression pattern of a set of 5 miRNAs (miR-21, miR-7, miR-124, miR-137 and miR-128) that are specifically over- or under-expressed in high grade glioma cells [18,26] in a series of 9 pediatric GBMs (pGBMs) and 4 adult GBMs (aGBMs) and in three glioblastoma cell lines (U87MG, A172, T98G). Since miR-21 was found to be over-expressed, we investigated its potential role in the response to Dox treatment in an in vitro GBM model.
Materials and methods
Patients
All patients with GBM (WHO-grade IV) seen between 2008 and 2013 at the Meyer Children’s University Hospital in Florence were eligible for this study. Histological diagnosis and tumor grading were carried out based on 2007-World Health Organization (2007-WHO) criteria [27]. The study was approved by the institutional Ethical Committee. Informed consent was obtained from the parents or legal guardians in all cases. Nine patients were enrolled in the study. Their main clinical characteristics are summarized in Table 1. Diagnosis was confirmed by the review of the CNS national panel of pathologists. Median age at the time of diagnosis was 8 ± 4.6 years (range, 1-15 years). All had been treated with chemotherapy and/or radiotherapy according to current front-line therapeutic studies of the Associazione Italiana Ematologia Oncologia Pediatrica (AIEOP). All underwent surgery for resection of disease, which turned to be complete in 3 of 9 cases. The median follow up was 10 ± 6.1 months (range, 3-24 months).
Table 1.
Clinical characteristics of pediatric Glioblastoma Multiforme (pGBM)
| ID | Gender | Age at diagnosis (years) | Surgery | First-line Treatment | Response | FU (months) | Status |
|---|---|---|---|---|---|---|---|
| P1 | F | 12 | PTR | TMZ + RT | PR | 8 | DOD |
| P2 | F | 1 | GTR | HDCT - RT | CR | 14 | DOD |
| P3 | M | 9 | PTR | Vinorelbine + RT | PR | 12 | DOD |
| P4 | F | 1 | GTR | HDCT + RT | CR | 24 | DOD |
| P5 | M | 4 | PTR | HDCT - RT | PR | 3 | DOD |
| P6 | F | 8 | PTR | Vinorelbine + RT | PR | 19 | DOD |
| P7 | M | 15 | PTR | Vinorelbine + RT | PR | 8 | DOD |
| P8 | M | 12 | By | TMZ + RT | PR | 7 | DOD |
| P9 | F | 7 | PTR | TMZ + RT | PR | 10 | DOD |
GTR: gross total removal; PTR: partial total removal; By: biopsy; HDCT: high-dose chemotherapy; RT: radiotherapy; TMZ: temozolomide; CR: complete response; PR: partial response; FU: follow-up; AWD: alive with disease; DOD: dead of disease.
Five adult GBM samples were obtained by the Pathology Unit of the Careggi University Hospital in Florence.
Cell lines
Three human GBM cell lines, A172, U87MG and T98G, were employed in this study (American Type Culture Collection; ATCC). U87MG and T98G were grown in Eagle’s Minimum Essential Medium, while A172 was grown in Dulbecco’s Modified Eagle Medium. Each medium was supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. All cell lines were maintained in a humidified atmosphere of 5% CO2-95% air at 37°C. Cells from exponentially growing cultures were used for all experiments.
Drug
Doxorubicin (Adriblastina 50 mg) was obtained from commercial sources (Pfizer) and tested at various plasma peak concentrations (PPC; 10×, 1×, and 0.1× PPC) [28] for miRNA expression studies. Doses of 0.1 and 0.5 µg/ml were used for MTT and TUNEL assay. The drug was prepared immediately prior to use.
In vitro drug assay
U87MG, A172 and T98G cell lines were seeded in a volume of 3 ml at 1 × 105 cells/well in 6-well plates, allowed to adhere for 24 hours and subsequently exposed to the drug for 12, 24 and 48 hours. The following Dox concentrations were used: 8.34 μg/ml for T98G and U87MG, and 0.834 μg/ml for A172. Untreated cells were used as a negative control. All experiments were conducted in triplicate.
Expression study
miRNAs were extracted with RecoverAllä Total Nucleic Acid Isolation (Ambion) from paraffin-embedded tissues (3-5 slices of tissue with a thick ≥ 10 μm) and with mirVanaä miRNA Isolation Kit (Ambion) from pellets of cell lines.
miR-21, miR-7, miR-124, miR-137 and miR-128 expression levels were determined using commercial assays (Assay on demand, Applied Biosystems) on a 7700 ABI PRISM Sequence Detector (Applied Biosystems).
Real-Time PCR was performed on cDNAs synthesized using the TaqManâ MicroRNA Reverse Transcription Kit (Applied Biosystems). All assays were performed in triplicate. For each miRNA, the expression levels, normalized to RNU48 (Applied Biosystems), were calculated using 2-∆∆Ct [29]. Adult and pediatric GBMs were subsequently normalized compared to FirstChoice® Human Brain Reference Total (Life Technologiesä). The expression results in pGBMs were also compared with those from a sample of pediatric non-tumoral cerebral cortex processed with the same experimental procedure, whereas the treated cell lines were normalized compared to the corresponding untreated cells.
MTT assay
Cytotoxicity was measured using the MTT assay (in vitro toxicology assay kit MTT based, Sigma). The key component of this assay is (3-[4,5-dimethylthiazol-2yl]-2,5-diphenyl tetrazolium bromide), a yellow salt that mitochondrial dehydrogenases of viable cells convert into purple formazan crystals, whose concentration is measured spectrophotometrically. We have conducted preliminary experiments to determine the best seeding concentration for U87MG, T98G and A172. Consequently, cells were seeded at the following densities: A172, 6 × 104 cells/well, U87MG, 4 × 104 cells/well and T98G, 2 × 104 cells/well in 24-well plates. After 24 hours, the cells were treated with 0.1 and 0.5 µg/ml Dox for 24 and 48 hours. The MTT assay was performed following the manufacturer’s instructions. The plates were placed on a shaker for 10 min to enhance solubilization of the precipitate. The absorbance (OD) of each well was then measured on a MULTISKAN FC (Thermo Scientific) microplate reader at a test wavelength of 550 nm. All experiments were performed in triplicate.
TUNEL assay
Apoptotic cells were detected by the terminal deoxynucleotidyl-transferase-mediated dUTP nick end-labeling (TUNEL) assay using in situ cell death detection kit, fluorescein (Roche). The best seeding concentration was preliminarily established at the following densities: A172, 18 × 104 cells/well, U87MG, 18 × 104 cells/well, and T98G, 3 × 104 cells/well in 6-well plates containing cover slips. After 24 hours, the cells were treated with 0.1 and 0.5 µg/ml Dox for 30 hours. After treatment, the cover slips were washed three times with PBS 1% and fixed in 4% paraformaldehyde solution for 30 minutes at room temperature. Then, cells were permeabilized by using sodium citrate 0.1%, TRITON × 100 0,1% for 5 min on ice. Finally, the TUNEL assay was performed, following the manufacturer’s instructions. The results were analyzed by fluorescence microscopy (Leitz, Type 307-148002, Wetzlar, Germany), equipped with E4 and N2.1 filters (Leica, Milan, Italy) using an oil immersion 100× magnification objective. Images were captured by a Canon digital camera using Remote Capture software (provided by Canon, Japan). All experiments were performed in triplicate.
miR-21 inhibition
Anti-miR-21 (AMI17000, id No. AM10206) was introduced into T98G cells at a final concentration of 10, 30, 60, 90, 120, 160 nM [30]. T98G cells were plated in 24-well plates (2 × 104 cells/well) and transfected 48 hours later using LipofectamineTM RNAi Max Transfection Agent (Invitrogen). The lowest possible concentration that achieved the most significant inhibition was chosen as the optimal dosage. Cells were transfected with the optimal dosage of 60 nM. To study the concomitant effects of miR-21 inhibition and Dox, cells were seeded into 6 well-plates at 4 × 104 cells/well and then transfected with anti-miR-21 at 60 nM, with or without Dox treatment at 0.5 µg/ml, for 72 hours [30].
MTT analysis on T98G cells transfected with anti-miR-21
T98G cells transfected with anti-miR-21 at 60 nM were subjected to MTT assay. Cells were seeded into 24 well-plates at 3 × 104 cells/well, transfected with anti-miR-21 at 60 nM for 24 hours and then treated with Dox. MTT analysis was performed after 72 hours of Dox treatment, following the manufacturer’s instructions. The OD of each well was measured on a MULTISKAN FC (Thermo Scientific) microplate reader at a test wavelength of 550 nm. All experiments were performed in triplicate.
TUNEL analysis on T98G cells transfected with anti-miR-21
T98G cells transfected with anti-miR-21 at 60 nM were subjected to TUNEL assay. Cells were seeded into 6 well-plates containing cover slips at 4 × 104 cells/well, transfected with anti-miR-21 at 60 nM for 24 hours and then treated with Dox. TUNEL analysis was performed after 72 hours of Dox treatment and the results were analyzed by fluorescence microscopy, as described above. All experiments were performed in triplicate.
Statistical analysis
Data were expressed as mean ± SD. Statistical analysis of in vitro drug assays was performed by using the one-way ANOVA test and post hoc Bonferroni-corrected t-test on the software version Graph Pad Prism 5.00. A level of p < 0.05 was accepted as statistically significant.
Statistical analysis of expression studies of adult and pediatric GBMs was performed by using the t-test and/or Levene test and the U Mannwhite test on the software package SPSS19.
Results
miRNA expression in pediatric and adult glioblastomas
Analysis of the series of 9 pGBMs and 4 aGBMs showed an overlapping expression pattern for all 5 miRNAs investigated, characterized by overexpression of miR-21 and reduced or lack of expression of miR-7, miR-124, miR-128 and miR-137.
The results of miRNA assays of the 9 pGBMs were compared to two different reference samples, FirstChoice® Human Brain Reference Total and pediatric non-tumoral cerebral cortex. Both analyzes yielded the same results. Differences in expression levels of 4/5 miRNAs were observed among the pGBM sample. This is likely due to cell heterogeneity in the samples (Figure 1A). No expression of miR-128 was detected.
Figure 1.
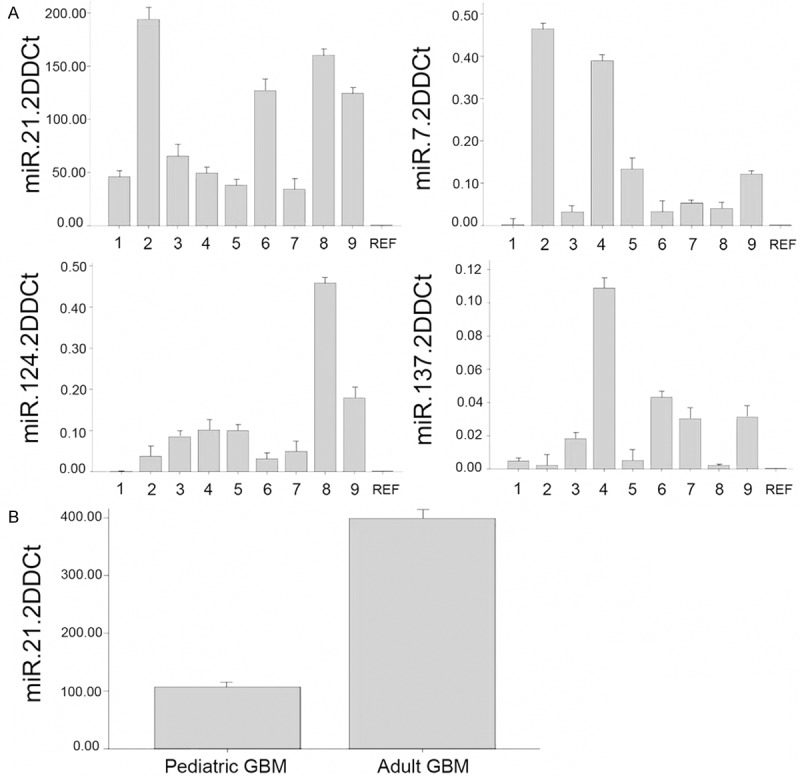
A: Expression of miR-21, miR-7, miR-124 e miR-137 in 9 pGBMs, calculated as 2-∆∆Ct; REF, FirstChoice® Human Brain Reference Total. B: Expression of miR-21 in adult and pediatric GBMs.
We found that the expression of miR-21 was significantly different between pGBMs and aGBMs, with 4× lower values in the pediatric than in the adult tumors (P < 0.001, t-test) relative to calibrator FirstChoice ® Human Brain Reference Total (Figure 1B).
miRNA expression in U87MG, A172 and T98G cell lines
In order to evaluate the effect of Dox treatment on miR-21, miR-7, miR-124, miR-128 and miR-137 expression, we treated U87MG, A172 and T98G cell lines with the chemotherapeutic agent at 0.834 µg/ml (0.1 × PPC for A172) and 8.34 µg/ml (1 × PPC, for T98G and U87MG). Subsequently, we measured miRNA levels at different time points (12, 24 and 48 hours) after treatment.
We observed low endogenous or no expression of miR-7, miR-124, miR-128 and miR-137 in A172, U87MG, and T98G treated and untreated cells (data not shown). In contrast, our data demonstrated that there was a dynamic change in miR-21 expression after Dox treatment in all cell lines. miR-21 was overexpressed in untreated A172, U87MG and T98G cells, while only low basal expression was found after Dox treatment (Figure 2).
Figure 2.
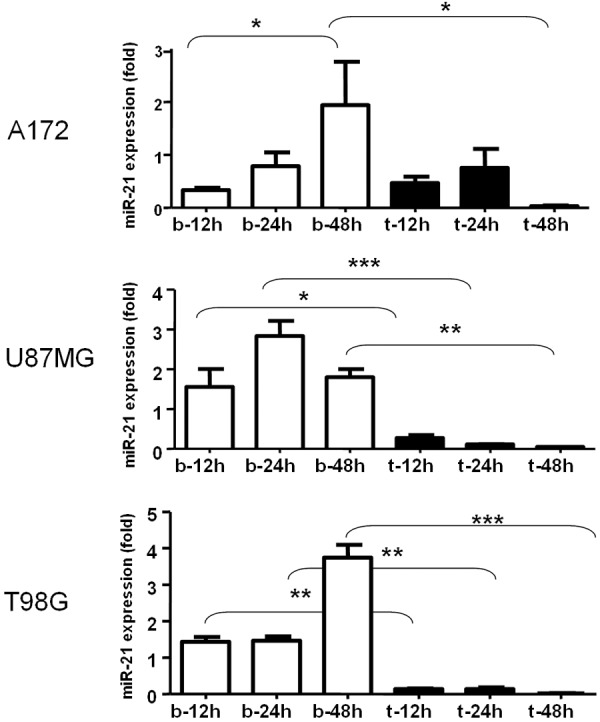
qRT-PCR analysis of miR-21 expression levels (mean ± SD) in A172, U87MG and T98G cell lines treated with Dox (12, 24 and 48 h). b: untreated cells; t: treated cells. *p < 0.05; **p < 0.001; ***p < 0.0001.
Dox sensitivity of GBM lines
Chemosensitivity was evaluated in three GBM cell lines treated for 24 and 48 hours with 0.1 and 0.5 µg/ml of Dox (Figure 3). The lowest possible concentrations that achieved the most significant results were chosen as the optimal dosage. No statistically significant difference was observed after 24 hours of treatment between either U87MG or T98G vs control, while A172 exhibited a statistically significant difference at both Dox concentrations used (OD values 0.6 ± 0.23 for 0.1 µg/ml Dox, and 0.6 ± 0.3 for 0.5 µg/ml Dox, respectively) vs control (OD 0.8 ± 0.2) (P < 0.05) (Figure 3A). Figure 3B illustrates the cytotoxic effect of Dox at 48 hours of treatment and demonstrates that A172 and U87MG were sensitive at 0.1 and 0.5 µg/ml concentrations, while T98G appeared to be resistant. OD values in A172 were 0.7 ± 0.1 and 0.14 ± 0.06, respectively, vs control 1.11 ± 0.1 (P < 0.01); OD values in U87MG were 0.65 ± 0.1 and 0.3 ± 0.005, respectively, vs control 0.8 ± 0.02 (P < 0.001). OD values did not significantly differ between T98G treated and untreated cells. Our MTT experiments demonstrated that the most effective cytotoxic Dox dosage was 0.5 µg/ml. Based on these results, we performed a TUNEL assay to investigate the pro-apoptotic effects of 0.5 µg/ml of Dox. This dosage induced apoptosis in A172 and U87MG but not in T98G, confirming that T98G is a drug resistant cell line (Figure 3C).
Figure 3.
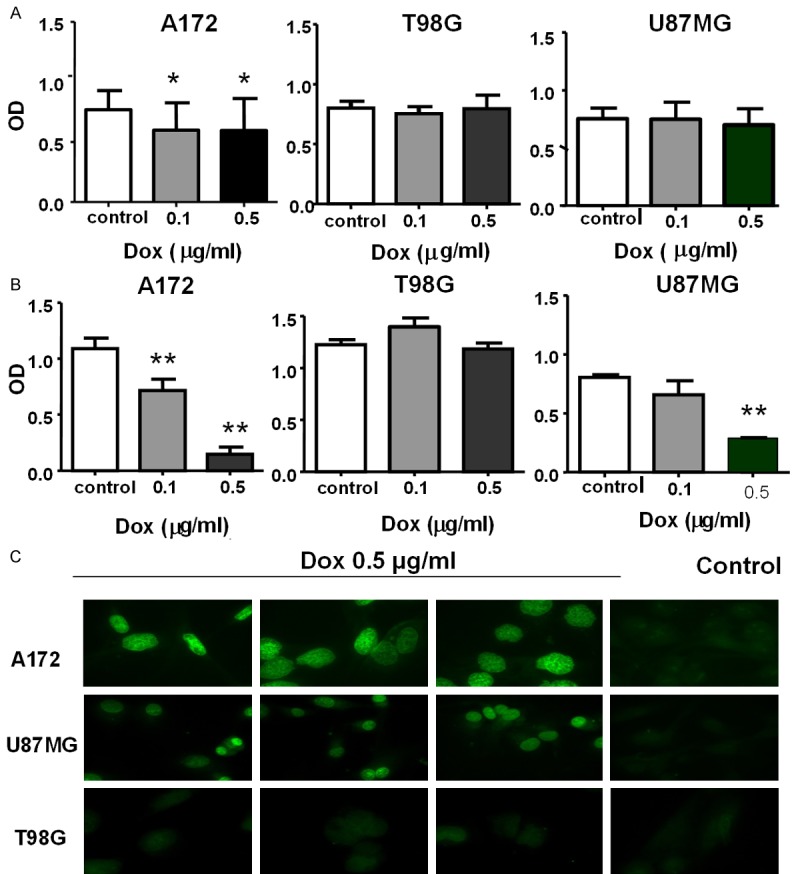
Cytotoxicity responses of A172, U87MG and T98G, after 24 (A) and 48 h (B) of Dox treatment, measured as OD by the MTT assay. C: TUNEL assay on A172, U87MG and T98G cell lines treated with 0.5 µg/ml Dox.
miR-21 inhibition in T98G cells
To investigate whether miR-21 regulation influences the drug resistant phenotype, we transfected anti-miR-21 into the T98G resistant cell line. To achieve the optimal effect of silencing with minimal off-target effects, different inhibitor concentrations (from 10 to 160 nM) were tested. We found that transfection with miR-21 inhibitor reduced miR-21 expression in a dose dependent manner (Figure 4A). The 60 nM concentration gave the maximal effect and resulted in approximately 80% decrease in miR-21 expression.
Figure 4.
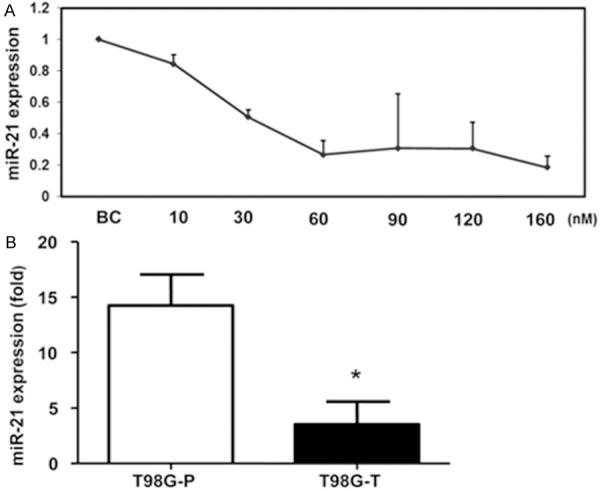
A: Transfection with miR-21 inhibitor reduces miR-21 expression in T98G cells in a dose dependent manner. BC is a blank transfection control used in comparative CT-fold change analysis. B: Levels (mean ± SD) of miR-21 expression in the T98G cell line before (T98G-P) and after transfection (T98G-T) with miR-21 inhibitor assessed by qRT-PCR. *p < 0.05.
The inhibitory effect of anti-miR-21 at 60 nM was assessed by qRT-PCR 48 hours after transfection. Our data showed that the expression level of miR-21 was significantly lower in T98G transfected cells (T98G-T) than in the parental cells (T98G-P) (Figure 4B).
Effects of concomitant Dox treatment and miR-21 inhibition
In order to test whether miR-21 inhibition improves the response of Dox-resistant cells to Dox re-challenge, T98G cells were transfected with anti-miR-21 for 24 hours and then treated with Dox 0.5 µg/ml. TUNEL analysis was performed after 72 hours of Dox treatment.
The data show that miR-21 inhibition in association with Dox treatment leads to increased apoptosis in T98G Dox-resistant cells (Figure 5A). In contrast, few apoptotic cells were observed in the corresponding cells treated with miR-21 inhibitor or Dox alone (Figure 5B, 5C). Figure 5D demonstrates that the treatment with miR-21 inhibitor (overall 96 hours) plus Dox (for 72 hours) resulted in a significant 23% increase in the apoptotic rate when compared with Dox treatment alone (Figure 5D).
Figure 5.
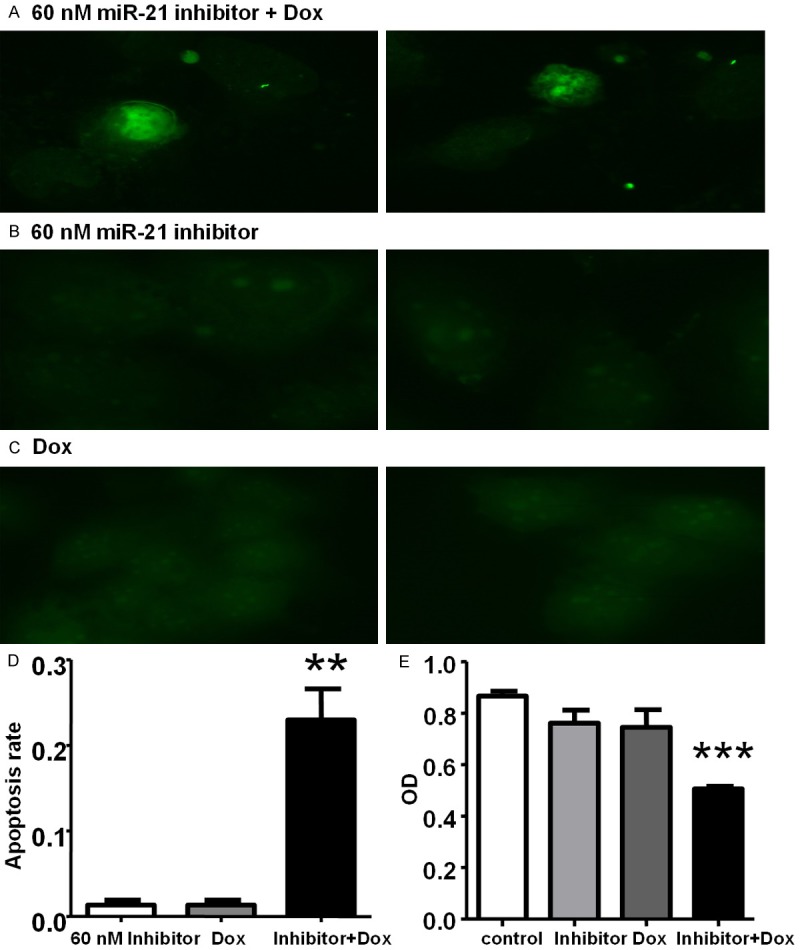
TUNEL assay, showing apoptotic nuclei in green. A: miR-21 inhibitor plus Dox. B: miR-21 inhibitor alone. C: Dox alone. D: Increased apoptotic rate after concomitant treatment with miR-21 inhibitor 60 nM and Dox 0.5 µg/ml (**p < 0.01). Bar chart shows the percentage (mean ± SD) of apoptotic cells counted from three randomly selected view fields. E: Cytotoxicity response of T98G cells transfected with miR-21 inhibitor 60 nM (96 h) and treated with Dox 0.5 ug/ml (72 h). Data are presented as mean ± SD (error bar). ***p < 0.001 inhibitor plus Dox vs control.
Based on these results, we tested the chemosensitivity of GBM cells treated with miR-21 inhibitor plus Dox by the MTT assay (Figure 5E). The data show that only the concomitant use of miR-21 inhibitor and Dox is effective in inhibiting the growth of T98G cells. While treatment with miR-21 inhibitor or Dox alone has no effect, the viability of T98G cells was largely reduced when the two substances were used in combination.
Our data suggest that combined Dox and miR-21 inhibitor therapy can sensitize GBM resistant cells to anthracyclines by enhancing apoptosis.
Discussion
In this study, in order to identify miRNAs that are involved in the pathogenesis and development of pediatric and adult high grade gliomas, we first used quantitative RT-PCR to analyze the expression of 5 miRNAs that had previously been found to be dysregulated in GBMs samples [18,26]. miR-21 was overexpressed in all 9 pGBM samples investigated, while the expression of miR-7, miR-124, miR-128 and miR-137 was either reduced or absent. Interestingly, we detected a significant difference between pGBMs and aGBMs, since the former showed a 4X lower degree of miR-21 expression compared to the latter (p = 0.001).
Anthracyclines such as Dox are commonly used in the treatment of a wide range of tumors, including hematological malignancies, many types of carcinoma and soft tissue sarcomas [31]. Data on in vitro and in vivo malignant glioma models suggest that Dox can be effective for these tumors [32-34]. Eramo and co-workers have demonstrated that Dox displays high antitumor activity against GBM stem cells [35].
In order to evaluate the cellular response to anthracyclines in glioblastoma we used three GBM cell lines (A172, U87MG and T98G), identifying a different behaviors among them. In fact, a cytotoxic effect was observed in A172 and U87MG after 48 hours of Dox treatment while T98G cells appeared to be resistant to the drug. This was confirmed by the finding of apoptosis induction following Dox treatment in A172 and U87MG, but not in T98G.
On the same cells we analyzed the expression of the 5 miRNAs described above; only miR-21 exhibited a different expression level in all cell lines with or without Dox treatment.
To validate the possible association of miR-21 with drug resistance of T98G cells, we investigated the effects of a specific miR-21 inhibition on the cells. The data showed that the expression level of miR-21 was significantly lower in T98G transfected cells (T98G-T) than in the parental control cells (T98G-P); moreover, the transfected cell line showed a high apoptotic rate compared to the control after Dox treatment.
Our results suggest that combined Dox and miR-21 inhibitor therapy may regulate anthracyclines resistance via augmented apoptotic pathway.
Different signal transduction pathways seem to be involved in the chemoresistance mechanisms of GBMs. miRNAs are candidate modulators of the response to antineoplastic agents of these malignant tumors. miRNA expression has been investigated in pediatric malignant brain tumors, including GBM, using microarray expression profiling. Pediatric CNS tumors exhibited tumor-specific miRNA signatures and differential miRNA expression (miR-129, miR-142-5p, and miR-25) compared to normal brain [19,20]. Moreover, some miRNAs have been shown to down-regulate the levels of proteins involved in key pathways downstream of tyrosine kinase signaling in adult GBM [18].
miR-21, an oncomiR, plays a crucial role in a plethora of biological functions and diseases including development, cancer and inflammation. miR-21 is one of the most investigated miRNAs in human cells, and its expression is significantly up-regulated in several tumors. miR-21 expression has been correlated with tumor grade and has been proposed as a marker of tumor progression [36,37]. miR-21 regulates a nuclear complex including AP-1/b-catenin [38] and also plays a role in the response to anti-EGFR treatment through modulation of the EGFR signaling pathway [38].
Original experiments demonstrated that miR-21 is frequently up-regulated in human GBM and that its inhibition leads to caspase stimulation and associated apoptotic cell death in different GBM cell lines [39]. In addition, miR-21 seems to contribute to tumorigenicity and invasion of glioma cells through targeting the components of the p53 and TGF-b pathways, mitochondrial apoptosis-related genes, as well as RECK and TIMP3 [22,40].
Importantly, it has been shown that down-regulation of miR-21 can inhibit the growth of GBM cell lines and induce apoptosis independently of PTEN status. These observations were confirmed in in vivo xenografts, highlighting the potential clinical relevance of miR-21-targeting agents [41]. Recently, miR-21 was also shown to regulate PDCD4, a tumor suppressor gene, in GBM cells [42]. GBM-derived cell lines treated with anti-miR-21 had reduced proliferation and also exhibited enhanced apoptosis, compared with untreated controls. In addition, cell lines in which miR-21 levels were inhibited displayed decreased anchorage-independent growth, whereas GBM-derived cell lines expressing PDCD4 showed increased apoptosis and diminished anchorage-independent growth [43].
miR-21 seems to regulate drug resistance in various cancers, and therefore the use of miR-21 inhibitors may function as an effective approach for reversing drug resistance in cancer cells. High levels of miR-21 are a common feature of GBM, and it has been shown that it can function as an anti-apoptotic factor in cultured GBM cells [39]. It has already been demonstrated that up-regulation of miR-21 can induce Dox chemoresistance in T24 human bladder cancer cells while down-regulation of miR-21 sensitized T24 cells to the drug [44]. Likewise, miR-21 blockage increased the chemosensitivity of human GBM cells to Taxol. Furthermore, down-regulation of miR-21 increased the chemosensitivity of glioma cells to Etoposide [45]. The concomitant use of miR-21 inhibitors and antineoplastic drugs could therefore be an effective therapeutic strategy for suppressing the growth of GBM, regardless of PTEN status [46].
Treatment of glioma cells with anti-miR-21 did not cause any change in proliferative capacity, although it resulted in significant reduction of the ability to form colonies in soft agar, further suggesting that this miRNA might play an important role in neoplastic transformation [45].
In conclusion, our study provides evidence that miR-21 inhibition in addition to pharmacological treatment could pave the way to new effective approaches for reversing drug resistance in GBM. Further studies are warranted to better understand how miRNAs can be explored as a potential chemotherapy adjunct in the treatment of resistant brain tumors.
Acknowledgements
This work was supported by: Associazione Italiana per la Ricerca sul Cancro (AIRC), grant IG-12799; “Amicodivalerio” Onlus; “Noi per Voi” Onlus, Fondazione Tommasino Bacciotti.
Disclosure of conflict of interest
None.
References
- 1.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–53. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci. 2009;100:2235–41. doi: 10.1111/j.1349-7006.2009.01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubuc AM, Northcott PA, Mack S, Witt H, Pfister S, Taylor MD. The genetics of pediatric brain tumors. Curr Neurol Neurosci Rep. 2010;10:215–23. doi: 10.1007/s11910-010-0103-9. [DOI] [PubMed] [Google Scholar]
- 4.Pfister S, Janzarik WG, Remke M, Ernst A, Werft W, Becker N, Toedt G, Wittmann A, Kratz C, Olbrich H, Ahmadi R, Thieme B, Joos S, Radlwimmer B, Kulozik A, Pietsch T, Herold-Mende C, Gnekow A, Reifenberger G, Korshunov A, Scheurlen W, Omran H, Lichter P. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118:1739–49. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollack IF, Hamilton RL, Finkelstein SD, Lieberman F. Molecular abnormalities and correlations with tumor response and outcome in glioma patients. Neuroimaging Clin N Am. 2002;12:627–39. doi: 10.1016/s1052-5149(02)00030-8. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–8. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O’brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–41. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623–31. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, Liu J, Cui Y, Bian X, Bie P, Qian C. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310:160–9. doi: 10.1016/j.canlet.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem Biophys Res Commun. 2003;304:184–90. doi: 10.1016/s0006-291x(03)00554-0. [DOI] [PubMed] [Google Scholar]
- 13.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9:180–6. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–81. doi: 10.1261/rna.5980303. Erratum in: RNA 2004; 10: 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogaev EI. Small RNAs in human brain development and disorders. Biochemistry (Mosc) 2005;70:1404–7. doi: 10.1007/s10541-005-0276-z. [DOI] [PubMed] [Google Scholar]
- 16.Pang JC, Kwok WK, Chen Z, Ng HK. Oncogenic role of microRNAs in brain tumors. Acta Neuropathol. 2009;117:599–611. doi: 10.1007/s00401-009-0525-0. [DOI] [PubMed] [Google Scholar]
- 17.Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386:1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 18.Lawler S, Chiocca EA. Emerging functions of microRNAs in glioblastoma. J Neurooncol. 2009;92:297–306. doi: 10.1007/s11060-009-9843-2. [DOI] [PubMed] [Google Scholar]
- 19.Birks DK, Barton VN, Donson AM, Handler MH, Vibhakar R, Foreman NK. Survey of MicroRNA expression in pediatric brain tumors. Pediatr Blood Cancer. 2011;56:211–6. doi: 10.1002/pbc.22723. [DOI] [PubMed] [Google Scholar]
- 20.Miele E, Buttarelli FR, Arcella A, Begalli F, Garg N, Silvano M, Po A, Baldi C, Carissimo G, Antonelli M, Spinelli GP, Capalbo C, Donofrio V, Morra I, Nozza P, Gulino A, Giangaspero F, Ferretti E. High-throughput microRNA profiling of pediatric high-grade gliomas. Neuro Oncol. 2014;16:228–40. doi: 10.1093/neuonc/not215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lou Y, Yang X, Wang F, Cui Z, Huang Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int J Mol Med. 2010;26:819–827. doi: 10.3892/ijmm_00000530. [DOI] [PubMed] [Google Scholar]
- 22.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 24.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ZX, Lu BB, Wang H, Cheng ZX, Yin YM. MicroRNA-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch Med Res. 2011;42:281–90. doi: 10.1016/j.arcmed.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Møller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion. Mol Neurobiol. 2013;47:131–44. doi: 10.1007/s12035-012-8349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis DL, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu CC, Zhou FM, Wang YQ, Zhong P, Jiang GF, Miao A, Lou H. Chemotherapy of cerebral gliomas directed by the chemosensitivity test in vitro: a clinical study. Chinese-German Journal of Clinical Oncology. 2007;6:269–273. [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Wong ST, Zhang XQ, Zhuang JT, Chan HL, Li CH, Leung GK. MicroRNA-21 inhibition enhances in vitro chemosensitivity of temozolomide-resistant glioblastoma cells. Anticancer Res. 2012;32:2835–41. [PubMed] [Google Scholar]
- 31.Benjamin RS, Riggs CE Jr, Bachur NR. Pharmacokinetics and metabolism of adriamycin in man. Clin Pharmacol Ther. 1973;14:592–600. doi: 10.1002/cpt1973144part1592. [DOI] [PubMed] [Google Scholar]
- 32.Abe T, Hasegawa S, Taniguchi K, Yokomizo A, Kuwano T, Ono M, Mori T, Hori S, Kohno K, Kuwano M. Possible involvement of multidrug-resistance-associated protein (MRP) gene expression in spontaneous drug resistance to vincristine, etoposide and adriamycin in human glioma cells. Int J Cancer. 1994;58:860–864. doi: 10.1002/ijc.2910580619. [DOI] [PubMed] [Google Scholar]
- 33.Wolff JE, Trilling T, Mölenkamp G, Egeler RM, Jürgens H. Chemosensitivity of glioma cells in vitro: a Meta-analysis. J Cancer Res Clin Oncol. 1999;125:481–486. doi: 10.1007/s004320050305. [DOI] [PubMed] [Google Scholar]
- 34.Steiniger SC, Kreuter J, Khalansky AS, Skidan IN, Bobruskin AI, Smirnova ZS, Severin SE, Uhl R, Kock M, Geiger KD, Gelperina SE. Chemotherapy of glioblastoma in rats using doxorubicin-loaded nanoparticles. Int J Cancer. 2004;109:759–767. doi: 10.1002/ijc.20048. [DOI] [PubMed] [Google Scholar]
- 35.Eramo A, Ricci-Vitiani L, Zeuner A, Pallini R, Lotti F, Sette G, Pilozzi E, Larocca LM, Peschle C, De Maria R. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 2006;13:1238–1241. doi: 10.1038/sj.cdd.4401872. [DOI] [PubMed] [Google Scholar]
- 36.Cho WC. OncomiRs: the discovery and the progress of microRNAs in cancer. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berthois Y, Delfino C, Metellus P, Fina F, Nanni-Metellus I, Al Aswy H, Pirisi V, Ouafik L, Boudouresque F. Differential expression of miR200a-3p and miR21 in grade II-III and grade IV gliomas: Evidence that miR200a-3p is regulated by O 6-methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biol Ther. 2014;15:938–50. doi: 10.4161/cbt.28920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang KL, Han L, Chen LY, Shi ZD, Yang M, Ren Y, Chen LC, Zhang JX, Pu PY, Kang CS. Blockage of a miR-21/EGFR regulatory feedback loop augments anti-EGFR therapy in glioblastomas. Cancer Lett. 2014;342:139–49. doi: 10.1016/j.canlet.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 39.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 40.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 41.Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Wang B, Wang G, Jia Z, Pu P, Zhang W, Kang C. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–55. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Liu W, Chao T, Zhang Y, Yan X, Gong Y, Qiang B, Yuan J, Sun M, Peng X. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 43.Gaur AB, Holbeck SL, Colburn NH, Israel MA. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol. 2011;13:580–90. doi: 10.1093/neuonc/nor033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao J, Lu Q, Wu D, Li P, Xu B, Qing W, Wang M, Zhang Z, Zhang W. microRNA-21 modulates cell proliferation and sensitivity to doxorubicin in bladder cancer cells. Oncol Rep. 2011;25:1721–9. doi: 10.3892/or.2011.1245. [DOI] [PubMed] [Google Scholar]
- 45.Rao SA, Santosh V, Somasundaram K. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol. 2010;23:1404–17. doi: 10.1038/modpathol.2010.135. [DOI] [PubMed] [Google Scholar]
- 46.Ren Y, Zhou X, Mei M, Yuan XB, Han L, Wang GX, Jia ZF, Xu P, Pu PY, Kang CS. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer. 2010;10:27. doi: 10.1186/1471-2407-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]


