Abstract
Approximately 35% of breast cancers exhibit PIK3CA activating mutation. Since PIK3CA hotspot mutation is the most frequently mutated gene in human breast cancers and primarily overlaps in HER2+ as well as ER+ breast cancers, the subset of patients bearing PIK3CA activating mutation does not get fullest benefit from either anti-HER2 or anti-hormonal agents. Literature also suggests that these patients may have chemotherapy resistance. Indeed, multiple clinical trials are currently evaluating the efficacy of over 30 drugs targeting different nodal points in the PI3K-AKT-mTOR pathway in breast and other cancers. However, to date, responses of solid tumors to PI3K pathway inhibitor monotherapy remains modest with an accompanied rapid emergences of drug resistance. MYC elevation represents one of the potential modes of actions by which breast tumors develop resistance to the PI3K pathway-specific targeted therapies. As products of oncogenes, both MYC and PIK3CA are well-established onco-proteins which contribute to breast oncogenesis. However, their similarities out number their dissimilarities in the context of their specific oncogenic cellular signals. In this review we will describe the specific cellular signals initiated following alteration in the MYC gene and PIK3CA gene in breast cancers. We will interrogate how MYC gene alterations influence the action of PI3K pathway targeted drugs in the context of PIK3CA mutation towards the development PI3K inhibitor induced drug-resistance in breast cancers.
Keywords: Breast tumors, MYC, PIK3CA, PI3K inhibitors, resistance
Introduction
Breast cancers like other solid cancers are characterized by gene mutations and chromosomal aberrations [1,2]. Breast cancer is the most common cancer of women in the U.S. and other western countries, with an accumulated life time incidence rate of about 11%. About 10% of breast cancers are inherited, mostly caused by mutations in BRCA1 and BRCA2. The rest are sporadic breast cancers caused by somatic mutations and chromosome instability in the breast tissue [3]. Both MYC and PIK3CA are among frequently amplified genes in addition to other well-known oncogenes including ERBB2, FGFR1, and CCND1 in breast cancers. Many tumor types (if not all) exhibit survival and/or growth dependence on a mutationally activated particular gene, commonly a kinase through a process termed “oncogene addiction”. This principle of targeted kinase inhibition has provided clinical success in treating diverse cancer types [4-9]. However, the single most concern that impedes the sustained clinical benefits of targeted therapies is the observed emergence of acquired drug resistance. As we are evolving in clinics to target an organ site cancer with genomic-data-driven pathway targeted drugs, the problem of drug induced resistance is becoming a formidable challenge. In this review we will try to understand the role of specific cellular signals those are brought into action following alteration(s) in the MYC gene and PIK3CA gene in breast cancers. We will also cross-examine how MYC gene alterations influence the action of the PI3K pathway targeted drugs in the context of PIK3CA mutation towards the development of PI3K pathway-specific inhibitor induced drug-resistance and how oncogenic mutation of PIK3CA synergistically interacts with MYC functions in breast cancers.
Alterations of MYC and PIK3CA genes in breast cancers
MYC is a proto-oncogene that transcribes its protein product containing a basic helix-loop-helix domain. As a transcription factor MYC protein regulates up to 15% of all human genes. Hence MYC gene product is tightly regulated at multiple levels of cell signaling, and the protein acts as downstream effector of several signaling pathways related to all fundamental functions of a cell. MYC is one of the most commonly altered oncogenes in human cancers [10]. In breast cancer, MYC target genes are involved in cell growth, proliferation, transformation, immortalization, metastasis-associated phenotypes, DNA-damage response, angiogenesis and cell-cycle control. Hyperactivation of MYC in tumor cells sets the permissive stage for the oncogenic signals and actively participates in the cellular transformation. This action is achieved by the evolutionarily conserved function of MYC which modulates protein synthesis. The MYC oncogenic program enhances the protein synthesis capacity of cancer cells by directly contributing to their survival, proliferation, and genome instability. MYC has also been demonstrated to be an important component of the “oncogenic nexus” acting in concert with PP2A and CIP2A in achieving the tumorigenic transformation in cells [11].
From the time Bishop and his co-workers discovered the MYC gene in the late 1970s [12-14], an enormous volume of scientific literature has been accumulated to demonstrate its fundamental role towards the malignant transformation of human and animal cells [15,16]. Almost all types of human malignancies including breast cancer have amplification and/or overexpression of this gene. Correlation of amplification of the MYC oncogene and overexpression of the MYC protein in high-grade breast cancer has been reported [17]. A summary of the amplification, RNA or protein expression of MYC gene in human breast cancers has been elegantly presented in the review by Liao and Dickson in 2000 wherein authors have painstakingly described the profound roles of MYC in breast cancer and its relationship with actions of different hormones those are etiologically related to breast cancer [15]. In later years, role of genetic and epigenetic alterations of MYC gene was further expanded to field of the multistep process of disease progression in breast cancers [18]. Alterations of MYC gene and MYC protein levels have been found related to the disease progression in ductal and lobular breast cancer [19]. The involvement of MYC was established in different subtypes including ER+, HER2+ and BRCA1-associated of breast cancers [18,20-22]. Elevated MYC expression leads to a poor prognosis in sporadic breast cancer patients those are BRCA1-deficient [23]. BRCA1 has been linked to transcriptional regulation through interaction with MYC and its role in BRCA1-associated breast cancer makes it an important target in basal-like/triple-negative breast cancers [18]. MYC oncogene amplification is observed in BRCA1-associated breast cancers wherein the aggressive histopathological features of tumors are in part due to dysregulated MYC activity and MYC also contributes to tumor progression [21]. MYC amplification has also been identified as an important predictor of response to the targeted therapies like HER2-targeted therapies [18].
In 1985, Whitman and colleagues showed that the transforming ability of the polyoma virus middle-T antigen (PyMT), a membrane-bound tyrosine kinase, closely correlated with phosphatidylinositol (PI) kinase activity [24]. Although PyMT was later found to have independent oncogenic properties, its regulation of PI3K revealed that hyperactivation of this pathway can lead to uncontrolled proliferation, enhanced migration, and adhesion-independent growth [25]. Recent observations revealed further important insights into the mechanism of regulation and activation of the PI3K pathway in cancer. Amplifications in the genes encoding p110α, p85α and AKT have been described and there is recent evidence of an activating mutation in AKT1 in breast, colorectal and ovarian cancers [26]. Notably, alteration in PIK3CA, PTEN, AKT1 and PIK3R1 (the gene for the p85α regulatory subunit of PI3K) were generally non-overlapped in tumor samples. In addition to this direct evidence of deregulation of the PI3K-AKT-mTOR network, it is well documented that receptor tyrosine kinases (RTKs), such as trans-membrane receptors (via tyrosine kinases) can be aberrantly activated in various cancers, leading to activation of the PI3K-AKT-mTOR signaling pathway [27]. These tyrosine kinases are able to phosphorylate critical tyrosine residues within activation motifs, often located within the receptors themselves (e.g., autophosphorylation of the YXXM motif in PDGFR) or are present on protein adaptors (e.g., IRS, SHC, GRB, GAB, and CBL). As a multi-domain-containing protein, the regulatory subunit of PI3K (p85, p55, p50) binds to the phosphorylated pYXXM motif via their SH2 domains resulting in PI3K membrane recruitment and generates the second messenger PI (3, 4, 5) P3 from PIP2. In addition, the RBD domain of p110α catalytic subunit is recruited to the plasma membrane through direct binding to the GTP-bound active form of membrane-bound (myristylated) RAS. Indeed, many of the most novel biomarker/mechanism-based cancer drugs, such as Gefitinib, Cetuximab, Trastuzumab and Lapatinib act through the inhibition of aberrantly activated RTKs, reducing the signaling through the PI3K-AKT-mTOR pathway. The overwhelming evidence of deregulation of the PI3K-AKT-mTOR pathway in tumorigenesis means that inhibition of this signaling pathway is an attractive avenue of investigation for pharmacological intervention. There are several small molecule inhibitors (SMIs) of PI3Ks or mTORs that have been developed and historically used as pharmacological tools to study the effect of PI3K or mTOR inhibition. Multiple clinical trials are currently evaluating the efficacy of over 30 drugs targeting different nodes in the PI3K pathway in breast and other cancers (https://clinicaltrials.gov). Although, targeted inhibition of PIK3CA holds significant promise, there are worries that an eventual development of resistance to these drugs and a clonal survival advantage would ultimately limit the clinical efficacy.
An alteration summary for MYC and PIK3CA genes in BC and subtypes
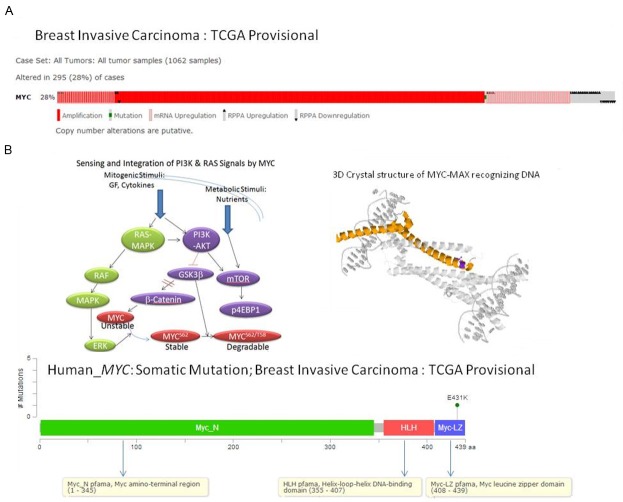
In order to know the extent of changes in MYC gene in breast cancer, we obtained an alteration-summary for MYC gene in breast cancer (Figure 1). Data mining was carried out using cBioPortal for Cancer Genomics, a data portal [cBioPortal for Cancer Genomics: [28,29]], available at http://www.cbioportal.org to measure the incidence of alterations in MYC, as per the criteria mentioned in the legends of figures. The database query was based on deregulation (mutation, copy number alterations and altered expression) of the MYC. We acknowledge the cBioPortal for Cancer Genomics site (http://cbioportal.org) which provides a web resource for exploring, visualizing, and analyzing multidimensional cancer genomics data. We acknowledge the TCGA Research Network for generating TCGA datasets. Figure 1A showed that the MYC was altered in 28% (295) of the cases (1062 samples) of the breast invasive carcinomas (Oncoprint data obtained using c-BioPortal; TCGA Provisional). The only somatic mutation occurring in the MYC gene in the selected samples was observed to be less than 5 in number of the only site of MYC mutation (E431K) (Figure 1B). Oncoprint data obtained using c-BioPortal (Figure 2) showed that there is a limited overlap between the 24% of altered MYC and the 37% of the altered PIK3CA of the breast invasive carcinomas (1062 samples; TCGA Provisional) (Figure 2: Upper Panel). The highest alteration (35%) in MYC was observed in basal-like types of BC (Figure 2: Upper Panel).
Figure 1.
Alterations (A) and somatic mutations (B) of human MYC gene in Breast Invasive Carcinoma (TCGA, Provisional). (A) Alterations (amplification, mutation, mRNA upregulation, RPPA upregulation and RPPA downregulation) of MYC gene in Breast Invasive Carcinoma (TCGA, Provisional). Data was obtained using c-BioPortal. Unaltered cases and white spaces between values were removed. The case set contained 1062 tumor samples. Following Genomic Profiles were selected: (1) mutations, (2) putative copy-number alteration (CNA) from GISTIC, (3) mRNA expression Z-scores (RNA Seq V2 RSEM) with Z-score thresholds ± 2.0, and (4) protein/phospho-protein level (RPPA) with Z-score thresholds ± 2.0. (B) The number of somatic mutations of MYC gene at different sites (domains) have been plotted in the selected samples. The somatic mutation rate of MYC gene in the selected samples has been presented in the text. Different domains/regions are presented in different colors. The left inset shows the schematic representation of interactions between mitogen mediated PI3K and RAS signaling pathways and role of MYC transcription factor as a “Signal Integrator”. Mitogenic stimulation with growth factors (GF)/cytokines leads to activation of the RAS-MAPK pathway and the PI3K-mTOR pathway. MYC protein level, its transcription, its synthesis as well as post translational modification, stability and degradation are either in parallel and/or serially regulated by the RAS-MAPK pathway and the PI3K-mTOR pathway. The synthesized nascent MYC protein is unstable in nature and has short half-life. MYC protein stability is increased via the post-translational phosphorylation at S62 by activated ERK. ERK can be activated downstream of RAS activation. Active RAS induces activation of its downstream effector pathways: the MAPK and PI3K pathways. While ERK activation following RAS activation stabilizes newly synthesized MYC protein, PI3K activation blocks MYC protein degradation by inhibiting phosphorylation at T58 by GSK3beta. GSK3beta initiates its degradation by phosphorylation of MYC protein at T58. Phosphorylation at T58 by GSK3beta requires prior phosphorylation at S62. Phosphorylation at T58 induces subsequent dephosphorylation at S62 by PP2A. Activated PI3K pathway (either following RAS activation or GF induction) inhibits GSK3beta and stabilizaes beta-catenin to induce MYC transcription in the nucleus. Thus RAS-mediated activation of ERK and PI3K-mediated inhibition of GSK3beta determine the amount of MYC protein in a tumor cell at a particular point of time by controlling both its stability and degradation via post-translational phosphorylation. The right inset shows the 3D crystal structure of chain A of MYC onco-protein (PDB 1nkp : crystal structure of myc-max recognizing dna) as obtained from c-BioPortal. Bound molecules are displayed in the cartoon (inset). The color represents the protein by secondary structures. Alpha helics is represented in yellow, beta sheets in blue and loops in light grey. The side chain atoms for every mapped residue are displayed. All mutated residues are represented in a single violet color. We acknowledge the cBioPortal for Cancer Genomics site (http://cbioportal.org) which provides a Web resource for exploring, visualizing, and analyzing multi-dimensional cancer genomics data. The portal reduces molecular profiling data from cancer tissues and cell lines into readily understandable genetic, epigenetic, gene expression and proteomic events [28]. We acknowledge works of Cerami et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data [28,29]. We acknowledge the TCGA Research Network for generating TCGA datasets.
Figure 2.
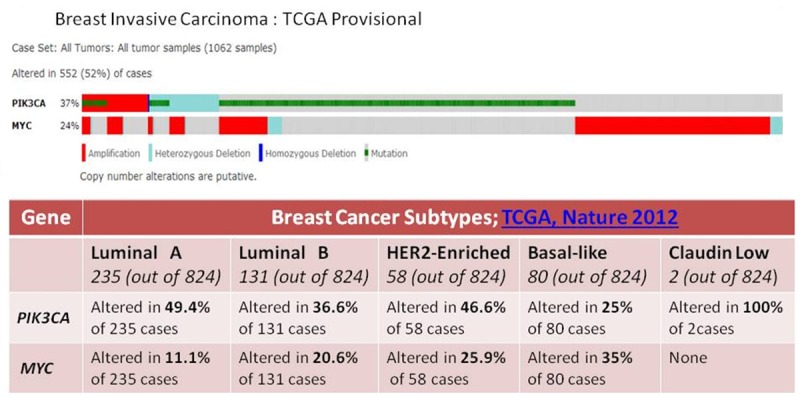
Overlap between alterations of human MYC and PIK3CA genes in Breast Invasive Carcinoma (TCGA, Provisional) (upper panel) and differential alterations of human MYC and PIK3CA genes in subtypes of Breast Invasive Carcinoma (TCGA, Nature 2012) (lower panel). Upper panel: Alterations (amplification, mutation, heterozygous deletion and homozygous deletion) of MYC and PIK3CA genes in Breast Invasive Carcinoma (TCGA, Provisional) are presented. Data was obtained using c-BioPortal. Unaltered cases and white spaces between values were removed. The case set contained 1062 tumor samples. Following Genomic Profiles were selected: (1) mutations, (2) putative copy-number alteration (CNA) from GISTIC, (3) mRNA expression Z-scores (RNA Seq V2 RSEM) with Z-score thresholds ± 2.0, and (4) protein/phospho-protein level (RPPA) with Z-score thresholds ± 2.0. Lower panel: Table shows changes in MYC and PIK3CA genes in five different subtypes of Breast Invasive Carcinoma (TCGA, Nature 2012) including Luminal A, Luminal B, HER2-enriched, Basal-like and Claudin-low. A custom case set was build using c-BioPortal (TCGA , Nature 2012 data set containing 825 cases; raw data at the NCI.) Following Genomic Profiles were selected: (1) mutations, (2) putative copy-number alteration (CNA) from GISTIC, (3) mRNA expression Z-scores (RNA Seq V2 RSEM) with Z-score thresholds ± 2.0, and (4) protein/phospho-protein level (RPPA) with Z-score thresholds ± 2.0. (Total 824 samples). The total numbers of samples in the respective subtypes are shown in the parenthesis.
In contrast to alteration in MYC gene (Figure 1A) in 28% (295) of the cases (1062 samples) of the breast invasive carcinomas (TCGA Provisional), the PIK3CA was altered in 37% of the cases (390 of cases) in the same selected samples (Figure 3A). Out of this 28% of the MYC altered cases, the predominant form of alteration was found to be the amplification of the gene followed by the upregulation of the mRNA. This pattern of the alteration of gene is characteristically different from that observed in PIK3CA gene in the same set of patients’ samples. Figure 3A shows that the predominant form of alterations in the PIK3CA observed in the breast invasive carcinomas (TCGA Provisional) is the mutation. This pattern of the alteration of gene is characteristically different from that observed in MYC gene in the same set of patients’ samples. Figure 1A shows that the predominant form of alterations in the MYC observed in the breast invasive carcinomas (TCGA Provisional) is the amplification. In certain samples, the mutations of the gene were found to occur along with the upregulation of PIK3CA mRNA or the amplification of the PIK3CA gene. There is also a significant difference observed between the somatic mutation rates of the MYC gene and the somatic mutation rates of the PIK3CA gene in the selected samples as presented in the Figures 1B, 3B. The somatic mutation rate of the MYC gene was 0.1% as compared to 29.8% in the PIK3CA gene in the selected samples (Figures 1B, 3B). Not only the rate of somatic mutation was higher in PIK3CA gene but also the distribution patterns of the somatic mutations were much wider between different domains of the corresponding protein of the PIK3CA gene. In addition the number of individual somatic mutation occurring in PIK3CA gene far exceeded the number of individual somatic mutation occurring in MYC gene. The most predominant somatic mutation occurring in PIK3CA gene, H1047L/R/Y was found in nearly 140 times as compared to less than 5 number of the only mutation (E431K) occurring in the MYC gene (Leucine Zipper domain) in the same selected samples (Figures 1B, 3B). The highest % of alteration in PIK3CA gene was observed in luminal A subtype (49.4%) while basal-like subtype exhibited the highest alteration in the MYC gene (35%) (Figure 2: Lower Panel). Interestingly, this differential alteration of MYC gene, specially mRNA upregulation can be explained by the reported activation of Wnt-beta-catenin pathway in this subset of breast cancer as reported from our group and others [30,31]. Our study also demonstrated that the upregulation of this pathway is associated with the metastatic behavior of the disease [32] in line with other reported observations that Wnt/β-catenin signaling pathway can be a potential therapeutic target in the treatment of triple negative breast cancer [33].
Figure 3.
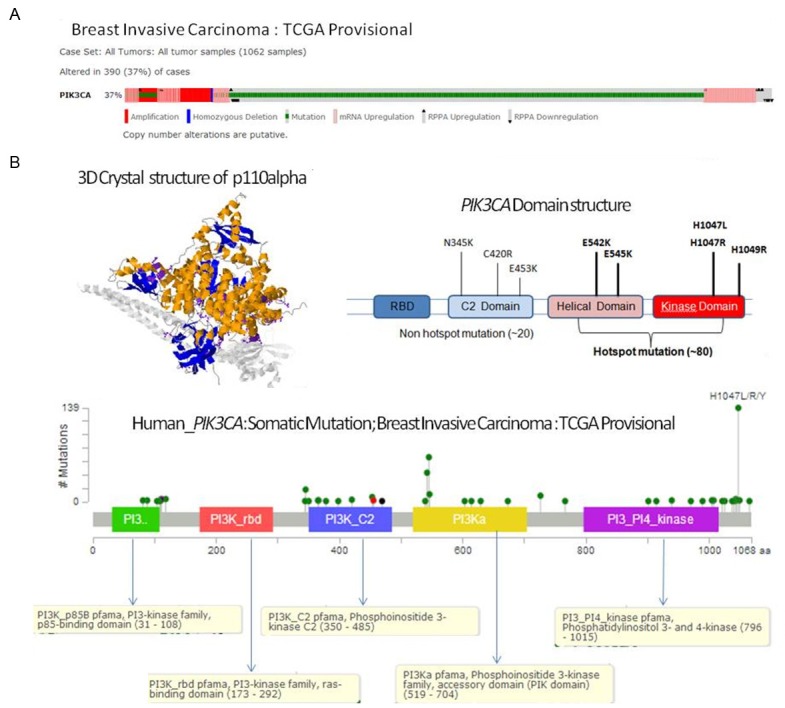
Alterations (A) and somatic mutations (B) of human PIK3CA gene in Breast Invasive Carcinoma (TCGA, Provisional). (A) Alterations (amplification, mutation, mRNA upregulation, RPPA upregulation and RPPA downregulation) of PIK3CA gene in Breast Invasive Carcinoma (TCGA, Provisional). Data was obtained using c-BioPortal. Unaltered cases and white spaces between values were removed. The case set contained 1062 tumor samples. Following Genomic Profiles were selected: (1) mutations, (2) putative copy-number alteration (CNA) from GISTIC, (3) mRNA expression Z-scores (RNA Seq V2 RSEM) with Z-score thresholds ± 2.0, and (4) protein/phospho-protein level (RPPA) with Z-score thresholds ± 2.0. (B) The numbers of somatic mutations of PIK3CA gene at different sites (domains) have been plotted in the selected samples. The somatic mutation rate of PIK3CA gene in the selected samples has been presented in the text. Different domains/regions are presented in different colors. The left inset shows the 3D crystal structure of chain A of phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (PDB 4/23: crystal structure of p110alpha complexed withnish2 of p85alpha and pi-103) as obtained from c-BioPortal. Bound molecules are displayed in the cartoon (inset). The color represents the protein by secondary structures. Alpha helics is represented in yellow, beta sheets in blue and loops in light grey. The side chain atoms for every mapped residue are displayed. All mutated residues are represented in a single violet color. The right inset shows commonly identified hotspot and non-hotspot mutations of PIK3CA in different domains as reported in various cancers which can be used as a reference. We acknowledge the cBioPortal for Cancer Genomics site (http://cbioportal.org) which provides a Web resource for exploring, visualizing, and analyzing multi-dimensional cancer genomics data. The portal reduces molecular profiling data from cancer tissues and cell lines into readily understandable genetic, epigenetic, gene expression and proteomic events [28]. We acknowledge works of Cerami et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data [29]. We acknowledge the TCGA Research Network for generating TCGA datasets.
Targeting the PI3K-AKT-mTOR signals in BC
The PI3K-AKT-mTOR signaling network has been reported to control tumorigenesis. Numerous signaling molecules of the PI3K-AKT-mTOR network are altered (mutated, up-regulated or down-regulated) in several tumor types, leading to increased PI3K signaling. The major oncogenic mutations in PI3K cluster occurs in two separate regions of its p110α catalytic subunit, the helical (E542K and E545K) and the kinase (H1047R) domains; both types yield constitutive lipid kinase activity [34,35]. According to Professor Vogelstein oncogenes (unlike tumor suppressor genes) are recurrently mutated at the same amino acid positions [36]. Except the mutation in the TP53 suppressor gene, PIK3CA hotspot mutation is the most frequently mutated gene in human breast cancers and primarily overlaps in HER2+ as well as ER+ breast cancers [37-39]. Presence of these mutations in cancer cells confer resistance to targeted therapies in both HER2+ and ER+ breast cancers [40-43] and demands pharmacological interventions of PI3K catalytic subunit (p110) inhibitor to overcome the resistance to PIK3CA-mediated therapy [8,9,44]. In addition a loss of function mutation (deletion or mutation or epigenetic alteration) of tumor suppressor PTEN, the antagonistic phosphatase to class I PI3K has been reported which leads to an increase in levels of PtdIns (3,4,5) P3 and consequently activation of AKT [45,46]. More recently activating mutations have been discovered in PIK3CA, the gene that encodes the class IA catalytic subunit p110α, in numerous cancers including lung, breast and colorectal cancer (CRC) [35]. PIK3CA mutations are thought to arise early in breast cancer development and are known to be selected for the progression as this mutation can be found in ductal carcinoma in situ as well as in invasive breast cancers/metastatic samples [47].
Agents targeting the PI3K-AKT-mTOR signaling pathways either alone or in combination have shown promise in early phase clinical trials and are currently being studied in phases I/II stages of clinical trials in various cancers including breast cancers. Several drugs targeting multiple levels of the PI3K network (that is PI3K, AKT, mTOR) have been developed. A number of ATP-mimetics that bind competitively and reversibly to the ATP-binding pocket of p110 are in early clinical development. These include the pan-PI3K inhibitors (BKM120, XL-147, PX-866, PKI-587, and GDC-0941), the p110α- specific inhibitors (BYL719, GDC-0032, and MLN1117), p110β- specific inhibitors (GSK2636771, SAR260301), the p110δ-specific inhibitor (CAL-101, Idelalisib; very recently approved by FDA for CLL patients [48]), the dual PI3K/mTOR inhibitors (BEZ235, BGT226, PF-4691502, GDC-0980, and XL-765), the allosteric inhibitors of mTOR (FDA approved everolimus, temsirolimus, and ridaforolimus) and the mTORC1/2 kinase inhibitors (MLN0128, AZD8055 and OSI-027). The pan-PI3K and p110α-specific inhibitors are equally potent against oncogenic p110α mutants. The development of isozyme-specific antagonists has permitted the delivery of significantly higher doses of the drugs (anti-p110α and anti-p110β) with appreciably lower side effects as compared to pan-PI3K inhibitors. Everolimus was the first one to be evaluated in clinical trials. BOLERO-2 trial, a randomized, double-blind, placebo-controlled phase III study of exemestane with or without everolimus (Afinitor; Novartis) in ER+/HER2- postmenopausal advanced breast cancer refractory to non-steroidal aromatase inhibitor (AI), demonstrated a median PFS of 10.6 months with combination therapy, compared with 4.1 months with exemestane alone (HR = 0.36; 95% CI Z 0.27e0.47; P < 0.001) [49], leading to U.S. Food and Drug Administration (FDA) approval for its application in the AI-resistant population. Next-generation sequencing identified almost 1,500 sequence alterations, 24 rearrangements, and about 550 copy number variations. The average tumor sample contained 4.1 genetic alterations, and at least one known somatic alteration was identified in 219 patients. The pathways harboring the most genetic alterations were PIK3CA (48%), CCND1 (31%), TP53 (23%), and FGFR1 (18%). Patients with a single alteration in one of these pathways had a median progression-free survival (PFS) of 214 days with everolimus, compared to 77 days with placebo (HR = 0.26). For those with multiple alterations, median PFS was 138 and 128 days, respectively (HR = 0.28) (2013 ASCO Annual Meeting reported by Prof. Gabriel N. Hortobagyi). The pan-PI3K inhibitor BKM120 (Novartis) is in phase III trials in combination with fulvestrant in patients with ER+ metastatic breast cancer resistant to prior AI therapy [(BELLE 2); NCT01610284] [36] or resistant also to an mTOR inhibitor [(BELLE 3); NCT01633060)]. In addition, a p110α inhibitor of PI3K has shown some promise in PIK3CA mutant breast cancer in phase I studies. In the preliminary report of this phase I study of the p110α inhibitor BYL719 (Novartis), 6/18 (33%) patients with heavily pretreated metastatic breast cancer with PIK3CA mutation achieved tumor shrinkage >20%, and two of these six patients achieved a partial response [50]. Ma et al., recently showed that the pan-PI3K inhibitor BKM120 plus fulvestrant produced partial response or disease stabilization for over 6 months in 10 of 18 patients with metastatic ER+ positive breast cancer. The results of this trial led to the ongoing phase III trials of this combination for patients with ER+ breast cancer who had disease progression on an aromatase inhibitor [51]. Another phase Ib international study showed that the combination of a p110α-specific inhibitor GDC-0032 with fulvestrant yielded a 73% response rate [52]. Emerging research showed that the combination approach of HER2 and PI3K targeting agents was required for tumor regression in preclinical model of HER2 amplification and PIK3CA mutation [9,53,54]. Dual mTORC1/2 and HER2 blockade resulted in antitumor activity in the anti-HER2 therapy resistant preclinical models. The simultaneous blockade of both the PI3K/AKT/mTOR and the MEK-ERK pathways obtained by combining lapatinib with INK-128 acts synergistically in inducing cell death and tumor regression in breast cancer models refractory to anti-HER2 therapy [54].
It has been also reported that PIK3CA mutations partially uncouple PI3K from upstream RTKs (e.g. HER2) to allow the development and maintenance of resistance to tyrosine kinase-targeted therapies [9]. The presence of PIK3CA mutations has been shown to determine clinical outcomes in different trials. Women bearing HER2+/HR+ tumors with mutations in the PI3K-AKT pathway respond poorly to neoadjuvent therapy in the German GeparSixto study (reported in San Antonio Breast Cancer symposium 2013). The combined analysis from both GeparQuino and GeparSixto studies demonstrated that in HER2+ patients, pathological complete response rates after dual HER2 blockade were significantly lower in the PIK3CA mutant group compared to patients with wild type PIK3CA (17% vs 27%). In HER2+/HR+ patients who harbored PIK3CA mutations, only 6.3% achieved pathological complete response compared with 30.8% for those without a PIK3CA mutation [55].
MYC cross-talks with PI3K-mTOR pathway
MYC is located upstream of PI3K-related DDR kinases as reported by Reimann et al., explaining the upregulation of a DDR following MYC activation and the MYC-evoked DNA damage response accounted for treatment resistance in vivo [56]. The functional involvement of MYC gene regulated transcriptional network with the PI3K-mTOR pathway has been reported to control different oncogenic cellular signals including proliferation, anti-apoptosis, nutrient response, RAS-MAPK pathway activation and DNA damage response. The PI3K family includes ataxia telangiectasia-mutated (ATM), ataxia telangiectasia and RAD3-related (ATR) and DNA-dependent protein kinase (DNA-PK). These kinases are all activated by DNA damage [57]. MYC (1) induces DNA damage, (2) increase reactive oxygen species and (3) mitigate p53 function leading to oncogene-induced genetic instability. The presence of deregulated MYC partially disables the p53-mediated DNA damage response, enabling cells with damaged genomes to enter the cycle [58].
Data from the “Basket” trials strongly advocates that the organ-site location of the tumor such as breast or lung or prostate is becoming less important than the genomic/proteomic information of the tumor (“Basket” trials; David B. Solit; Memorial Sloan Kettering Cancer center; Preannual meeting seminar, “Genetics and Genomics for the Practicing Clinicians” held immediately before the ASCO Annual meeting 2014). An undeniable piece of evidence of the oncogenic functional interaction between MYC and the PI3K-mTOR pathway can be presented from the studies in Burkitt lymphoma in which the immunoglobulin (IG) promoter-c-MYC translocation is a hallmark of the disease [59,60] and in the experimental models of the disease activation of mTOR is observed [61,62]. Synergy between PI3K signaling and MYC function has been reported by Sander et al., in Burkitt lymphomagenesis [63]. Their finding of recurrent PI3K pathway activation in human Burkitt lymphoma indicated that deregulated c-MYC and PI3K activity cooperate in Burkitt lymphoma pathogenesis. In line with this observed synergy, an impaired cytotoxic response was observed due to upregulation of NOTCH-MYC signaling following PI3K/mTOR inhibition in T-cell acute lymphoblastic leukaemia [64]. Results of the study by Shepherd et al., showed that drugs targeting PI3K-mTOR can upregulate NOTCH-MYC activity, have implications for the use of PI3K inhibitors for the treatment of other malignancies with activated NOTCH. PI-103 (PI3K and mTOR dual inhibitor) addition led to upregulation of c-MYC, which was blocked by co-incubation with a γ-secretase inhibitor (GSI; NOTCH inhibition), indicating that PI3K-mTOR inhibition resulted in activation of the NOTCH-MYC pathway as suggested by their microarray studies showing a global increase in NOTCH target gene expression upon PI3K-mTOR inhibition. NOTCH-MYC-induced resistance to PI3K-mTOR inhibition was supported by synergistic induction of cell death by PI-103 and a small molecule c-MYC inhibitor and by reduction of the cytotoxic effect of PI-103 + GSI by c-MYC overexpression. In other hematological malignancies like B-cell lymphomas, a combined inhibition of PI3K-related DNA damage response kinases and mTORC1 induced apoptosis indicating that combined DDR and mTORC1 inhibition may represent a novel therapeutic strategy for the management of MYC-driven cancers [60]. Gene expression profiling revealed elevated NOTCH1 mRNA expression in triple negative breast cancers, both basaloid and claudin-low subtypes. It has been reported that NOTCH ligands, Jagged1 and Jagged2, are correlated with poor prognosis in TNBC and AKT is activated downstream of NOTCH in breast cancer cell lines. Interestingly, Zhu et al demonstrated a correlation of NOTCH1, pAKT and nuclear NF-κB expression in triple negative breast cancer [65]. NOTCH-1 signaling pathway has been reported to be involved in mediating the effect of Genistein in the inhibition of growth of MDA-MB-231 triple-negative breast cancer cells [66]. Recently Stoeck et al., used Next-generation sequencing to identify NOTCH mutations in a large collection of diverse solid tumors including triple negative breast cancer [67]. NOTCH1 and NOTCH2 rearrangements leading to constitutive receptor activation were observed only in triple-negative breast cancers. Accordingly, they observed that TNBC cell lines with NOTCH1 rearrangements associated with high levels of activated NOTCH1 (N1-ICD) were sensitive to the gamma-secretase inhibitor (GSI) in vitro and in vivo, whereas cell lines with NOTCH2 rearrangements were resistant to GSI. In their study, immunohistochemical staining of N1-ICD in TNBC xenografts correlated with responsiveness, and expression levels of the direct NOTCH target gene HES4 was found to be correlated with outcome in patients with TNBC. These observations become relevant in the context of the involvement of MYC in the disease because of the fact that MYC is an downstream effector of NOTCH and NOTCH induced mammary tumorogenesis [68]. The above facts indicate that it is possible to target MYC-regulated pathways in combination with inhibitors of other oncogenic pathways in the basal-like subtype of breast cancer.
Another important link of the functional convergence between MYC gene and mammalian target of rapamycin (mTOR) is initiation factor 4E binding protein-1 (4EBP1), a master regulator of protein synthesis [69]. Pourdehnad et al., found that mTOR-dependent phosphorylation of 4EBP1, an important mTOR substrate is required for cancer cell survival in MYC gene-dependent tumor initiation and maintenance. The clinical implications of this critical link between MYC and mTOR-dependent phosphorylation of 4EBP1 was tested in clinics by observing the therapeutic response in human lymphomas. Clinically active mTOR inhibitor, which is capable of blocking mTOR-dependent 4EBP1 phosphorylation had remarkable therapeutic efficacy in MYC-driven hematological cancers. Studies by Li et al., on the nutrient-responsive signaling network that controls metabolic gene transcription in Drosophila larvae following amino acid (AA) starvation have provided the genetic proof of interaction between MYC and PI3K. Their data show that the widespread changes in metabolic gene expression in Drosophila larvae following AA starvation are mediated in large part through the mTOR signaling pathway and MYC is one of the important transcriptional mechanisms by which these changes occurred [70].
The unique post-translational control of MYC dynamics by sequential phosphorylation at S62 and T58 allows MYC protein to integrate upstream signals from both the RAS-MEK-ERK and the PI3K-AKT-mTOR pathways both in parallel or in series (RAS activation activates downstream PI3K), critically regulating a wide range of cell fates (growth, proliferation, and programmed cell death), energy/amino acid metabolism and DNA damage repair. MYC senses and intergrates ERK and PI3K signals [71]. In their study Lee et al., analyzed a well-defined signaling module for MYC regulation using a kinetic model constrained by experimental data and observations. In this module they observed that MYC acts as an integrator of its upstream signals that differentially regulate its stability [71]. A diagrammatic representation of the PI3K-MYC interaction has been presented in the Figure 1B (left inset). Sears et al., reported that multiple RAS-dependent phosphorylation pathways regulate MYC protein stability [72]. Their result demonstrated that an activation of the RAS-RAF-ERK pathway extends the half-life of the MYC protein to enhance the MYC activity in tumor cells. Investigating mitogen stimulation regulated two N-terminal phosphorylation sites in MYC, T58 and S62, they showed that the S62 phosphorylation mediated via ERK is required for RAS induced stabilization of MYC. T58 phosphorylation mediated by GSK3beta (a downstream effctor molecule of the PI3K-AKT signaling pathway) in a prior S62 phosphorylation dependent manner is associated with degradation of MYC. Additionally, the RAS-dependent PI3K pathway also contributes to the MYC protein accumulation via GSK3beta activity. These observations thus define a synergistic role for multiple RAS-mediated phosphorylation pathways in the control of MYC protein accumulation during the initial stage of cell proliferation. Activation of MYC via RAS activation has also been reported in neuroblastomas where active RAS is needed to block N-MYC degradation, promoting cooperative RAS- and N-MYC-dependent cell cycle progression of the tumor cells [73]. In addition to the RAS-PI3K pathway mediated increase in the accumulation of MYC protein, the PI3K pathway activation can also contribute to the transcription of MYC gene via a signaling mediator of Wnt-beta-catenin pathway (Figure 1B; left inset), the involvement of which is well documented in breast cancer [30-32]. Figure 1B; left inset represents interactions between mitogen mediated PI3K and RAS signaling pathways and role of MYC transcription factor as a “Signal Integrator”. Mitogenic stimulation with growth factors (GF)/cytokines leads to activation of the RAS-MAPK and the PI3K-mTOR pathways. MYC protein level, its transcription, its synthesis as well as post translational modification, stability and degradation are either in parallel and or serially regulated by the RAS-MAPK and the PI3K-mTOR pathways. Patterns of RAS-mediated activation of ERK and PI3K-mediated inhibition of GSK3beta determine the amount of MYC protein in a tumor cell at a particular time by controlling both its stability and degradation via post-translational phosphorylation. In breast cancers, the interactive synergy between MYC and PI3K has been reported to be mediated through estrogen enhancing proliferation of mammary epithelial cells. Seventeen-beta-estradiol stimulated both a short-term transient and a sustained increase in MYC protein expression in MCF7 cells. The MYC dependent survival signal generated by E2, which have been strongly implicated in the development of ER+ breast cancers was dependent upon basal levels of mTOR and its upstream regulators PI3K. These data provide evidence that E2 promotes survival signals in breast cancer cells through an mTOR-dependent increase in MYC expression [74].
The data obtain from the above studies provide a rational basis for the use of drug combinations that target both the pathways. It was recently reported that the induction of MYC-regulated genes is associated with poor outcome in human cancers [75]. It has been also reported by other that MYC overexpression had no effect on cellular sensitivity to doxorubicin but rendered cells even more sensitive to paclitaxel, which are two classical chemotherapeutic agents [76].
Resistance to pathway targeted inhibitors in BC
Although MYC remains one of the most commonly altered oncogenes in human cancers, yet therapies directly targeting MYC hyperactivation (or its immediate downstream) are not successful in the clinic unlike targeted therapies against activated oncogenic kinases [77]. Recently, components of the protein translation machinery have been exploited as therapeutic targets for MYC -driven cancers which may represent a highly relevant strategy for the treatment of MYC -dependent human cancers. Guided by the tumor dependency framework, mechanism of resistance to anti-cancer agents have been analyzed extensively in recent years with special focus on small molecule kinase inhibitors. At the end, these efforts have given rise to 3 main categories of resistance to targeted therapies.
1. Secondary genetic alteration in the target gene itself is one of the most common drug resistance mechanisms for the targeted therapy. This mechanism has been most extensively described and studied for imatinib, which binds the inactive conformation of ABL [78]. Secondary somatic alterations have been detected in a wide variety of tumors from patients treated with variety of targeted kinase inhibitors [79-81].
2. Activation of bypass signaling mechanisms following the treatment of targeted therapies. An illustrate example of bypass-mediated resistance has been described in EGFR-mutant non small cell lung cancer. Here, reactivation of PI3K-AKT signaling leads to drug resistance [82]. It has been reported by other that the aberrant activation of the PI3K-AKT signaling pathway in the presence of a EGFR kinase inhibitor gefitinib can occur as a result of activation of MET or by its ligand hepatocyte growth factor (HGF) or IGF-1R signaling [83-86]. Another avenue through which bypass resistance mechanism may operate involves modulation of feedback loops. Examples include HER3 pathway up-regulation in the setting of AKT inhibition [87], PI3K-AKT activation by mTOR inhibitors [88] and augmentation of AKT signaling by MEK inhibitors [89].
3. Acquired drug resistance involves genomic alterations that dysreguate signaling components acting either upstream or downstream of the target protein. To date, this kind of drug resistance mechanism has been most extensively studied for MEK and RAF inhibitors. Amplification of BRAF has been described as an alternative upstream mechanism of resistance to the MEK inhibitor AZD6244. This mechanism leads to increased MEK phosphorylation and consequently ERK activation, which no longer can be inhibited by AZD6244 [90,91]. In addition, an oncogenic PIK3CA activating mutation was sufficient to cause gefitinib resistance in EGFR mutant cancer cells and has also been observed in an erlotinib-EGFR mutant tumor, thus indicating that downstream effectors may ultimately become relevant in this context as well [92,93].
MYC expression and resistance to PI3K pathway targeted inhibitors
MYC overexpression is sufficient to confer resistance to PI3K and mTOR inhibitors. Diversity for 8q24 (this chromosomal region harbors several important oncogenes including MYC) was consistently higher in HER2+ tumors compared to other subtype. Interestingly, diversity of 8q24 was lower in primary triple negative breast cancers and increased in both distant and lymph node metastases, whereas the opposite trend was observed for HER2 cases [94].
It was an initial general consensus that mutations in the oncogenic PI3K, possibly in the kinase domain (inhibitor binding domain), would yield resistance to catalytic PI3K inhibition. But the success of the PI3K pathway-specific inhibitors at least in preclinical settings and also in some extent in clinical settings with other targeted inhibitors, translational scientists thought and found that acquired drug resistance may involve alterations that deregulated signaling component acting downstream of the targeted protein. Using an integrated copy number and expression-based-approach within the dual PI3K/mTOR inhibitor (BEZ235) resistant cells, Thomas M. Roberts and colleagues determined that MYC, a commonly deregulated breast cancer oncogene, was responsible for the acquired BEZ235 resistance [76]. Following their study, Pixu Liu and group have demonstrated by using transgenic mouse models and a series of elegant experiments that MYC contributed to PIK3CA independent of tumor growth and resistance to PI3K-catalytic inhibition. They found that in some mice tumors rapidly and completely regressed to a non-palpable state within 1-2 months following inactivation of oncogenic PIK3CA with no re-growth indicating that these tumors remained addicted on PIK3CA for their maintenance. However, in another group of mice tumors partially regressed but then resumed growth in the absence of PIK3CA activation [95]. These studies clearly specify that some tumors although initiated with PIK3CA activating mutation but during the tumor progression they are no longer dependent on PIK3CA status and probably not respond to catalytic inhibition of PI3K. Their SNP array analyses of recurrent tumors revealed a common amplification on chromosome 15 spanning 1.48 Mb (Chromosome 15:61,271,320-62,750,432), which contains the coding sequence for a single gene, MYC and knockdown of MYC dramatically reduced tumor incidence and extended the time to tumor onset. A chemical genetic screen identified MYC and NOTCH pathway activation as mechanisms of resistance to PI3K inhibitors in breast cancer cell lines [96]. Molecular analyses revealed that both the mouse and human MYC genes are direct transcriptional target of NOTCH not only in breast cancer but also in T-cell acute lymphoblastic leukemia/lymphoma (as mentioned before) [97-99]. Klinakis and groups’ mouse genetic results clearly demonstrated that MYC was indispensable for the development of NOTCH regressing mammary carcinoma. They also demonstrated by using immune-staining that >90% of the cases (20/22) exhibited high MYC expression, NOTCH was also highly expressed [97]. It has been also reported by others that treatment of cells with dual PI3K/mTOR inhibitor (PI-103) led to upregulation of MYC, which was blocked by co-incubation with g-secretase inhibitor, indicating that dual PI3K/mTOR inhibition resulted in activation of the NOTCH-MYC pathway. The NOTCH-MYC pathway-induced resistance to PI3K/mTOR inhibition was supported by synergistic cell death induction by PI-103 plus MYC inhibition, and by reduction of cytotoxic effect of PI-103 plus g-secretase inhibitor by MYC overexpression [64]. Experiments carried out by Muellner MK et al showed that cells transduced with MYC displayed high level of resistance to dual PI3K/mTOR inhibitor or PI3K inhibitors. Similarly, knockdown of MYC to levels comparable to non-transformed control cells completely reversed the resistance to dual PI3K/mTOR inhibitor. Interestingly, overexpression of the NOTCH canonical target genes HES1, HEY1 or HEY2 did not confer PI3K pathway inhibitor resistance [96]. Furthermore, there are several human breast cancer data bases confirming increased MYC expression in PIK3CA mutant breast cancers with frequencies ranging from 27% to 47% [38,100].
It has been known for quite some time that MYC is one of the downstream target molecules of the PI3K-AKT signaling pathway in AKT-mediated phosphorylation and inhibition of GSK3β prevents the phosphorylation and degradation of MYC protein [101]. MYC is also a downstream effector molecule of the mTOR pathway. The stimulation of translation of MYC through the induction of eukaryotic initiation factor 4E (a major mediator of cap-dependent mRNA translation and tightly regulated by mTOR pathway) is one of the known mechanisms MYC drives protein translation and is implicated in MYC-mediated tumorigenesis [102,103]. Recently, Ilic et al reported that eIF4E gene amplification/overexpression developed as a compensatory resistance mechanism in cells initially sensitive to the combined PI3K/mTOR inhibition. Considering that the gene encoding eIF4E is an established MYC-regulated target, cooperation between MYC and eIF4E in regulating resistance mechanism is a possibility [76,104]. These findings suggest that aberrant elevation of MYC represents a potential mechanism by which tumors develop resistance to PI3K inhibition, and thus combination therapies targeting both PI3K and MYC (or its upstream affector molecule, like g-secretase since MYC lacks critical hydrophobic pockets it is highly challenging to target by small molecule compounds, [105,106] may be necessary and sufficient to circumvent resistance to PI3K-targeted therapy. Recently, Elkabrts and colleagues have reported that despite the presence of activating PIK3CA mutations, not all patients benefited from BYL719 (p110α-specfic inhibitor) treatment, suggesting that their tumors may be intrinsically resistant to p110α-specfic inhibitor [107]. Their cell line-based mechanistic data revealed that breast cancer cells resistant to BYL719 had persistent activation of mTORC1 signaling, although AKT phosphorylation was dampened. Here, we can argue that mTOR-dependent upregulation in MYC expression may be the cause for BYL719 resistant in PIK3CA mutated breast cancer cells [please see Rodrik, V. et al., 2005 [74]].
Beside breast cancer, in anaplastic thyroid carcinoma model MYC cooperates with PI3K activation to induce more aggressive features, including pan-PI3K drug (BKM120) resistance and also enhanced metastatic behavior[108]. In addition to activating PI3K-AKT and MEK-ERK signaling, mTOR inhibition by rapamycin can also induce MYC protein phosphorylation and accumulation in colorectal cancer cells. Functional analysis indicates that rapamycin-induced MYC phosphorylation is dependent on PDK1 but independent of PI3K and AKT activity [109]. They also reported that rapamycin-induced MYC activation is associated with the loss of PPP2R2B, which encodes for the B55β regulatory subunit of the serine/threonine protein phosphatase PP2A and loss of PPP2R2B results in aberrant activation of PDK1. They further demonstrated that pharmacological or genetic inhibition of PDK1 markedly inhibited MYC phosphorylation, leading to enhance sensitivity to rapamycin in colon cancer cells.
Factors other than MYC activation are also responsible for PI3K inhibitor resistance in PIK3CA mutated conditions. Multiple studies have described clinically relevant and potentially targetable adaptive responses as well as pre-existing or acquired genetic resistance to PI3K inhibitor, including downstream activation of mTOR (via variety of mechanisms independent of AKT activation) or the RAS-RAF-MEK-ERK pathway activation or increased expression of anti-apoptotic BCL2 family members [101,110]. Recently, Vora et al developed PI3K inhibitor resistant breast cancer cell lines and demonstrated PI3K inhibitor independent CyclinD1/CDK4-dependent RB phosphorylation leading to E2F dependent cell survival and proliferation. They also demonstrated that a combination of CDK4/6 inhibitor plus PI3K inhibitor synergistically reduced in vitro cell viability and in vivo tumor regression in BYL719-resistant/PIK3CA mutant xenografts [111].
Treatment strategy and future perspective
The MYC is involved in many critical processes in malignant cells, including proliferation, growth, differentiation, and metabolism [112]. Since MYC activation/amplification is an important mechanism underlying resistance to the PI3K-AKT-mTOR pathway inhibitors, developing an effective therapeutic strategy for targeting MYC may be necessary to overcome this pathway-targeted drug-resistance. Furthermore, MYC also plays an important role for cancer stem cells initiation and maintenance and its association with tumor recurrence following treatment indicate that MYC induction following the PI3K-AKT-mTOR pathway-specific inhibition may be a serious issue in the clinic. Although MYC is a valid candidate for cancer target, direct inhibition of MYC has not successful yet in the clinics. Thus, alternative strategies include targeting MYC by (a) context dependent upstream of MYC affector candidates like NOTCH, PDK1 and (b) interfering with key MYC downstream target gene(s). Future clinical trial design for PI3K-pathawy specific inhibitor should focus on (1) continued efforts to select appropriate patients following genomic profiling, (2) continued efforts for the tissue collection in post-neoadjuvent as well as metastatic settings for better understanding of the significant pathway(s) alterations that are associated with tumor pathogenesis and development of therapeutic resistance and (3) combination of complementary pathway inhibitor to maximize the clinical efficacy and minimize resistance to the therapy.
Conclusion
In an ideal world, a resistance will be predictable based on (1) the genomic background preexisting within the tumor and (2) the nature of signals the drug impedes. Our collective goal will be to establish a knowledge base that can be utilized to predict possible mechanisms of resistance to these inhibitors, especially if such mechanisms may preexist within the tumors. It would be crucial to obtain such information beforehand because this would render clinicians the advantage of “first strike” in their war against drug-induced resistance in breast cancers; choosing patients who might benefit maximally from this pharmacological intervention. Like other targeted therapies, understanding the diversity of mechanisms that give rise to PI3K inhibitor resistance utilizing laboratory-based preclinical models is expected to help clinical hypothesis testing and ultimately provide the rational of right combination strategy that can overcome resistance mechanism(s).
Acknowledgements
We acknowledge the TCGA Research Network for generating TCGA datasets which were used in this study. We used cBioPortal for Cancer Genomics which provides visualization, analysis and download of large-scale cancer genomics data sets. We acknowledge works of Gao et al. Sci. Signal. 2013 & Cerami et al. Cancer Discov. 2012 as we used cBioPortal. Authors acknowledge Avera Research Institute, Sioux Falls, SD and Dept. of Internal Medicine, SSOM, USD, Sioux Falls, SD.
Disclosure of conflict of interest
The author(s) confirm that this article content has no conflicts of interest.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 3.Kadota M, Sato M, Duncan B, Ooshima A, Yang HH, Diaz-Meyer N, Gere S, Kageyama S, Fukuoka J, Nagata T, Tsukada K, Dunn BK, Wakefield LM, Lee MP. Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. Cancer Res. 2009;69:7357–7365. doi: 10.1158/0008-5472.CAN-09-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, Capdeville R, Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 5.Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, Fletcher JA, Silverman SG, Silberman SL, Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker BJ, Corless C, Fletcher CD, Joensuu H. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 6.Inoue A, Suzuki T, Fukuhara T, Maemondo M, Kimura Y, Morikawa N, Watanabe H, Saijo Y, Nukiwa T. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J. Clin. Oncol. 2006;24:3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez CG, Ma CX, Crowder RJ, Guintoli T, Phommaly C, Gao F, Lin L, Ellis MJ. Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer. Breast Cancer Res. 2011;13:R21. doi: 10.1186/bcr2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rexer BN, Chanthaphaychith S, Dahlman K, Arteaga CL. Direct inhibition of PI3K in combination with dual HER2 inhibitors is required for optimal antitumor activity in HER2+ breast cancer cells. Breast Cancer Res. 2014;16:R9. doi: 10.1186/bcr3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 11.De P, Carlson J, Leyland-Jones B, Dey N. Oncogenic nexus of cancerous inhibitor of protein phosphatase 2A (CIP2A): An oncoprotein with many hands. Oncotarget. 2014;5:4581–602. doi: 10.18632/oncotarget.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop JM. Oncogenes. Sci Am. 1982;246:80–92. doi: 10.1038/scientificamerican0382-80. [DOI] [PubMed] [Google Scholar]
- 13.Bishop JM. Viruses, genes, and cancer. Harvey Lect. 1982;78:137–172. [PubMed] [Google Scholar]
- 14.Bishop JM. Retroviruses and cancer genes. Adv Cancer Res. 1982;37:1–32. doi: 10.1016/s0065-230x(08)60880-5. [DOI] [PubMed] [Google Scholar]
- 15.Liao DJ, Dickson RB. c-Myc in breast cancer. Endocr Relat Cancer. 2000;7:143–164. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- 16.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blancato J, Singh B, Liu A, Liao DJ, Dickson RB. Correlation of amplification and overexpression of the c-myc oncogene in high-grade breast cancer: FISH, in situ hybridisation and immunohistochemical analyses. Br J Cancer. 2004;90:1612–1619. doi: 10.1038/sj.bjc.6601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Olopade OI. MYC in breast tumor progression. Expert Rev Anticancer Ther. 2008;8:1689–1698. doi: 10.1586/14737140.8.10.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chrzan P, Skokowski J, Karmolinski A, Pawelczyk T. Amplification of c-myc gene and overexpression of c-Myc protein in breast cancer and adjacent non-neoplastic tissue. Clin Biochem. 2001;34:557–562. doi: 10.1016/s0009-9120(01)00260-0. [DOI] [PubMed] [Google Scholar]
- 20.Musgrove EA, Sergio CM, Loi S, Inman CK, Anderson LR, Alles MC, Pinese M, Caldon CE, Schutte J, Gardiner-Garden M, Ormandy CJ, McArthur G, Butt AJ, Sutherland RL. Identification of functional networks of estrogen- and c-Myc-responsive genes and their relationship to response to tamoxifen therapy in breast cancer. PLoS One. 2008;3:e2987. doi: 10.1371/journal.pone.0002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grushko TA, Dignam JJ, Das S, Blackwood AM, Perou CM, Ridderstrale KK, Anderson KN, Wei MJ, Adams AJ, Hagos FG, Sveen L, Lynch HT, Weber BL, Olopade OI. MYC is amplified in BRCA1-associated breast cancers. Clin Cancer Res. 2004;10:499–507. doi: 10.1158/1078-0432.ccr-0976-03. [DOI] [PubMed] [Google Scholar]
- 22.Adem C, Soderberg CL, Hafner K, Reynolds C, Slezak JM, Sinclair CS, Sellers TA, Schaid DJ, Couch F, Hartmann LC, Jenkins RB. ERBB2, TBX2, RPS6KB1, and MYC alterations in breast tissues of BRCA1 and BRCA2 mutation carriers. Genes Chromosomes Cancer. 2004;41:1–11. doi: 10.1002/gcc.20057. [DOI] [PubMed] [Google Scholar]
- 23.Ren J, Jin F, Yu Z, Zhao L, Wang L, Bai X, Zhao H, Yao W, Mi X, Wang E, Olopade OI, Wei M. MYC overexpression and poor prognosis in sporadic breast cancer with BRCA1 deficiency. Tumour Biol. 2013;34:3945–3958. doi: 10.1007/s13277-013-0983-9. [DOI] [PubMed] [Google Scholar]
- 24.Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 25.Liu S, Knapp S, Ahmed AA. The structural basis of PI3K cancer mutations: from mechanism to therapy. Cancer Res. 2014;74:641–646. doi: 10.1158/0008-5472.CAN-13-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 27.Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dey N, Young B, Abramovitz M, Bouzyk M, Barwick B, De P, Leyland-Jones B. Differential activation of Wnt-beta-catenin pathway in triple negative breast cancer increases MMP7 in a PTEN dependent manner. PLoS One. 2013;8:e77425. doi: 10.1371/journal.pone.0077425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, Natrajan R, Reis-Filho JS. beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol. 2011;24:209–231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 32.Dey N, Barwick BG, Moreno CS, Ordanic-Kodani M, Chen Z, Oprea-Ilies G, Tang W, Catzavelos C, Kerstann KF, Sledge GW Jr, Abramovitz M, Bouzyk M, De P, Leyland-Jones BR. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer. 2013;13:537. doi: 10.1186/1471-2407-13-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King TD, Suto MJ, Li Y. The Wnt/beta-catenin signaling pathway: a potential therapeutic target in the treatment of triple negative breast cancer. J Cell Biochem. 2012;113:13–18. doi: 10.1002/jcb.23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuels Y, Diaz LA Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 36.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 38.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, Symmans WF, Pusztai L, Nolden LK, Horlings H, Berns K, Hung MC, van de Vijver MJ, Valero V, Gray JW, Bernards R, Mills GB, Hennessy BT. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, Sampath D, Sliwkowski MX. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 41.De P, Miskimins K, Dey N, Leyland-Jones B. Promise of rapalogues versus mTOR kinase inhibitors in subset specific breast cancer: old targets new hope. Cancer Treat Rev. 2013;39:403–412. doi: 10.1016/j.ctrv.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Ma CX, Crowder RJ, Ellis MJ. Importance of PI3-kinase pathway in response/resistance to aromatase inhibitors. Steroids. 2011;76:750–752. doi: 10.1016/j.steroids.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 43.Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. J. Clin. Oncol. 2011;29:4452–4461. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S, Maira M, Garcia-Echeverria C, Parra JL, Arribas J, Baselga J. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 46.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 47.Kalinsky K, Heguy A, Bhanot UK, Patil S, Moynahan ME. PIK3CA mutations rarely demonstrate genotypic intratumoral heterogeneity and are selected for in breast cancer progression. Breast Cancer Res Treat. 2011;129:635–643. doi: 10.1007/s10549-011-1601-4. [DOI] [PubMed] [Google Scholar]
- 48.Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, Barrientos JC, Zelenetz AD, Kipps TJ, Flinn I, Ghia P, Eradat H, Ervin T, Lamanna N, Coiffier B, Pettitt AR, Ma S, Stilgenbauer S, Cramer P, Aiello M, Johnson DM, Miller LL, Li D, Jahn TM, Dansey RD, Hallek M, O’Brien SM. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370:997–1007. doi: 10.1056/NEJMoa1315226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baselga J, Campone M, Piccart M, Burris HA 3rd, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, Beck JT, Ito Y, Yardley D, Deleu I, Perez A, Bachelot T, Vittori L, Xu Z, Mukhopadhyay P, Lebwohl D, Hortobagyi GN. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Juric D, Argiles G, Burris HA, Gonzalez-Angulo AM, Saura C, Quadt C, Douglas M, Demanse D, De Buck S, Baselga J. Phase I study of BYL719, an alpha-specific PI3K inhibitor, in patients with PIK3CA mutant advanced solid tumors: preliminary efficacy and safety in patients with PIK3CA mutant ER-positive (ER+) metastatic breast cancer (MBC) San Antonio Breast Cancer Symposium. 2012 Abstract # P6-10-07. [Google Scholar]
- 51.Ma CX, Wang J, Luo J, et al. A phase I study of BKM120 and fulvestrant in postmenopausal women with estrogen receptor-positive metastatic breast cancer. San Antonio Breast Cancer Symposium. 2013 Abstract # PD1-4. [Google Scholar]
- 52.Juric D, Saura C, Cervantes A, et al. Ph1b study of the PI3K inhibitor GDC-0032 in combination with fulvestrant in patients with hormone receptor- positive advanced breast cancer. San Antonio Breast Cancer Symposium. 2013 Abstract # PD1-3. [Google Scholar]
- 53.Eichhorn PJ, Gili M, Scaltriti M, Serra V, Guzman M, Nijkamp W, Beijersbergen RL, Valero V, Seoane J, Bernards R, Baselga J. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia-Garcia C, Ibrahim YH, Serra V, Calvo MT, Guzman M, Grueso J, Aura C, Perez J, Jessen K, Liu Y, Rommel C, Tabernero J, Baselga J, Scaltriti M. Dual mTORC1/2 and HER2 blockade results in antitumor activity in preclinical models of breast cancer resistant to anti-HER2 therapy. Clin Cancer Res. 2012;18:2603–2612. doi: 10.1158/1078-0432.CCR-11-2750. [DOI] [PubMed] [Google Scholar]
- 55.Loibl S, Denkert C, Schneeweis A, et al. PIK3CA mutation predicts resistance to anti-HER2/chemotherapy in primary HER2-positive/hormone receptor-positive breast cancer—1(1):prospective analysis of 737 participants of the GeparSixto and GeparQuinto studies. San Antonio Breast Cancer Symposium. 2013 Abstract # S4-06. [Google Scholar]
- 56.Reimann M, Loddenkemper C, Rudolph C, Schildhauer I, Teichmann B, Stein H, Schlegelberger B, Dorken B, Schmitt CA. The Myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood. 2007;110:2996–3004. doi: 10.1182/blood-2007-02-075614. [DOI] [PubMed] [Google Scholar]
- 57.Al-Ejeh F, Kumar R, Wiegmans A, Lakhani SR, Brown MP, Khanna KK. Harnessing the complexity of DNA-damage response pathways to improve cancer treatment outcomes. Oncogene. 2010;29:6085–6098. doi: 10.1038/onc.2010.407. [DOI] [PubMed] [Google Scholar]
- 58.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 59.Klapproth K, Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. Br J Haematol. 2010;149:484–497. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- 60.Shortt J, Martin BP, Newbold A, Hannan KM, Devlin JR, Baker AJ, Ralli R, Cullinane C, Schmitt CA, Reimann M, Hall MN, Wall M, Hannan RD, Pearson RB, McArthur GA, Johnstone RW. Combined inhibition of PI3K-related DNA damage response kinases and mTORC1 induces apoptosis in MYC-driven B-cell lymphomas. Blood. 2013;121:2964–2974. doi: 10.1182/blood-2012-08-446096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT, Kogan SC, Nadon R, Housman DE, Lowe SW, Pelletier J. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci U S A. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ravitz MJ, Chen L, Lynch M, Schmidt EV. c-myc Repression of TSC2 contributes to control of translation initiation and Myc-induced transformation. Cancer Res. 2007;67:11209–11217. doi: 10.1158/0008-5472.CAN-06-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sander S, Calado DP, Srinivasan L, Kochert K, Zhang B, Rosolowski M, Rodig SJ, Holzmann K, Stilgenbauer S, Siebert R, Bullinger L, Rajewsky K. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell. 2012;22:167–179. doi: 10.1016/j.ccr.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shepherd C, Banerjee L, Cheung CW, Mansour MR, Jenkinson S, Gale RE, Khwaja A. PI3K/mTOR inhibition upregulates NOTCH-MYC signalling leading to an impaired cytotoxic response. Leukemia. 2013;27:650–660. doi: 10.1038/leu.2012.285. [DOI] [PubMed] [Google Scholar]
- 65.Zhu H, Bhaijee F, Ishaq N, Pepper DJ, Backus K, Brown AS, Zhou X, Miele L. Correlation of Notch1, pAKT and nuclear NF-kappaB expression in triple negative breast cancer. Am J Cancer Res. 2013;3:230–239. [PMC free article] [PubMed] [Google Scholar]
- 66.Pan H, Zhou W, He W, Liu X, Ding Q, Ling L, Zha X, Wang S. Genistein inhibits MDA-MB-231 triple-negative breast cancer cell growth by inhibiting NF-kappaB activity via the Notch-1 pathway. Int J Mol Med. 2012;30:337–343. doi: 10.3892/ijmm.2012.990. [DOI] [PubMed] [Google Scholar]
- 67.Stoeck A, Lejnine S, Truong A, Pan L, Wang H, Zang C, Yuan J, Ware C, MacLean J, Garrett-Engele PW, Kluk M, Laskey J, Haines BB, Moskaluk C, Zawel L, Fawell S, Gilliland G, Zhang T, Kremer BE, Knoechel B, Bernstein BE, Pear WS, Liu XS, Aster JC, Sathyanarayanan S. Discovery of Biomarkers Predictive of GSI Response in Triple-Negative Breast Cancer and Adenoid Cystic Carcinoma. Cancer Discov. 2014;4:1154–1167. doi: 10.1158/2159-8290.CD-13-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu J, Chen Y, Olopade OI. MYC and Breast Cancer. Genes Cancer. 2010;1:629–640. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci U S A. 2013;110:11988–11993. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L, Edgar BA, Grewal SS. Nutritional control of gene expression in Drosophila larvae via TOR, Myc and a novel cis-regulatory element. BMC Cell Biol. 2010;11:7. doi: 10.1186/1471-2121-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee T, Yao G, Nevins J, You L. Sensing and integration of Erk and PI3K signals by Myc. PLoS Comput Biol. 2008;4:e1000013. doi: 10.1371/journal.pcbi.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yaari S, Jacob-Hirsch J, Amariglio N, Haklai R, Rechavi G, Kloog Y. Disruption of cooperation between Ras and MycN in human neuroblastoma cells promotes growth arrest. Clin Cancer Res. 2005;11:4321–4330. doi: 10.1158/1078-0432.CCR-04-2071. [DOI] [PubMed] [Google Scholar]
- 74.Rodrik V, Zheng Y, Harrow F, Chen Y, Foster DA. Survival signals generated by estrogen and phospholipase D in MCF-7 breast cancer cells are dependent on Myc. Mol Cell Biol. 2005;25:7917–7925. doi: 10.1128/MCB.25.17.7917-7925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolfer A, Wittner BS, Irimia D, Flavin RJ, Lupien M, Gunawardane RN, Meyer CA, Lightcap ES, Tamayo P, Mesirov JP, Liu XS, Shioda T, Toner M, Loda M, Brown M, Brugge JS, Ramaswamy S. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci U S A. 2010;107:3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A. 2011;108:E699–708. doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 78.Sherbenou DW, Druker BJ. Applying the discovery of the Philadelphia chromosome. J Clin Invest. 2007;117:2067–2074. doi: 10.1172/JCI31988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 80.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 81.Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H, Ishikawa Y, Kimura H, Mitsudomi T, Tanio Y, Mano H ALK Lung Cancer Study Group. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 82.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 83.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 84.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang WC, Yu CJ, Gazdar A, Pass H, Rusch V, Gerald W, Huang SF, Yang PC, Miller V, Ladanyi M, Yang CH, Pao W. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, Lindeman NI, Murphy C, Akhavanfard S, Yeap BY, Xiao Y, Capelletti M, Iafrate AJ, Lee C, Christensen JG, Engelman JA, Janne PA. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T, Ogino H, Kakiuchi S, Hanibuchi M, Nishioka Y, Uehara H, Mitsudomi T, Yatabe Y, Nakamura T, Sone S. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 87.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, Baselga J, Rosen N. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mirzoeva OK, Das D, Heiser LM, Bhattacharya S, Siwak D, Gendelman R, Bayani N, Wang NJ, Neve RM, Guan Y, Hu Z, Knight Z, Feiler HS, Gascard P, Parvin B, Spellman PT, Shokat KM, Wyrobek AJ, Bissell MJ, McCormick F, Kuo WL, Mills GB, Gray JW, Korn WM. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res. 2009;69:565–572. doi: 10.1158/0008-5472.CAN-08-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, Edwards PA, Smith PD, Cook SJ. Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Sci Signal. 2011;4:ra17. doi: 10.1126/scisignal.2001752. [DOI] [PubMed] [Google Scholar]
- 91.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, Salton M, Dahlman KB, Tadi M, Wargo JA, Flaherty KT, Kelley MC, Misteli T, Chapman PB, Sosman JA, Graeber TG, Ribas A, Lo RS, Rosen N, Solit DB. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, Naumov GN, Yeap BY, Jarrell E, Sun J, Tracy S, Zhao X, Heymach JV, Johnson BE, Cantley LC, Janne PA. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, Akhavanfard S, Heist RS, Temel J, Christensen JG, Wain JC, Lynch TJ, Vernovsky K, Mark EJ, Lanuti M, Iafrate AJ, Mino-Kenudson M, Engelman JA. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Almendro V, Kim HJ, Cheng YK, Gonen M, Itzkovitz S, Argani P, van Oudenaarden A, Sukumar S, Michor F, Polyak K. Genetic and phenotypic diversity in breast tumor metastases. Cancer Res. 2014;74:1338–1348. doi: 10.1158/0008-5472.CAN-13-2357-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, Yang J, Semaan DJ, Chen C, Fox EA, Gray NS, Monahan J, Schlegel R, Beroukhim R, Mills GB, Zhao JJ. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muellner MK, Uras IZ, Gapp BV, Kerzendorfer C, Smida M, Lechtermann H, Craig-Mueller N, Colinge J, Duernberger G, Nijman SM. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat Chem Biol. 2011;7:787–793. doi: 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klinakis A, Szabolcs M, Politi K, Kiaris H, Artavanis-Tsakonas S, Efstratiadis A. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci U S A. 2006;103:9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP, Aster JC, Sigaux F, Soulier J, Look AT, Young RA, Califano A, Ferrando AA. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VE, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 101.Klempner SJ, Myers AP, Cantley LC. What a tangled web we weave: emerging resistance mechanisms to inhibition of the phosphoinositide 3-kinase pathway. Cancer Discov. 2013;3:1345–1354. doi: 10.1158/2159-8290.CD-13-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lin CJ, Malina A, Pelletier J. c-Myc and eIF4F constitute a feedforward loop that regulates cell growth: implications for anticancer therapy. Cancer Res. 2009;69:7491–7494. doi: 10.1158/0008-5472.CAN-09-0813. [DOI] [PubMed] [Google Scholar]
- 103.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 104.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 105.Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83:1688–1695. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 107.Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, Tao J, Campos AB, Rodon J, Ibrahim YH, Serra V, Rodrik-Outmezguine V, Hazra S, Singh S, Kim P, Quadt C, Liu M, Huang A, Rosen N, Engelman JA, Scaltriti M, Baselga J. mTORC1 inhibition is required for sensitivity to PI3K p110alpha inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med. 2013;5:196ra199. doi: 10.1126/scitranslmed.3005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Russo MA, Cristofano AD, et al. MYC amplification and overexpression Is associated with metastasis and drug resistance in a mouse model of anaplastic thyroid carcinoma. 104th Annual Meeting of the American Association for Cancer Research. 2013 Abstract # 5192. [Google Scholar]
- 109.Tan J, Lee PL, Li Z, Jiang X, Lim YC, Hooi SC, Yu Q. B55beta-associated PP2A complex controls PDK1-directed myc signaling and modulates rapamycin sensitivity in colorectal cancer. Cancer Cell. 2010;18:459–471. doi: 10.1016/j.ccr.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 110.Bagrodia S, Smeal T, Abraham RT. Mechanisms of intrinsic and acquired resistance to kinase-targeted therapies. Pigment Cell Melanoma Res. 2012;25:819–831. doi: 10.1111/pcmr.12007. [DOI] [PubMed] [Google Scholar]
- 111.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, Lockerman EL, Pollack SF, Liu M, Li X, Lehar J, Wiesmann M, Wartmann M, Chen Y, Cao ZA, Pinzon-Ortiz M, Kim S, Schlegel R, Huang A, Engelman JA. CDK 4/6 Inhibitors Sensitize PIK3CA Mutant Breast Cancer to PI3K Inhibitors. Cancer Cell. 2014;26:136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]