Abstract
Background: Radiation resistance poses a major clinical challenge in treatment of esophageal squamous cell carcinoma (ESCC). However, the mechanisms of radioresistance has not been fully elucidated. Since accumulating evidence demonstrates that aberrant expression of microRNAs (miRNAs) contributes to cancer sensitivity to radiation, we aimed to identify miRNAs associated with radioresistance of ESCC. Methods: In this study, we used GeneChip miRNA Array to perform an comparison of miRNAs expression in tissues from primary ESCC and recurrent ESCC in situ after radiotherapy. Differential expressions of miRNAs were comfirmed by quantitative Real-Time PCR in tissues and six ESCC cell lines. Cell radiosensitivity were determined by colony formation assay. Functional analyses of miRNA-381 in ESCC cells growth and metastasis were performed by MTT and Transwell Assays. In vivo assays of the functions of miRNA-381 were performed in tumor xenografts. Results: One miRNA candidate, miRNA-381, was found to be downregulated in radiation resistance tissues and cells. Enforced expression of miRNA-381 increased radiosensitivity of ESCC cells and promoted nonaggressive phenotype including decreased cellular proliferation and migration. In contrast, inhibition of miRNA-381 in ESCC cells promoted radiation resistance and development of an aggressive phenotype. In vivo assays extended the significance of these results, showing that miRNA-381 overexpression decreased the tumor growth and the resistance to radiation treatment in tumor xenografts. Conclusions: Together, our work reveals miRNA-381 expression as a critical determinant of radiosensitivity in esophageal cancer cells.
Keywords: microRNA, esophageal squamous cell carcinoma, radioresistance, aberrant expression
Introduction
Esophageal carcinoma (EC) remains one of the leading causes of death due to cancer [1], with a 5-year survival rate of < 20% [2]. Esophageal cancer usually occurs as either adenocarcinoma or squamous cell carcinoma (ESCC) which dominantly occurred in East Asia and accounts for 95% of all Chinese EC patients [3]. Currently, the radiotherapy has been considered as an effective, well-established treatment for ESCC. However, patients can still develop recurrent cancer which tend to display a more aggressive phenotype because of the inherent ability of ESCC cells to become radioresistant [4]. Thus, understanding the molecular mechanisms of ESCC underlying radiation sensitivity or resistance may ultimately improve the effectiveness of radiation treatment in killing cancer cells and minimize the risk of recurrence [5].
Previous studies have identified an association between radioresistance and the aberrant expression of many genes, making these genes as potential therapeutic targets for treatment of cancer. However, direct alteration of single gene usually insufficiently affect cellular radiosensitivity due to multiple regulated signaling of cancer progress [6]. At present, the microRNAs (miRNAs), a class of small noncoding RNA molecules, is becoming a promising and emerging research topic in cancer radioresistance because of their potential ability to simultaneously regulate multiple oncogenic pathways that may influence radiation response [7,8]. By completely or incompletely binding to target mRNAs, miRNAs can block translation or lead to degradation of mRNAs, resulting in the inhibited expression of various proteins [9]. Accumulating evidence reveals that differential expression of miRNAs may contribute to many human diseases, including cancer [10]. Depending on their potential target genes, miRNAs can act both onco- and anti-onco effects and regulate many cancer-associated behaviors, such as metastasis, invasion, angiogenesis, and chemoresistant phenotype [11]. In addition, several miRNAs have been correlated with patient survival and may be involved in cancer radioresistance [12]. However, there are little data on the miRNAs expression profile of radioresistance in ESCC, and the potential functions and underlying mechanisms of these miRNAs remain unclear.
In this study, we show that miRNA-381 is differential expression in ESCC tissues and cells with different radiosensitivity and can sensitize ESCC cells to radiation treatment. We also characterize the functions of miRNA-381 in tumour progression, which show a negative regulation in cell proliferation, migration, and invasion. Thus, we have identified a novel microRNA as a key regulator of radiosensitivity in ESCC.
Materials and method
Human sample collection
Three pairs of specimens of primary ESCC and recurrent ESCC in situ after radiotherapy from the same patient were collected from Tangdu Hospiatl of Forth Military Medical University. This study was approved by the IRB of Forth Military Medical University. All tissues were pathologically examined. Written informed consent forms were obtained from all subjects and all clinical investigation had been conducted according to the principles expressed in the Declaration of Helsinki.
Cell lines and cell culture
The human ESCC cell lines TE1, ECA109, EC9706, KYSE30, KYSE150, and KYSE450 were obtained from American Type Culture Collection (Manassas, VA) and grown in complete growth medium as recommended by the manufacturer. These cells were maintained in a humidified 5% CO2 atmosphere at 37°C. Each cell line was regularly authenticated by verifying its morphology and testing to confirm the absence of mycoplasma contamination.
Colony formation assay
Cells were seeded at various densities (1 × 102-2 × 104) of cells in six-well tissue culture plates with complete medium. Twenty-four hours later, the cells were treated with a single dose of X irradiation (0, 2, 4, 6, or 8 Gy) and then incubated for additional 14 days. The cultures were fixed with methanol and stained with crystal violet. The number of colonies with > 50 cells was counted under a dissecting microscope. Plating efficiencies (PE) were calculated as the number of colonies divided by the number of cells seeded. The survival fraction (SF) was calculated using the following equation: SF = Colonies Counted/Cells Seeded× (PE/100). Three independent experiments were performed. The data were analyzed using SigmaPlot 12.0 software (Systat Software, Inc., CA, USA) with the linear-quadratic (LQ) model to calculate the parameters SF2, D0, Dq, and N.
RNA isolation, microarrays, and quantitative Real-Time PCR
Total RNA was extracted from the ESCC tissues or cells using TRIzol reagent (Invitrogen, San Diego, CA) according to the manufacturer’s instructions. The Affymetrix GeneChip miRNA Array (Santa Clara, CA) is composed of 6703 probe sets for miRNAs that are registered in the Sanger miRBase database version 19 (http://www.mirbase.org/). The arrays were performed at KangChen Bio-Tech (Shanghai, China) as described both on the KangChen website and in previous reports.
Quantitative real-time PCR (qRT-PCR) was performed on a BioRad iQ5 Real-Time PCR Detection System with a SYBR Green I Master Mix (TAKARA, Japan). miRNA abundance was normalized to U6. RNA from three separate cell pellets pretreatment was analyzed. Relative gene expression was calculated using the method given in Applied Biosystems User Bulletin No. 2. (P/N 4303859B), with non-targeting miRNA-treated cells acting as the control in each data set. Primer pairs used in this study were: miR-381, F, 5’-AGTCTATACAAGGGCAAGCTCTC-3’/R, 5’-ATCCATGACAGATCCCTACCG-3’; U6, F, 5’-CTCGCTTCGGCAGCACA-3’/R, 5’-AACGCTTCACGAATTTGCGT-3’.
Transfection of the pre-miR-381 or inhibitor
6-well plates were seeded with 5 × 104 cell/well in 2 mL media 24 hr before transfection; cells were 80%-90% confluent. Cells were transfected with pre-miR-381 (mimic, 50 nM) or antisense of miRNA-381 (inhibitor, 100 nM) (Ribobio, China) using Lipofectamine 2000 Reagent (Life Technologies, Grand Island, NY) according to manufacturer’s instruction. After 48 hr of transfection, cells were used for qRT-PCR or migration and invasion assays. All miRNA vector were purchased from SBI (System Biosciences, USA). To generate stable miR-381 overexpress cell lines, cells were infected with lentiviral transduction particles containing pre-miR-381 plasmid or miRNA scramble control lentiviral vector. Cells were grown in the presence of 4 μg/mL puromycin for selection of stably transfected clones.
MTT assays
The cells were seeded at a density of 5 × 103 cells/well in 96-well plates at a final volume of 180 µl in incubation, at 37°C, with 5% CO2. After different time incubation, 20 µl of 5 mg/ml solution of MTT (Sigma, St. Louis, MO, USA) in PBS was added to each well. The plates were then incubated for 4 h at 37°C. The reaction was then solubilized in 100% dimethylsulfoxide (Sigma, St. Louis, MO, USA) 20 µl /well, and shaken for 15 min. Absorbance of each well was measured on a multidetection microplate reader (BMG LABTECH, Durham, NC, USA) at a wavelength of 570 nm.
Cell migration assays
Migration assays were conducted using Transwell plates with 8 μm pore size membranes (Corning Inc., Corning, NY) as described in previously literature [13]. After incubation for 4 hr, cells remaining in the upper side of the filter were removed with cotton swabs. The cells attached on the lower surface were fixed and stained using crystal violet and washed with water. Cells were counted with five high power fields per membrane and results were presented as the mean number of cells migrated per field per membrane. All experiments were conducted in triplicate.
Tumor xenograft experiments
All experiments involving mice were approved by Institutional Research Committee of Forth Military Medical University. All mice received humane care in compliance with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. 2.5 million cancer cells were mixed in a 1:1 (v:v) ratio with Growth Factor Reduced Matrigel (Becton, Dickinson and Company), and the mixture was injected subcutaneously into the right flanks of 6- to 7-week-old BALB/c nu/nu nude mice. Tumor volume (in mm3) was determined by caliper measurements performed every 2 days and calculated by using the modified ellipse formula: (volume = length × width2/2) [8]. When the tumor volumes reached approximately 300 mm3, mice were randomly assigned to mock ionizing radiation or a 5 Gy dose of ionizing radiation. 40 days after tumor initiation, the mice were killed by cervical dislocation, and their tumors were excised and measured.
Statistical analyses
The results of the quantitative data in this study are expressed as the mean ± standard deviation (SD). Data were analyzed by paired or independent t-test between paired samples or two independent groups. One way ANOVA was used for within-group comparison. The correlation between miRNA expression and radiosensitivity of ESCC cells was analyzed by Spearman correlation analysis. Data analysis was performed using IBM® SPSS Statistics (version 19.0). A P value of less than 0.05 was considered significant.
Results
Differential miRNA expression profiles between primary ESCC and recurrent ESCC in situ after radiotherapy
An miRNA expression array indicated that a total of 19 miRNAs were differentially expressed (changed in expression by > 1.5-fold) in the recurrent ESCC in situ after radiotherapy when compared with paired primary ESCC, that including 9 up-regulated and 11 down-regulated miRNAs (Figure 1A). Furthermore, using TaqMan quantitative real-time PCR, we confirmed that the relative expression levels of 2 up-regulated and 3 down-regulated miRNAs were consistent with the microarray data (Figure 1B).
Figure 1.
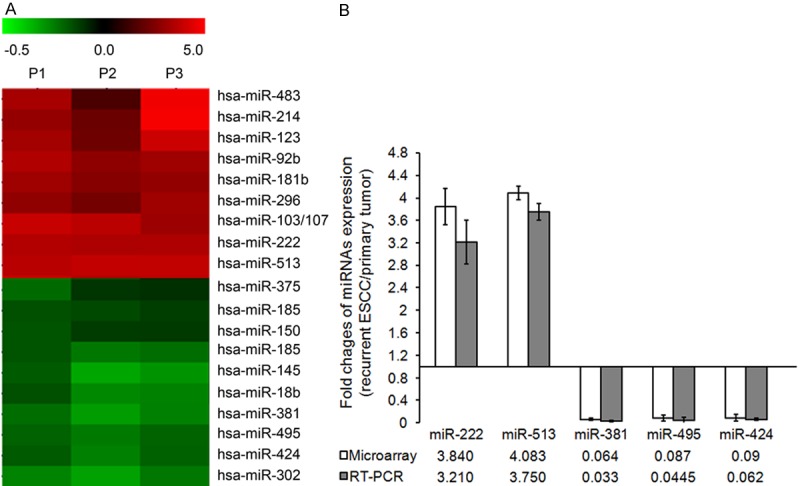
miRNA expression profiles of ESCC between primary tumor and paired recurrent tumor after radiotherapy. A: An miRNA expression array was performed to test miRNAs expression between primary tumor and recurrent tumor after radiotherapy. The red color represented a high expression level of miRNAs in recurrent ESCC when compared with primary tumor. The green color represented a low expression level of miRNAs in recurrent cancer. B: Differential expression miRNAs were confirmed by qRT-PCR. U6 served as an internal control. The results were presented as the means ± SD of fold changes of miRNA expression in recurrent ESCC when compared with the primary ESCC.
Higher expression of miRNA-381 indicates higher radiosensitivity of ESCC cells
In order to confirm the correlation between differential expression miRNAs and radiosensitivity of ESCC, we further evaluate the expression of above 5 miRNAs in 6 ESCC cell lines with differential profiles of radiosensitivity. The radiosensitivity of ESCC cell lines was evaluated by Colony formation assay after exposed to gradually increasing doses of irradiation (0, 2, 4, 6 and 8 Gy). The data demonstrated the decreasing survival of all ESCC cells following increasing doses of irradiation (data not shown). The fraction of surviving cells was calculated and compared between six different cell lines. As shown in Figure 2A, TE1 cells presented with the highest radiosensitivity, while KYSE450 showed more resistance to irradiation than other five cell lines (ANOVA test, P < 0.05). The trend of radiosensitivity in these six cell lines was TE1>ECA109>EC9706>KYSE30>KYSE150>KYSE450. We further characterized the parameters associated with radiobiology by using a modified linear-quadratic model programme (Sigmaplot V12.2). The indices including 37% dose slope (D0), quasi threshold dose (Dq), and survival fraction at 2 Gy (SF2), corresponding to radioresistance were lower in TE1 cells and higher in KYSE450 cells (Table 1).
Figure 2.
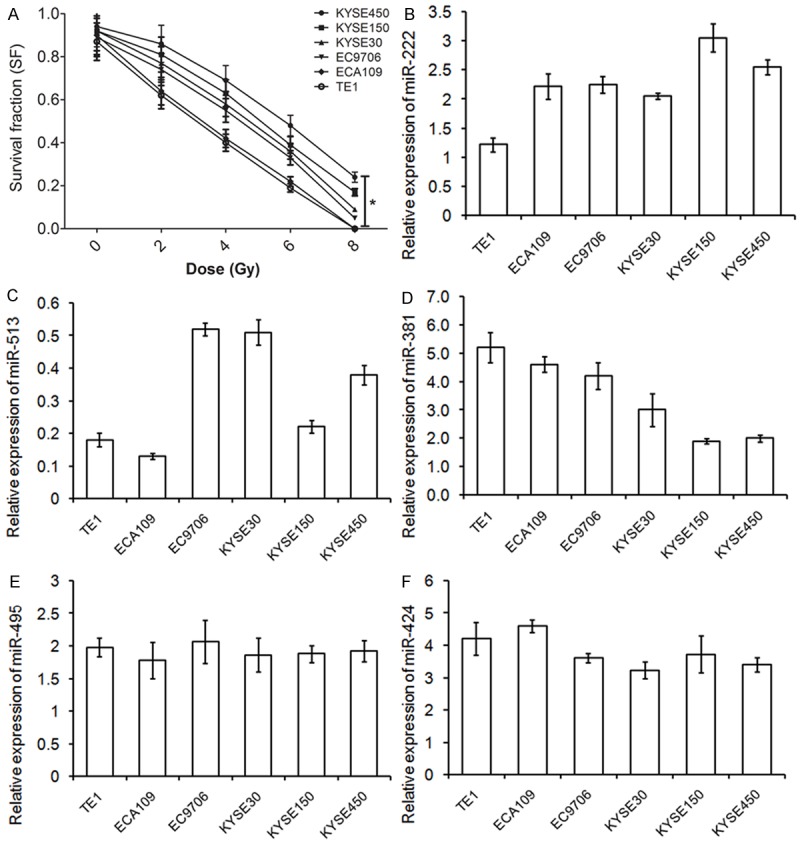
miRNA-381 is higher expressed in radiosensitive ESCC cells. A: Radiosensitivity of ESCC cell lines. Six ESCC cell lines TE1, ECA109, EC9706, KYSE30, KYSE150, and KYSE450 were irradiated with 0, 2, 4, 6, or 8 Gy and conducted to Colony formation assay. The results are presented as the means ± SD of values obtained in 3 independent experiments. *ANOVA test, P < 0.05. B-F: miRNA-381 expression in six ESCC cell lines was detected using qRT-PCR. U6 served as an internal control. An ANOVA test was used to determine the statistical significance of differences among the groups. The results are presented as the means ± SD of values obtained in 3 independent experiments.
Table 1.
Related parameters of cell survival curve standard model
| Dose (Gy) | KYSE450 | KYSE150 | KYSE30 | EC9706 | ECA109 | TE1 | P value |
|---|---|---|---|---|---|---|---|
| D0 | 3.78 ± 0.031 | 3.48 ± 0.061 | 3.48 ± 0.009 | 3.39 ± 0.176 | 2.81 ± 0.008 | 2.73 ± 0.016 | 0.047 |
| Dq | 6.00 ± 0.146 | 4.62 ± 0.010 | 3.55 ± 0.009 | 3.02 ± 0.073 | 1.85 ± 0.003 | 1.65 ± 0.010 | 0.012 |
| SF2 | 0.86 ± 0.008 | 0.81 ± 0.109 | 0.77 ± 0.012 | 0.74 ± 0.008 | 0.64 ± 0.003 | 0.62 ± 0.023 | 0.034 |
D0: 37% dose slope; Dq: quasithreshold dose; SF2: survival fraction at 2 Gy.
The expression of miRNAs was tested in each cell lines by qRT-PCR (Figure 2B-F). Interesting, when these 5 qRT-PCR-verified miRNAs was generated based on the trend of radiosensitivity within six ESCC cell lines, the expression of miRNA-381 was decreasing as cells in resistance to irradiation. Compared to the most radioresistant KYSE450 cell line, the radiosensitive TE1 cells expressed approximately 2.5 fold of miRNA-381 (Figure 2E, P < 0.001). The Spearman correlation analysis verified a negative correlation with radioresistant capacity and expression of miRNA-381 in ESCC cells (r 2 = -0.974, P = 0.001).
miRNA-381 increases radio sensitivity of ESCC cells and promotes nonaggressive phenotype
Based upon the above-verified differential expression of miRNA-381, we aimed to examine the potential role of miRNA-381 in radiobiology of ESCC cells by overexpression of miRNA-381 in radioresistant KYSE450 and KYSE150 cells. A miRNA-381 precursor vector and a control vector were used to transfect KYSE450 and KYSE150 cells in vitro. The transfection efficiency was demonstrated by qRT-PCR that miRNA-381 was successfully up-regulated 6.3 ± 0.82 times and 5.5 ± 0.74 times respectively in above two cell lines (P < 0.001; Figure 3A). After overexpression of miRNA-381, radiation clonogenic survival assays were performed. The miRNA-381 precursor-transfected cells displayed significantly decreased survival fraction (SF) compared with control cells (AUC 1.34 vs. 1.75, P < 0.05 in KYSE450 cell line; AUC 1.06 vs. 1.82, P < 0.01; Figure 3B), reflecting on increased radiosensitivity.
Figure 3.
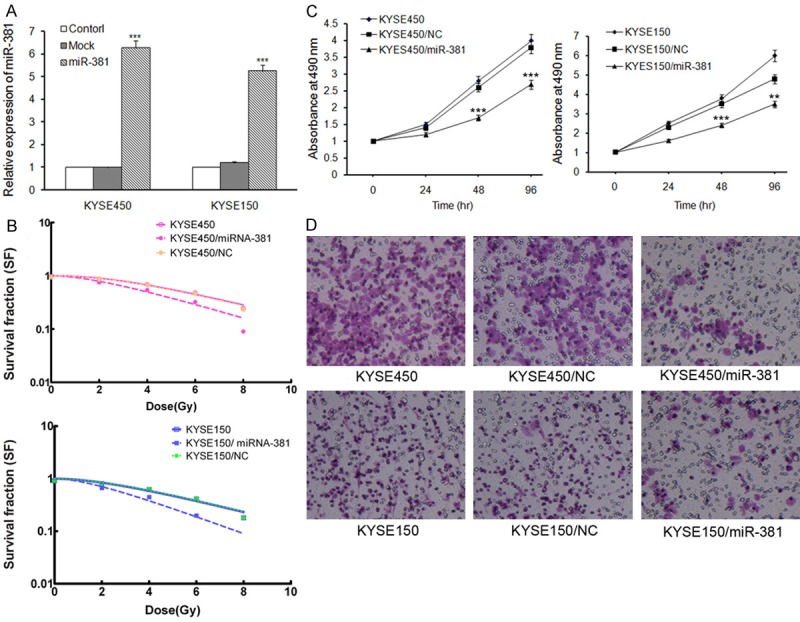
miRNA-381 over-expression in radioresistant KYSE450 and KYSE150 cells results in increased sensitivity to irradiation. A: miRNA-381 over-expression was verified by qRT-PCR analysis. KYSE450 and KYSE150 cells were transfected with miRNA-381 precursor vector or a control vector using Lipofectamine. miRNA-381 expression was confirmed 48 hr after transfection using qRT-PCR. U6 served as an internal control. ***P < 0.001. B: Radiation clonogenic survival assays were performed, and surviving fraction fitted to the linear quadratic equation for KYSE450 and KYSE150 cells transfected with control or miRNA-381 precursor vector. C: The effect of miRNA-381 on ESCC cells growth, as measured using the MTT assay. The results are presented as the means ± SD of the OD values obtained in 3 independent experiments. **P < 0.01, ***P < 0.001. D: Cell Transwell assays were conducted to investigate the role of miRNA-381 on ESCC cells migration.
In addition, we also conducted several parallel studies to further investigate the function of miRNA-381 in cell aggression. MTT analysis revealed that miRNA-381 overexpressed KYSE450 and KYSE150 cells proliferated significantly slower than control cells (P < 0.01, Figure 3C). In cell Transwell assays, migration (Figure 3D) was significantly reduced by overexpression of miRNA-381 in both KYSE450 and KYSE150 cell lines, supporting the negative functional role of miRNA-381 in aggression of ESCC cells.
Inhibition of miRNA-381 of ESCC cells promotes radiation resistance and development of an aggressive phenotype
Following overexpression of miRNA-381 in the radioresistant cells, we then suppressed the expression of miRNA-381 in radiosensitive TE1 and ECA109 cells to investigate its role in ESCC radioresistance. Our data revealed that the miRNA-381 was successfully inhibited by transfection of antisense of miRNA-381 in TE1 and ECA109 cells (P < 0.01, Figure 4A). Inhibition of miRNA-381 led to an increased survival capacity of TE1 and ECA109 cells following irradiation (P < 0.01, Figure 4B). Moreover, down regulation of miRNA-381 also promoted TE1 and ECA109 to develop an aggressive phenotype with increased capability of proliferation, and migration (Figure 4C and 4D).
Figure 4.
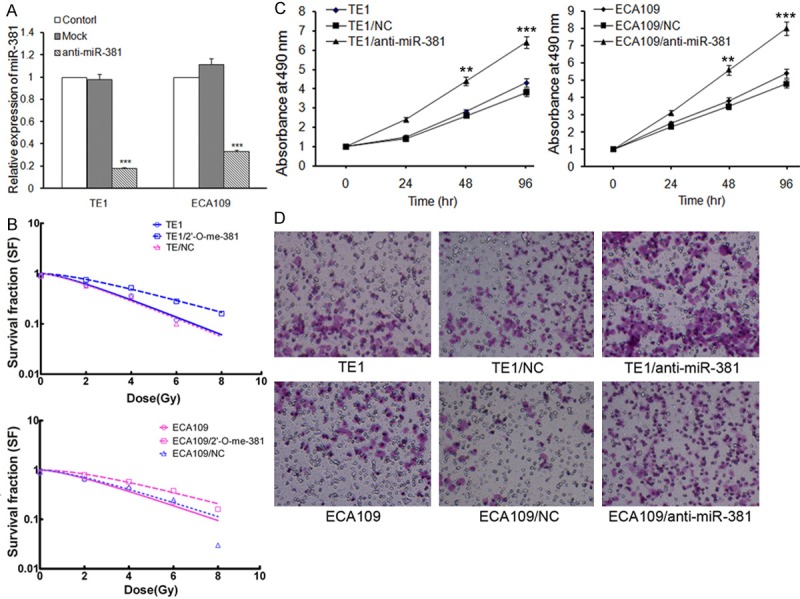
miRNA-381 suppression in radiosensitive TE1 and ECA109 cells leads to decreased sensitivity to irradiation. A: Inhibition of miRNA-381 was verified by qRT-PCR analysis. TE1 and ECA109 cells were transfected with antisense of miRNA-381 or control RNA using Lipofectamine. miRNA-381 expression was confirmed 48 hr after transfection using qRT-PCR. U6 served as an internal control. ***P < 0.001. B: Radiation clonogenic survival assays were performed, and surviving fraction fitted to the linear quadratic equation for TE1 and ECA109 cells transfected with control RNA or antisense of miRNA-381. C: The effect of miRNA-381 on ESCC cells growth, as measured using the MTT assay. The results are presented as the means ± SD of the OD values obtained in 3 independent experiments. **P < 0.01, ***P < 0.001. D: Cell Transwell assays were conducted to investigate the role of miRNA-381 on ESCC cells migration.
Overexpression of miRNA-381 increases radiation sensitivity in vivo
To investigate the in vivo functions of miRNA-381 in tumors on growth and radiation response, we established the stable miRNA-381 over-expressed KYSE450 cell line (KYSE450-miR-3811) and negative control KYSE450 cell line (KYSE450-NC), and generated subcutaneous tumors in BALB/c nu/nu nude mice using KYSE450-NC and KYSE450-miR-381 cells. Consist with in vitro results, KYSE450-miR-381 tumors grew slower than KYSE450-control tumors (Figure 5A). Following by irradiation of the tumors with a dose of 5 Gy, the KYSE450-miR-381 tumors showed less radiation resistant with a shorter growth delay (Figure 5B). 40 days after tumor initiation, tumors were removed and macroscopically measured. As shown in Figure 5C, irradiation could significantly suppress tumor growth and this effect was promoted by over-expression of miRNA-381. Moreover, over-expression of miRNA-381 also inhibited tumor growth, although the difference was not statistically significant (P = 0.126).
Figure 5.
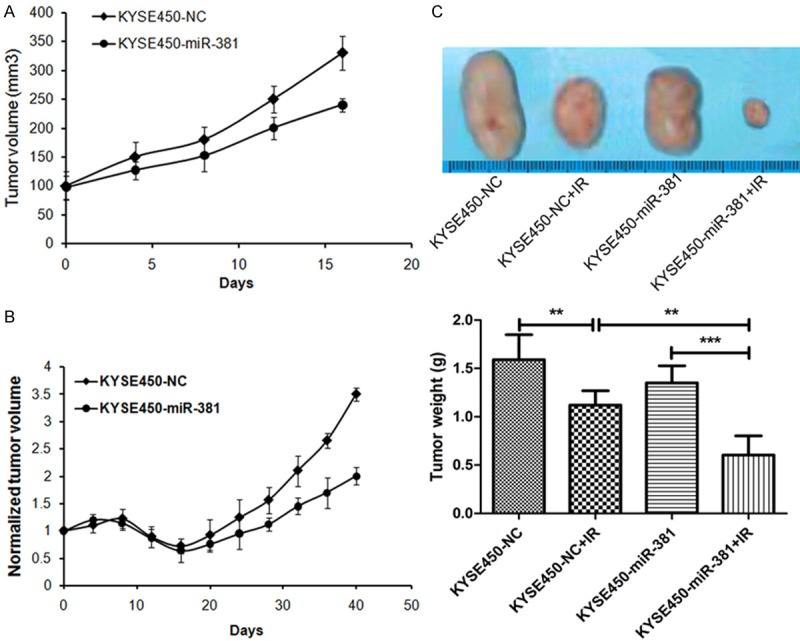
miR-381 reduces tumor growth and radiation resistance in vivo. A: Nude mice were subcutaneously injected with 107 KYSE450 cells infected with a control vector or pre-miR-381 vector, tumor volume was recorded and presented as mean ± SD. Each group was composed of 8 mice. *P < 0.05. B: When the average tumor volume reached approximately 300 mm3, the tumors were irradiated with a single 5 Gy dose of IR. Mean tumor volumes normalized to starting volume and SD were presented as tumor growth curves. *P < 0.05, **P < 0.01. C: Representative images of the tumors from mice 40 days after irradiation and a quantitative summary of the tumor weights (n = 6 mice per group). *P < 0.05, **P < 0.01.
Discussion
Radioresistance is a challenging obstacle in treatment of ESCC and research addressing this problem is essential. Previous studies have demonstrated that the process of developing radioresistance is complicated, which involves multiple molecular mechanisms [11]. Recently, the aberrant expression of miRNAs has been functionally analyzed in tumors, and miRNAs were found to be associated with various malignant behaviors of tumor cells including radioresistance [14]. However, there are little data on the miRNAs expression profile of radioresistance in ESCC. In this present study, we found differential expression profiles of miRNAs in ESCC with different irradiation sensitivity and identified that miRNA-381 was associated with ESCC radiosensitivity, which significantly increased in radiosensitive TE1 cells and decreased in radioresistant KYSE450 cells. In vitro functional studies further demonstrated that overexpression of miRNA-381 increases radiosensitivity of ESCC cells, promotes non aggressive phenotype, and overexpression of miRNA-381 confers a radiosensitivity and grow inhibition in radioresistant KYSE450 cells.
Results from several other groups have confirmed about forty-five miRNAs involved in the radioresistance of human cancers (see review by M. Ahmad Chaudhry [15], Chanatip Metheetrairut and Frank J Slack [14]). In esophageal carcinoma, only three miRNAs, including miRNA-21 [16], miRNA-22 [17], and miRNA-31 [18], were found to contribute to the radioresistance by targeting different down-stream genes. Su et al. recently found a set of miRNAs aberrantly expressed in acquired radioresistant ESCC cell and identified miRNA-301a as a promotor in sensibilization of radiotherapy through wnt/-catenin signal pathway [19]. These previous reports indicated the potential role of miRNAs in radioresistance of ESCC. Based on our differential expression profiles of miRNAs associated with ESCC cells radioresistance, we confirmed the important roles of miRNA in radioresistance and revealed that inhibition of miRNA-381 could promote radioresistance of ESCC both in vitro and in vivo.
Previous researches also showed other biological functions of miRNA-381 in both cancerous and noncancerous conditions. Lee et al. found that forced expression of miRNA-381 inhibited colony-forming capacity of normal and malignant mast cell lines [20]. This effect was also observed in renal cancer cells, which miRNA-381 could suppressed the proliferation of cancer cells [21]. Moreover, miRNA-381 further exerted the inhibiting effects in cell migration and invasion of lung adenocarcinoma [22], indicated the carcinostasis of miRNA-381. However, in glioma, miRNA-381 may act as an oncomiR to stimulate tumor growth [23,24]. Our data revealed that miRNA-381 could inhibit ESCC cell proliferation and migration. Of note, miRNA-381 can also sensitize several cancer cells to chemotherapy, such as leukemia and renal cancer [25,26]. Whether miRNA-381 has the similar function in ESCC needs more investigations.
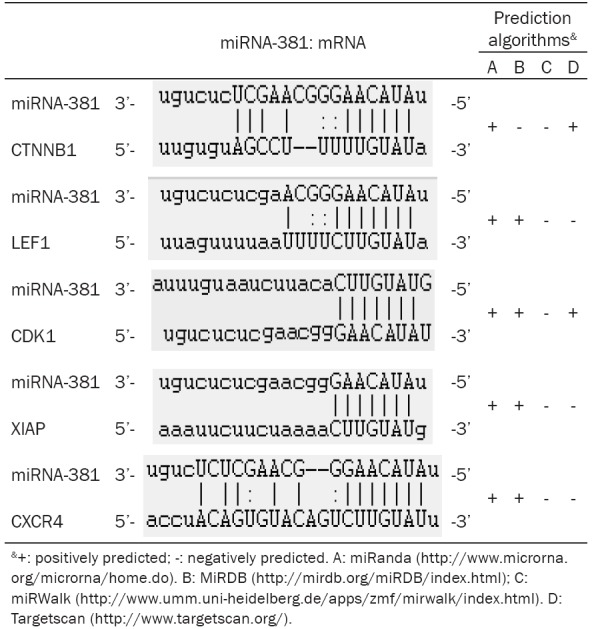
miRNA-381 exerts its biological functions through the regulation of various target genes, such as MITF [20], LRRC4 [23], ID1 [22], MDR1 [25], BRD7 [24], and WEE1 [26]. Herein, we used bioinformatic algorithms to predict another five genes, including CTNNB1, LEF1, CDK1, XIAP, and CXCR4, as downstream of miRNA-381. As shown in Table 2, the miRNA-381 has potential capability to directly bind the 3’-UTR of these five mRNAs that may degrade these genes or inhibit transcription. As each gene mentioned above can regulate various cell behaviors and involved in carcinostasis or carcinogenesis of ESCC [27-31], it is very possible that one or all of them have important roles in miRNA-381 related radioresistance of ESCC. Thus, a number of initiatives are needed to undergo to understand the particular mechanism.
Table 2.
Potential targets of miR-381 by different prediction algorithms
|
In conclusion, the results of our present study have strongly showed that the miRNA-381 sensitizes ESCC cells to irradiation and inhibits cell proliferation, migration, and invasion, indicating that it is a valuable radioresistance-associated biomarker and a promising therapeutic target for treatment of ESCC. However, the clinical significance of miRNA-381 has not been verified in this study and therefore requires further investigation. Furthermore, it remains unclear exact downstream pathway of miRNA-381 and how the miRNA-381 signalling pathway participates in regulation of radioresistance in ESCC. Our laboratory recently discovered that miRNA-381 was significantly down-regulated and the predicted target genes CTNNB1, LEF1, CDK1, XIAP, and CXCR4 were correspondingly up-regulated in radioresistant ESCC cells compared to radiosensitive cells. This discovery suggests that miRNA-381 regulate radioresistance may via above signalling pathway, which is the direction of our future research.
Acknowledgements
This work was supported by Chinese National Science Foundation Projects (NSFC 81301922 and 81302055).
Disclosure of conflict of interest
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, He Y, Zheng R, Zhang S, Zeng H, Zou X, He J. Esophageal cancer incidence and mortality in China, 2009. J Thorac Dis. 2013;5:19–26. doi: 10.3978/j.issn.2072-1439.2013.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JJ, Tannock IF. Repopulation of cancer cells during therapy: an important cause of treatment failure. Nat Rev Cancer. 2005;5:516–525. doi: 10.1038/nrc1650. [DOI] [PubMed] [Google Scholar]
- 5.Wang P, Zhang J, Zhang L, Zhu Z, Fan J, Chen L, Zhuang L, Luo J, Chen H, Liu L, Chen Z, Meng Z. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–1143. e1112. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 6.Chinnaiyan P, Allen GW, Harari PM. Radiation and new molecular agents, part II: targeting HDAC, HSP90, IGF-1R, PI3K, and Ras. Semin Radiat Oncol. 2006;16:59–64. doi: 10.1016/j.semradonc.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang X, Taeb S, Jahangiri S, Emmenegger U, Tran E, Bruce J, Mesci A, Korpela E, Vesprini D, Wong CS, Bristow RG, Liu FF, Liu SK. miRNA-95 mediates radioresistance in tumors by targeting the sphingolipid phosphatase SGPP1. Cancer Res. 2013;73:6972–6986. doi: 10.1158/0008-5472.CAN-13-1657. [DOI] [PubMed] [Google Scholar]
- 9.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 10.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G, Liu Y, Su Z, Ren S, Zhu G, Tian Y, Qiu Y. MicroRNA-324-3p regulates nasopharyngeal carcinoma radioresistance by directly targeting WNT2B. Eur J Cancer. 2013;49:2596–2607. doi: 10.1016/j.ejca.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Wu Q, Yang Z, Shi Y, Fan D. MiRNAs in Human Cancers: The Diagnostic and Therapeutic Implications. Curr Pharm Des. 2014;20:5336–47. doi: 10.2174/1381612820666140128204914. [DOI] [PubMed] [Google Scholar]
- 13.Cai H, Xu Y. The role of LPA and YAP signaling in long-term migration of human ovarian cancer cells. Cell Commun Signal. 2013;11:31. doi: 10.1186/1478-811X-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr Opin Genet Dev. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhry MA. Radiation-induced microRNA: Discovery, functional analysis, and cancer radiotherapy. J Cell Biochem. 2014;115:436–449. doi: 10.1002/jcb.24694. [DOI] [PubMed] [Google Scholar]
- 16.Huang S, Li XQ, Chen X, Che SM, Chen W, Zhang XZ. Inhibition of microRNA-21 increases radiosensitivity of esophageal cancer cells through phosphatase and tensin homolog deleted on chromosome 10 activation. Dis Esophagus. 2013;26:823–831. doi: 10.1111/j.1442-2050.2012.01389.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang XC, Zhang ZB, Wang YY, Wu HY, Li DG, Meng AM, Fan FY. Increased miRNA-22 expression sensitizes esophageal squamous cell carcinoma to irradiation. J Radiat Res. 2013;54:401–408. doi: 10.1093/jrr/rrs113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynam-Lennon N, Reynolds JV, Marignol L, Sheils OM, Pidgeon GP, Maher SG. MicroRNA-31 modulates tumour sensitivity to radiation in oesophageal adenocarcinoma. J Mol Med (Berl) 2012;90:1449–1458. doi: 10.1007/s00109-012-0924-x. [DOI] [PubMed] [Google Scholar]
- 19.Su H, Jin X, Zhang X, Xue S, Deng X, Shen L, Fang Y, Xie C. Identification of microRNAs involved in the radioresistance of esophageal cancer cells. Cell Biol Int. 2014;38:318–325. doi: 10.1002/cbin.10202. [DOI] [PubMed] [Google Scholar]
- 20.Lee YN, Brandal S, Noel P, Wentzel E, Mendell JT, McDevitt MA, Kapur R, Carter M, Metcalfe DD, Takemoto CM. KIT signaling regulates MITF expression through miRNAs in normal and malignant mast cell proliferation. Blood. 2011;117:3629–3640. doi: 10.1182/blood-2010-07-293548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen B, Duan L, Yin G, Tan J, Jiang X. Simultaneously expressed miR-424 and miR-381 synergistically suppress the proliferation and survival of renal cancer cells---Cdc2 activity is up-regulated by targeting WEE1. Clinics (Sao Paulo) 2013;68:825–833. doi: 10.6061/clinics/2013(06)17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothschild SI, Tschan MP, Jaggi R, Fey MF, Gugger M, Gautschi O. MicroRNA-381 represses ID1 and is deregulated in lung adenocarcinoma. J Thorac Oncol. 2012;7:1069–1077. doi: 10.1097/JTO.0b013e31824fe976. [DOI] [PubMed] [Google Scholar]
- 23.Tang H, Liu X, Wang Z, She X, Zeng X, Deng M, Liao Q, Guo X, Wang R, Li X, Zeng F, Wu M, Li G. Interaction of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res. 2011;1390:21–32. doi: 10.1016/j.brainres.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 24.Tang H, Wang Z, Liu Q, Liu X, Wu M, Li G. Disturbing miR-182 and -381 inhibits BRD7 transcription and glioma growth by directly targeting LRRC4. PLoS One. 2014;9:e84146. doi: 10.1371/journal.pone.0084146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Ohms SJ, Li Z, Wang Q, Gong G, Hu Y, Mao Z, Shannon MF, Fan JY. Changes in the expression of miR-381 and miR-495 are inversely associated with the expression of the MDR1 gene and development of multi-drug resistance. PLoS One. 2013;8:e82062. doi: 10.1371/journal.pone.0082062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen B, Duan L, Yin G, Tan J, Jiang X. miR-381, a novel intrinsic WEE1 inhibitor, sensitizes renal cancer cells to 5-FU by up-regulation of Cdc2 activities in 786-O. J Chemother. 2013;25:229–238. doi: 10.1179/1973947813Y.0000000092. [DOI] [PubMed] [Google Scholar]
- 27.Li HZ, Gao XS, Xiong W, Zhao J, Zhang H, Zhou DM. Identification of differentially expressed genes related to radioresistance of human esophageal cancer cells. Chin J Cancer. 2010;29:882–888. doi: 10.5732/cjc.010.10148. [DOI] [PubMed] [Google Scholar]
- 28.Cloos CR, Daniels DH, Kalen A, Matthews K, Du J, Goswami PC, Cullen JJ. Mitochondrial DNA depletion induces radioresistance by suppressing G2 checkpoint activation in human pancreatic cancer cells. Radiat Res. 2009;171:581–587. doi: 10.1667/RR1395.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S, Ding F, Luo A, Chen A, Yu Z, Ren S, Liu Z, Zhang L. XIAP is highly expressed in esophageal cancer and its downregulation by RNAi sensitizes esophageal carcinoma cell lines to chemotherapeutics. Cancer Biol Ther. 2007;6:973–980. doi: 10.4161/cbt.6.6.4195. [DOI] [PubMed] [Google Scholar]
- 30.Koishi K, Yoshikawa R, Tsujimura T, Hashimoto-Tamaoki T, Kojima S, Yanagi H, Yamamura T, Fujiwara Y. Persistent CXCR4 expression after preoperative chemoradiotherapy predicts early recurrence and poor prognosis in esophageal cancer. World J Gastroenterol. 2006;12:7585–7590. doi: 10.3748/wjg.v12.i47.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gockel I, Schimanski CC, Heinrich C, Wehler T, Frerichs K, Drescher D, von Langsdorff C, Domeyer M, Biesterfeld S, Galle PR, Junginger T, Moehler M. Expression of chemokine receptor CXCR4 in esophageal squamous cell and adenocarcinoma. BMC Cancer. 2006;6:290. doi: 10.1186/1471-2407-6-290. [DOI] [PMC free article] [PubMed] [Google Scholar]


