Abstract
Survivin, the smallest member of IAP (inhibitor of apoptosis) family, is a dual functional protein acting as a critical apoptosis inhibitor and key cell cycle regulator. Survivin is usually expressed in embryonic tissues during development and undetectable in most terminally differentiated tissues. Numerous studies demonstrate that survivin is selectively upregulated in almost all types of human malignancies and its overexpression positively correlates with poor prognosis, tumor recurrence, and therapeutic resistance. This differential expression of survivin in tumors and normal tissues draws a great interest to develop survivin-targeted therapy for cancer treatment. Nonetheless, the molecular mechanisms controlling survivin expression in malignant tumor cells have not been fully understood. While aberrant activation of receptor tyrosine kinases (RTKs) and the downstream signaling, such as PI-3K/Akt, MEK/MAPK, mTOR, and STAT pathways, have frequently been shown to upregulate survivin, recent data suggest that a class of noncoding RNAs, microRNAs (miRNAs) also play an important role in survivin dysregulation in human cancers. Here, we focus on survivin expression-regulated by specific miRNAs binding to the 3’-UTR of survivin mRNA, and summarize the latest advances on survivin-targeted therapy in clinical trials and the therapeutic potential of survivin-targeting miRNAs in cancer.
Keywords: Survivin, miRNA, targeted therapy, cancer
Introduction
Survivin (encoded by gene BIRC5) is the smallest member of inhibitor of apoptosis (IAP) protein family. As endogenous inhibitors of caspases, this family has eight known members, including NAIP/BIRC1, cIAP1/BIRC2, cIAP2/BIRC3, XIAP/BIRC4, Survivin/BIRC5, BRUCE/BIRC6, livin/BIRC7, and ILP2/BIRC8 [1,2]. All IAPs contain at least one BIR (Baculovirus Inhibitor of apoptosis protein Repeat) domain, which is required for their classification as IAP proteins. Survivin has only one BIR domain. The coding gene of survivin, BIRC5 locates at chromosome 17q25 in humans, consisting of three introns and four exons [1,2]. Alternative splicing of BIRC5 pre-mRNA generates five transcripts, translating into five different proteins, which are wild-type survivin [3], survivin-2B, survivin-Ex3 [4], survivin-2 [5] and survivin-3B [6,7]. Although survivin was initially discovered as an anti-apoptotic protein, it also serves as an indispensable regulator of cell mitosis [8].
Dysregulation of survivin is a key signature of many cancers. A number of studies demonstrate that survivin is selectively overexpressed in almost all human cancers, but undetectable in normal differentiated tissues [9,10]. Its expression is associated with aberrant activation of several receptor tyrosine kinases (RTKs), such as the epidermal growth factor receptor (EGFR), erbB2 (Her2), the insulin-like growth factor-1 (IGF-1) receptor (IGF-1R), and various cell survival signaling cascades, including PI-3K/Akt, mitogen-activated protein kinase (MEK)/mitogen-activated protein kinase (MAPK), mammalian target of rapamycin (mTOR), signal transducer and activator of transcription (STAT), hypoxia-inducible factor-1 (HIF-1), and etc [9,10]. Survivin carries out its anti-apoptotic function through inhibition of caspase-9 with the help of a cellular protein named hepatitis B X-interacting protein (HBXIP) [11]. It can also inhibit apoptosis via stabilizing XIAP from ubiquitination/proteasomal destruction by forming a survivin-XIAP complex [12]. In cancer cells, overexpression of survivin is correlated with poor clinical outcome, tumor recurrence, and therapeutic resistance [13,14]. For instance, we have shown that elevated expression of erbB3 confers paclitaxel resistance in erbB2-overexpressing breast cancer cells via PI-3K/Akt-dependent upregulation of survivin [15], and targeting of erbB3 with a therapeutic antibody re-sensitizes the breast cancer cells to paclitaxel mainly through downregulation of survivin [16]. In addition to the survival signaling pathways upstream of survivin, recent studies suggest that epigenetic mechanisms also contribute to survivin dysregulation in human cancers. For example: at transcriptional level, aberrant hypo- or hyper-methylation of survivin promoter could lead to upregulation of survivin [17,18]; at post-transcriptional level, microRNAs (miRNAs) bind to the 3’-untranslated region (UTR) of survivin mRNA and thereby alter survivin protein translation or induce its mRNA degradation. In this review, we summarize our recent understanding on miRNA regulation of survivin in human cancers, and discuss the current and future approaches targeting of survivin for cancer therapy.
MicroRNA regulation of survivin expression
MiRNAs are short noncoding RNAs of ~22 nucleotides that negatively regulate gene expression at the post-transcriptional level. In general, miRNAs bind to the 3’-UTR of its target mRNA via sequence-guided recognition to trigger mRNA degradation or translational repression [19]. A mature miRNA forms a RNA-protein complex called RNA-induced silencing complex (RISC) to carry out its gene silencing function. Although the 3’-UTRs of mRNAs are the most frequent regions of miRNA targeting, the open reading frames (ORFs) could also be targeted. The frequency and effect of ORF targeting by miRNAs are less than that of 3’-UTR targeting, whereas 5’-UTR targeting is a rare event [19,20]. One miRNA may have multiple mRNA targets; likewise, one mRNA can be regulated by multiple miRNAs. It has been shown that each cancer owns unique pattern of miRNA expression profiles [21]. Several critical survivin-targeting miRNAs have been identified in human cancers.
MiR-203
MiR-203 is one of the most studied survivin-targeting miRNAs. MiR-203 directly targets survivin mRNA and plays a pivotal role in prostate cancer (PCa) progression and metastasis. Saini and colleagues found that in advanced PCa, expression of miR-203 was specifically attenuated in the metastatic bone tissues and prostate cancer cell lines derived from bone metastasis [22]. Restoration of miR-203 in PCa cell line PC3 dramatically decreased bone metastasis in a mouse model. Ectopic expression of miR-203 in PC3 cells suppressed epithelial-mesenchymal transition (EMT), invasion, and motility, and induced cell cycle G1 arrest and apoptosis in vitro. The anti-metastatic effect of miR-203 was partially due to downregulation of survivin. Meanwhile, miR-203 also regulated a cohort of pro-metastatic genes, including ZEB2, Bmi1, Smad4, and the bone-specific transcriptional regulators Runx2 and Dlx5 [22]. Its capability to exhibit multi-layer metastatic repression makes miR-203 an intriguing tool in the treatment of advanced PCa [22]. In addition, the expression levels of miR-203 and survivin are inversely correlated in laryngeal cancer patient samples [23]. Forced expression of miR-203 in laryngeal carcinoma cell line Hep-2 decreased cell growth, which was caused by cell cycle G1 arrest [23]. The tumor suppressive activity of miR-203 has also been reported in the progression of hepatocellular carcinoma [24], lung cancer cells [25], and pancreatic cancer cells [26]. Reintroducing miR-203 to these cancer cells inhibited cell proliferation by reduction of survivin [24-26]. One study with estrogen responsive breast cancer MCF7 cells found that estradiol (E2) treatment downregulated survivin-targeting miRNAs - miR-203, miR-16 and miR-143, which were responsible for estradiol-induced cell growth and survivin upregulation [27]. A recent report provided a detailed explanation of miR-203 regulation of survivin in human Merkel cell carcinoma (MCC). A human polyomavirus called Merkel cell polyomavirus (MCV) can integrate, with its tumor-specific T-antigen mutation, into the DNA of tumor cells. MCC can be divided into two subtypes based upon the detectability of MCV T-antigens, MCV-positive (MCV+) MCCs and MCV-negative (MCV-) MCCs. It has been demonstrated that miR-203 induces cell growth inhibition via downregulation of survivin only in MCV-MCCs, but not in MCV+MCCs. In contrast, the expression of survivin in MCV+MCCs is enhanced by MCV T-antigens [28]. Thus, both MCV+MCCs and MCV-MCCs share the same trait of Survivin dysregulation, providing a strong rationale to develop survivin-targeted therapy for MCCs. Nonetheless, different mechanisms upregulating survivin should be taken into consideration when designing therapeutic strategies.
MiR-34a
MiR-34a has been shown to reduce expression of survivin. The mechanism of miR-34a action in controlling survivin expression varies upon different types of cancer. Current literature regarding miR-34a regulation of survivin can be classified as the following: 1) Indirect regulation: with ectopic expression of miR-34a or drug-induced upregulation of miR-34a in cancer cells, survivin is reduced through downregulation of its upstream activators or transcriptional factors, which are targets of miR-34a [29-32]; 2) Direct regulation [33-35]: these studies haven’t confirmed with luciferase reporter assays whether miR-34a directly binds to the 3’-UTR of survivin mRNA and functions, although TargetScan (http://www.targetscan.org/) predicts a seed sequence for miR-34a at position 1940-1946 of survivin 3’-UTR. MiR-34a was significantly downregulated in head and neck squamous cell carcinoma (HNSCC) tissues and cell lines [29]. Studies with tumor samples from HNSCC patients revealed an inverse correlation between miR-34a and survivin or E2F3 levels. Overexpression of miR-34a markedly decreased E2F3 and survivin in HNSCC cell line UM-SCC-74A. In the rescue experiments, survivin expression was completely restored by a miR-34a-resistant isoform of E2F3a, whereas E2F3b only partially restored survivin. These data support a crucial role for the miR-34a-E2F3a-survivin axis in mediating miR-34a’s tumor suppressive function in HNSCCs [29]. In non-small cell lung cancer (NSCLC) cells, tocotrienol-induced miR-34a inhibited Notch-1 and subsequently reduced survivin, as survivin was one of the downstream targets of Notch-1 [30]. Ectopic expression of miR-34a in laryngeal squamous cell carcinoma cell lines, a gastric cancer cell line and murine melanoma cells was also reported to inhibit cell growth by downregulation of survivin [33-35]. Thus, the precise mechanisms of miR-34a regulation of survivin need to be quested with consideration of cancer types.
miRNAs with multiple binding sites at 3’-UTR of survivin mRNA
Among the reported miRNAs that target survivin mRNA (Figure 1), some have multiple binding sites on its 3’-UTR. One of them is miR-542-3p, which has three putative binding sites [36]. Luciferase assays using site-specific mutant constructs have verified that miR-542-3p directly regulates survivin expression primarily through its binding at the 2nd predicted binding site. Since the studies were only conducted in a NSCLC cell line A549, one cannot rule out the possibility that miR-542-3p may simultaneously bind to all three sites to regulate survivin in other tumor types. Transfection of miR-542-3p mimic in cancer cells resulted in downregulation of survivin and growth inhibition due to cell cycle arrest at both G1 and G2/M phases [36]. The role of miR-708 in targeting survivin, inducing apoptosis and suppressing tumorigenicity in renal cancer cells has been reported [37]. The expression of miR-708 was widely attenuated in human renal cell carcinomas (RCCs). Forced expression of miR-708 in RCC cells decreased cell proliferation, clonality, invasion and migration, and increased apoptosis. Intratumoral delivery of miR-708 led to regression of RCC murine tumor xenografts. Survivin was identified as an important target of miR-708 in carrying out its antitumor activity. Specific knockdown of survivin by siRNA partially phenocopied miR-708 overexpression. It is predicted that miR-708 has three binding sites on 3’-UTR of survivin, which was confirmed by luciferase reporter assays [37]. However, it remains unknown which binding site plays a more critical role or miR-708 equally utilizes all three of them to downregulate survivin. It will be interesting to test whether the miRNAs with multiple binding sites on 3’-UTR of survivin are more effective than those with single binding site to inhibit survivin. We have recently reported that three miRNAs, miR-125a, miR-125b and miR-205 act in concert to downregulate erbB2/erbB3 receptors in human breast cancer cells [38]. It is conceivable to hypothesize that functional cooperation may also exist among the multiple binding sites of one miRNA. Thus, the miRNAs with multiple binding sites may be more promising tools in miRNA-replacement therapy. Since the amount of the miRNAs with multiple-binding sites used in therapy would be less, the effect of such miRNAs could be more specific and their side effects should be minimized.
Figure 1.
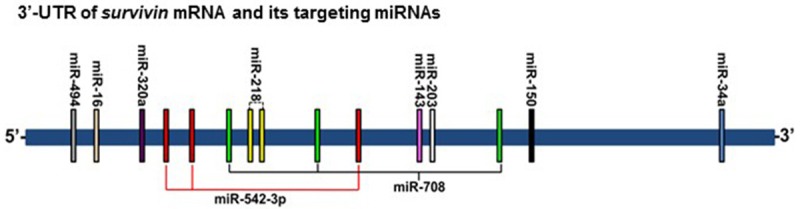
Schematic representation of survivin-targeting miRNAs and their relative binding sites on 3’-UTR of survivin mRNA. Vertical bars indicate the miRNAs’ binding sites, and the bars with identical color represent the same miRNA. Both miR-542-3p and miR-708 have three binding sites on the 3’-UTR of survivin mRNA.
Regulation of survivin-targeting miRNAs
As aforementioned, the miRNAs targeting survivin are widely attenuated in different types of cancer (Table 1). This raises the questions: how those miRNAs are regulated and what are the causes of their downregulation. TEL-AML1 fusion protein is the most frequently identified in childhood leukemia. Two survivin-targeting miRNAs, miR-494 and miR-320a were found to be upregulated upon TEL-AML1 silencing [39]. The expression levels of miR-494 and miR-320a were lower in TEL-AML1 positive leukemia as compared with the immunophenotype-matched TEL-AML1 negative ones. Within TEL-AML1 positive leukemia, the expression of the two miRNAs was inversely correlated with the expression of survivin. It has been concluded that TEL-AML1 may exert its anti-apoptotic function in part by suppressing miR-494 and miR-320a, and thereby resulting in enhanced expression of survivin [39]. In nasopharyngeal carcinoma (NPC), miR-218 is frequently downregulated in primary NPC tissues and cell lines [40]. Luciferase assays have confirmed that survivin, ROBO1 and connexin43 are direct targets of miR-218. Primary miR-218 is transcribed from two chromosomal locations, which are embedded in the intronic regions of SLIT2 (4p15.31) and SLIT3 (5q35.1). The promoter regions of SLIT2 and SLIT3 are hypermethylated in NPC. Thus, the reduction of miR-218 is due to epigenetic silencing of SLIT2 and SLIT3 promoters. Survivin and ROBO1, two of the most important targets of miR-218, are upregulated in NPC, leading to NPC cell survival and migration [40]. In addition, the regulatory axis, miR-27a-ZBTB10-Sp-survivin is intensively studied by Safe and colleagues [41-43]. They have tried a variety of chemicals and combinations of drugs in human cancer cells and found that the oncogenic miR-27a can be downregulated by different treatments. As a result, one of the targets of miR-27a, ZBTB10, a repressor of Sp transcriptional factor family members, is upregulated, leading to downregulation of the Sp-dependent genes, including survivin [41-43]. Furthermore, both miR-203 and miR-542-3p have been identified as tumor suppressive miRNAs, and are frequently downregulated due to promoter methylation in cancers of breast, prostate, and liver as well as gastric B-cell lymphoma [44-50]. To date, there is no report on promoter hypermethylation of miR-203 and miR-542-3p in NSCLC. Although the expression of miR-203 is much lower in lung cancer cell lines than that in normal bronchial epithelial cells [25], it is unclear if the reduced miR-203 is also attributed to its promoter methylation. Our recent data show that the expression levels of both miR-203 and miR-542-3p are significantly decreased in the majority of NSCLC tumors as compared to the adjacent normal lung tissues (Wang and Liu, unpublished data). We are currently testing whether promoter methylation and/or other epigenetic alterations play an important role in downregulation of the survivin-targeting miRNAs in NSCLC. Hence, when considering restoring survivin-targeting miRNAs in human cancers, elucidating the molecular mechanisms through which the miRNAs are reduced will provide us new avenues to bring back the “good” miRNAs.
Table 1.
Reported survivin-targeting miRNAs and their expression status in human cancers
| miRNAs | Expression Status |
|---|---|
| miR-494 | Downregulated by TEL-AML1 in ALL [39]. |
| miR-16 | Downregualted by E2 in MCF-7 cells [27]. |
| Downregulated in colorectal cancers [90]. | |
| miR-320a | Downregulated by TEL-AML1 in ALL [39]. |
| miR-542-3p | The 2nd binding site of miR-542-3p on 3’-UTR of survivin was confirmed to be the primary site in A549 cells [36]. |
| miR-708 | Downregualted in renal cell carcinoma [37]. |
| miR-218 | Downregulated by promoter methylation in nasopharyngeal carcinoma. Binding on 2nd site was confirmed by mutation with luciferase assay [40]. |
| miR-143 | Downregualted by E2 in MCF-7 cells [27]. |
| miR-203 | Specifically attenuated in advanced metastatic prostate cancer [22]. |
| Downregualted in laryngeal cancer [23], hepatocellular carcinoma [24], lung cancer [25], and pancreatic cancer cells [26]. | |
| It regulates survivin expression in MCV-MCCs [28]. | |
| Downregualted by E2 in MCF-7 cells [27]. | |
| miR-150 | Reduces survivin levels in Burkitt’s lymphoma cell line DG75 [91]. |
| miR-34a | E2F3a was downregulated by miR-34a in HNSCCs. Expression of survivin was reduced as the result of E2F3a downregulation [29]. |
| Induced by delta-tocotrienol in NSCLC, and thereby downregulates survivin by targeting Notch-1 [30]. | |
| Induced by CDK1 inhibitor purvalanol A in neuroblastoma, and therebydownregulates survivin by targeting the transcriptional factor MYCN [31]. | |
| Overexpression in laryngeal squamous cell carcinoma [35], gastric cancer [32,33], and murine melanoma cell lines [34]. |
Abbreviations: ALL: acute lymphoblastic leukemia; E2: 17β-estradiol; MCV: Merkel cell polyomavirus; MCC: Merkel cell carcinoma; HNSCC: head neck squamous cell carcinoma; NSCLC: non-small cell lung cancer.
Targeting of survivin in cancer treatment
Survivin has long been recognized as an important target because of its selective expression in tumor, but not normal tissues [9,10]. The differential expression of survivin in tumor and normal tissues inspires people to develop survivin-targeted therapy. Although several strategies, including the transcriptional inhibitor YM155, antisense oligonucleotide (ASO), immunotherapy, and gene therapy have been designed to target survivin [14,51-53], and some of them are actively under clinical trials in a wide variety of human cancers (http://www.clinicaltrials.gov), there is currently no survivin-targeted therapy approved for cancer treatment.
Clinical evaluation of survivin-targeted strategy for cancer therapy
LY2181308 (Eli Lilly, Indianapolis, IN) is a 2’-O-methoxymethyl modified ASO targeting survivin mRNA [54]. It has been tested in phase II clinical trials in patients with relapsed or refractory AML (as single agent or combined with idarubicin and cytarabine) [55], NSCLC (combined with docetaxel), and castration-resistant prostate cancer (CRPC) (combined with docetaxel/prednisone) [56]. In the treatment of AML, LY2181308 was well tolerated as single agent, and showed some clinical benefits and no additional toxicity in combination with chemotherapy [55]. More patients are needed to validate the data. However, in combination with docetaxel/prednisone, LY2181308 showed no improvement as compared to docetaxel/prednisone alone in the treatment of CRPC [56]. Another survivin-targeting ASO, EZN-3042 (Enzon Pharmaceuticals, Inc., Piscataway, NJ) [57] exhibited very good pre-clinical activity in acute lymphoblastic leukemia (ALL) cells and mouse models [58,59], leading to phase I clinical trial in children with relapsed ALL. Unfortunately, severe dose-limiting toxicities, grade 3 γ-glutamyl transferase elevation and grade 3 gastrointestinal bleeding, were seen at dose level 1 (2.5 mg/kg). It was not tolerated when EZN-3042 was combined with chemotherapy. The trial was terminated, and the company ended the development of this agent [60]. Immunotherapies with dendritic cell- or prime-based vaccination targeting survivin are ongoing in various human cancers [61,62].
Terameprocol (EM-1421, Erimos Pharmaceuticals, Houston, TX) and YM155 (Astellas Pharma US, Inc., Northbrook, IL) are transcriptional repressors of survivin promoter. The suppressive effect of Terameprocol on survivin is based upon its selective inhibition of Sp1. Thus, Terameprocol is a global transcriptional inhibitor rather than a gene specific inhibitor [63]. It could inhibit the replication of Sp1 regulated viruses [64,65] and downregulate CDK1 and vascular endothelial growth factor (VEGF) via inhibition of Sp1, resulting in cell cycle inhibition, decreased angiogenesis and enhanced apoptosis in tumor cells [66]. Clinical trials have been initiated to test its antitumor activity in leukemia, cervical intraepithelial neoplasia and recurrent or refractory solid tumors. YM155 is an agent standing out from chemical library screening with a potent suppressive activity against survivin promoter [67]. It is the only small molecule that has been claimed to specifically block survivin in humans [68]. In the following section, we briefly discuss the mechanism of YM155 action and the current clinical studies of its antitumor activity.
The transcriptional repressor YM155 of survivin
YM155 (also called Sepantronium Bromide) is a small molecule suppressant of survivin promoter. It is an imidazolium-based compound, identified from high-throughput screening of a chemical library with luciferase assays [67]. The Drug Discovery Research group of Astellas Pharma, Inc. has published several articles indicating that YM155 acts as a transcriptional suppressant of survivin. They show that YM155 binds to interleukin enhancer-binding factor 3 (ILF3), leading to dissociation of p54nrb from ILF3 and resulting in distinct subcellular localization of ILF3 and p54nrb [69]. Since the ILF3/p54nrb transcriptional complex is required for survivin expression, thus YM155 downregulates survivin via its capability to disrupt the ILF3/p54nrb complex [69,70]. It has also been reported that YM155 disrupts the binding of Sp1 to survivin core promoter region, leading to suppressed survivin promoter activity and consequent survivin reduction [71]. YM155 inhibits survivin expression and induces cell death of various cancer cells at nanomolar level in vitro [67,72]. As a leading inhibitor of survivin being tested in clinical trials (http://www.clinicaltrial.gov/ct2/results?term=YM155&Search=Search), YM155 has become the first choice in survivin-related studies. Meanwhile, questions have been raised regarding the mechanisms and specificity of YM155 [68]. Trevor Glaros and colleagues provide the first evidence suggesting that YM155 is a DNA damage agent rather than a transcriptional suppressor of survivin, because the concentrations of YM155 required inducing rH2AX and P-KAP1, two hallmarks of DNA damage, were much lower than that needed to inhibit survivin [73]. Therefore, downregulation of survivin-induced by YM155 is likely a secondary event following DNA damage. Data from a human Merkel cell carcinoma cell line MKL-1 showed that YM155 did not induce G2/M arrest, but reduced all early DNA synthesis. In addition, YM155 only decreased the growth rate of tumor xenografts-established from MKL-1 cells and the tumors grew back when YM155 was withdrawn [74]. Our recent studies with the acute myeloid leukemia (AML) cell line Kasumi-1 reveal similar results of the “off-target” effects of YM155. Lower concentrations of YM155 that promotes Kasumi-1 cells undergoing apoptosis fail to downregulate survivin, rather significantly enhance rH2AX. Furthermore, YM155 does not induce cell cycle G2/M arrest in Kasumi-1 cells, whereas specific knockdown of survivin expression with shRNAs does (Huang and Liu, unpublished data).
Despite the controversies of the mechanism of action, YM155 has being tested in several phase II clinical studies, as monotherapy or combined with traditional chemotherapeutics or monoclonal antibodies. In a study of 41 patients with refractory diffuse large B-cell lymphoma (DLBCL), only 1 patient had a complete remission and 2 patients responded with a median progression-free survival of 58 days, indicating a very limited activity of YM155 as single agent in DLBCL [75]. Phase II studies with YM155 as monotherapy have also been carried out in unresectable stage III or IV melanoma [76], advanced refractory NSCLC [77], castration-resistant taxane-pretreated prostate cancer [78]. Although the adverse events of YM155 as single therapy was well tolerated, its antitumor activity was fairly modest [75-78]. Thus, the combinatorial strategies of YM155 with other therapeutics were sought in clinical trials. Currently, YM155 in combination with docetaxel is being tested in patients with stage III or IV melanoma (NCT01009775), advanced hormone refractory prostate cancer (NCT00514267), and Her2 negative metastatic breast cancer (NCT01038804). Recent studies reveal that the combinations of YM155 and monoclonal antibody Rituximab (anti-CD20) [79] or alemtuzumab/Campath-1H (anti-CD25) [80] show significant improvements in tumor regression and survival in mouse models with human B-Cell non-Hodgkin lymphoma or adult T-cell leukemia, respectively. Corresponding clinical studies are ongoing in CD20-positive B cell non-Hodgkin’s lymphoma patients (NCT01007292) and adult T-Cell leukemia/lymphoma patients (NCT00061048). Nonetheless, a recent phase I/II study indicates that YM155 is safe, but it fails to show an improvement in response rates to paclitaxel and carboplatin in patients with advanced NSCLC [81]. This failure is likely due to limitations in the specificity of YM155 and its insufficient inhibition of survivin in patients [51]. It is believed that novel therapies which can effectively downregulate survivin in vivo are required to enhance the efficacy of chemotherapy, and thereby reduce the risk of relapse and improve the survival of patients with cancers, including NSCLC.
Therapeutic potential of survivin-targeting miRNAs in human cancers
One of the main goals in studying survivin-targeting miRNAs is to integrate those miRNAs in treating cancer patients. MiRNA-based therapeutic strategies can be divided by their expression levels: 1) miRNA replacement therapy is applied to bring back downregulated miRNAs (for cancer: tumor suppressive miRNAs); 2) miRNA inhibition therapy is applied to suppress overexpressed miRNAs (for cancer: oncogenic miRNAs) [82]. Since stability of miRNAs, uptake efficiency, toxicity and targeting specificity issues are big concerns in in vivo conditions, most ongoing miRNA-related clinical trials use miRNAs as biomarkers, other than therapeutic tools [83,84]. MiR-122 is the first miRNA reached clinical trials as targeted therapy. MiR-122 in HCV (Hepatitis C virus) patients is hepatic-specifically inhibited by an anti-miR122 drug (a locked nucleic acid (LNA)-modified miR-122 antagonist), named miravirsen/ SPC3649 (Santaris Pharma A/S, Copenhagen, Denmark), study of which has already reached phase II clinical trial (http://clinicaltrials.gov/ct2/results?term=mir122&Search=Search) [83]. In May 2013, the first clinical trial using miR-34 mimics (trade name: MRX34, Mirna Therapeutics, Austin, TX) as replacement therapy (ClinicalTrials.gov Identifier: NCT01829971) was initiated. It is worth noting that, this is the first and only miRNA replacement therapy carried out under clinical trial up until now. In addition to survivin, the other targets of miR-34a also involve in G1/S transition (c-MYC, E2F, CDK4, CDK6), apoptosis inhibition (Bcl2, SIRT1) and tumor cell invasion (c-MET) [85], thus render a wide range benefit for miR-34 replacement therapy. Now the ongoing clinical trial of MRX34 is carried out in liver-base cancers and hematologic malignancies.
miRNA delivery methods
To use miRNAs in vivo for treatment, proper delivery methods are required to transport the miRNAs to their targets. The delivery systems of miRNA can be divided as viral-based systems and non-viral systems. Toxicity and immunogenicity are major drawbacks of viral-base vectors, limiting their clinical usage [82]. Non-viral delivery systems, including PEI (Polyethylenimine) and lipid-based systems are more favorable for clinical use. The materials and methods of nonviral delivery systems are reviewed by Zhang et al in detail [82]. The nanocarriers synthesized from different materials with distinct surface modifications are also tested for miRNA delivery [82,85].
It is believed that efficient, low toxic and specific delivery of survivin-targeting miRNAs to cancer cells is important for effective cancer therapy. Antibody-coated nanoparticles have been studied as a delivery system for survivin-targeting miRNAs [34,86,87]. Chen et al [34] used a liposome-polycation-hyaluronic acid nanoparticle formulation modified with tumor-targeting single-chain antibody fragment (scFv) for systemic delivery of miR-34a in lung metastasis of murine B16F10 melanoma. Delivered miR-34a downregulated survivin expression in the metastatic tumor and reduced tumor burden in the lung. Gaca et al [87] developed a human serum albumin-based nanoparticulate carrier system for delivery of an engineered miRNA, whose sequence was based on a siRNA targeting at the ORF of survivin mRNA. The nanoparticle enhanced sensitivity of colorectal cancer SW480 cells to the treatment of ionizing radiation [87]. In a following study, the human serum albumin nanoparticle was coupled with a membrane heat shock protein Hsp70-specific antibody (cmHsp70.1). The cmHsp70.1-conjugated survivin-targeting miRNA nanoparticle exhibited an enhanced tumor cell uptake and increased the efficacy of radiation therapy in vitro, suggesting that development of antibody-based tumor specific targeting systems for miRNAs would be beneficial [86].
Future direction
To effectively treat cancer patients, one of the optimal strategies is to find targets which can distinguish tumor cells from normal tissues. The goal is to aim at these targets to kill cancer cells while minimize the hazards to healthy cells. Because of its selective expression in tumor tissues, survivin is considered as one of those targets and has been studied over a decade since its identification in 1997 [3]. Survivin has been consistently confirmed to play a vital role in tumor cell survival. A number of cancer survival-related signaling pathways converge on survivin and the expression of survivin is highly correlated with therapeutic resistance, advanced tumor, and poor prognosis. Survivin is not an enzyme or a cell-surface molecule, excluding it from traditional drug targets. Nonetheless, it possesses nodal properties and involves in multiple signaling mechanisms in tumor maintenance. Thus, ensuring interventions on survivin can cause devastating results to cancer cells [9]. Although the expression of survivin is high during embryonic development and undetectable in most terminally differentiated tissues, it does express in some normal adult tissues and cells, such as hematopoietic progenitor cells, vascular endothelial cells, T cells, erythroid cells, ovary, testes, and liver [88]. Functions of these normal tissues should be taken into consideration when using survivin as a target for cancer therapy [88].
Studies on understanding the molecular basis of survivin dysregulation in human cancers provide us new avenues to identify novel therapeutic strategy inhibiting survivin. The miRNAs binding to 3’-UTR of survivin mRNA emerge as important players for regulation of survivin expression. The specific miRNA expression profiles [21] in cancers present unique opportunities to manipulate survivin epigenetically. We believe that miRNAs, particular those with multiple binding sites on 3’-UTR of survivin mRNA, are novel, exciting tools to effectively inhibit survivin for cancer treatment. As “sister” miRNAs work cooperatively to downregulate their common targets [89], it is likely that the multiple binding sites of one miRNA may exhibit synergistic activity to inhibit the target. The two miRNAs, miR-542-3p and miR-708, each of which has three binding sites on 3’-UTR of survivin mRNA, should be more specific and effective than other miRNAs with single or two binding sites to downregulate survivin. It is worth testing whether miR-542-3p and/or miR-708 may be developed as promising agents targeting of survivin to enhance the efficacy of those commonly used therapeutic agents in cancer treatment.
Acknowledgements
This work was supported in part by the NIH/NCI grant 1R03CA181918-01, the National Natural Science Foundation of China (NSFC grant No. 81472763), and a research award from Elsa U. Pardee Foundation (BL).
Disclosure of conflict of interest
The authors declare that they have no competing interests.
References
- 1.Fulda S. Inhibitor of apoptosis proteins in hematological malignancies. Leukemia. 2009;23:467–476. doi: 10.1038/leu.2008.329. [DOI] [PubMed] [Google Scholar]
- 2.Hunter AM, LaCasse EC, Korneluk RG. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12:1543–1568. doi: 10.1007/s10495-007-0087-3. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 4.Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD. Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999;59:6097–6102. [PubMed] [Google Scholar]
- 5.Caldas H, Honsey LE, Altura RA. Survivin 2alpha: a novel Survivin splice variant expressed in human malignancies. Mol Cancer. 2005;4:11. doi: 10.1186/1476-4598-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F. Role of survivin and its splice variants in tumorigenesis. Br J Cancer. 2005;92:212–216. doi: 10.1038/sj.bjc.6602340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badran A, Yoshida A, Ishikawa K, Goi T, Yamaguchi A, Ueda T, Inuzuka M. Identification of a novel splice variant of the human anti-apoptopsis gene survivin. Biochem Biophys Res Commun. 2004;314:902–907. doi: 10.1016/j.bbrc.2003.12.178. [DOI] [PubMed] [Google Scholar]
- 8.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 9.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 10.Kanwar JR, Kamalapuram SK, Kanwar RK. Survivin signaling in clinical oncology: a multifaceted dragon. Med Res Rev. 2013;33:765–789. doi: 10.1002/med.21264. [DOI] [PubMed] [Google Scholar]
- 11.Marusawa H, Matsuzawa S, Welsh K, Zou H, Armstrong R, Tamm I, Reed JC. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003;22:2729–2740. doi: 10.1093/emboj/cdg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohi T, Okada K, Xia F, Wilford CE, Samuel T, Welsh K, Marusawa H, Zou H, Armstrong R, Matsuzawa S, Salvesen GS, Reed JC, Altieri DC. An IAP-IAP complex inhibits apoptosis. J Biol Chem. 2004;279:34087–34090. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- 13.Zaffaroni N, Daidone MG. Survivin expression and resistance to anticancer treatments: perspectives for new therapeutic interventions. Drug Resist Updat. 2002;5:65–72. doi: 10.1016/s1368-7646(02)00049-3. [DOI] [PubMed] [Google Scholar]
- 14.Coumar MS, Tsai FY, Kanwar JR, Sarvagalla S, Cheung CH. Treat cancers by targeting survivin: just a dream or future reality? Cancer Treat Rev. 2013;39:802–811. doi: 10.1016/j.ctrv.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Huang X, Lee CK, Liu B. Elevated expression of erbB3 confers paclitaxel resistance in erbB2-overexpressing breast cancer cells via upregulation of Survivin. Oncogene. 2010;29:4225–4236. doi: 10.1038/onc.2010.180. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Huang J, Lyu H, Cai B, Yang X, Li F, Tan J, Edgerton SM, Thor AD, Lee CK, Liu B. Therapeutic targeting of erbB3 with MM-121/SAR256212 enhances antitumor activity of paclitaxel against erbB2-overexpressing breast cancer. Breast Cancer Res. 2013;15:R101. doi: 10.1186/bcr3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nabilsi NH, Broaddus RR, Loose DS. DNA methylation inhibits p53-mediated survivin repression. Oncogene. 2009;28:2046–2050. doi: 10.1038/onc.2009.62. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka C, Uzawa K, Shibahara T, Yokoe H, Noma H, Tanzawa H. Expression of an inhibitor of apoptosis, survivin, in oral carcinogenesis. J Dent Res. 2003;82:607–611. doi: 10.1177/154405910308200807. [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 22.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y, Dahiya R. Regulatory Role of mir-203 in Prostate Cancer Progression and Metastasis. Clin Cancer Res. 2011;17:5287–5298. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 23.Bian K, Fan J, Zhang X, Yang XW, Zhu HY, Wang L, Sun JY, Meng YL, Cui PC, Cheng SY, Zhang J, Zhao J, Yang AG, Zhang R. MicroRNA-203 leads to G1 phase cell cycle arrest in laryngeal carcinoma cells by directly targeting survivin. FEBS Lett. 2012;586:804–809. doi: 10.1016/j.febslet.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 24.Wei W, Wanjun L, Hui S, Dongyue C, Xinjun Y, Jisheng Z. miR-203 inhibits proliferation of HCC cells by targeting survivin. Cell Biochem Funct. 2013;31:82–85. doi: 10.1002/cbf.2863. [DOI] [PubMed] [Google Scholar]
- 25.Jin J, Deng J, Wang F, Xia X, Qiu T, Lu W, Li X, Zhang H, Gu X, Liu Y, Cao W, Shao W. The expression and function of microRNA-203 in lung cancer. Tumour Biol. 2013;34:349–357. doi: 10.1007/s13277-012-0556-3. [DOI] [PubMed] [Google Scholar]
- 26.Xu D, Wang Q, An Y, Xu L. MiR203 regulates the proliferation, apoptosis and cell cycle progression of pancreatic cancer cells by targeting Survivin. Mol Med Rep. 2013;8:379–384. doi: 10.3892/mmr.2013.1504. [DOI] [PubMed] [Google Scholar]
- 27.Yu X, Zhang X, Dhakal IB, Beggs M, Kadlubar S, Luo D. Induction of cell proliferation and survival genes by estradiol-repressed microRNAs in breast cancer cells. BMC Cancer. 2012;12:29. doi: 10.1186/1471-2407-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie H, Lee L, Caramuta S, Hoog A, Browaldh N, Bjornhagen V, Larsson C, Lui WO. MicroRNA Expression Patterns Related to Merkel Cell Polyomavirus Infection in Human Merkel Cell Carcinoma. J Invest Dermatol. 2014;134:507–517. doi: 10.1038/jid.2013.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar B, Yadav A, Lang J, Teknos TN, Kumar P. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PLoS One. 2012;7:e37601. doi: 10.1371/journal.pone.0037601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji X, Wang Z, Geamanu A, Goja A, Sarkar FH, Gupta SV. Delta-tocotrienol suppresses Notch-1 pathway by upregulating miR-34a in nonsmall cell lung cancer cells. Int J Cancer. 2012;131:2668–2677. doi: 10.1002/ijc.27549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Tsai YH, Tseng SH. Inhibition of cyclin-dependent kinase 1-induced cell death in neuroblastoma cells through the microRNA-34a-MYCN-survivin pathway. Surgery. 2013;153:4–16. doi: 10.1016/j.surg.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Cao W, Yang W, Fan R, Li H, Jiang J, Geng M, Jin Y, Wu Y. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour Biol. 2013;35:1287–95. doi: 10.1007/s13277-013-1171-7. [DOI] [PubMed] [Google Scholar]
- 33.Cao W, Fan R, Wang L, Cheng S, Li H, Jiang J, Geng M, Jin Y, Wu Y. Expression and regulatory function of miRNA-34a in targeting survivin in gastric cancer cells. Tumour Biol. 2013;34:963–971. doi: 10.1007/s13277-012-0632-8. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Z, Zhan G, Ye D, Ren Y, Cheng L, Wu Z, Guo J. MicroRNA-34a affects the occurrence of laryngeal squamous cell carcinoma by targeting the antiapoptotic gene survivin. Med Oncol. 2012;29:2473–2480. doi: 10.1007/s12032-011-0156-x. [DOI] [PubMed] [Google Scholar]
- 36.Yoon S, Choi YC, Lee S, Jeong Y, Yoon J, Baek K. Induction of growth arrest by miR-542-3p that targets survivin. FEBS Lett. 2010;584:4048–4052. doi: 10.1016/j.febslet.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 37.Saini S, Yamamura S, Majid S, Shahryari V, Hirata H, Tanaka Y, Dahiya R. MicroRNA-708 induces apoptosis and suppresses tumorigenicity in renal cancer cells. Cancer Res. 2011;71:6208–6219. doi: 10.1158/0008-5472.CAN-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Huang J, Lyu H, Lee CK, Tan J, Wang J, Liu B. Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis. 2013;4:e556. doi: 10.1038/cddis.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diakos C, Zhong S, Xiao Y, Zhou M, Vasconcelos GM, Krapf G, Yeh RF, Zheng S, Kang M, Wiencke JK, Pombo-de-Oliveira MS, Panzer-Grumayer R, Wiemels JL. TEL-AML1 regulation of survivin and apoptosis via miRNA-494 and miRNA-320a. Blood. 2010;116:4885–4893. doi: 10.1182/blood-2009-02-206706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alajez NM, Lenarduzzi M, Ito E, Hui AB, Shi W, Bruce J, Yue S, Huang SH, Xu W, Waldron J, O’Sullivan B, Liu FF. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011;71:2381–2391. doi: 10.1158/0008-5472.CAN-10-2754. [DOI] [PubMed] [Google Scholar]
- 41.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 42.Pathi SS, Jutooru I, Chadalapaka G, Sreevalsan S, Anand S, Thatcher GR, Safe S. GT-094, a NO-NSAID, inhibits colon cancer cell growth by activation of a reactive oxygen species-microRNA-27a: ZBTB10-specificity protein pathway. Mol Cancer Res. 2011;9:195–202. doi: 10.1158/1541-7786.MCR-10-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mertens-Talcott SU, Noratto GD, Li X, Angel-Morales G, Bertoldi MC, Safe S. Betulinic acid decreases ER-negative breast cancer cell growth in vitro and in vivo: role of Sp transcription factors and microRNA-27a:ZBTB10. Mol Carcinog. 2013;52:591–602. doi: 10.1002/mc.21893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhatnagar N, Li X, Padi SK, Zhang Q, Tang MS, Guo B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010;1:e105. doi: 10.1038/cddis.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, Huang MY, Hou JH, Wu QL, Zeng MS, Huang WL, Zeng YX, Shao JY. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552–3562. doi: 10.1158/0008-5472.CAN-10-2435. [DOI] [PubMed] [Google Scholar]
- 46.Boll K, Reiche K, Kasack K, Morbt N, Kretzschmar AK, Tomm JM, Verhaegh G, Schalken J, von Bergen M, Horn F, Hackermuller J. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene. 2012;32:277–85. doi: 10.1038/onc.2012.55. [DOI] [PubMed] [Google Scholar]
- 47.Craig VJ, Cogliatti SB, Rehrauer H, Wundisch T, Muller A. Epigenetic silencing of microRNA-203 dysregulates ABL1 expression and drives Helicobacter-associated gastric lymphomagenesis. Cancer Res. 2011;71:3616–3624. doi: 10.1158/0008-5472.CAN-10-3907. [DOI] [PubMed] [Google Scholar]
- 48.Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, Croce N, Vandesompele J, Mestdagh P, Finazzi-Agro E, Levine AJ, Melino G, Bernardini S, Candi E. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2012;32:127–34. doi: 10.1038/onc.2012.14. [DOI] [PubMed] [Google Scholar]
- 49.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Zhang B, Li W, Fu L, Zhu Z, Dong JT. Epigenetic Silencing of miR-203 Upregulates SNAI2 and Contributes to the Invasiveness of Malignant Breast Cancer Cells. Genes Cancer. 2011;2:782–791. doi: 10.1177/1947601911429743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rauch A, Hennig D, Schafer C, Wirth M, Marx C, Heinzel T, Schneider G, Kramer OH. Survivin and YM155: how faithful is the liaison? Biochim Biophys Acta. 2014;1845:202–220. doi: 10.1016/j.bbcan.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Carter BZ, Milella M, Altieri DC, Andreeff M. Cytokine-regulated expression of survivin in myeloid leukemia. Blood. 2001;97:2784–2790. doi: 10.1182/blood.v97.9.2784. [DOI] [PubMed] [Google Scholar]
- 53.Kelly RJ, Lopez-Chavez A, Citrin D, Janik JE, Morris JC. Impacting tumor cell-fate by targeting the inhibitor of apoptosis protein survivin. Mol Cancer. 2011;10:35. doi: 10.1186/1476-4598-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molckovsky A, Siu LL. First-in-class, first-in-human phase I results of targeted agents: highlights of the 2008 American society of clinical oncology meeting. J Hematol Oncol. 2008;1:20. doi: 10.1186/1756-8722-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erba HP, Sayar H, Juckett M, Lahn M, Andre V, Callies S, Schmidt S, Kadam S, Brandt JT, Van Bockstaele D, Andreeff M. Safety and pharmacokinetics of the antisense oligonucleotide (ASO) LY2181308 as a single-agent or in combination with idarubicin and cytarabine in patients with refractory or relapsed acute myeloid leukemia (AML) Invest New Drugs. 2013;31:1023–1034. doi: 10.1007/s10637-013-9935-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiechno P, Somer BG, Mellado B, Chlosta PL, Cervera Grau JM, Castellano D, Reuter C, Stockle M, Kamradt J, Pikiel J, Duran I, Wedel S, Callies S, Andre V, Hurt K, Brown J, Lahn M, Heinrich B. A Randomised Phase 2 Study Combining LY2181308 Sodium (Survivin Antisense Oligonucleotide) with First-line Docetaxel/Prednisone in Patients with Castration-resistant Prostate Cancer. Eur Urol. 2013;65:516–20. doi: 10.1016/j.eururo.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 57.Sapra P, Wang M, Bandaru R, Zhao H, Greenberger LM, Horak ID. Down-modulation of survivin expression and inhibition of tumor growth in vivo by EZN-3042, a locked nucleic acid antisense oligonucleotide. Nucleosides Nucleotides Nucleic Acids. 2010;29:97–112. doi: 10.1080/15257771003597733. [DOI] [PubMed] [Google Scholar]
- 58.Park E, Gang EJ, Hsieh YT, Schaefer P, Chae S, Klemm L, Huantes S, Loh M, Conway EM, Kang ES, Hoe Koo H, Hofmann WK, Heisterkamp N, Pelus L, Keerthivasan G, Crispino J, Kahn M, Muschen M, Kim YM. Targeting survivin overcomes drug resistance in acute lymphoblastic leukemia. Blood. 2011;118:2191–2199. doi: 10.1182/blood-2011-04-351239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morrison DJ, Hogan LE, Condos G, Bhatla T, Germino N, Moskowitz NP, Lee L, Bhojwani D, Horton TM, Belitskaya-Levy I, Greenberger LM, Horak ID, Grupp SA, Teachey DT, Raetz EA, Carroll WL. Endogenous knockdown of survivin improves chemotherapeutic response in ALL models. Leukemia. 2012;26:271–279. doi: 10.1038/leu.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raetz EA, Morrison D, Romanos-Sirakis E, Gaynon P, Sposto R, Bhojwani D, Bostrom BC, Brown P, Eckroth E, Cassar J, Malvar J, Buchbinder A, Carroll WL. A Phase I Study of EZN-3042, a Novel Survivin Messenger Ribonucleic Acid (mRNA) Antagonist, Administered in Combination With Chemotherapy in Children With Relapsed Acute Lymphoblastic Leukemia (ALL): A Report From the Therapeutic Advances in Childhood Leukemia and Lymphoma (TACL) Consortium. J Pediatr Hematol Oncol. 2013;36:458–63. doi: 10.1097/MPH.0b013e3182a8f58f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srivastava AK, Sharma RK, Yolcu ES, Ulker V, MacLeod K, Dinc G, Shirwan H. Prime-boost vaccination with SA-4-1BBL costimulatory molecule and survivin eradicates lung carcinoma in CD8+ T and NK cell dependent manner. PLoS One. 2012;7:e48463. doi: 10.1371/journal.pone.0048463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hobo W, Strobbe L, Maas F, Fredrix H, Greupink-Draaisma A, Esendam B, de Witte T, Preijers F, Levenga H, van Rees B, Raymakers R, Schaap N, Dolstra H. Immunogenicity of dendritic cells pulsed with MAGE3, Survivin and B-cell maturation antigen mRNA for vaccination of multiple myeloma patients. Cancer Immunol Immunother. 2013;62:1381–1392. doi: 10.1007/s00262-013-1438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 64.Chen H, Teng L, Li JN, Park R, Mold DE, Gnabre J, Hwu JR, Tseng WN, Huang RC. Antiviral activities of methylated nordihydroguaiaretic acids. 2. Targeting herpes simplex virus replication by the mutation insensitive transcription inhibitor tetra-O-methyl-NDGA. J Med Chem. 1998;41:3001–3007. doi: 10.1021/jm980182w. [DOI] [PubMed] [Google Scholar]
- 65.Park R, Giza PE, Mold DE, Huang RC. Inhibition of HSV-1 replication and reactivation by the mutation-insensitive transcription inhibitor tetra-O-glycyl-nordihydroguaiaretic acid. Antiviral Res. 2003;58:35–45. doi: 10.1016/s0166-3542(02)00165-1. [DOI] [PubMed] [Google Scholar]
- 66.Smolewski P. Terameprocol, a novel site-specific transcription inhibitor with anticancer activity. IDrugs. 2008;11:204–214. [PubMed] [Google Scholar]
- 67.Nakahara T, Kita A, Yamanaka K, Mori M, Amino N, Takeuchi M, Tominaga F, Hatakeyama S, Kinoyama I, Matsuhisa A, Kudoh M, Sasamata M. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67:8014–8021. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- 68.Holmes D. Cancer drug’s survivin suppression called into question. Nat Med. 2012;18:842–843. doi: 10.1038/nm0612-842b. [DOI] [PubMed] [Google Scholar]
- 69.Nakamura N, Yamauchi T, Hiramoto M, Yuri M, Naito M, Takeuchi M, Yamanaka K, Kita A, Nakahara T, Kinoyama I, Matsuhisa A, Kaneko N, Koutoku H, Sasamata M, Yokota H, Kawabata S, Furuichi K. Interleukin enhancer-binding factor 3/NF110 is a target of YM155, a suppressant of survivin. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.013243. 013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamauchi T, Nakamura N, Hiramoto M, Yuri M, Yokota H, Naitou M, Takeuchi M, Yamanaka K, Kita A, Nakahara T, Kinoyama I, Matsuhisa A, Kaneko N, Koutoku H, Sasamata M, Kobori M, Katou M, Tawara S, Kawabata S, Furuichi K. Sepantronium bromide (YM155) induces disruption of the ILF3/p54(nrb) complex, which is required for survivin expression. Biochem Biophys Res Commun. 2012;425:711–716. doi: 10.1016/j.bbrc.2012.07.103. [DOI] [PubMed] [Google Scholar]
- 71.Cheng Q, Ling X, Haller A, Nakahara T, Yamanaka K, Kita A, Koutoku H, Takeuchi M, Brattain MG, Li F. Suppression of survivin promoter activity by YM155 involves disruption of Sp1-DNA interaction in the survivin core promoter. Int J Biochem Mol Biol. 2012;3:179–197. [PMC free article] [PubMed] [Google Scholar]
- 72.Nakahara T, Kita A, Yamanaka K, Mori M, Amino N, Takeuchi M, Tominaga F, Kinoyama I, Matsuhisa A, Kudou M, Sasamata M. Broad spectrum and potent antitumor activities of YM155, a novel small-molecule survivin suppressant, in a wide variety of human cancer cell lines and xenograft models. Cancer Sci. 2011;102:614–621. doi: 10.1111/j.1349-7006.2010.01834.x. [DOI] [PubMed] [Google Scholar]
- 73.Glaros TG, Stockwin LH, Mullendore ME, Smith B, Morrison BL, Newton DL. The “survivin suppressants” NSC 80467 and YM155 induce a DNA damage response. Cancer Chemother Pharmacol. 2012;70:207–212. doi: 10.1007/s00280-012-1868-0. [DOI] [PubMed] [Google Scholar]
- 74.Arora R, Shuda M, Guastafierro A, Feng H, Toptan T, Tolstov Y, Normolle D, Vollmer LL, Vogt A, Dömling A, Brodsky JL, Chang Y, Moore PS. Survivin is a therapeutic target in merkel cell carcinoma. Sci Transl Med. 2012;4:133ra156. doi: 10.1126/scitranslmed.3003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheson BD, Bartlett NL, Vose JM, Lopez-Hernandez A, Seiz AL, Keating AT, Shamsili S, Papadopoulos KP. A phase II study of the survivin suppressant YM155 in patients with refractory diffuse large B-cell lymphoma. Cancer. 2012;118:3128–3134. doi: 10.1002/cncr.26510. [DOI] [PubMed] [Google Scholar]
- 76.Lewis KD, Samlowski W, Ward J, Catlett J, Cranmer L, Kirkwood J, Lawson D, Whitman E, Gonzalez R. A multi-center phase II evaluation of the small molecule survivin suppressor YM155 in patients with unresectable stage III or IV melanoma. Invest New Drugs. 2011;29:161–166. doi: 10.1007/s10637-009-9333-6. [DOI] [PubMed] [Google Scholar]
- 77.Giaccone G, Zatloukal P, Roubec J, Floor K, Musil J, Kuta M, van Klaveren RJ, Chaudhary S, Gunther A, Shamsili S. Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non-small-cell lung cancer. J. Clin. Oncol. 2009;27:4481–4486. doi: 10.1200/JCO.2008.21.1862. [DOI] [PubMed] [Google Scholar]
- 78.Tolcher AW, Quinn DI, Ferrari A, Ahmann F, Giaccone G, Drake T, Keating A, de Bono JS. A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxane-pretreated prostate cancer. Ann Oncol. 2012;23:968–973. doi: 10.1093/annonc/mdr353. [DOI] [PubMed] [Google Scholar]
- 79.Kita A, Mitsuoka K, Kaneko N, Nakata M, Yamanaka K, Jitsuoka M, Miyoshi S, Noda A, Mori M, Nakahara T, Sasamata M. Sepantronium bromide (YM155) enhances response of human B-cell non-Hodgkin lymphoma to rituximab. J Pharmacol Exp Ther. 2012;343:178–183. doi: 10.1124/jpet.112.195925. [DOI] [PubMed] [Google Scholar]
- 80.Chen J, Pise-Masison CA, Shih JH, Morris JC, Janik JE, Conlon KC, Keating A, Waldmann TA. Markedly additive antitumor activity with the combination of a selective survivin suppressant YM155 and alemtuzumab in adult T-cell leukemia. Blood. 2013;121:2029–2037. doi: 10.1182/blood-2012-05-427773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelly RJ, Thomas A, Rajan A, Chun G, Lopez-Chavez A, Szabo E, Spencer S, Carter CA, Guha U, Khozin S, Poondru S, Van Sant C, Keating A, Steinberg SM, Figg W, Giaccone G. A phase I/II study of sepantronium bromide (YM155, survivin suppressor) with paclitaxel and carboplatin in patients with advanced non-small-cell lung cancer. Ann Oncol. 2013;24:2601–2606. doi: 10.1093/annonc/mdt249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93:98–104. doi: 10.1038/clpt.2012.192. [DOI] [PubMed] [Google Scholar]
- 84.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121–1126. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gulla A, Tagliaferri P, Tassone P, Caraglia M. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaca S, Reichert S, Multhoff G, Wacker M, Hehlgans S, Botzler C, Gehrmann M, Rodel C, Kreuter J, Rodel F. Targeting by cmHsp70.1-antibody coated and survivin miRNA plasmid loaded nanoparticles to radiosensitize glioblastoma cells. J Control Release. 2013;172:201–206. doi: 10.1016/j.jconrel.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 87.Gaca S, Reichert S, Rodel C, Rodel F, Kreuter J. Survivin-miRNA-loaded nanoparticles as auxiliary tools for radiation therapy: preparation, characterisation, drug release, cytotoxicity and therapeutic effect on colorectal cancer cells. J Microencapsul. 2012;29:685–694. doi: 10.3109/02652048.2012.680511. [DOI] [PubMed] [Google Scholar]
- 88.Fukuda S, Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther. 2006;5:1087–1098. doi: 10.1158/1535-7163.MCT-05-0375. [DOI] [PubMed] [Google Scholar]
- 89.Wahdan-Alaswad R, Liu B. “Sister” miRNAs in cancers. Cell Cycle. 2013;12:3703–3704. doi: 10.4161/cc.26875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma Q, Wang X, Li Z, Li B, Ma F, Peng L, Zhang Y, Xu A, Jiang B. microRNA-16 represses colorectal cancer cell growth in vitro by regulating the p53/survivin signaling pathway. Oncol Rep. 2013;29:1652–1658. doi: 10.3892/or.2013.2262. [DOI] [PubMed] [Google Scholar]
- 91.Tan LP, Wang M, Robertus JL, Schakel RN, Gibcus JH, Diepstra A, Harms G, Peh SC, Reijmers RM, Pals ST, Kroesen BJ, Kluin PM, Poppema S, van den Berg A. miRNA profiling of B-cell subsets: specific miRNA profile for germinal center B cells with variation between centroblasts and centrocytes. Lab Invest. 2009;89:708–716. doi: 10.1038/labinvest.2009.26. [DOI] [PubMed] [Google Scholar]


