Abstract
PI4KIIIβ recruitment to Golgi membranes relies on GBF1/Arf and ACBD3. Enteroviruses such as poliovirus and coxsackievirus recruit PI4KIIIβ to their replication sites via their 3A proteins. Here, we show that human rhinovirus (HRV) 3A also recruited PI4KIIIβ to replication sites. Unlike other enterovirus 3A proteins, HRV 3A failed to bind GBF1. Although HRV 3A was previously shown to interact with ACBD3, our data suggest that PI4KIIIβ recruitment occurred independently of both GBF1 and ACBD3.
INTRODUCTION
Enteroviruses (family Picornaviridae), such as poliovirus (PV) and coxsackievirus B3 (CVB3), rely on host factor phosphatidylinositol-4-kinase IIIβ (PI4KIIIβ) for genome replication (1, 2). The recruitment of PI4KIIIβ to the replication sites is mediated by the 3A viral nonstructural protein (1). PI4KIIIβ is normally recruited to the Golgi membranes by the small GTPase Arf1 (3) and the Arf1 activator guanine nucleotide exchange factor (GEF) GBF1 (4). ACBD3 (acyl-coenzyme A [CoA]-binding protein domain 3) also interacts with PI4KIIIβ to recruit it to the Golgi membranes (5, 6). CVB3 3A and PV 3A both interact with the N terminus of GBF1 (7, 8) as well as ACBD3 (9). However, we and others recently showed that the PI4KIIIβ recruitment by 3A of PV and CVB3 seems to occur independently of both GBF1/Arf1 and ACBD3 (9, 10). We studied this by individually expressing mutant 3A proteins of CVB3 that no longer interact with GBF1 (9). Unfortunately, these 3A mutations render CVB3 unviable (F. J. M. van Kuppeveld, unpublished data); thus, we have not yet been able to study the role of GBF1 in PI4KIIIβ recruitment in infected cells. Yeast two-hybrid analysis suggests that the 3A proteins of human rhinoviruses (HRV), which also belong to the Enterovirus genus, do not interact with the N terminus of GBF1 (8). In this study, we set out to investigate the consequences of the inability of HRV 3A proteins to interact with GBF1, focusing on the possible role of GBF1 and ACBD3 in PI4KIIIβ recruitment and virus replication.
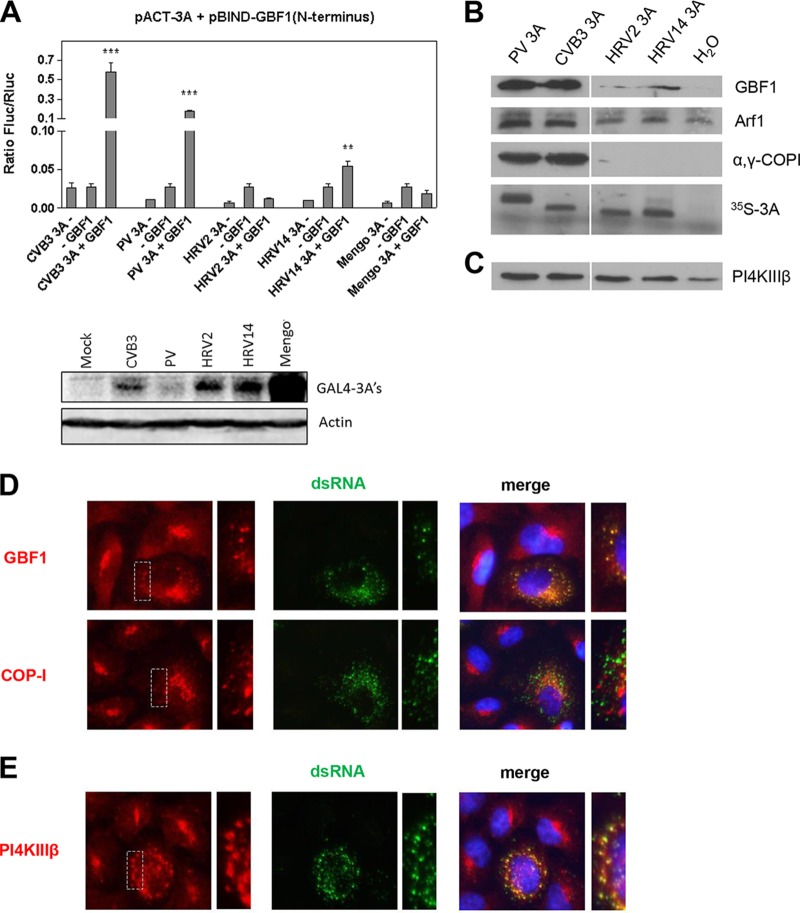
First, we studied the interaction between the N terminus of GBF1 and the 3A proteins of HRV2 and HRV14, which are members of group A and B rhinoviruses, respectively, in a mammalian environment, employing the mammalian two-hybrid (M2H) assay (Promega) as described previously (9). We used the 3As of CVB3, PV, and mengovirus, belonging to the Cardiovirus genus of the Picornaviridae, as controls. Expression of all 3A proteins was verified by Western blot analysis (Fig. 1A, lower half). Importantly, all enterovirus 3A proteins were previously tested and validated to be competent in interacting with other proteins in the M2H assay (9). Both CVB3 3A and PV 3A interacted with the N terminus of GBF1, while no interaction was detected for mengovirus 3A (Fig. 1A, upper panel). HRV2 3A did not bind the N terminus of GBF1, while HRV14 3A interacted to only a limited extent. We considered the possibility that the HRV 3A proteins bind to the C terminus of GBF1. Therefore, we studied whether 3A expression induced recruitment of full-length GBF1 to membranes in HeLa S10 cell extracts as described elsewhere (11). In line with previous results (9, 11), PV and CVB3 3A expression induced accumulation of GBF1, Arf1, and the Arf1 effector COP-I on membranes (Fig. 1B). HRV14 3A showed a low level of GBF1/Arf1 recruitment and no COP-I recruitment, whereas HRV2 3A accumulated little, if any, GBF1/Arf1 and COP-I on membranes. Taken together, these results demonstrate that the 3A proteins of HRVs poorly interact with GBF1 compared to those of PV and CVB3.
FIG 1.
Interaction of HRV 3A with GBF1. (A) Upper half: interaction of different picornavirus 3A proteins with the N terminus of GBF1 in the mammalian two-hybrid system (M2H). Bars show the means of the results from three samples with standard deviations (SD). Significant differences compared to the highest control determined with a Student's t test are indicated as follows: ** = P < 0.01; *** = P < 0.001. Lower half: Western blotting of picornaviral GAL4-3A proteins tested in M2H. Mengo, mengovirus. (B and C) In vitro HeLa S10 cell extract assay. RNA coding for various enterovirus 3A proteins was translated in HeLa S10 cell extracts. Membranes were isolated by centrifugation and subjected to Western blot analysis to detect the membrane-associated proteins using antibodies against GBF1, Arf1, and α,γ-COPI (B) or PI4KIIIβ (C). The efficiency of the translation reaction was assessed by [35S]methionine labeling. (D and E) BGM cells were infected with HRV2 for 8 h, followed by staining of dsRNA, an infection marker, together with GBF1 or COP-I (D) or PI4KIIIβ (E). Nuclei were visualized with DAPI (4′,6-diamidino-2-phenylindole). The dashed areas are enlarged on the right to show the overlap between signals.
Next, we investigated whether GBF1 is present at replication sites in HRV2-infected cells. BGM cells were infected with HRV2, fixed after 8 h, and stained with antibodies against GBF1 (provided by K. Nakayama, Kyoto University, Japan) and COP-I (provided by F. Wieland, Biochemie-Zentrum, Heidelberg, Germany). In uninfected cells, both GBF1 and COP-I displayed a perinuclear Golgi pattern (Fig. 1D). In HRV2-infected cells, GBF1 and COP-I showed a dispersed pattern similar to those seen in PV- and CVB3-infected cells (1, 12). Moreover, the signal of GBF1, but not that of COP-I, overlapped with that of double-stranded RNA (dsRNA), a marker of viral replication (Fig. 1D), as also observed in PV- and CVB3-infected cells (1, 12). Thus, GBF1 localizes at replication sites in HRV2-infected cells despite the lack of a direct interaction with the 3A protein.
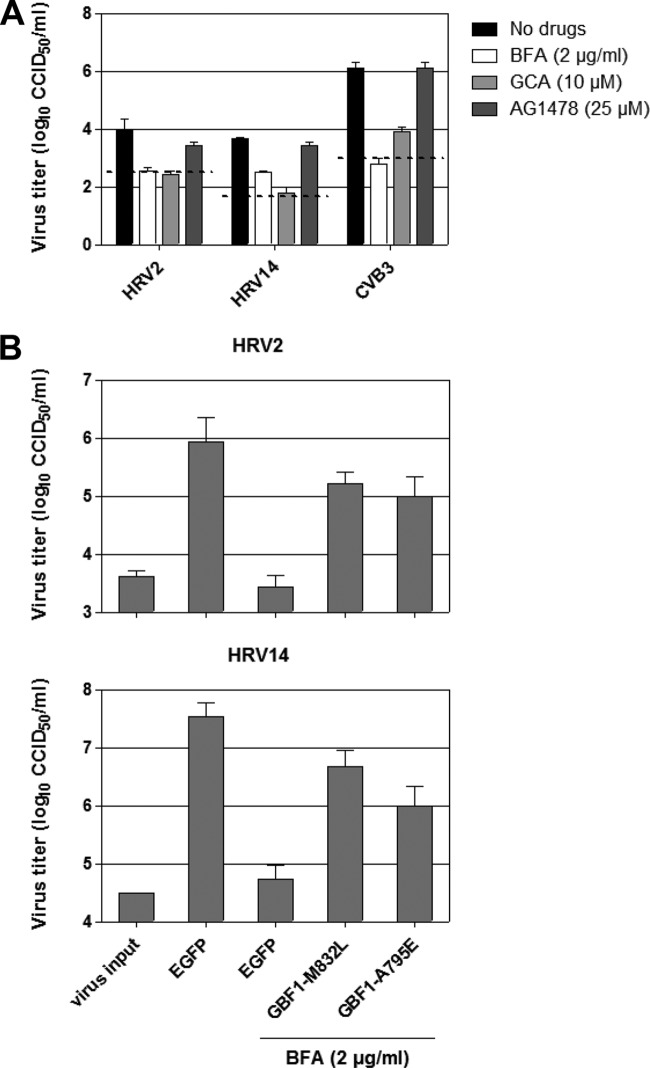
Localization of GBF1 to the HRV replication sites implies, but does not prove, that it is essential for replication. HRV replication is sensitive to brefeldin A (BFA) (13), an inhibitor of the ArfGEFs GBF1, BIG1, and BIG2, but it remained to be determined which of these ArfGEFs is important for HRV. To test whether GBF1 is indeed crucial to HRV replication, we determined the sensitivity of HRV2 and HRV14 to the specific GBF1 inhibitor Golgicide A (GCA). Figure 2A shows that both HRV2 and HRV14 were sensitive to GCA. HRV replication was largely insensitive to treatment with AG1478, a compound that disperses the Golgi membranes without directly targeting GBF1 (14), showing that mere disruption of the secretory pathway is not detrimental to HRV replication. Together, these results suggest that inhibition of GBF1 impairs HRV replication. To corroborate this finding, we examined whether expression of a BFA-resistant GBF1 (11, 15) could restore HRV replication in the presence of BFA as described elsewhere (15). Figure 2B shows that BFA-resistant GBF1-M832L and GBF1-A795E markedly protected HRV replication against the inhibitory effects of BFA. This result confirms that GBF1, and not BIG1 or BIG2, is important for HRV replication. Importantly, this result also indicates that a direct interaction between 3A and GBF1 is not essential for rescue of virus replication, as was previously suggested (11).
FIG 2.
HRV replication depends on GBF1. (A) Sensitivity of HRV2 and HRV14 to GBF1 inhibitors. HeLa R19 cells were infected with virus at a low multiplicity of infection (MOI) for 30 min. After removal of the inoculum, fresh compound-containing medium was added to the cells. After 8 h, cells were freeze-thawed to release intracellular viruses. Samples were titrated on HeLa R19 cells. CVB3 was used as a positive control. Virus input levels (determined at t = 0.5 h) are indicated with a dashed line. CCID50, 50% cell culture infective dose. (B) Replication rescue experiment. HeLa R19 cells were transfected with plasmids encoding EGFP, BFA-resistant GBF1-M832L, or BFA-resistant GBF1-A795E. One day later, cells were infected with virus in the presence of BFA. After 24 h, cells were freeze-thawed and samples were titrated on HeLa R19 cells.
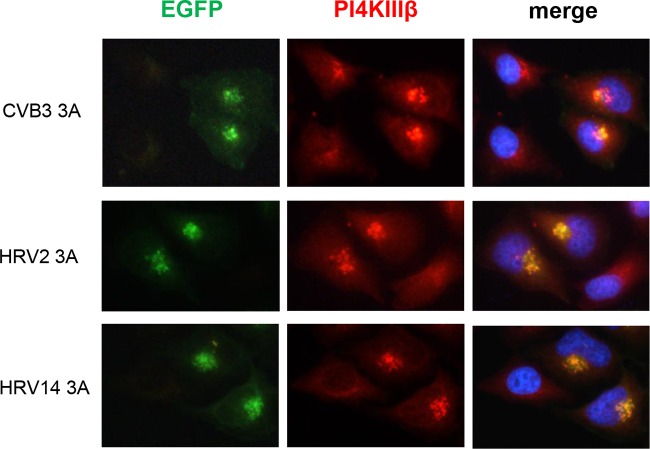
For PV and CVB3, PI4KIIIβ recruitment to replication sites is mediated by their 3A protein (1). Recently, we and others found that replication of all enteroviruses, including HRV, is abolished in the presence of PI4KIIIβ inhibitors (2, 16). Although this indicates that the kinase is also a crucial host factor for HRV, it is as yet unknown whether HRV also actively recruits PI4KIIIβ to replication sites and whether this is mediated via its 3A protein, since the absence of a 3A-GBF1 interaction for HRV could affect recruitment of downstream factor PI4KIIIβ. Immunofluorescence analysis showed that PI4KIIIβ localized to replication sites in HRV2-infected cells (Fig. 1E) in a manner similar to that observed for GBF1 (Fig. 1D). To test whether PI4KIIIβ is recruited by 3A, the HeLa S10 cell extracts expressing HRV 3A proteins were also analyzed for the presence of PI4KIIIβ on membranes. Figure 1C shows that the 3A proteins of both HRVs induce an accumulation of PI4KIIIβ on membranes similar to that seen with CVB3 and PV 3As. We corroborated this finding in intact cells by immunofluorescence. HeLa cells expressing enhanced green fluorescent protein (EGFP)-tagged 3A of CVB3, HRV2, and HRV14 were stained for endogenous GBF1 and PI4KIIIβ (Millipore). Basal levels of GBF1 were localized to 3A-containing membranes, but we did not observe an active recruitment (data not shown). The presence of GBF1 on 3A-containing membranes is most likely a consequence of HRV 3A localizing to and modifying Golgi membranes, where GBF1 (and also other Golgi proteins) normally resides. In contrast, the signal of PI4KIIIβ was substantially increased on 3A-containing membranes compared to the Golgi membranes in untransfected cells, indicating that all 3A proteins induced an active recruitment of PI4KIIIβ to membranes (Fig. 3).
FIG 3.
HRV 3A recruits PI4KIIIβ to membranes. EGFP-tagged 3A proteins were expressed in HeLa R19 cells 1 day prior to fixation. Cells were stained for endogenous PI4KIIIβ. Nuclei were visualized with DAPI.
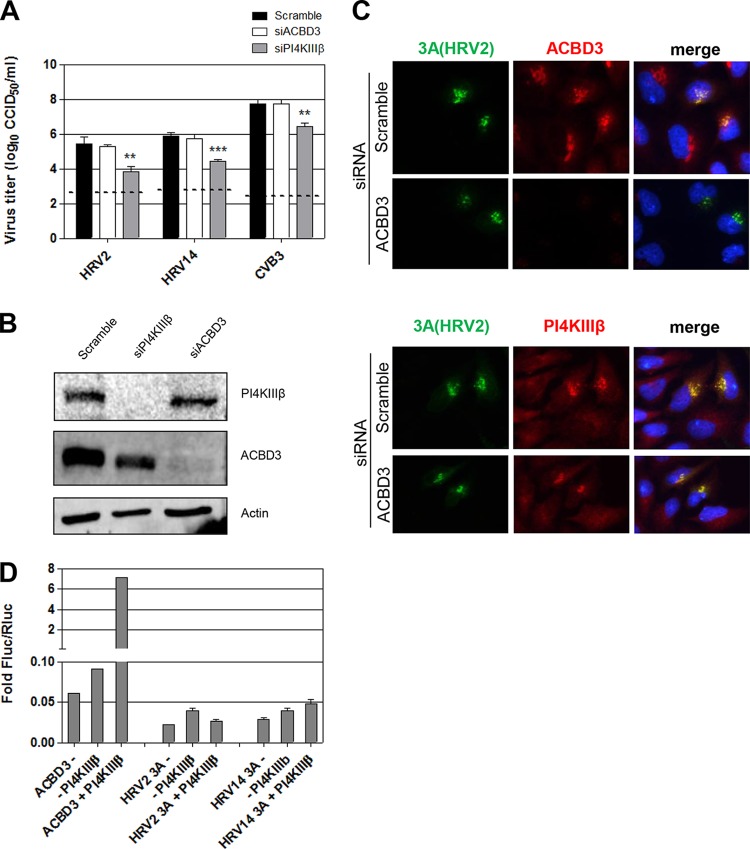
PI4KIIIβ was recently shown to be recruited to replication sites of Aichi virus (family Picornaviridae, genus Kobuvirus) via ACBD3 (5). PV and CVB3 also interact with ACBD3, but this interaction seems dispensable for kinase recruitment (9, 10, 17). Moreover, ACBD3 depletion does not inhibit PV and CVB3 replication (9, 10). Since HRV 3A proteins lack the ability to bind GBF1, we considered the possibility that they recruit PI4KIIIβ via ACBD3. Recently, we showed that HRV 3A proteins interact with ACBD3 in a M2H assay (9). Here, we investigated whether ACBD3 is essential for HRV replication and kinase recruitment. To this end, HeLa cells were treated for 3 days with Scramble small interfering RNA (siRNA) (9), siRNA targeting hACBD3 (5, 9), or siRNA targeting PI4KIIIβ (9). Efficient knockdown was verified with Western blot analysis (Fig. 4B). Depletion of PI4KIIIβ significantly reduced HRV2 and HRV14 replication in a manner similar to that seen with CVB3, whereas knockdown of ACBD3 had no effect on replication (Fig. 4A). ACBD3 depletion also did not affect the ability of HRV2 3A to recruit PI4KIIIβ (Fig. 4C). Thus, ACBD3 is not a crucial host factor for HRV replication and does not contribute to PI4KIIIβ recruitment.
FIG 4.
GBF1-independent recruitment of PI4KIIIβ by HRV2 3A does not rely on ACBD3. (A) Sensitivity of HRV2 and HRV14 to depletion of ACBD3 or PI4KIIIβ by siRNA treatment. HeLa R19 cells (2,000 cells) were reverse transfected with 2 pmol siRNA and 0.1 μl Lipofectamine 2000 mixed in 20 μl Opti-MEM in a 96-well format. Two days posttransfection, cells were infected with virus for 30 min, after which the medium was replaced with fresh medium. Samples were either immediately frozen to determine virus input levels (indicated with a dashed line) or incubated for 24 h. Significant differences compared to the results of treatment with Scramble siRNA were determined with a Student's t test (** = P < 0.01; *** = P < 0.001). (B) Western blot analysis of cell lysates confirming efficient knockdown by siRNA treatments. (C) HRV2 3A recruits PI4KIIIβ in ACBD3-depleted cells. HeLa R19 cells were reverse transfected with Scramble siRNA or siRNA against ACBD3 as described for panel A. After 48 h, cells were transfected with EGFP-tagged HRV2 3A. At 24 h after DNA transfection, cells were fixed and stained with antibodies against endogenous ACBD3 or PI4KIIIβ. (D) Interaction of HRV 3A proteins with PI4KIIIβ in the mammalian two-hybrid system. Bars show the means of the results from three samples with SD.
Having found that GBF1 and ACBD3 are dispensable for PI4KIIIβ recruitment by HRV 3A, we tested the interaction between 3A and PI4KIIIβ in the M2H assay. In line with our previous results, PI4KIIIβ was interaction competent in this assay, as shown by the signal detected together with ACBD3 (Fig. 4D). No signal was detected for both HRV 3A proteins with PI4KIIIβ, which suggests that there is no direct interaction between HRV 3A and PI4KIIIβ. Together, these results indicate that HRV 3A proteins indirectly induce PI4KIIIβ recruitment.
In conclusion, we have shown that HRV replication depends on GBF1 and PI4KIIIβ but not on ACBD3. The recruitment of the lipid kinase is mediated by the 3A protein in a GBF1- and ACBD3-independent manner. These findings are in agreement with our recent observations of ectopic expression of mutant CVB3 3As that were unable to bind GBF1 but that still recruited the kinase in ACBD3-depleted cells (9). It remains to be elucidated by which viral protein GBF1 is recruited to HRV replication sites. PI4KIIIβ recruitment is mediated indirectly by HRV 3A but possibly also by other viral proteins. It was recently shown for PV that, besides 3A, 2BC also recruits the kinase (18), which may also be the case for HRVs. Together, these results show that the replication requirements and recruitment strategies of rhinoviruses are more similar to those of other enteroviruses than had been initially believed based on the lack of a 3A-GBF1 interaction. Furthermore, these findings suggest that GBF1 binding, ACBD3 interaction, and PI4KIIIβ recruitment are three separate functions of enterovirus 3A proteins.
ACKNOWLEDGMENTS
We are very grateful to K. Nakayama and H.-W. Shin (Graduate School of Pharmaceutical Sciences, Kyoto University, Japan) for generously providing an excellent polyclonal rabbit serum against GBF1.
This work was supported by research grants from The Netherlands Organization for Scientific Research (NWO-VENI-863.12.005 to H.M.V.D.S., NWO-ALW-820.02.018 to F.J.M.V.K., and NWO-VICI-91812628 to F.J.M.V.K.) and the European Union 7th Framework (EUVIRNA Marie Curie Initial Training Network; grant agreement number 264286 to F.J.M.V.K.) and by start-up funds of the University of Maryland to G.A.B.
REFERENCES
- 1.Hsu N-Y, Ilnytska O, Belov G, Santiana M, Chen Y-H, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJM, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Schaar HM, Leyssen P, Thibaut HJ, de Palma A, van der Linden L, Lanke KHW, Lacroix C, Verbeken E, Conrath K, Macleod AM, Mitchell DR, Palmer NJ, van de Poël H, Andrews M, Neyts J, van Kuppeveld FJM. 2013. A novel, broad-spectrum inhibitor of enterovirus replication that targets host cell factor PI4KIIIβ. Antimicrob Agents Chemother 57:4971–4981. doi: 10.1128/AAC.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. 2004. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol 6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 4.Donaldson JG, Jackson CL. 2000. Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol 12:475–482. doi: 10.1016/S0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki J, Ishikawa K, Arita M, Taniguchi K. 2012. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J 31:754–766. doi: 10.1038/emboj.2011.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sohda M, Misumi Y, Yamamoto A, Yano A, Nakamura N, Ikehara Y. 2001. Identification and characterization of a novel Golgi protein, GCP60, that interacts with the integral membrane protein giantin. J Biol Chem 276:45298–45306. doi: 10.1074/jbc.M108961200. [DOI] [PubMed] [Google Scholar]
- 7.Wessels E, Duijsings D, Niu T-K, Neumann S, Oorschot VM, de Lange F, Lanke KHW, Klumperman J, Henke A, Jackson CL, Melchers WJG, van Kuppeveld FJM. 2006. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev Cell 11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Wessels E, Duijsings D, Lanke KHW, van Dooren SHJ, Jackson CL, Melchers WJG, van Kuppeveld FJM. 2006. Effects of picornavirus 3A proteins on protein transport and GBF1-dependent COP-I recruitment. J Virol 80:11852–11860. doi: 10.1128/JVI.01225-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorobantu CM, van der Schaar HM, Ford LA, Strating JRPM, Ulferts R, Fang Y, Belov G, van Kuppeveld FJM. 2014. Recruitment of PI4KIIIβ to coxsackievirus B3 replication organelles is independent of ACBD3, GBF1, and Arf1. J Virol 88:2725–2736. doi: 10.1128/JVI.03650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Téoulé F, Brisac C, Pelletier I, Vidalain P-O, Jégouic S, Mirabelli C, Bessaud M, Combelas N, Autret A, Tangy F, Delpeyroux F, Blondel B. 2013. The Golgi protein ACBD3, an interactor for poliovirus protein 3A, modulates poliovirus replication. J Virol 87:11031–11046. doi: 10.1128/JVI.00304-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog 4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol 81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irurzun A, Perez L, Carrasco L. 1992. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology 191:166–175. doi: 10.1016/0042-6822(92)90178-R. [DOI] [PubMed] [Google Scholar]
- 14.van der Linden L, van der Schaar HM, Lanke KHW, Neyts J, van Kuppeveld FJM. 2010. Differential effects of the putative GBF1 inhibitors Golgicide A and AG1478 on enterovirus replication. J Virol 84:7535–7542. doi: 10.1128/JVI.02684-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanke KHW, van der Schaar HM, Belov GA, Feng Q, Duijsings D, Jackson CL, Ehrenfeld E, van Kuppeveld FJM. 2009. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J Virol 83:11940–11949. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spickler C, Lippens J, Laberge M-K, Desmeules S, Bellavance E, Garneau M, Guo T, Hucke O, Leyssen P, Neyts J, Vaillancourt FH, Décor A, O'Meara J, Franti M, Gauthier A. 2013. Phosphatidylinositol 4-kinase III beta is essential for the replication of human rhinovirus and its inhibition causes a lethal phenotype in vivo. Antimicrob Agents Chemother 57:3358–3368. doi: 10.1128/AAC.00303-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greninger AL, Knudsen GM, Betegon M, Burlingame AL, Derisi JL. 2012. The 3A protein from multiple picornaviruses utilizes the Golgi adaptor protein ACBD3 to recruit PI4KIIIβ. J Virol 86:3605–3616. doi: 10.1128/JVI.06778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arita M. 2014. Phosphatidylinositol-4 kinase III beta and oxysterol-binding protein accumulate unesterified cholesterol on poliovirus-induced membrane structure. Microbiol Immunol 58:239–256. doi: 10.1111/1348-0421.12144. [DOI] [PubMed] [Google Scholar]