Abstract
Rabies virus replicates in the cytoplasm of host cells, but rabies virus phosphoprotein (P-protein) undergoes active nucleocytoplasmic trafficking. Here we show that the largely nuclear P-protein isoform P3 can localize to nucleoli and forms specific interactions with nucleolin. Importantly, depletion of nucleolin expression inhibits viral protein expression and infectious virus production by infected cells. This provides the first evidence that lyssaviruses interact with nucleolin and that nucleolin is important to lyssavirus infection.
TEXT
Lyssaviruses, including rabies virus (RABV) and Australian bat lyssavirus, cause over 55,000 human deaths worldwide annually (1). The RABV phosphoprotein (P-protein) has critical roles in genome transcription/replication and antagonism of interferon (IFN)-dependent antiviral responses (2–9). In infected cells, the P gene produces full-length P-protein (P1) and several N-terminally truncated isoforms, predominantly P2 and P3, via a ribosomal leaky scanning mechanism (7, 10). Although RABV replication occurs in the cytoplasm, the nucleocytoplasmic distribution of P-protein in infected cells is dependent on cellular nuclear export mechanisms, indicating that P-protein undergoes active nuclear trafficking during infection (7).
P-protein isoforms have distinct nuclear trafficking properties whereby P1 and P2 localize mainly in the cytoplasm, while P3 can accumulate in the nucleus (11–13) (Fig. 1). This depends on specific nuclear localization (NLS) and nuclear export (NES) sequences, including an N-terminal NLS (N-NLS) (Fig. 1) that is active only in P3, indicative of specific intranuclear functions (7, 11–13). Notably, a number of viruses with cytoplasmic replication cycles express proteins able to enter the nucleus and to interact with nucleoli and core nucleolar proteins, such as nucleolin (NCL) and fibrillarin (FBL), suggestive of key roles in infection potentially related to functions in ribosome biogenesis, RNA processing, and regulation of cell cycle progression and apoptosis (14–18). Possible roles for nucleoli/nucleolar proteins in lyssavirus infection, however, have not been investigated.
FIG 1.
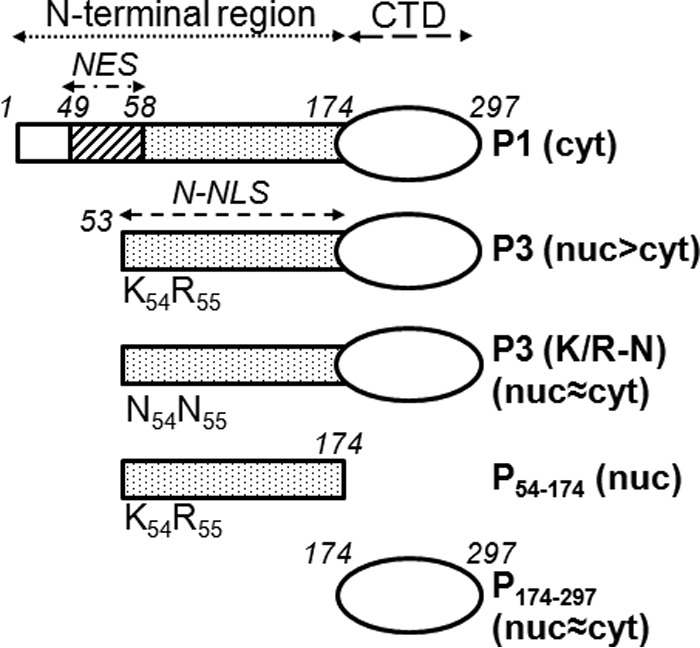
RABV P-protein constructs used in the study. P-proteins and derivatives thereof used in this study are shown schematically; constructs used to express P-proteins/derivatives from CVS RABV fused to GFP were generated by cloning into pEGFP-C1 as described previously (13) or have been described elsewhere (11). Residue positions corresponding to full-length P-protein are in italics, and regions corresponding to the N-terminal region, C-terminal domain (CTD), N-terminal NES (residues 49 to 58), and N-NLS (residues 54 to 174) are shown. The N-terminal NES mediates nuclear export of P1 but is truncated and inactivated in P3 concomitant with activation of the N-NLS. Substitution of the K and R residues at positions 54 and 55 for asparagine inhibits N-NLS function. The reported subcellular distribution of the proteins is indicated (cyt, cytoplasmic; nuc, nuclear).
To identify proteins that interact with P-protein, we immunoprecipitated total P-protein from SK-N-BE cells infected with challenge virus standard (CVS) RABV for mass-spectroscopic analysis using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF). NCL was identified among the proteins coprecipitated (data not shown). To verify this interaction, we performed immunoprecipitation (IP) analysis using HEK293T cells transfected to express P1, P3, or P3(KR-N), in which the N-NLS residues K54 and R55 are replaced by asparagine to inhibit nuclear localization (11), fused to green fluorescent protein (GFP) (Fig. 1). As previously described (19), we observed that nucleolin was present as several forms in cell lysates (Fig. 2A and 3E, Input) with apparent molecular masses between ca. 55 and 110 kDa; this includes NCL modified posttranslationally, including through extensive phosphorylation, methylation, and acetylation (20–22), as well as NCL autocleavage products (19, 23, 24). NCL coprecipitated with GFP-P3 but not GFP alone (Fig. 2A). NCL also coprecipitated with P1 and P3(KR-N), despite the fact that NCL is strongly nuclear/nucleolar while P1 is cytoplasmic and P3(KR-N) is inhibited for nuclear import compared with wild-type P3 (Fig. 2A) (11). Notably, we also found that in all cases, P-protein coprecipitated specific forms of NCL (ca. 110 kDa and ca. 70 kDa or less) without precipitating forms between ca. 70 and 100 kDa (Fig. 2A; see also Fig. 3E); thus it appears that P-protein forms selective interactions with specific modified and/or cleaved forms of NCL. We additionally analyzed IPs of GFP-P1 and P3 by mass spectroscopy, identifying NCL as an interactor (data not shown); however, the core nucleolar protein FBL was not detected in this analysis, indicating that interaction with NCL is specific.
FIG 2.
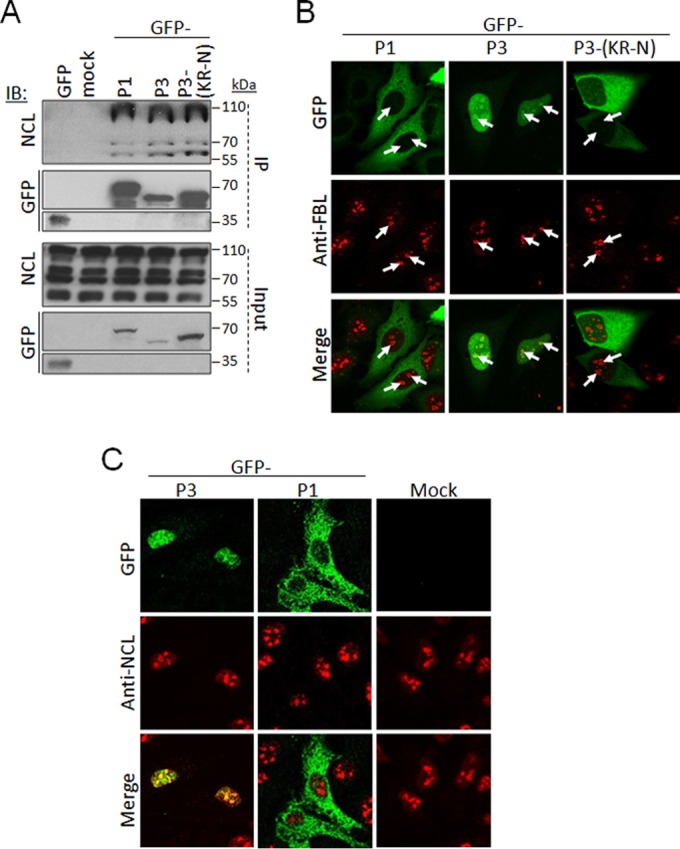
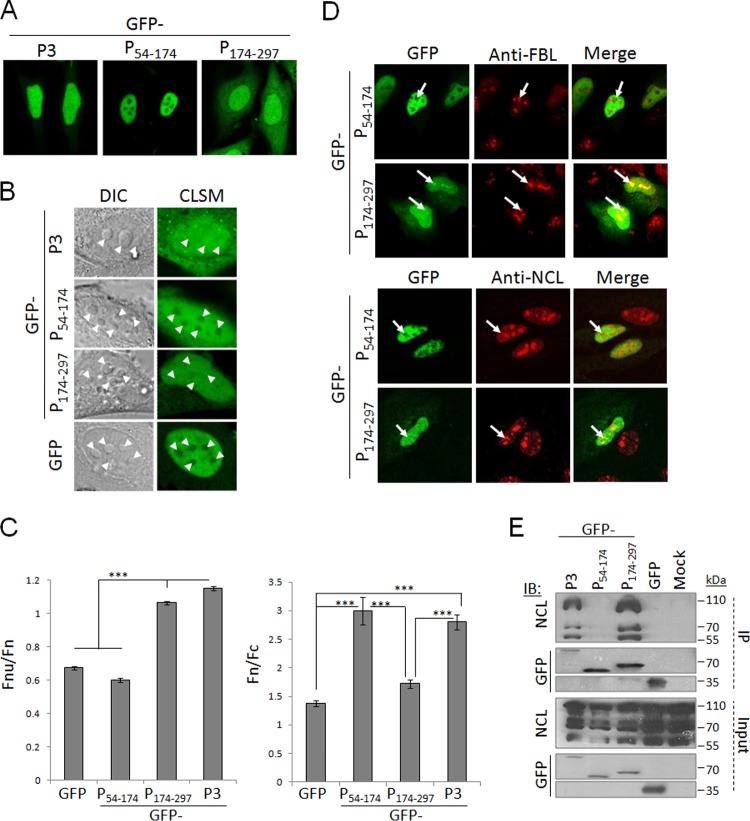
P-protein can interact with NCL and localize to nucleoli. (A) HEK293T cells were transfected to express the indicated GFP-fused P-proteins or GFP alone or mock transfected, using Lipofectamine 2000. At 18 h posttransfection, cells were lysed, and immunoprecipitation (IP) of GFP-fused proteins was performed using the GFP-trap system (ChromoTek) (8). Lysates (input) and IPs were analyzed by immunoblotting (IB) using antibodies against NCL or GFP. (B and C) HeLa cells transfected to express the indicated proteins were fixed and immunostained with anti-FBL (B) or anti-NCL (C) antibody (Abcam) followed by Alexa Fluor-568-conjugated secondary antibody, and analyzed by CLSM using an Olympus FV1000 microscope with 100× oil immersion objective. The white arrows in FBL-stained samples indicate nucleoli.
FIG 3.
The P-protein CTD is necessary and sufficient to mediate nucleolar localization and NCL interaction. (A and B) HeLa cells expressing the indicated proteins were analyzed live using CLSM, with DIC microscopy to identify nucleoli (B), using a Leica SP5 microscope with 63× oil immersion objective. (C) Images such as those in panel B were analyzed to determine the Fnu/Fn (left; n ≥ 40 nucleoli from 2 independent assays) and Fn/Fc (right; n ≥ 15 cells from 2 independent assays) as described previously (11, 28). Data are means ± standard errors of the means (SEM), and statistical analysis used Student's t test (***, P < 0.0001). (D) HeLa cells transfected to express the indicated proteins were fixed and immunostained for FBL or NCL before CLSM analysis. (E) HEK293T cells transfected to express the indicated proteins or mock transfected were subjected to immunoprecipitation followed by IB analysis.
To examine the nucleolar interaction of P1, P3, and P3(KR-N) in intact cells, we transfected HeLa cells to express the GFP-fused proteins, before fixation and immunostaining with antibodies against the nucleolar marker FBL and analysis by confocal laser scanning microscopy (CLSM). This revealed that P3 localized throughout the nucleus, including within FBL-labeled nucleoli; this localization was substantially reduced for P3(KR-N) (Fig. 2B). Due to its exclusion from the nucleus, P1 did not colocalize with FBL/nucleoli (Fig. 2B). NCL can shuttle between the nucleus and cytoplasm but localizes principally in nucleoli (24). Immunostaining for NCL confirmed this distribution and indicated that P3 but not P1 can localize within NCL-labeled nucleoli (Fig. 2C). Expression of P1 or P3 did not appear to substantially affect the localization of NCL compared with mock-transfected cells. This suggests that P1 does not cause substantial relocalization of NCL to the cytoplasm such that their coimmunoprecipitation is likely to include associations formed postlysis; however, since cellular NCL includes a cytoplasmic pool (25, 26), it is also likely that P1 interacts with cytoplasmic NCL in intact cells. Clearly, however, the data indicate that NCL interaction requires a sequence within residues 56 to 297 of P-protein.
To determine the region of P3 required for nucleolar localization and NCL interaction, we analyzed the localization of GFP-fused P54–174 (N-terminal region) and P174–297 (C-terminal domain [CTD]) (Fig. 1) in living cells (Fig. 3A and B) or cells immunostained for FBL or NCL (Fig. 3D), using CLSM. GFP-P54–174 was strongly nuclear due to N-NLS activity (11) but was excluded from nucleoli, consistent with the requirement for specific signals for nucleolar targeting (16) and suggesting that the nucleolar localization signal of P3 is within the CTD (Fig. 3A). Consistent with this GFP-P174–297, although less nuclear due to deletion of the N-NLS, clearly could localize in nucleoli (Fig. 3A). To quantify the level of nucleolar localization of each of these proteins, we analyzed the nucleus of living cells expressing the different GFP-fused P-proteins or GFP alone, using differential interference contrast (DIC) microscopy to identify nucleoli and using CLSM to assess GFP localization within the nucleus/nucleoli (Fig. 3B). DIC typically identified 3 to 6 characteristic nucleolar structures within each nucleus. As expected, GFP was excluded from the nucleolus, and fusion to P3 or P174–297, but not P54–174, conferred the capacity to localize within the nucleolus (Fig. 3B). Calculation of the ratio of nucleolar to nuclear fluorescence corrected for background fluorescence (Fnu/Fn) in living cells was performed as previously described (27, 28); we also calculated the nuclear to cytoplasmic fluorescence ratio (Fn/Fc) (11) to determine the level of protein accumulation in the nucleus (Fig. 3C). This confirmed that GFP and GFP-P54–174 were excluded from nucleoli even though P54–174 conferred a significant (P < 0.0001) increase in nuclear accumulation. GFP-P3 and GFP-P174–297 showed significantly (P < 0.0001) increased nucleolar accumulation compared with GFP and GFP-P54–174, consistent with the presence of a nucleolar localization sequence (Fig. 3C). Nucleolar localization of GFP-P3 and GFP-P174–297 was also observed in fixed cells immunostained for NCL or FBL (Fig. 3D), and IP analysis identified coprecipitation of NCL with GFP-P174–297 but not GFP-P54–174 (Fig. 3E), indicating that the CTD is necessary and sufficient to target nucleoli and to interact with NCL. This suggests that in P3 the N-NLS mediates efficient nuclear import, and a CTD-localized nucleolar localization/NCL interaction signal mediates nucleolar association.
To examine the role of NCL in infection, we transfected U-373-MG cells with scrambled small interfering RNA (Scr-siRNA) or NCL-targeting siRNA using Dharmafect-1 reagent, according to the manufacturer's instructions (Thermo Scientific). To exclude any off-target effects, we tested two siRNAs targeting distinct sequences of NCL (NCL1 [5′-UCCAAGGUAACUUUAUUUCUU-3′] and NCL2 [5′-UUCUUUGACAGGCUCUUCCUU-3′]) (29) (Fig. 4 and data not shown). U373-MG cells (2 × 105) were transfected with siRNA (25 nM) on days 1 and 2, before infection with CVS RABV on day 3. Cells were lysed 16, 24, or 28 h later, and the expression of NCL, tubulin, and viral P- and N (nucleo)-proteins was analyzed by Western blotting (Fig. 4A and B and data not shown). NCL1 and NCL2, but not Scr-siRNA, depleted NCL in infected and mock-infected cells, but tubulin expression was unaffected, indicating that NCL depletion had no major effect on cell viability; we also confirmed directly that cell viability was unaffected by NCL siRNA transfection using a trypan blue exclusion assay (data not shown). P-protein was clearly detectable in infected Scr-siRNA-transfected cells, but in cells transfected with NCL1 or NCL2 siRNA, P-protein expression was strongly reduced (Fig. 4A and data not shown). Comparable effects on N-protein were observed (Fig. 4B). Inhibition of P-protein expression was observed at 16, 24, and 28 h postinfection and at multiplicities of infection (MOI) of 0.5, 2, and 5 (Fig. 4A and B and data not shown). To determine whether these effects correlated with altered infectious virus production, we collected supernatants from cells transfected with NCL1 or NCL2 and infected at MOI of 5 for 24 h, and determined viral titers by immunofluorescence using the Poisson distribution (30). NCL depletion by NCL1 or NCL2 resulted in a ca. 6-10-fold decrease in titer (Fig. 4C and data not shown).
FIG 4.
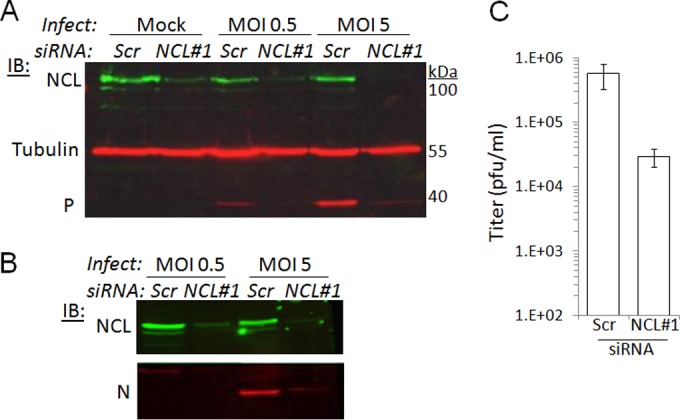
NCL is required for RABV infection. (A) U-373-MG cells transfected with Scr (control) or NCL-targeting siRNA (NCL1 [29]) were infected with CVS11 RABV at an MOI of 0.5 or 5, or mock infected, for 24 h. Cell lysates were then analyzed by IB using antibodies against NCL (rabbit antibody ab22758; Abcam), P-protein (mouse antibody 25C2 [39]), and tubulin (mouse antibody N3656; Amersham) and fluorescently conjugated anti-rabbit and anti-mouse IgG secondary antibodies. The blot shown is representative of 2 assays. (B) Cell lysates from the assay represented in panel A were analyzed by IB using antibody against RABV N protein (62B5) (40). (C) Viral titers were determined in supernatants from RABV-infected cells transfected with Scr or NCL1 siRNA (means ± standard deviations [n = 2] from 2 assays).
In conclusion, our data show for the first time that RABV can interact with nucleoli and NCL via the P-protein and that NCL expression is important to infection. This identifies a novel host interface in lyssavirus infection, providing a potential new target for the development of attenuated vaccine strains and antivirals against these highly pathogenic viruses. Previous data have indicated roles for the nucleolus and nucleolin in the replication of viruses with nuclear replication cycles (e.g., retroviruses and DNA viruses), including direct roles for nucleoli as sites of genome replication (31–33). However, a number of studies have also indicated that certain cytoplasmic RNA viruses can interact with the nucleolus/nucleolar proteins with importance to infection (16, 18); our study now extends these observations to lyssaviruses. These interactions are likely to relate to viral modulation of the multifunctional nucleolus/nucleolin, potentially including effects on ribosome biogenesis, nucleocytoplasmic transport, apoptosis, cell cycle and immunity (14, 34). P-protein has well established roles in viral modulation of host cell biology, including innate immune responses (3, 4, 35–38), suggesting that it might target the nucleolus to facilitate these functions; this possibility will form the basis of future studies in our laboratory.
ACKNOWLEDGMENTS
This research was supported by the National Health and Medical Research Council Australia project grant 1003244 to G.W.M., Australian Research Council Discovery project grant DP110101749 to G.W.M. and D.A.J., National Health and Medical Research Council Australia Senior Principal Research Fellowship 1002486 to D.A.J., and Grimwade Fellowship (Miegunyah Trust) to G.W.M.
We acknowledge the facilities of Monash Micro Imaging (Monash University, Victoria, Australia) and the Melbourne Advanced Microscopy Facility (Melbourne University, Victoria, Australia).
REFERENCES
- 1.Schnell MJ, McGettigan JP, Wirblich C, Papaneri A. 2010. The cell biology of rabies virus: using stealth to reach the brain. Nat Rev Microbiol 8:51–61. doi: 10.1038/nrmicro2260. [DOI] [PubMed] [Google Scholar]
- 2.Brzózka K, Finke S, Conzelmann KK. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J Virol 79:7673–7681. doi: 10.1128/JVI.79.12.7673-7681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chelbi-Alix MK, Vidy A, Bougrini JE, Blondel D. 2006. Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. J Interferon Cytokine Res 26:271–280. doi: 10.1089/jir.2006.26.271. [DOI] [PubMed] [Google Scholar]
- 4.Vidy A, Chelbi-Alix M, Blondel D. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J Virol 79:14411–14420. doi: 10.1128/JVI.79.22.14411-14420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito N, Moseley GW, Blondel D, Shimizu K, Rowe CL, Ito Y, Masatani T, Nakagawa K, Jans DA, Sugiyama M. 2010. Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J Virol 84:6699–6710. doi: 10.1128/JVI.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieu KG, Brice A, Wiltzer L, Hirst B, Jans DA, Blondel D, Moseley GW. 2013. The rabies virus interferon antagonist P protein interacts with activated STAT3 and inhibits Gp130 receptor signaling. J Virol 87:8261–8265. doi: 10.1128/JVI.00989-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oksayan S, Ito N, Moseley G, Blondel D. 2012. Subcellular trafficking in rhabdovirus infection and immune evasion: a novel target for therapeutics. Infect Disord Drug Targets 12:38–58. doi: 10.2174/187152612798994966. [DOI] [PubMed] [Google Scholar]
- 8.Wiltzer L, Larrous F, Oksayan S, Ito N, Marsh GA, Wang LF, Blondel D, Bourhy H, Jans DA, Moseley GW. 2012. Conservation of a unique mechanism of immune evasion across the Lyssavirus genus. J Virol 86:10194–10199. doi: 10.1128/JVI.01249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiltzer L, Okada K, Yamaoka S, Larrous F, Kuusisto HV, Sugiyama M, Blondel D, Bourhy H, Jans DA, Ito N, Moseley GW. 2014. Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J Infect Dis 209:1744–1753. doi: 10.1093/infdis/jit829. [DOI] [PubMed] [Google Scholar]
- 10.Chenik M, Chebli K, Blondel D. 1995. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J Virol 69:707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oksayan S, Wiltzer L, Rowe CL, Blondel D, Jans DA, Moseley GW. 2012. A novel nuclear trafficking module regulates the nucleocytoplasmic localization of the rabies virus interferon antagonist, P protein. J Biol Chem 287:28112–28121. doi: 10.1074/jbc.M112.374694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasdeloup D, Poisson N, Raux H, Gaudin Y, Ruigrok RWH, Blondel D. 2005. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology 334:284–293. doi: 10.1016/j.virol.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Moseley GW, Filmer RP, DeJesus MA, Jans DA. 2007. Nucleocytoplasmic distribution of rabies virus P-protein is regulated by phosphorylation adjacent to C-terminal nuclear import and export signals. Biochemistry 46:12053–12061. doi: 10.1021/bi700521m. [DOI] [PubMed] [Google Scholar]
- 14.Boisvert FM, Van Koningsbruggen S, Navascués J, Lamond AI. 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 15.Matthews DE, Emmott E, Hiscox JA. 2011. Viruses and the nucleolus, p 321–345 InOlsen MOJ. (ed), The nucleolus. Springer, New York, NY. [Google Scholar]
- 16.Hiscox JA. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol 5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiscox JA, Whitehouse A, Matthews DA. 2010. Nucleolar proteomics and viral infection. Proteomics 10:4077–4086. doi: 10.1002/pmic.201000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salvetti A, Greco A. 2014. Viruses and the nucleolus: the fatal attraction. Biochim Biophys Acta 1842:840–847. doi: 10.1016/j.bbadis.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang SH, Yeh NH. 1993. The self-cleaving activity of nucleolin determines its molecular dynamics in relation to cell proliferation. Exp Cell Res 208:48–53. doi: 10.1006/excr.1993.1221. [DOI] [PubMed] [Google Scholar]
- 20.Belenguer P, Caizergues-Ferrer M, Labbe JC, Doree M, Amalric F. 1990. Mitosis-specific phosphorylation of nucleolin by p34(cdc2) protein kinase. Mol Cell Biol 10:3607–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das S, Cong R, Shandilya J, Senapati P, Moindrot B, Monier K, Delage H, Mongelard F, Kumar S, Kundu TK, Bouvet P. 2013. Characterization of nucleolin K88 acetylation defines a new pool of nucleolin colocalizing with pre-mRNA splicing factors. FEBS Lett 587:417–424. doi: 10.1016/j.febslet.2013.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Lischwe MA, Roberts KD, Yeoman LC, Busch H. 1982. Nucleolar specific acidic phosphoprotein C23 is highly methylated. J Biol Chem 257:14600–14602. [PubMed] [Google Scholar]
- 23.Chen CM, Chiang SY, Yeh NH. 1991. Increased stability of nucleolin in proliferating cells by inhibition of its self-cleaving activity. J Biol Chem 266:7754–7758. [PubMed] [Google Scholar]
- 24.Srivastava M, Pollard HB. 1999. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J 13:1911–1922. [PubMed] [Google Scholar]
- 25.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 26.Schwab MS, Dreyer C. 1997. Protein phosphorylation sites regulate the function of the bipartite NLS of nucleolin. Eur J Cell Biol 73:287–297. [PubMed] [Google Scholar]
- 27.Roth DM, Moseley GW, Glover D, Pouton CW, Jans DA. 2007. A microtubule-facilitated nuclear import pathway for cancer regulatory proteins. Traffic 8:673–686. doi: 10.1111/j.1600-0854.2007.00564.x. [DOI] [PubMed] [Google Scholar]
- 28.Roth DM, Moseley GW, Pouton CW, Jans DA. 2011. Mechanism of microtubule-facilitated “fast track” nuclear import. J Biol Chem 286:14335–14351. doi: 10.1074/jbc.M110.210302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P. 2007. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol 8:66. doi: 10.1186/1471-2199-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obiang L, Raux H, Ouldali M, Blondel D, Gaudin Y. 2012. Phenotypes of vesicular stomatitis virus mutants with mutations in the PSAP motif of the matrix protein. J Gen Virol 93:857–865. doi: 10.1099/vir.0.039800-0. [DOI] [PubMed] [Google Scholar]
- 31.Hiscox JA. 2002. The nucleolus—a gateway to viral infection? Arch Virol 147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pyper JM, Clements JE, Zink MC. 1998. The nucleolus is the site of Borna disease virus RNA transcription and replication. J Virol 72:7697–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greco A. 2009. Involvement of the nucleolus in replication of human viruses. Rev Med Virol 19:201–214. doi: 10.1002/rmv.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. 2008. Nucleolus: the fascinating nuclear body. Histochem Cell Biol 129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vidy A, El Bougrini J, Chelbi-Alix MK, Blondel D. 2007. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J Virol 81:4255–4263. doi: 10.1128/JVI.01930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rieder M, Brzózka K, Pfaller CK, Cox JH, Stitz L, Conzelmann KK. 2011. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: Inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J Virol 85:842–852. doi: 10.1128/JVI.01427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blondel D, Regad T, Poisson N, Pavie B, Harper F, Pandolfi PP, De Thé H, Chelbi-Alix MK. 2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21:7957–7970. doi: 10.1038/sj.onc.1205931. [DOI] [PubMed] [Google Scholar]
- 38.Marschalek A, Drechsel L, Conzelmann KK. 2012. The importance of being short: the role of rabies virus phosphoprotein isoforms assessed by differential IRES translation initiation. Eur J Cell Biol 91:17–23. doi: 10.1016/j.ejcb.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Raux H, Iseni F, Lafay F, Blondel D. 1997. Mapping of monoclonal antibody epitopes of the rabies virus P protein. J Gen Virol 78:119–124. [DOI] [PubMed] [Google Scholar]
- 40.Lahaye X, Vidy A, Pomier C, Obiang L, Harper F, Gaudin Y, Blondel D. 2009. Functional characterization of negri bodies (NBs) in rabies virus-infected cells: evidence that NBs are sites of viral transcription and replication. J Virol 83:7948–7958. doi: 10.1128/JVI.00554-09. [DOI] [PMC free article] [PubMed] [Google Scholar]