ABSTRACT
The HIV-1 Gag polyprotein precursor composed of the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains orchestrates virus assembly via interactions between MA and the cell plasma membrane (PM) on one hand and NC and the genomic RNA on the other hand. As the Gag precursor can adopt a bent conformation, a potential interaction of the NC domain with the PM cannot be excluded during Gag assembly at the PM. To investigate the possible interaction of NC with lipid membranes in the absence of any interference from the other domains of Gag, we quantitatively characterized by fluorescence spectroscopy the binding of the mature NC protein to large unilamellar vesicles (LUVs) used as membrane models. We found that NC, either in its free form or bound to an oligonucleotide, was binding with high affinity (∼107 M−1) to negatively charged LUVs. The number of NC binding sites, but not the binding constant, was observed to decrease with the percentage of negatively charged lipids in the LUV composition, suggesting that NC and NC/oligonucleotide complexes were able to recruit negatively charged lipids to ensure optimal binding. However, in contrast to MA, NC did not exhibit a preference for phosphatidylinositol-(4,5)-bisphosphate. These results lead us to propose a modified Gag assembly model where the NC domain contributes to the initial binding of the bent form of Gag to the PM.
IMPORTANCE The NC protein is a highly conserved nucleic acid binding protein that plays numerous key roles in HIV-1 replication. While accumulating evidence shows that NC either as a mature protein or as a domain of the Gag precursor also interacts with host proteins, only a few data are available on the possible interaction of NC with lipid membranes. Interestingly, during HIV-1 assembly, the Gag precursor is thought to adopt a bent conformation where the NC domain may interact with the plasma membrane. In this context, we quantitatively characterized the binding of NC, as a free protein or as a complex with nucleic acids, to lipid membranes and showed that the latter constitute a binding platform for NC. Taken together, our data suggest that the NC domain may play a role in the initial binding events of Gag to the plasma membrane during HIV-1 assembly.
INTRODUCTION
The nucleocapsid protein (NC) of human immunodeficiency virus type 1 (HIV-1) is a small basic nucleic acid binding protein with two strictly conserved CCHC motifs that strongly bind zinc (1). Mature NC results from the protease-mediated cleavage of the Gag polyprotein precursor and plays key roles in HIV-1 replication (2–5). During the early steps, NC acts as a cofactor of the reverse transcriptase (RT), to promote the initiation of reverse transcription (6–8), as well as the two obligatory strand transfers (9, 10). Moreover, NC also reduces RT pauses at the initiation step (11–13), increases the overall RT processivity (14, 15), promotes synthesis through pause sites (16–19), helps to remove the RNA fragments resulting from the RT RNase H activity (20), and may contribute to the protection of the viral DNA (vDNA) during its nuclear import (21). Therefore, NC is thought to play a central role in the production of full-length vDNA as well as in its integration into the host cell genome (22–25).
The late steps of the viral life cycle are orchestrated by the Gag polyprotein precursor, formed of four major structural domains, namely, the matrix (MA), capsid (CA), nucleocapsid (NC), and p6. While the MA domain (GagMA) (26) targets Gag to the inner leaflet of the plasma membrane (PM) through a myristoyl group at its N terminus and a highly basic region (HBR) (27–31), the NC domain (GagNC) specifically interacts with the genomic RNA (gRNA) among the large excess of cellular RNAs. This GagNC/gRNA recognition constitutes a prerequisite for gRNA packaging (32–40) and dimerization (41–43). Interaction of GagNC with gRNA is also important for the CA-mediated multimerization of Gag (44–50), albeit cellular mRNAs can replace gRNA in the formation of virus-like particles (51).
Deleting GagNC or replacing all its basic residues with neutral ones was reported to partially impair the anchoring and the multimerization of Gag at the PM (47, 52). This phenotype was observed when the mutated Gag was expressed either alone or coexpressed with wild-type Gag in the same cell. Though the role of GagNC in Gag oligomerization could be a consequence of its binding to RNAs, which act as a scaffold to direct Gag-Gag interactions, a nonexclusive hypothesis is that GagNC possibly contributes to the binding of Gag to the PM. Along this line, recombinant Gag molecules were found to adopt a bent globular conformation (53–55) and were proposed to interact with the gRNA through both their MA and NC domains (56–60). Moreover, in the absence of RNAs, Gag proteins in their bent conformation were shown to bind to negatively charged model membranes, suggesting that in addition to the MA domain, the NC domain may bind to membranes. Interestingly, RNAs have been shown to decrease the affinity of MA for lipid membranes (61, 62). Thus, RNAs and notably gRNA may act as negative regulators of nonspecific membrane binding, likely by reducing the electrostatic interactions of the HBR with acidic lipids and preventing myristate exposure (62, 63). Phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] and probably other phosphoinositides (64, 65), which are essentially found on the cytoplasmic leaflet of the PM, can outcompete gRNA (61, 66) and promote the anchoring of the myristoylated MA domain to the PM (62, 67). The PI(4,5)P2-induced release of GagMA from the gRNA is also thought to promote Gag trimerization and a structural switch of Gag to its rod-shaped conformation that is observed in virus particles (68–70).
In this context, as the gRNA decreases the affinity of GagMA for the membrane, it can be hypothesized that GagNC participates at least transiently, together with GagMA, to the initial binding of the Gag/gRNA complex to the PM. In order to further characterize the possible interaction of GagNC with lipid membranes, we used the mature NC protein, to avoid any interfering effects brought by the other domains of Gag, and large unilamellar vesicles (LUVs) as lipid membrane models. By using a combination of fluorescence spectroscopy techniques, we found a strong interaction of NC, in its free form and bound to nucleic acids, with negatively charged lipid membranes, but with no preferential binding to PI(4,5)P2. Taken together, our data support a possible role of NC in the initial binding of Gag to the inner leaflet of the PM.
MATERIALS AND METHODS
Materials.
All chemicals and solvents for spectroscopic measurements were from Sigma-Aldrich. Dioleoylphosphatidylcholine (DOPC), dioleoylphosphatidylethanolamine (DOPE), dioleoylphosphatidylserine (DOPS), dioleoylphosphatidylglycerol (DOPG), and phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] were from Sigma-Aldrich. Rhodamine-DOPE (Rh-DOPE) was purchased from Avanti Polar Lipids (Birmingham, AL). The concentration of phospholipid stock solutions in chloroform was determined by dry weight. The HIV-1 nonlabeled dTAR sequence and dTAR labeled with a fluorescein at its 3′ end were synthesized and purified by high-pressure liquid chromatography (HPLC) by IBA GmbH (Germany). The concentration of dTAR was calculated from its absorbance measured with a Cary 400 spectrophotometer (Varian), using a molar extinction coefficient at 260 nm of 515,070 M−1 · cm−1 and at 488 nm of 80,000 M−1 · cm−1 for dTAR and dTAR–6-carboxyfluorescein (dTAR-FAM), respectively.
Preparation of peptides.
NC peptides were prepared by solid-phase peptide synthesis on a 433A synthesizer (ABI, Foster City, CA), HPLC purified, and characterized by ion spray mass spectrometry, as previously described (71, 72). The MFL probe, a functionalized derivative of 4′-(dimethylamino)-3-hydroxyflavone, was prepared and coupled to the N terminus of the NC peptides, as previously described (73). The lissamine rhodamine dye (LRh) was synthesized as described previously (74) and coupled to the N terminus of the NC peptides in the same way as the MFL probe. To get the zinc-bound form of NC peptides, 2.2 molar equivalents of ZnSO4 was added to the peptide and pH was raised to 7.4. Noticeably, in the case of MFL-labeled peptides, a large excess of Zn2+ ions should be avoided since this ion could affect its fluorescence. Peptide concentration was determined using an extinction coefficient of 5,700 M−1 cm−1 at 280 nm for nonlabeled peptides, 33,000 M−1 cm−1 at 400 nm for the MFL-labeled peptides, and 80,000 M−1 cm−1 at 555 nm for LRh-labeled NC. Note that the nucleic acid chaperone properties (75, 76) of the labeled NC peptide were tested as described previously (72) and found to be close to those of the unlabeled NC, indicating that the MFL label marginally perturbs the peptide structure and activity (data not shown).
Preparation of LUVs.
Large unilamellar vesicles (LUVs) as well as LUVs labeled with 1% Rh-DOPE (LUVs-Rh), were prepared in 20 mM phosphate buffer, 150 mM NaCl, pH 7.4, by the classical extrusion method (77). Suspensions of multilamellar vesicles were extruded with a Lipex Biomembranes extruder (Vancouver, Canada), using first filters of 0.2 μm (7 passages) and thereafter 0.1 μm (10 passages). This protocol leads to LUVs with a mean diameter of 0.11 μm, as measured by dynamic light scattering (DLS) with a Malvern Zetamaster 300 (Malvern, United Kingdom). To calculate the vesicle concentration, the extended radius of the vesicles (R) was considered to be 535 Å, as determined by DLS measurements. The thickness of the lipid bilayer (t) and the average lipid density (d) were assumed to be 40 Å and 70 Å2/lipid, respectively (78). The number of lipids per vesicle was determined as n = 4π[R2 + (R − t)2]/d = 9.54 × 104 lipids/vesicle, so that the vesicle concentration corresponds to C(vesicles) = C(lipids)/n.
Fluorescence spectroscopy.
Fluorescence spectra were recorded at 20°C on a FluoroMax or Fluorolog (Jobin Yvon Horiba) spectrofluorometer equipped with a thermostated cuvette holder. All spectra were corrected for the emission of the corresponding blank solution (neat solvent, lipid vesicles). Quantum yields (QY) were calculated, using 4′-(dialkylamino)-3-hydroxyflavone in ethanol (QY = 0.52 [79]), as a reference. To calculate the insertion depth of the MFL label in the lipid bilayer when the MFL-labeled NC peptides were bound to LUVs, the parallax quenching method using nitroxide lipids was used, as described previously (80–82). To determine the NC/LUV binding parameters, 0.1 μM MFL-NC was titrated with increasing concentrations of LUVs at 20°C. To determine the binding parameters of the MFL-NC/dTAR (3:1) complexes to LUVs, the complexes were prepared first by mixing 0.1 μM MFL-NC with 33 nM dTAR and then titrated with increasing concentrations of LUVs at 20°C.
The average number ν of moles of MFL-NC bound per mole of LUVs is calculated from the fluorescence intensities using ν = (I0 − I)/(I0 − It)Pt/Lt, where Pt and Lt designate the total concentration of MFL-NC and LUVs (calculated as indicated above), respectively, and It designates the fluorescence at the plateau when all the peptide is bound, whereas I0 and I correspond to the fluorescence intensities of the peptide in the absence and in the presence of a given concentration of LUVs, respectively. Furthermore, the free peptide concentration P is deduced by P = Pt − νLt. The same calculations are used for the binding of MFL-NC/dTAR complexes to LUVs.
To further demonstrate the interaction between the binding partners in the ternary complex NC/dTAR/LUVs, fluorescence resonance energy transfer (FRET) experiments were performed. In the FRET approach, an excited fluorophore, acting as a FRET donor, transfers nonradiatively its energy to an acceptor through dipole-dipole coupling. The efficiency of this energy transfer is inversely proportional to the sixth power of the distance between the donor and acceptor, making FRET extremely sensitive to small changes in distance. Due to the short distances where FRET applies (<8 nm), this method is used to evidence the interaction between two molecular species. The interactions between our partners were monitored two at a time, so that three combinations were used, namely, MFL-NC with LUVs-Rh, dTAR-FAM with LRh-NC, and dTAR-FAM with LUVs-Rh. The NC/dTAR (3:1) complexes (0.3 μM NC added to 0.1 μM dTAR) were prepared first and then added to an equivalent volume of a LUV solution (100 μM in lipid concentration). Excitation wavelengths were 400 nm and 480 nm for MFL-NC and dTAR-FAM, respectively. Emission spectra were recorded between 420 nm and 700 nm for the MFL/Rh couple and between 500 and 700 nm for the FAM/Rh and FAM/LRh couples. All experiments were performed in 20 mM phosphate buffer in the presence of 150 mM NaCl, pH 7.4, to limit nonspecific electrostatic interactions.
RESULTS
Fluorescence assay to monitor the binding of NC to model membranes.
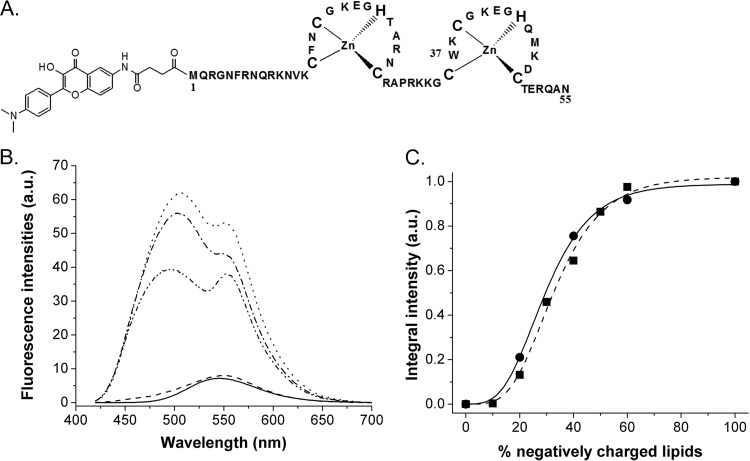
To show and characterize the binding of NC to membranes, we used a synthetic NC protein covalently labeled at its N terminus by MFL (73, 83), an environment-sensitive probe of the 3-hydroxyflavone family (Fig. 1A), and large unilamellar vesicles (LUVs) of 100 nm in diameter as membrane models. Due to an excited-state intramolecular proton transfer reaction, the MFL probe is characterized by two excited-state species, namely, the normal and tautomeric forms (N* and T*), showing two well-separated emission bands, differently sensitive to the polarity and hydration of the probe environment (73, 79, 84).
FIG 1.
Binding of NC-MFL to LUVs, taken as a membrane model. (A) Structure of the NC peptide N terminally labeled with the MFL probe. (B) Fluorescence spectra of MFL-NC in the absence (solid line) and in the presence of LUVs composed of either neutral DOPC (dashed line), negatively charged DOPS (line with alternating dashes and dots) or DOPG (dotted line), or LUVmix1, a lipid mixture composed of DOPC-DOPE-DOPS-SM-PI(4,5)P2 at a ratio of 16:46:25:8:5 (mol% of lipids) (line with dashes and double dots). Concentrations of NC and lipids were 0.25 μM and 100 μM, respectively. (C) Dependence of MFL-NC/LUV binding on the percentage of negatively charged DOPS (■) or DOPG (●) lipids in mixture with DOPC, as monitored by the integral fluorescence intensity of the label. The data points were fitted by a sigmoidal function (solid and dashed lines) to guide the eye. The concentration of peptide was 0.3 μM. The concentrations of DOPC/DOPS and DOPC/DOPG lipids were 200 and 100 μM, respectively. Experiments were performed in 20 mM phosphate buffer, pH 7.4, in the presence of 150 mM NaCl to limit nonspecific electrostatic interactions. Excitation wavelength was 400 nm.
While the labeled NC showed a single emission band (Fig. 1B) similar to that of the free MFL label in water (73), negatively charged LUVs, composed of either DOPS or DOPG phospholipids added at a molar ratio of 400 lipids per MFL-NC peptide, caused a dual emission (Fig. 1B) with a strong increase (∼ 8.5-fold) in quantum yield. These important changes in the MFL emission can be ascribed to the transfer of the MFL probe from the polar environment of the buffer to the aprotic environment of the hydrophobic membrane. These data clearly show that the MFL-NC peptide binds to negatively charged LUVs, irrespective of the nature of the lipid polar head group. In sharp contrast, neutral DOPC LUVs did not significantly affect the MFL-NC fluorescence spectrum (Fig. 1B), suggesting that NC does not bind to neutral lipid vesicles. To further correlate NC binding to LUVs with the presence of negatively charged lipids, we increased the molar fraction of DOPS to DOPC and monitored the binding of these LUVs to MFL-NC through the changes in the probe fluorescence intensity (Fig. 1C). While no change in MFL intensity was observed with a molar fraction of 10%, a sharp increase was observed when the molar ratio was increased from 20 to 60 mol%, where a plateau was reached. The dependence of NC binding on the molar fraction of PS (Fig. 1C) was similar to the one reported for the MA protein (85) or the Gag protein (86). This indicates that, similarly to MA or Gag (85, 86), electrostatic interactions play an important role in the binding of the positively charged NC protein to negatively charged lipids. This conclusion is further substantiated by the strong analogy of MFL-NC with MFL-labeled poly-l-lysine that was reported to bind through electrostatic interactions to negatively charged LUVs and to induce similar changes in MFL fluorescence parameters (73). Interestingly, LUVs composed of a mixture of lipids, DOPC-DOPE-DOPS-SM-PI(4,5)P2, in a ratio of 16:46:25:8:5 (mol% of lipids), which mimics the content in negatively charged phospholipids of the inner leaflet of the PM (87–90), led to changes in the MFL-NC spectrum, close to those observed upon addition of negatively charged LUVs (Fig. 1B). The only difference between these LUVs (called LUVmix1) and pure DOPS LUVs is their somewhat lower quantum yield, probably due to their lower content (30%) in negatively charged lipids. Thus, it appears that NC can probably bind to the inner leaflet of the PM through electrostatic interactions with negatively charged lipids.
Determination of the NC binding parameters to lipid membranes.
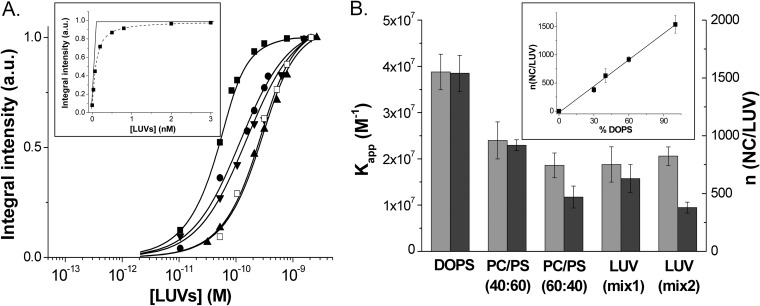
In order to determine the binding parameters of NC to LUVs, we titrated MLF-NC using increasing concentrations of LUVs with different lipid compositions (Fig. 2A). To fit the binding curves, we first used an equation that considers the binding of NC to lipid bilayers to be governed by a molar partition coefficient Km:
| (1) |
where Pm and Pt are the concentrations of NC protein bound to the membrane and total protein, respectively. I0, I, and If are the fluorescence intensities in the absence of lipid, in the presence of a given lipid concentration, and in the presence of a saturating lipid concentration, respectively. Assuming that NC can access only lipids at the external surface of the vesicles and that the concentration of bound NC is negligible compared to that of accessible lipids, the concentration of free lipids can be approximated by the equation [L′] = [L′]t/2, where [L′]t is the total concentration of lipids. Using this approximation, equation 1 was used to fit the binding curves of MA to lipid bilayers (85, 91). Using the same equation, we found that the Km value of NC for DOPS LUVs was (4.5 ± 0.7) × 105 M−1 (Table 1) and, thus, was close to the affinity [(3.5 ± 1) × 105 M−1] reported for the binding of MA to the same kind of LUVs (92). Due to this rather high affinity, it is likely that the concentration of bound peptide is probably not negligible in comparison to [L′]t, especially for the first points of the titration, where the concentration of peptide is higher than the concentration of LUVs, so that the approximation [L′] = [L′]t/2 is not fully valid. Moreover, equation 1 is formally equivalent to the equation describing a classical protein-ligand interaction with a one-to-one stoichiometry (93, 94). Due to its multiple charges and its large area compared to that of lipid head groups, it is unlikely that NC binds to only one lipid in a LUV. In fact, the real entities that bind NC are the lipid vesicles. Assuming that each vesicle can bind a number n of NC molecules with an apparent binding constant Kapp, we can use the equation that we derived for the binding of NC to oligonucleotides, which is based on the same binding model (95, 96):
| (2) |
where Pt is the total concentration of peptide and Lt is the total concentration of LUVs, calculated as indicated in Materials and Methods. This equation does not make any approximation on the concentration of the free species and allows a direct fitting of the fluorescence intensity I of the labeled protein. The number of binding sites n was determined from the intercept of the tangent to the initial points of the curve with the tangent to the plateau value (Fig. 2A, inset). For DOPS LUVs, we obtained a binding stoichiometry of 1,500 (±150) NC protein molecules per LUV, which corresponds to about 29 (±3) lipids per NC molecule. By taking 70 Å2 as the surface of a lipid, each peptide thus occupies about 20 nm2 on the vesicle surface. As this occupied surface is substantially larger than the ∼4-nm2 area calculated from the largest section (3.2 × 1.5 nm) of the three-dimensional (3D) structure of NC (PDB 1ESK), this suggests that NC molecules do not fully coat the LUV surface, probably as a consequence of the repulsive electrostatic forces between the nonneutralized charges of the NC molecules bound to the lipid surface. Using the stoichiometry value obtained from the inset of Fig. 2A, we determined from the fit of the binding curves with equation 2 a Kapp value of (3.9 ± 0.9) × 107 M−1 (Fig. 2B; Table 1) for each NC binding site on the DOPS vesicles. As this Kapp value was obtained through the same formalism as that applied for the binding of NC to nucleic acids (95, 96), a direct comparison is thus possible and shows that the affinity of NC for negatively charged lipids is of the same order as the affinity of NC to nucleic acids (97).
FIG 2.
Determination of the binding parameters of NC protein to model lipid membranes. (A) Titration of MFL-NC (0.1 μM) with LUVs of different lipid compositions: DOPS (■), DOPC-DOPS (40:60) (●), DOPC-DOPS (60:40) (▲), DOPC-DOPE-DOPS-SM-PI(4,5)P2 (16:46:25:8:5) (LUVmix1, ▼), and DOPC-DOPE-DOPS-SM (16:46:30:8) (LUVmix2, □). Solid lines correspond to the fit of the data points with equation 2, using the binding constant and the number of binding sites per LUVs indicated in panel B. (Inset) Determination of the number of binding sites from the intercept of the tangent to the first points of the titration with the tangent to the fluorescence plateau. (B) Binding affinity (light gray) and number of binding sites per LUV (dark gray) as a function of the LUV composition. The values of Kapp and n were determined from the binding curves in panel A. (Inset) Linear dependence of the number of binding sites as a function of the percentage of negatively charged lipids in the LUV composition. All experiments were in 20 mM phosphate buffer, 150 mM NaCl, pH 7.4. Excitation wavelength was 400 nm.
TABLE 1.
Comparison of Km and Kapp values obtained for the binding of NC derivatives to LUVs of different compositionsa
| Peptide | LUV composition | Km (10−5 M−1) | Kapp (10−5 M−1) |
|---|---|---|---|
| NC | DOPS | 4.5 (±0.7) | 390 (±20) |
| DOPC/DOPS (40:60) | 2.0 (±0.1) | 220 (±70) | |
| DOPC/DOPS (60:40) | 0.9 (±0.1) | 180 (±10) | |
| LUVmix1 | 1.4 (±0.2) | 170 (±30) | |
| LUVmix2 | 0.7 (±0.1) | 200 (±20) | |
| NCH23C | DOPC/DOPS (40:60) | 2.2 (±0.2) | 220 (±20) |
| NCSSHS | DOPC/DOPS (40:60) | 2.6 (±0.3) | 160 (±20) |
| NC(11–55) | DOPC/DOPS (40:60) | 0.30 (±0.02) | 20 (±1) |
Km and Kapp values were obtained by fitting the titration curves of MFL-labeled NC derivatives by the LUVs of indicated compositions, using equations 1 and 2, respectively. The compositions of LUVmix1 and LUVmix2 were DOPC-DOPE-DOPS-SM-PI(4,5)P2 (16:46:25:8:5) and DOPC-DOPE-DOPS-SM (16:46:30:8), respectively.
Interestingly, addition of an increasing proportion of DOPC to DOPS LUVs was found to sharply decrease the number of NC binding sites per LUV to 920 (±50), 470 (±90), and 630 (±100) for DOPC-DOPS (40:60), DOPC-DOPS (60:40), and LUVmix1, respectively (Fig. 2B). In contrast, the binding affinities were only slightly modified, since we observed only a 2-fold decrease between pure DOPS LUVs and LUVmix1, which corresponds to a negligible change of the binding energy (<0.5 kcal · mol−1). Since GagMA binds to the PM via an electrostatic interaction between its HBR domain and negatively charged lipids and also through specific recognition of PI(4,5)P2, we investigated whether NC could also specifically interact with this lipid. To this aim, we replaced 5% PI(4,5)P2 with 5% DOPS in the composition of LUVmix1, so that the total percentage of DOPS becomes 30%. Thus, by using LUVmix2, composed of DOPC-DOPE-DOPS-SM (16:46:30:8), the number of binding sites decreased to 380 (±50) NC/LUV with no significant change in the binding affinity (Fig. 2B). As PI(4,5)P2 molecules are characterized by 3 negative charges versus only one for DOPS, one PI(4,5)P2 molecule may be equivalent to 3 DOPS molecules in the binding process. Therefore, the real percentage of negative charges in LUVmix1 that contains 5% PI(4,5)P2 and 25% DOPS is 40%, explaining the similarities with the DOPC-DOPS (60:40) composition. Moreover, the negligible changes in the Kapp values upon the replacement of PI(4,5)P2 by DOPS further suggested that in contrast to myristylated MA (91), NC does not show a preferential binding for PI(4,5)P2. Together with the limited changes in the Kapp values, a linear correlation was observed between the number of binding sites and the percentage of negatively charged lipids in the LUVs (Fig. 2B, inset), strongly suggesting that NC can recruit negatively charged lipids to ensure a tight binding to LUVs.
Taken together, our data indicate that NC binds with high affinity to lipid membranes, supporting the notion that GagNC could participate, at least transiently, in the initial binding of Gag to the PM.
Determinants of NC binding to lipid membranes.
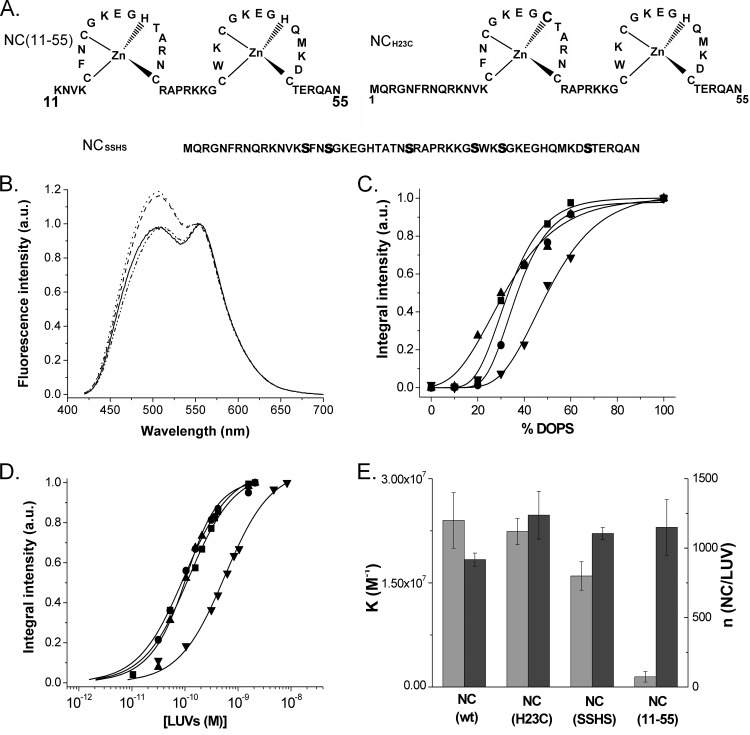
In order to determine which regions of NC are instrumental for membrane binding, we used three NC mutants (Fig. 3A). The role of the basic unfolded N-terminal (amino acids [aa] 1 to 10) domain was evaluated with the NC(11–55) derivative. The role of the central folded zinc finger domain, which is critical for a number of NC functions (5, 95, 98–100), was evaluated with the NCSSHS mutant, where all cysteine residues were replaced by serines to prevent the coordination of zinc ions and therefore the folding of the fingers (10). Finally, to examine the contribution of the hydrophobic plateau at the top of the central finger motif, we used the NCH23C mutant, where the His23 residue is replaced by a Cys residue. This substitution induces a misfolding of the proximal finger motif that prevents formation of the hydrophobic plateau (101).
FIG 3.
Interaction of NC mutants with LUVs. (A) Sequences of the NC mutants. Mutations are shown in bold. (B) Fluorescence spectra of MFL-labeled NC (dashed line), NCH23C (line with dashes and double dots), NCSSHS (solid line), and NC(11–55) (dotted line) peptides in the presence of DOPS LUVs. Concentrations of peptides and lipids were 0.25 and 200 μM, respectively. (C) Dependence of the binding of MFL-labeled NC (■), NCH23C (●), NCSSHS (▲), and NC(11–55) (▼) peptides on the percentage of negatively charged DOPS lipids in DOPC-DOPS LUVs. The data points were fitted by a sigmoidal function (solid lines) to guide the eye. Concentrations of peptides and LUVs were 0.25 and 200 μM, respectively. (D) Titrations of MFL-labeled NC, NCH23C, NCSSHS, and NC(11–55) peptides (0.1 μM) with DOPC-DOPS (40:60) LUVs. Symbols are as in panel C. The number of binding sites per LUV and the binding affinities were obtained as described for Fig. 2A. Solid lines correspond to the fit of the data points with equation 2, using the parameters given in panel E. (E) Binding parameters of NC mutants to DOPC-DOPS (40:60) LUVs (Kapp in light gray, n in dark gray). Buffer and excitation wavelength were as in Fig. 2.
To monitor their interaction with LUVs, the NC mutants were N terminally labeled with the MFL probe. As for the labeled native NC, addition of a large excess of negatively charged LUVs (200 μM in lipids) to all labeled mutants induced a two-band emission and strongly increased their quantum yield, indicating that all peptides were able to bind to these LUVs (Fig. 3B). Interestingly, the N*/T* ratio values were significantly lower with the NCH23C (IN*/IT* = 1.07) and NCSSHS (IN*/IT* = 1.14) mutants than with the native protein (IN*/IT* = 1.27), indicating that the MFL probe linked to the amino acid at position 1 experienced a less polar environment in these mutants than in native NC, upon binding to LUVs. This suggests that the position of the NC N-terminal (aa 1 to 10) domain on LUVs depends on the proper folding of the fingers. In contrast, the fluorescence parameters of the labeled NC(11–55) peptide (IN*/IT* = 1.24) were indistinguishable from those of MFL-NC, indicating that the amino acids at position 11 in NC(11–55) and at position 1 in NC(1–55) experience a similar environment at the membrane.
To further determine the localization of the NC probe in the membrane, parallax measurements were performed to determine the depth of the label from the center of the bilayer (73). This distance is recovered through the amount of quenching of the MFL emission observed with lipids carrying fluorescence quenchers at different depths in the bilayer. Using DOPC-DOPS (40:60) vesicles, we obtained average depths of 16.2 to 17.7 Å from the center of the bilayer for both MFL-NC and the three NC mutants (data not shown). These values pointed to a shallow location of the probe close to the lipid head groups for all four NC peptides. However, the accuracy of this technique is not sufficient to show binding depth differences that could explain the differences in the N*/T* ratio values observed for the NCH23C and NCSSHS mutants compared to the native protein.
Next, we compared the binding of the four labeled NC peptides to LUVs containing an increasing fraction of negatively charged lipids (Fig. 3C). The binding of the NC mutants to LUVs was found to strongly rely on the percentage of negatively charged lipids. In fact, as for the native NC, an optimal binding for NCH23C and NCSSHS peptides was reached with LUVs containing 60% negatively charged lipids. In contrast, NC(11–55) needs a higher percentage of negatively charged lipids, suggesting that it probably binds to LUVs with a lower affinity. By investigating the binding parameters of the NC mutants to DOPC-DOPS (40:60) LUVs, we found that the MFL-labeled NC, NCH23C, and NCSSHS showed nearly indistinguishable binding parameters. This indicates that the proper folding of the zinc fingers, as well as the hydrophobic plateau at the top of the zinc fingers, is not essential for the interaction of NC with the lipids. Nevertheless, the differences in polarity of the MFL probe observed in Fig. 3B suggest that the properly folded fingers may control the position of the N-terminal domain of NC in the membrane. Moreover, as the binding is thought to be mainly electrostatic, it can be speculated that the numerous positively charged amino acids distributed along the NC sequence indifferently intervene in the binding to the lipids, so that their number is more important than their localization in the sequence. In agreement with this conclusion, NC(11–55), which contains three basic residues fewer than NC(1–55), shows a >10-fold decrease in its binding constant (Fig. 3D and E).
Binding properties of NC/DNA complexes to lipid membranes.
Since the functions of NC and GagNC in virus replication are mediated by an interplay with viral nucleic acids (97), we studied the possible binding of NC/nucleic acid complexes to lipid membranes. As a nucleic acid model sequence, we used the dTAR 55-nucleotide (nt) sequence corresponding to the DNA version of the HIV-1 RNA transactivation response element. To show the binding of the NC/dTAR (3:1) complex to lipid membranes, we first performed a series of FRET experiments by monitoring the interactions between two partners at a time. As we have three partners, the FRET efficiencies of the three combinations of two labeled partners were determined in the presence or the absence of the third nonlabeled partner. As both the interactions of NC with LUVs (this work) and with nucleic acids (95, 102, 103) are dependent on electrostatic interactions, we systematically investigated the FRET efficiencies at two salt concentrations (30 mM and 150 mM NaCl) (Table 2). For the interactions between MFL-NC and DOPS LUVs labeled by 1% Rh-DOPE (LUV-Rh), we observed a strong FRET efficiency (83 to 90%) that was slightly affected by the addition of nonlabeled dTAR or by the salt concentration. These data indicate that NC is bound to LUVs both in the absence and in the presence of dTAR, being close enough to the Rh-DOPE molecules to give a high FRET efficiency. It can also be concluded that dTAR does not significantly outcompete NC from the LUVs to form binary dTAR/NC complexes.
TABLE 2.
FRET efficiencies of binary and ternary complexes between NC, dTAR, and LUVs
| Monitored interactiona | Nonlabeled partner | FRET efficiency for NaCl concn: |
|
|---|---|---|---|
| 150 mM | 30 mM | ||
| MFL-NC/LUV-Rh | − | 0.84 | 0.90 |
| dTAR | 0.83 | 0.85 | |
| dTAR-FAM/LUV-Rh | − | 0 | 0 |
| NC | 0.09 | 0.27 | |
| dTAR-FAM/LRh-NC | − | 0.72 | 0.80 |
| LUVs | 0.37 | 0.62 | |
FRET experiments were performed between two labeled partners in the absence (−) and in the presence of the third nonlabeled partner in 20 mM phosphate buffer with 150 mM or 30 mM NaCl (pH 7.4).
Next, we monitored the FRET efficiency of dTAR labeled with fluorescein at its 3′ end (dTAR-FAM) to LUV-Rh in the absence and the presence of nonlabeled NC. As expected, due to the inability of the negatively charged oligonucleotide to bind to negatively charged lipid vesicles (104), we did not observe any FRET in the absence of NC. In contrast, a significant FRET efficiency (9% at 150 mM NaCl and 27% at 30 mM NaCl) was observed when NC was added, revealing the formation of a ternary NC/dTAR/LUV complex. Finally, we observed a strong FRET efficiency (>70%) between dTAR-FAM and NC labeled at its N terminus by lissamine rhodamine (LRh-NC) in the binary NC/dTAR complex. Addition of nonlabeled LUVs to Rh-NC/dTAR-FAM was found to preserve a good FRET efficiency (37% at 150 mM NaCl and 62% at 30 mM NaCl). Taken together, our data showing a high FRET efficiency between MFL-NC and LUV-Rh in the presence of dTAR, as well as a high FRET efficiency between dTAR-FAM and LRh-NC in the presence of LUV, unambiguously confirmed the formation of NC/dTAR/LUV ternary complexes. Noticeably, the lower FRET efficiencies observed for dTAR-FAM/LUV-Rh and dTAR-FAM/LRh-NC in the ternary mixtures at 150 mM NaCl than at 30 mM NaCl suggest that fewer dTAR molecules may be bound to the LUVs, probably as a consequence of a limited coating of dTAR by NC molecules at the highest salt concentration. However, it cannot be excluded that the two salt concentrations induce different conformations of the NC/dTAR complexes bound to the LUVs.
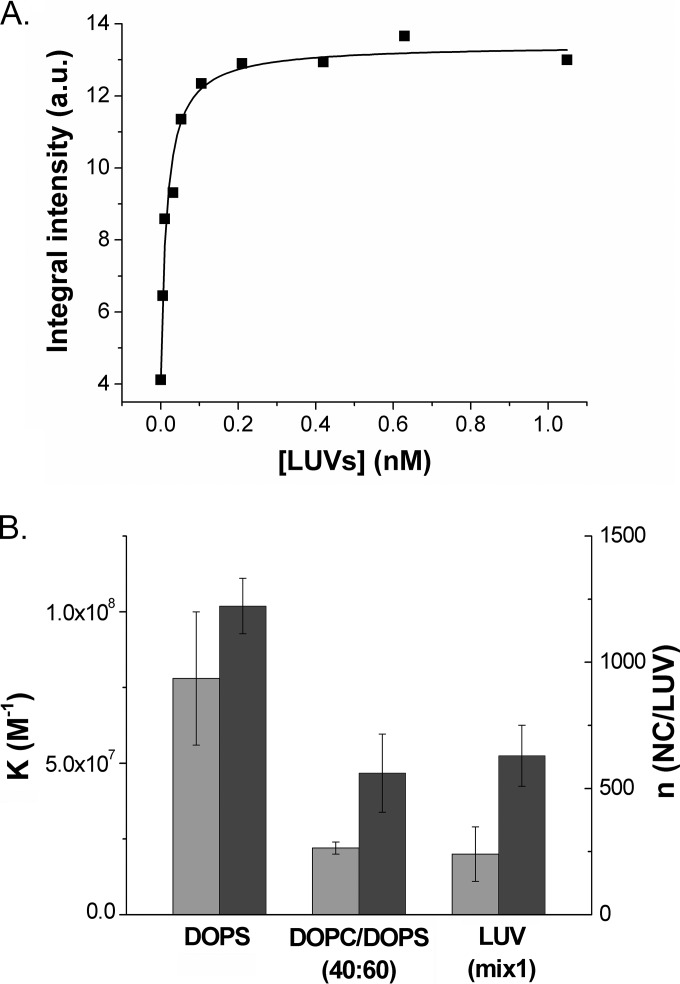
Next, we determined the binding parameters of the MFL-NC/dTAR complexes to LUVs of various compositions (DOPS, DOPC-DOPS [40:60], and LUVmix1) by titrating the complexes with increasing concentrations of LUVs in 20 mM phosphate buffer, 150 mM NaCl, pH 7.4 (Fig. 4A). For all three compositions of LUVs, the binding constants of the NC/dTAR complexes were found to be similar to those of NC but with a lower number of binding sites, as expected from the increased size of the NC/dTAR complexes compared to free NC (Fig. 4B). The reduction in the number of binding sites was limited to 20% for DOPS LUVs but up to 45% for LUVmix1. Since, by analogy with free NC, only the number of binding sites, but not the affinity of the NC/dTAR complexes for the LUVs, was found to vary with the percentage of negatively charged lipids, it can be concluded that NC/dTAR complexes are also able to recruit negatively charged lipids in LUVs in order to optimize their binding.
FIG 4.
Determination of the binding parameters of the NC/dTAR complexes to model lipid membranes. (A) Titration curve of MFL-NC/dTAR (3:1) complexes with DOPS LUVs. The number of binding sites per LUV and the binding affinities were obtained as described for Fig. 2A. The solid line corresponds to the fit of the data points with equation 2. Concentrations of NC and dTAR were 100 nM and 33 nM, respectively. (B) Binding parameters of NC/dTAR complexes to DOPS LUV, DOPC/DOPS (40:60) LUVs, and LUVmix1 (Kapp in light gray, n in dark gray). Buffer and excitation wavelength were as in Fig. 2.
Taken together, our data indicate that the NC/DNA complexes bind with high affinity to negatively charged lipid membranes, supporting the notion that the NC domain of Gag may interact with the inner leaflet of the PM and in so doing might participate in Gag assembly at the level of the PM.
DISCUSSION
The aim of the present work was to investigate the possible interactions of HIV-1 NC protein with lipid membranes. The possible binding of GagNC to membranes has already been evoked (58, 59, 105) or deduced from the cellular localization of Gag lacking the NC domain (52), but the evidence of this interaction together with its characterization was still missing. To that end, we investigated the binding of fluorescently labeled synthetic NC, free or bound to nucleic acids, to LUVs with different lipid compositions, taken as model membranes. This was carried out with the mature NC in order to avoid any influence from the other domains of Gag.
Our results show that NC, free or bound to an oligonucleotide, binds with high affinity to negatively charged LUVs but not to neutral ones. The NC-membrane interaction is thought to be mostly electrostatic, which was confirmed by the analysis of the binding of NC mutants to LUVs. Though it cannot be excluded that the MFL probe contributes to the binding of the labeled peptides to LUVs, its contribution is likely limited as it is not sufficient to promote the binding of the labeled peptides to neutral LUVs. Importantly, while the number of binding sites was found to decrease with the percentage of negatively charged lipids in the LUV composition, the binding constant value was nearly independent of this percentage, suggesting that NC and NC-oligonucleotide complexes can recruit negatively charged lipids to ensure optimal binding. Such a recruitment of negatively charged lipids by NC is in line with the recent hypothesis that Gag multimerization at the PM could induce the formation of acidic lipid-enriched microdomains (ALEM) (106, 107).
Interestingly, the binding constant of NC for negatively charged lipids is similar to that of the interaction of unmyristoylated MA and Gag proteins with DOPS-containing LUVs (91, 92), which is mainly governed by electrostatic interactions through the HBR sequence of MA (59, 60, 85, 106). However, in contrast to the myristoylated GagMA domain (67, 91, 108), which binds avidly and specifically to PI(4,5)P2 through a hydrophobic interaction with the exposed myristyl chain, no specific binding was found for the NC-PI(4,5)P2 interaction. Therefore, it is likely that the affinity of NC for negatively charged lipids is substantially lower than the final affinity of the myristoylated MA domain for PI(4,5)P2, following the myristyl switch. In agreement with this conclusion, mutating the N-terminal glycine of GagMA, to which the myristate is attached, impairs Gag-membrane association (27, 28, 109).
Another major difference between the MA and NC domains is their relative affinities for RNA and lipids. Indeed, due to its high affinity for the myristoylated Gag, PI(4,5)P2 can outcompete RNA bound to the myristoylated GagMA domain (58, 61, 63, 66). In contrast, results in Fig. 4B indicate that PI(4,5)P2 cannot dissociate the NC/nucleic acid complexes, suggesting that GagNC might be able to simultaneously bind RNA and the PM. This hypothesis is supported by the multivalent nature of NC, which allows it to bind at the same time two nucleic acid molecules (110). These differences between the relative stabilities of the different complexes, together with the ability of NC and NC/nucleic acid complexes to interact with negatively charged lipid membranes, lead us to revisit the HIV-1 assembly model, previously proposed by several groups (55, 61, 70, 111) (for reviews, see references 59, 60, and 63). In this revised model, we propose that the NC domain of Gag participates in a transient manner in the binding of Gag to the PM, allowing the recruitment of negatively charged lipids (Fig. 5).
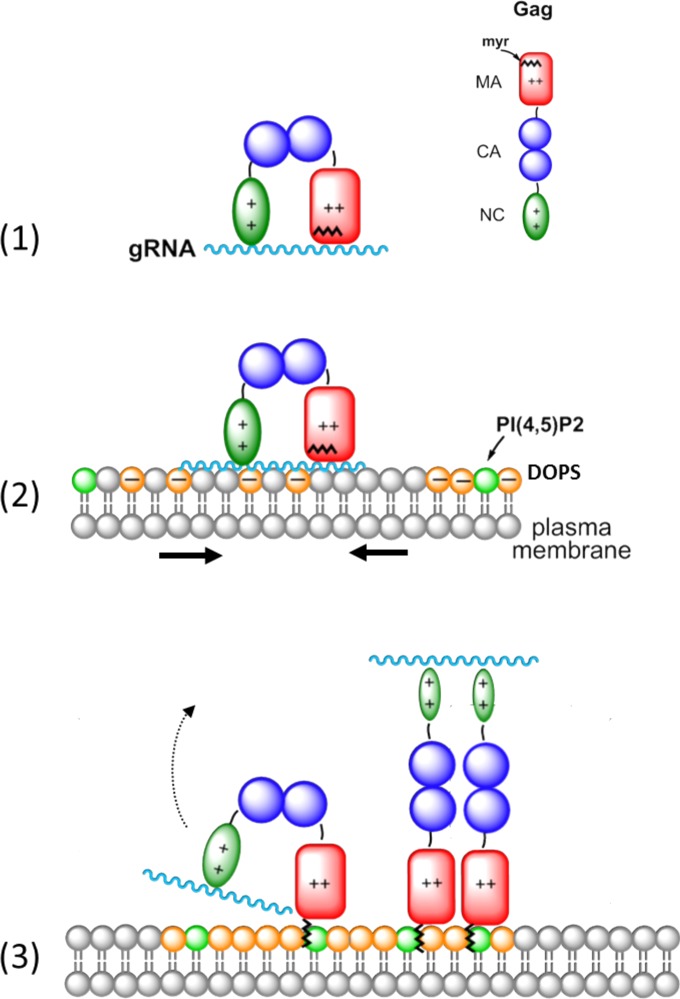
FIG 5.
Proposed role of the GagNC domain in the binding of Gag to the plasma membrane. (1) Newly made Gag polyprotein binds in its bent conformation to the gRNA via its NC and MA domains. (2) The Gag-gRNA complex interacts with the plasma membrane via its MA and NC domains. (3) The NC domain recruits negatively charged lipids, including PI(4,5)P2, that form an acidic lipid-enriched microdomain. This in turn leads to the binding of GagMA to PI(4,5)P2 through its previously sequestered myristyl group. This may finally cause the dissociation of GagMA from the gRNA and promote the rod-shape conformation of Gag.
This model stipulates that at the start of Gag assembly, the NC and MA domains are involved in the selection and recruitment of the gRNA (Fig. 5, step 1), which is facilitated by the globular or bent conformation of Gag (36, 55, 59, 61, 62, 69, 70, 112). At this stage, the binding of the gRNA to GagMA is thought to be mediated by the HBR sequence of Gag (61, 64), while the myristyl group is still buried in the GagMA core. Next, as NC bound to a nucleic acid can interact with negatively charged membranes (Fig. 4), the Gag-gRNA complex may bind to the cytoplasmic leaflet of the PM through its MA and NC domains. To strengthen its interaction with the PM, the gRNA-bound NC domain is thought to recruit negatively charged phospholipids, notably PI(4,5)P2 and PS, in order to form an ALEM (Fig. 5, step 2). This NC-induced recruitment of ALEM is consistent with the reported ability of GagNC to accelerate and stabilize the binding of Gag to cellular membranes (113). Once recruited, PI(4,5)P2 should induce the myristyl switch which stabilizes the anchoring of the GagMA into the PM. After the dissociation of the gRNA from the MA domain, the binding energy of the NC-gRNA complex for the PM is probably not sufficient to maintain the globular/bent conformation of Gag, so that the GagNC-gRNA complex is released from the membrane, allowing Gag to adopt an extended rod-shape conformation (Fig. 5, step 3) found in immature viral particles (114, 115).
This model proposes that the initial steps of HIV-1 assembly might result from binding reactions of the GagMA and GagNC domains with both the gRNA and the PM. These binding events associated with conformational changes could thus control the Gag assembly process at the PM. Such a dual role of the NC domain in the binding of Gag to the PM and the gRNA could explain, at least in part, the decrease in the accumulation of Gag at the PM, upon deleting the NC domain (52). This view may differ from previous reports showing that GagZip chimeric proteins (50, 116–118), where GagNC is replaced by a heterologous transcription factor leucine zipper, can undergo assembly with the release of immature virus-like particles resembling those formed by wild-type Gag (50, 116–119). However, these immature particles contain very little genomic or cellular RNA (116), indicating that the GagZip proteins probably do not interact with nucleic acids in the cellular context. As a consequence of the fused leucine zipper motif, the myristyl group of GagMA may be exposed very early during the assembly process, directly anchoring the GagZip proteins to the PM and bypassing the steps described in Fig. 5. Further validation of the possible role of GagNC, as proposed in Fig. 5, will be elucidated in due course by monitoring the assembly of Gag and Gag mutants in the cellular context.
ACKNOWLEDGMENTS
This work and V.P. were supported by ANR Fluoaant. This work was also supported by the ANR Femtostack and the European Project THINPAD “Targeting the HIV-1 Nucleocapsid Protein to fight Antiretroviral Drug Resistance” (FP7—grant agreement 601969). N.K. was supported by a fellowship from the French Ministère de la Recherche.
We thank Andrey Klymchenko for useful discussion. We are also indebted to Jean-Luc Darlix for critical reading of the manuscript.
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Mély Y, De Rocquigny H, Morellet N, Roques BP, Gérad D. 1996. Zinc binding to the HIV-1 nucleocapsid protein: a thermodynamic investigation by fluorescence spectroscopy. Biochemistry 35:5175–5182. doi: 10.1021/bi952587d. [DOI] [PubMed] [Google Scholar]
- 2.Thomas JA, Gorelick RJ. 2008. Nucleocapsid protein function in early infection processes. Virus Res 134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamson CS, Freed EO. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol 55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 4.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. 2010. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol 7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muriaux D, Darlix J-L. 2010. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol 7:744–753. doi: 10.4161/rna.7.6.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barraud P, Gaudin C, Dardel F, Tisné C. 2007. New insights into the formation of HIV-1 reverse transcription initiation complex. Biochimie 89:1204–1210. doi: 10.1016/j.biochi.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Hargittai MRS, Gorelick RJ, Rouzina I, Musier-Forsyth K. 2004. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol 337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Quan Y, Arts EJ, Li Z, Preston BD, De Rocquigny H, Roques BP, Darlix JL, Kleiman L, Parniak MA, Wainberg MA. 1996. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA(Lys3) in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol 70:4996–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. 2008. Strand transfer events during HIV-1 reverse transcription. Virus Res 134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 10.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol 74:8980–8988. doi: 10.1128/JVI.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang C, Rong L, Götte M, Li X, Quan Y, Kleiman L, Wainberg MA. 1998. Mechanistic studies of early pausing events during initiation of HIV-1 reverse transcription. J Biol Chem 273:21309–21315. [DOI] [PubMed] [Google Scholar]
- 12.Bampi C, Jacquenet S, Lener D, Décimo D, Darlix J-L. 2004. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Int J Biochem Cell Biol 36:1668–1686. doi: 10.1016/j.biocel.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 13.Rong L, Liang C, Hsu M, Guo X, Roques BP, Wainberg MA. 2001. HIV-1 nucleocapsid protein and the secondary structure of the binary complex formed between tRNA(Lys.3) and viral RNA template play different roles during initiation of (−) strand DNA reverse transcription. J Biol Chem 276:47725–47732. doi: 10.1074/jbc.M105124200. [DOI] [PubMed] [Google Scholar]
- 14.Bampi C, Bibillo A, Wendeler M, Divita G, Gorelick RJ, Le Grice SFJ, Darlix J-L. 2006. Nucleotide excision repair and template-independent addition by HIV-1 reverse transcriptase in the presence of nucleocapsid protein. J Biol Chem 281:11736–11743. doi: 10.1074/jbc.M600290200. [DOI] [PubMed] [Google Scholar]
- 15.Grohmann D, Godet J, Mély Y, Darlix J-L, Restle T. 2008. HIV-1 nucleocapsid traps reverse transcriptase on nucleic acid substrates. Biochemistry 47:12230–12240. doi: 10.1021/bi801386r. [DOI] [PubMed] [Google Scholar]
- 16.Wu T, Guo J, Bess J, Henderson LE, Levin JG. 1999. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J Virol 73:4794–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji X, Klarmann GJ, Preston BD. 1996. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry 35:132–143. [DOI] [PubMed] [Google Scholar]
- 18.Drummond JE, Mounts P, Gorelick RJ, Casas-Finet JR, Bosche WJ, Henderson LE, Waters DJ, Arthur LO. 1997. Wild-type and mutant HIV type 1 nucleocapsid proteins increase the proportion of long cDNA transcripts by viral reverse transcriptase. AIDS Res Hum Retroviruses 13:533–543. [DOI] [PubMed] [Google Scholar]
- 19.Klasens BI, Huthoff HT, Das AT, Jeeninga RE, Berkhout B. 1999. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim Biophys Acta 1444:355–370. [DOI] [PubMed] [Google Scholar]
- 20.Roda RH, Balakrishnan M, Hanson MN, Wöhrl BM, Le Grice SF, Roques BP, Gorelick RJ, Bambara RA. 2003. Role of the reverse transcriptase, nucleocapsid protein, and template structure in the two-step transfer mechanism in retroviral recombination. J Biol Chem 278:31536–31546. doi: 10.1074/jbc.M304608200. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamoorthy G, Roques B, Darlix J-L, Mély Y. 2003. DNA condensation by the nucleocapsid protein of HIV-1: a mechanism ensuring DNA protection. Nucleic Acids Res 31:5425–5432. doi: 10.1093/nar/gkg738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckman JS, Bosche WJ, Gorelick RJ. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J Virol 77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carteau S, Gorelick RJ, Bushman FD. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol 73:6670–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao K, Gorelick RJ, Johnson DG, Bushman F. 2003. Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. J Virol 77:1598–1603. doi: 10.1128/JVI.77.2.1598-1603.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas JA, Gagliardi TD, Alvord WG, Lubomirski M, Bosche WJ, Gorelick RJ. 2006. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology 353:41–51. doi: 10.1016/j.virol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol 244:198–223. [DOI] [PubMed] [Google Scholar]
- 27.Bryant M, Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A 87:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Göttlinger HG, Sodroski JG, Haseltine WA. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 86:5781–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou W, Parent LJ, Wills JW, Resh MD. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol 68:2556–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan X, Yu X, Lee TH, Essex M. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol 67:6387–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono A, Orenstein JM, Freed EO. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J Virol 74:2855–2866. doi: 10.1128/JVI.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aldovini A, Young RA. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol 64:1920–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cimarelli A, Sandin S, Höglund S, Luban J. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol 74:3046–3057. doi: 10.1128/JVI.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384–388. [DOI] [PubMed] [Google Scholar]
- 35.Mizuno A, Ido E, Goto T, Kuwata T, Nakai M, Hayami M. 1996. Mutational analysis of two zinc finger motifs in HIV type 1 nucleocapsid proteins: effects on proteolytic processing of Gag precursors and particle formation. AIDS Res Hum Retroviruses 12:793–800. [DOI] [PubMed] [Google Scholar]
- 36.Ott DE, Coren LV, Gagliardi TD. 2005. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J Virol 79:13839–13847. doi: 10.1128/JVI.79.22.13839-13847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott DE, Coren LV, Shatzer T. 2009. The nucleocapsid region of human immunodeficiency virus type 1 gag assists in the coordination of assembly and gag processing: role for RNA-Gag binding in the early stages of assembly. J Virol 83:7718–7727. doi: 10.1128/JVI.00099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poon DTK, Chertova EN, Ott DE. 2002. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55Gag: functional linkage between translation and RNA packaging. Virology 293:368–378. doi: 10.1006/viro.2001.1283. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Barklis E. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol 71:6765–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Barklis E. 1995. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol 69:5716–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y-X, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. 1999. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol 73:4251–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jalalirad M, Laughrea M. 2010. Formation of immature and mature genomic RNA dimers in wild-type and protease-inactive HIV-1: differential roles of the Gag polyprotein, nucleocapsid proteins NCp15, NCp9, NCp7, and the dimerization initiation site. Virology 407:225–236. doi: 10.1016/j.virol.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 43.Kafaie J, Song R, Abrahamyan L, Mouland AJ, Laughrea M. 2008. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology 375:592–610. doi: 10.1016/j.virol.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Alfadhli A, Dhenub TC, Still A, Barklis E. 2005. Analysis of human immunodeficiency virus type 1 Gag dimerization-induced assembly. J Virol 79:14498–14506. doi: 10.1128/JVI.79.23.14498-14506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowzard JB, Bennett RP, Krishna NK, Ernst SM, Rein A, Wills JW. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol 72:9034–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol 73:8527–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogue IB, Hoppe A, Ono A. 2009. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding. J Virol 83:7322–7336. doi: 10.1128/JVI.02545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee E-G, Linial ML. 2004. Basic residues of the retroviral nucleocapsid play different roles in Gag-Gag and Gag-Psi RNA interactions. J Virol 78:8486–8495. doi: 10.1128/JVI.78.16.8486-8495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandefur S, Smith RM, Varthakavi V, Spearman P. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J Virol 74:7238–7249. doi: 10.1128/JVI.74.16.7238-7249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol 72:1782–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rulli SJ Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol 81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grigorov B, Decimo D, Smagulova F, Pechoux C, Mougel M, Muriaux D, Darlix JL. 2007. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology 4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shkriabai N, Datta SAK, Zhao Z, Hess S, Rein A, Kvaratskhelia M. 2006. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- 54.Datta SAK, Zhao Z, Clark PK, Tarasov S, Alexandratos JN, Campbell SJ, Kvaratskhelia M, Lebowitz J, Rein A. 2007. Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. J Mol Biol 365:799–811. doi: 10.1016/j.jmb.2006.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datta SAK, Curtis JE, Ratcliff W, Clark PK, Crist RM, Lebowitz J, Krueger S, Rein A. 2007. Conformation of the HIV-1 Gag protein in solution. J Mol Biol 365:812–824. doi: 10.1016/j.jmb.2006.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alfadhli A, McNett H, Tsagli S, Bächinger HP, Peyton DH, Barklis E. 2011. HIV-1 matrix protein binding to RNA. J Mol Biol 410:653–666. doi: 10.1016/j.jmb.2011.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lochrie MA, Waugh S, Pratt DG Jr, Clever J, Parslow TG, Polisky B. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res 25:2902–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones CP, Datta SAK, Rein A, Rouzina I, Musier-Forsyth K. 2011. Matrix domain modulates HIV-1 Gag's nucleic acid chaperone activity via inositol phosphate binding. J Virol 85:1594–1603. doi: 10.1128/JVI.01809-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rein A, Datta SAK, Jones CP, Musier-Forsyth K. 2011. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem Sci 36:373–380. doi: 10.1016/j.tibs.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamard-Peron E, Muriaux D. 2011. Retroviral matrix and lipids, the intimate interaction. Retrovirology 8:15. doi: 10.1186/1742-4690-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chukkapalli V, Oh SJ, Ono A. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A 107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chukkapalli V, Ono A. 2011. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J Mol Biol 410:512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olety B, Ono A. 3July2014. Roles played by acidic lipids in HIV-1 Gag membrane binding. Virus Res doi: 10.1016/j.virusres.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chukkapalli V, Hogue IB, Boyko V, Hu W-S, Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag membrane binding. J Virol 82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan J, Dick RA, Vogt VM. 2011. Rous sarcoma virus gag has no specific requirement for phosphatidylinositol-(4,5)-bisphosphate for plasma membrane association in vivo or for liposome interaction in vitro. J Virol 85:10851–10860. doi: 10.1128/JVI.00760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alfadhli A, Still A, Barklis E. 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol 83:12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A 103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fuller SD, Wilk T, Gowen BE, Kräusslich HG, Vogt VM. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol 7:729–738. [DOI] [PubMed] [Google Scholar]
- 69.Datta SAK, Heinrich F, Raghunandan S, Krueger S, Curtis JE, Rein A, Nanda H. 2011. HIV-1 Gag extension: conformational changes require simultaneous interaction with membrane and nucleic acid. J Mol Biol 406:205–214. doi: 10.1016/j.jmb.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Munro JB, Nath A, Färber M, Datta SAK, Rein A, Rhoades E, Mothes W. 2014. A conformational transition observed in single HIV-1 Gag molecules during in vitro assembly of virus-like particles. J Virol 88:3577–3585. doi: 10.1128/JVI.03353-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Rocquigny H, Ficheux D, Gabus C, Fournié-Zaluski MC, Darlix JL, Roques BP. 1991. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem Biophys Res Commun 180:1010–1018. [DOI] [PubMed] [Google Scholar]
- 72.Shvadchak VV, Klymchenko AS, de Rocquigny H, Mély Y. 2009. Sensing peptide-oligonucleotide interactions by a two-color fluorescence label: application to the HIV-1 nucleocapsid protein. Nucleic Acids Res 37:e25. doi: 10.1093/nar/gkn1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Postupalenko VY, Shvadchak VV, Duportail G, Pivovarenko VG, Klymchenko AS, Mély Y. 2011. Monitoring membrane binding and insertion of peptides by two-color fluorescent label. Biochim Biophys Acta 1808:424–432. doi: 10.1016/j.bbamem.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 74.Bonnet D, Ilien B, Galzi J-L, Riché S, Antheaune C, Hibert M. 2006. A rapid and versatile method to label receptor ligands using “click” chemistry: validation with the muscarinic M1 antagonist pirenzepine. Bioconjug Chem 17:1618–1623. doi: 10.1021/bc060140j. [DOI] [PubMed] [Google Scholar]
- 75.Bernacchi S, Stoylov S, Piémont E, Ficheux D, Roques BP, Darlix JL, Mély Y. 2002. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J Mol Biol 317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- 76.Godet J, de Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix J-L, Mély Y. 2006. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J Mol Biol 356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 77.Hope MJ, Bally MB, Webb G, Cullis PR. 1985. Production of large unilamellar vesicles by a rapid extrusion procedure: characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim Biophys Acta 812:55–65. [DOI] [PubMed] [Google Scholar]
- 78.Lewis BA, Engelman DM. 1983. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol 166:211–217. [DOI] [PubMed] [Google Scholar]
- 79.Chou PT, Martinez ML, Clements JH. 1993. Reversal of excitation behavior of proton-transfer vs. charge-transfer by dielectric perturbation of electronic manifolds. J Phys Chem 97:2618–2622. [Google Scholar]
- 80.Chattopadhyay A, London E. 1987. Parallax method for direct measurement of membrane penetration depth utilizing fluorescence quenching by spin-labeled phospholipids. Biochemistry 26:39–45. [DOI] [PubMed] [Google Scholar]
- 81.Abrams FS, London E. 1993. Extension of the parallax analysis of membrane penetration depth to the polar region of model membranes: use of fluorescence quenching by a spin-label attached to the phospholipid polar headgroup. Biochemistry 32:10826–10831. [DOI] [PubMed] [Google Scholar]
- 82.Kaiser RD, London E. 1998. Location of diphenylhexatriene (DPH) and its derivatives within membranes: comparison of different fluorescence quenching analyses of membrane depth. Biochemistry 37:8180–8190. [DOI] [PubMed] [Google Scholar]
- 83.Pivovarenko VG, Zamotaiev OM, Shvadchak VV, Postupalenko VY, Klymchenko AS, Mély Y. 2012. Quantification of local hydration at the surface of biomolecules using dual-fluorescence labels. J Phys Chem A 116:3103–3109. doi: 10.1021/jp2101732. [DOI] [PubMed] [Google Scholar]
- 84.Klymchenko AS, Demchenko AP. 2003. Multiparametric probing of intermolecular interactions with fluorescent dye exhibiting excited state intramolecular proton transfer. Phys Chem Chem Phys 5:461–468. doi: 10.1039/b210352d. [DOI] [Google Scholar]
- 85.Dalton AK, Ako-Adjei D, Murray PS, Murray D, Vogt VM. 2007. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J Virol 81:6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dick RA, Goh SL, Feigenson GW, Vogt VM. 2012. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc Natl Acad Sci U S A 109:18761–18766. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Op den Kamp JAF. 1979. Lipid asymmetry in membranes. Annu Rev Biochem 48:47–71. [DOI] [PubMed] [Google Scholar]
- 88.Hanshaw RG, Smith BD. 2005. New reagents for phosphatidylserine recognition and detection of apoptosis. Bioorg Med Chem 13:5035–5042. doi: 10.1016/j.bmc.2005.04.071. [DOI] [PubMed] [Google Scholar]
- 89.Van Meer G, Voelker DR, Feigenson GW. 2008. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vácha R, Berkowitz ML, Jungwirth P. 2009. Molecular model of a cell plasma membrane with an asymmetric multicomponent composition: water permeation and ion effects. Biophys J 96:4493–4501. doi: 10.1016/j.bpj.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hamard-Peron E, Juillard F, Saad JS, Roy C, Roingeard P, Summers MF, Darlix J-L, Picart C, Muriaux D. 2010. Targeting of murine leukemia virus Gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the matrix. J Virol 84:503–515. doi: 10.1128/JVI.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ehrlich LS, Fong S, Scarlata S, Zybarth G, Carter C. 1996. Partitioning of HIV-1 Gag and Gag-related proteins to membranes. Biochemistry 35:3933–3943. [DOI] [PubMed] [Google Scholar]
- 93.Ben-Tal N, Honig B, Miller C, McLaughlin S. 1997. Electrostatic binding of proteins to membranes. Theoretical predictions and experimental results with charybdotoxin and phospholipid vesicles. Biophys J 73:1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Didier P, Sharma KK, Mély Y. 2011. Fluorescence techniques to characterise ligand binding to proteins, p 156–199 InPodjarny A, Dejaegere AP, Kieffer B (ed), RSC biomolecular sciences, vol 22 Biophysical approaches determining ligand binding to biomolecular targets: detection, measurement and modelling. Royal Society of Chemistry, Cambridge, United Kingdom. [Google Scholar]
- 95.Beltz H, Clauss C, Piémont E, Ficheux D, Gorelick RJ, Roques B, Gabus C, Darlix J-L, de Rocquigny H, Mély Y. 2005. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J Mol Biol 348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 96.Egelé C, Schaub E, Ramalanjaona N, Piémont E, Ficheux D, Roques B, Darlix J-L, Mély Y. 2004. HIV-1 nucleocapsid protein binds to the viral DNA initiation sequences and chaperones their kissing interactions. J Mol Biol 342:453–466. doi: 10.1016/j.jmb.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 97.Darlix J-L, Godet J, Ivanyi-Nagy R, Fossé P, Mauffret O, Mély Y. 2011. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J Mol Biol 410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 98.Post K, Kankia B, Gopalakrishnan S, Yang V, Cramer E, Saladores P, Gorelick RJ, Guo J, Musier-Forsyth K, Levin JG. 2009. Fidelity of plus-strand priming requires the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res 37:1755–1766. doi: 10.1093/nar/gkn1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Godet J, Ramalanjaona N, Sharma KK, Richert L, de Rocquigny H, Darlix J-L, Duportail G, Mély Y. 2011. Specific implications of the HIV-1 nucleocapsid zinc fingers in the annealing of the primer binding site complementary sequences during the obligatory plus strand transfer. Nucleic Acids Res 39:6633–6645. doi: 10.1093/nar/gkr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol 80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 101.Déméné H, Dong CZ, Ottmann M, Rouyez MC, Jullian N, Morellet N, Mely Y, Darlix JL, Fournié-Zaluski MC, Saragosti S. 1994. 1H NMR structure and biological studies of the His23→Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for virus infectivity. Biochemistry 33:11707–11716. [DOI] [PubMed] [Google Scholar]
- 102.Mély Y, de Rocquigny H, Sorinas-Jimeno M, Keith G, Roques BP, Marquet R, Gérard D. 1995. Binding of the HIV-1 nucleocapsid protein to the primer tRNA(3Lys), in vitro, is essentially not specific. J Biol Chem 270:1650–1656. [DOI] [PubMed] [Google Scholar]
- 103.Vuilleumier C, Bombarda E, Morellet N, Gérard D, Roques BP, Mély Y. 1999. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry 38:16816–16825. [DOI] [PubMed] [Google Scholar]
- 104.Clamme JP, Bernacchi S, Vuilleumier C, Duportail G, Mély Y. 2000. Gene transfer by cationic surfactants is essentially limited by the trapping of the surfactant/DNA complexes onto the cell membrane: a fluorescence investigation. Biochim Biophys Acta 1467:347–361. doi: 10.1016/S0005-2736(00)00230-3. [DOI] [PubMed] [Google Scholar]
- 105.Sandefur S, Varthakavi V, Spearman P. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol 72:2723–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kerviel A, Thomas A, Chaloin L, Favard C, Muriaux D. 2013. Virus assembly and plasma membrane domains: which came first? Virus Res 171:332–340. doi: 10.1016/j.virusres.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 107.Mariani C, Desdouits M, Favard C, Benaroch P, Muriaux DM. 2014. Role of Gag and lipids during HIV-1 assembly in CD4(+) T cells and macrophages. Front Microbiol 5:312. doi: 10.3389/fmicb.2014.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bouamr F, Scarlata S, Carter C. 2003. Role of myristylation in HIV-1 Gag assembly. Biochemistry 42:6408–6417. doi: 10.1021/bi020692z. [DOI] [PubMed] [Google Scholar]
- 109.Pal R, Reitz MS Jr, Tschachler E, Gallo RC, Sarngadharan MG, Veronese FD. 1990. Myristoylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res Hum Retroviruses 6:721–730. [DOI] [PubMed] [Google Scholar]
- 110.Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, McNitt KL, Johnson DG, Hixson CV, Gorelick RJ, Fabris D, Henderson LE, Rein A. 2006. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res 34:472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nanda H, Datta SAK, Heinrich F, Lösche M, Rein A, Krueger S, Curtis JE. 2010. Electrostatic interactions and binding orientation of HIV-1 matrix studied by neutron reflectivity. Biophys J 99:2516–2524. doi: 10.1016/j.bpj.2010.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chukkapalli V, Inlora J, Todd GC, Ono A. 2013. Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. J Virol 87:7155–7159. doi: 10.1128/JVI.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ono A, Freed EO. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A 98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Briggs JAG, Simon MN, Gross I, Kräusslich H-G, Fuller SD, Vogt VM, Johnson MC. 2004. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 115.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Crist RM, Datta SAK, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J Virol 83:2216–2225. doi: 10.1128/JVI.02031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Accola MA, Strack B, Göttlinger HG. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol 74:5395–5402. doi: 10.1128/JVI.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Johnson MC, Scobie HM, Ma YM, Vogt VM. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J Virol 76:11177–11185. doi: 10.1128/JVI.76.22.11177-11185.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Klein KC, Reed JC, Tanaka M, Nguyen VT, Giri S, Lingappa JR. 2011. HIV Gag-leucine zipper chimeras form ABCE1-containing intermediates and RNase-resistant immature capsids similar to those formed by wild-type HIV-1 Gag. J Virol 85:7419–7435. doi: 10.1128/JVI.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]