ABSTRACT
The envelope of influenza A viruses contains two large antigens, hemagglutinin (HA) and neuraminidase (NA). Conventional influenza virus vaccines induce neutralizing antibodies that are predominantly directed to the HA globular head, a domain that is subject to extensive antigenic drift. Antibodies directed to NA are induced at much lower levels, probably as a consequence of the immunodominance of the HA antigen. Although antibodies to NA may affect virus release by inhibiting the sialidase function of the glycoprotein, the antigen has been largely neglected in past vaccine design. In this study, we characterized the protective properties of monospecific immune sera that were generated by vaccination with recombinant RNA replicon particles encoding NA. These immune sera inhibited hemagglutination in an NA subtype-specific and HA subtype-independent manner and interfered with infection of MDCK cells. In addition, they inhibited the sialidase activities of various influenza viruses of the same and even different NA subtypes. With this, the anti-NA immune sera inhibited the spread of H5N1 highly pathogenic avian influenza virus and HA/NA-pseudotyped viruses in MDCK cells in a concentration-dependent manner. When chickens were immunized with NA recombinant replicon particles and subsequently infected with low-pathogenic avian influenza virus, inflammatory serum markers were significantly reduced and virus shedding was limited or eliminated. These findings suggest that NA antibodies can inhibit virus dissemination by interfering with both virus attachment and egress. Our results underline the potential of high-quality NA antibodies for controlling influenza virus replication and place emphasis on NA as a vaccine antigen.
IMPORTANCE The neuraminidase of influenza A viruses is a sialidase that acts as a receptor-destroying enzyme facilitating the release of progeny virus from infected cells. Here, we demonstrate that monospecific anti-NA immune sera inhibited not only sialidase activity, but also influenza virus hemagglutination and infection of MDCK cells, suggesting that NA antibodies can interfere with virus attachment. Inhibition of both processes, virus release and virus binding, may explain why NA antibodies efficiently blocked virus dissemination in vitro and in vivo. Anti-NA immune sera showed broader reactivity than anti-HA sera in hemagglutination inhibition tests and demonstrated cross-subtype activity in sialidase inhibition tests. These remarkable features of NA antibodies highlight the importance of the NA antigen for the development of next-generation influenza virus vaccines.
INTRODUCTION
Hemagglutinin (HA) is the most abundant antigen of the influenza A virus envelope. Antibodies that interfere with receptor-binding or fusion activity of HA usually have virus-neutralizing activity (1). However, immune pressure by the host permanently selects for virus escape mutants that are no longer neutralized, a phenomenon known as antigenic drift. Moreover, influenza A viruses acquiring a new HA subtype from a different influenza A virus through reassortment may easily escape from preexisting immunity (antigenic shift). Presently, 18 subtypes of HA are known and can be genetically and serologically discriminated. Subtypes H1 to H16 were detected in avian influenza viruses (AIV), while subtypes H17 and H18 were recently detected in bat influenza viruses. So far, only subtypes H1 to H3 have been found in human influenza viruses.
Neuraminidase (NA), the second major antigen of the viral envelope, is also subject to antigenic drift and shift, indicating that the antigen is under immune pressure, as well (3–6). Currently, 11 NA subtypes are known. Subtypes N1 to N9 have been detected in AIV, subtypes N1 and N2 are found in human influenza viruses, and subtypes N10 and N11 were recently discovered in bat influenza viruses (7). The different NAs of influenza A viruses can be phylogenetically classified into 3 groups, with group 1 comprising N1, N4, N5, and N8; group 2 comprising N2, N3, N6, N7, and N9; and group 3 containing N10 and N11 (7, 8).
The classical NA (subtypes N1 to N9) is a tetrameric glycoprotein that disposes of sialidase activity. The enzyme removes sialic acid residues from cellular and viral glycoproteins, thereby facilitating budding and release of progeny viruses from the host cell (9). Apart from its role in virus egress, NA may help initiate infection of respiratory epithelial cells (10). The current opinion is that antibodies to NA do not have neutralizing activity in terms of preventing influenza virus infection. Rather, they restrict virus spread in the infected host by inhibiting NA sialidase activity (11, 12). This “permissive” immunity can be quite effective in reducing virus titers, alleviating clinical symptoms, and reducing and shortening virus shedding (13–20).
Although there is evidence for a beneficial role of NA in immunity against influenza viruses (21, 22), currently used vaccines do not make use of these properties (11, 23). In fact, conventional inactivated human influenza virus vaccines are split and standardized according to HA quantities (24, 25), whereas the amount of NA often varies among different manufacturers and production lots (11). The NA antigen is underrepresented in vaccine preparations, as there are 4-fold to 5-fold fewer NA than HA molecules present in the viral envelope (26). Not surprisingly, the rate of seroconversion to NA has been reported to be quite low in the vaccinated human population (27, 28). There is experimental evidence that HA dominates NA in priming of B cells, explaining the low immunogenicity of NA (29, 30). Interestingly, antigen competition is eliminated if the two antigens are dissociated from each other and used separately for vaccination (31).
The NA antigen has been used for experimental vaccination employing plasmid vectors (32–34), viral vectored vaccines (35), virus-like particles (36, 37), and recombinant subunit vaccines (38, 39). In mammalian hosts, these vaccines conferred partial or even complete protection from infection with human or avian influenza viruses (15, 19, 32, 33, 35–39). In contrast, NA vaccines based on recombinant Newcastle disease virus (NDV) or infectious laryngotracheitis virus (ILTV) failed to protect chickens from lethal infection with highly pathogenic avian influenza virus (HPAIV), although the survival times of the animals were prolonged (40–42). Immunization of chickens with NA recombinant baculovirus, plasmid DNA, or alphaviral replicon particles provided partial protection from lethal infection with homologous HPAIV (43).
In the present study, recombinant virus replicon particles (VRPs) were used to generate monospecific immune sera directed to NA. These immune sera were characterized for the ability to inhibit hemagglutination, infection of Madin-Darby canine kidney (MDCK) cells, sialidase activity, and virus spread in vitro. In addition, the capacity of the NA antigen to protect chickens against infection with low-pathogenic avian influenza viruses (LPAIVs) was investigated. Our data highlight the potential of the NA antigen to induce protective immunity against influenza viruses.
(This work was performed by Stefan Halbherr in partial fulfillment of the requirements for a Ph.D. degree from the University of Bern [Graduate School for Cellular and Biomedical Sciences], Bern, Switzerland.)
MATERIALS AND METHODS
Cells.
BHK-21 cells were obtained from the German Cell Culture Collection (DSZM) (Braunschweig, Germany) and grown in Earle's minimal essential medium (MEM) (Life Technologies, Carlsbad, CA) supplemented with 5% fetal bovine serum (FBS) (Biowest). BHK-G43, a transgenic BHK-21 cell clone expressing the vesicular stomatitis virus (VSV) G protein in a regulated manner, was maintained as described previously (44). MDCK cells (type I) were provided by Georg Herrler (TiHo, Hannover, Germany) and cultured with MEM and 5% FBS. Vero-E6 cells were obtained from ATCC and cultured in Glasgow MEM containing 5% FBS.
Viruses.
Recombinant modified vaccinia virus Ankara expressing the RNA polymerase of the T7 phage (MVA-T7) was a kind gift from Gerd Sutter (LMU, Munich, Germany) (45). Recombinant VSV*ΔG(HAH5) and VSV*ΔG(HAH1) expressing the HA antigen of A/Yamaguchi/7/04 (H5N1) and A/duck/Italy/1447/05 (H1N1), respectively, have been reported previously (46). The HPAIV A/chicken/Yamaguchi/7/04 (H5N1) (47) and the LPAIV A/duck/Hokkaido/Vac-1/04 (H5N1) (48) were kindly provided by Yoshihiro Sakoda (Hokkaido University, Sapporo, Japan). The LPAIVs A/duck/Italy/1447/05 (H1N1), A/turkey/Italy/3675/99 (H7N1), and A/teal/Italy/1398/06 (H10N7) were kindly provided by William Dundon (Istituto Zooprofilattico Sperimentale delle Venezie [IZSV], Venice, Italy). A/swan/Potsdam/62/81 (H7N7) was obtained from Timm Harder (FLI, Riems, Germany). A/Puerto Rico/8/34 (H1N1) and A/swine/Belzig/2/01 (H1N1) were kindly provided by Paul Digard (University of Cambridge, Cambridge, United Kingdom) and Jürgen Stech (FLI, Riems, Germany), respectively. A/WSN/33 (H1N1), A/New Caledonia/20/99 (H1N1), and A/California/7/09 (H1N1) were kindly provided by Veronika von Messling (Paul-Ehrlich-Institut, Langen, Germany), W. Wunderli, (University Hospital of Geneva, Geneva, Switzerland), and the National Institute for Biological Standards and Control (NIBSC) (Potters Bar, United Kingdom), respectively. A/swine/USA/1976/31 (H1N1) was purchased from the American Type Culture Collection (Manassas, VA, USA). All influenza viruses were propagated in the allantoic cavities of 10-day-old embryonated specific-pathogen-free (SPF) chicken eggs for 2 days at 37°C.
Influenza viruses were titrated on MDCK cells either in the absence (for titration of HPAIV) or presence (for titration of LPAIV) of 1 μg/ml of acetylated trypsin (Sigma-Aldrich, St. Louis, MO). At 72 h postinfection (p.i.) with HPAIV, the cells were washed with phosphate-buffered saline (PBS) and fixed with 10% formalin containing 0.1% (wt/vol) crystal violet. The plates were washed with tap water to remove excess crystal violet and dried. In the case of LPAIV, infected cells were detected by immunostaining with a monoclonal antibody directed to the NP antigen (49). Virus titers were calculated according to the Spearman-Kärber method and expressed as 50% tissue culture infectious doses (TCID50)/ml (50).
Construction of plasmids.
The cDNAs encoding segments 4 and 6 of A/swine/Belzig/2/01 (H1N1), segment 4 of A/turkey/Italy/3675/99 (H7N1), and segment 6 of A/swan/Potsdam/62/81 (H7N7) were generated by reverse transcription of total RNA isolated from either virus-infected MDCK cells or the allantoic fluid of infected chicken embryos (46). The cDNAs encoding all eight RNA segments of A/chicken/Yamaguchi/7/04 (H5N1) and A/WSN/33 (H1N1) were kindly provided by Yoshihiro Sakoda (Hokkaido University, Sapporo, Japan) and Gabriele Neumann (University of Wisconsin—Madison, Madison, WI, USA), respectively. The open reading frames of influenza virus antigens were amplified by PCR and cloned into the pVSV* plasmid using the MluI and BstEII endonuclease restriction sites placed upstream and downstream of the fourth transcription unit of the VSV genome (51). A cDNA encoding enhanced green fluorescent protein (eGFP) was inserted into an additional transcription unit located in the intergenic region between the original G and L genes. The resulting plasmids were designated pVSV*ΔG(NA) and pVSV*ΔG(HA), in which the asterisk denotes the presence of the eGFP gene and ΔG indicates the absence of the glycoprotein G gene. For generation of pseudotype virus, a VSV vector backbone with 7 transcription units was used (52). The HA and NA genes of A/chicken/Yamaguchi/7/04 (H5N1) were inserted into the fourth and fifth transcription units, taking advantage of single MluI and XhoI restriction sites, respectively. The gene encoding a secreted version of the small (19-kDa) NanoLuc luciferase (Nluc) from the deep-sea shrimp Oplophorus gracilirostris (Promega, Madison, WI, USA) was inserted into the NheI site of the sixth transcription unit. The resulting plasmid was designated pVSVΔG(HA,NA,NLuc). Any mutations in the recombinant vector constructs were excluded by DNA sequencing.
Generation of recombinant VSV.
Recombinant VSV lacking the envelope glycoprotein G was generated as described previously (51, 53). Briefly, BHK-G43 cells were infected with recombinant MVA-T7 expressing T7 RNA polymerase (45) and subsequently transfected with a plasmid carrying a VSV antigenomic cDNA, along with three plasmids encoding the VSV proteins N, P, and L. All the genes were placed under the control of the T7 promoter. Expression of the VSV G protein was induced in BHK-G43 cells by adding 10−9 M mifepristone (Sigma) to the cell culture medium. At 24 h posttransfection, the cells were trypsinized and seeded into T-75 cell culture flasks (Corning B.V., Amsterdam, The Netherlands), along with an equal number of fresh BHK-G43 cells. The cells were incubated at 37°C for 24 h in the presence of mifepristone. The cell culture supernatant was clarified by low-speed centrifugation and passed through a 0.20-μm-pore-size filter. Recombinant VSV*ΔG, VSV*ΔG(NA), and VSV*ΔG(HA) particles were propagated on mifepristone-induced BHK-G43 cells and titrated on BHK-21 cells. Infectious titers were expressed as fluorescent focus-forming units (FFU) per milliliter. VSVΔG(HA,NA,Nluc) pseudotype virus was propagated and titrated on MDCK cells. For titration, the cells were infected with serial virus dilutions and incubated with MEM containing 0.9% (wt/vol) methylcellulose. The cells were fixed 24 h postinfection, permeabilized with 0.25% (vol/vol) Triton X-100, and incubated with rabbit polyclonal anti-VSV serum and subsequently with goat anti-rabbit IgG serum conjugated with horseradish peroxidase (Dako, Hamburg, Germany). Infected cells were visualized with AEC peroxidase substrate (0.05% [wt/vol] 3-amino-9-ethylcarbazole, 0.015% [vol/vol] H2O2, 0.05 M sodium acetate buffer, pH 5.5). Virus titers were calculated and expressed as PFU per ml.
Assay for sialidase activity on infected cells.
Vero-E6 cells were seeded into 96-well cell culture plates (2 × 104 cells/well). The following day, the confluent cells were infected with either VSV*ΔG, VSV*ΔG(NA), A/chicken/Yamaguchi/7/04 (H5N1), or A/swan/Potsdam/62/81 (H7N7). At 12 h postinfection, the cells were washed with PBS and incubated for 20 min at 37°C with 100 μl of PBS per well containing 20 μM the fluorogenic sialidase substrate 4-methylumbelliferyl-alpha-d-N-acetylneuraminic acid (MU-NANA) (Sigma-Aldrich). The supernatants were transferred to black 96-well plates, and fluorescence was detected with a SpectraMax Gemini plate reader (Molecular Devices) using 355 nm for excitation and 460 nm for emission wavelengths.
Immunization and infection of animals.
SPF chickens (5 weeks old) and piglets (8 weeks old) were obtained from the breeding unit of the Institute of Virology and Immunology (IVI) (Mittelhäusern, Switzerland). The animals were immunized by intramuscular (i.m.) injections of cell culture supernatant (twice with 0.25 ml for chickens and twice with 2.5 ml for pigs) containing recombinant VRPs (2 × 108 FFU/ml). Adjuvants were not employed. Three weeks after the first application, the animals were immunized a second time using the same VRPs, doses, and route. Immune sera were prepared 3 to 4 weeks after the boost and heated at 56°C for 30 min in order to inactivate components of the complement system. For hemagglutination inhibition (HI) tests, serum was treated for 18 h at 37°C with 50 mU of Vibrio cholerae neuraminidase (VCNA) (Sigma) per ml of serum. The VCNA was subsequently inactivated by incubating the samples for 30 min at 56°C.
In order to assess the protective capacity of NA antigen, SPF chickens were immunized twice with recombinant VRPs as described above. Three weeks after the second immunization, the animals were infected with the low-pathogenic avian influenza virus A/swan/Potsdam/62/81 (H7N7) via the intratracheal (i.t.) route using 107 TCID50 in 250 μl PBS. All animal experiments were performed in compliance with the Swiss animal protection law and approved by the animal welfare committee of the canton of Berne (authorization numbers 76/10 and 07/12).
Inhibition of virus entry.
Serial 2-fold diIutions of immune sera (50 μl) were incubated in quadruplicate for 1 h at room temperature with 50 μl of MEM containing 100 TCID50 of A/duck/Hokkaido/Vac-1/04 (H5N1). The serum-virus mixture was transferred to confluent MDCK cells grown in 96-well tissue culture plates (4 × 104 cells/well) and incubated for 1 h at 37°C. The cells were washed three times with PBS and incubated with MEM at 37°C in the absence of immune serum. At 24 h postinfection, the cells were washed with PBS, fixed with 3% paraformaldehyde, and permeabilized with 0.25% (vol/vol) Triton X-100. Influenza virus NP antigen was detected by incubating the cells with the anti-NP monoclonal antibody HB-65 and subsequently with goat anti-mouse IgG serum conjugated to horseradish peroxidase (Dako, Hamburg, Germany). Infected cells were visualized using AEC peroxidase substrate. The number of plaques per well was estimated using an automated spot analyzer (Cellular Technology Ltd., Bonn, Germany).
To analyze the inhibition of virus entry by immunofluorescence, MDCK cells were grown on 12-mm glass coverslips and incubated for 1 h at 4°C with 0.5 TCID50/cell of A/chicken/Yamaguchi/7/04 (H5N1). Subsequently, the cells were washed with PBS, incubated with MEM for 5 h at 37°C, fixed, and permeabilized as described above. The NP antigen was detected by incubating the cells with monoclonal antibody HB-65 and subsequently with anti-mouse IgG conjugated with Alexa Fluor 488 (1:500; Life Technologies, Carlsbad, CA, USA). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma).
For analysis of virus binding to the cell surface, MDCK cells were grown in 24-well cell culture plates and inoculated with A/chicken/Yamaguchi/7/04 (H5N1) for 1 h at 4°C in the absence or presence of immune sera. The cells were washed three times with ice-cold PBS, and total RNA was extracted using TRIzol reagent (Life Technologies) according to the manufacturer's protocol. Influenza virus RNA segment 7 was detected by quantitative reverse transcription (qRT)-PCR as described previously (54, 55).
Inhibition of virus spread in vitro.
MDCK cells grown in 96-well tissue culture plates (4 × 104 cells/well) were infected for 1 h at 37°C with 100 TCID50 of A/chicken/Yamaguchi/7/04 (H5N1) per well. The cells were washed twice with PBS and maintained for 24 h at 37°C with 100 μl/well of MEM containing serially diluted chicken immune sera or oseltamivir carboxylate (Toronto Research Chemicals). The cells were fixed with 3% paraformaldehyde, and influenza virus NP antigen was detected as described above.
For inhibition of pseudotype virus spread, MDCK cells were grown in 96-well microplates and infected for 1 h with VSVΔG(HA,NA,NLuc) using a multiplicity of infection (MOI) of 0.0025 PFU/cell. The cells were washed and incubated with 200 μl of MEM cell culture medium supplemented with serially diluted immune sera. The cell culture supernatant was harvested at 48 h postinfection. Cell culture supernatant (10 μl) was mixed with H2O (40 μl) and Nano-Glo luciferase substrate (50 μl; Promega) and transferred to white 96-well microplates. Luminescence was recorded with a Centro LB 960 microplate luminometer (Berthold Technologies, Bad Wildbad, Germany).
Sialidase inhibition assay.
Human and porcine influenza A viruses and LPAIV were propagated on MDCK cells in the presence of 1 μg/ml of acetylated trypsin. At 40 h postinfection, the cell culture supernatant was harvested and cell debris was removed by low-speed centrifugation. Virus was pelleted from the clarified supernatant by ultracentrifugation (105,000 × g; 60 min; 4°C) and suspended in PBS (pH 7.2) containing 1 mM CaCl2 and 0.5 mM MgCl2. Suspended virus (50 μl) corresponding to 0.04 mU of sialidase was incubated for 1 h at 21°C with 50 μl of serially diluted chicken immune sera or oseltamivir carboxylate in the presence of 0.1% (wt/vol) bovine serum albumin (BSA) (Sigma). One unit of sialidase was defined as the amount of enzyme required to produce 1 μmol of methylumbelliferone in 1 min at 37°C, pH 7.2. The mixtures were transferred to Nunc MaxiSorp 96-well plates (Thermo Scientific, Waltham, MA, USA) coated with 1 μg of glycophorin A (Sigma) per well and blocked with 1% (wt/vol) BSA (Sigma). The plates were incubated for 16 h at 37°C and washed with PBS containing 0.05% (vol/vol) Tween 20 (PBS-T). Each well received 50 μl of PBS containing 0.1% (wt/vol) BSA and 0.5 μg/ml of biotinylated peanut agglutinin (PNA) (Sigma). Immobilized glycophorin A was incubated with PNA for 1 h at room temperature, washed with PBS-T, and then incubated for 30 min at 21°C with streptavidin-peroxidase conjugate (Dako; 1:5,000 in PBS). The plates were rinsed several times with PBS-T and once with PBS before 50 μl/well of TMB peroxidase substrate (Life Technologies) was added. The reaction was stopped by adding 50 μl/well of 0.16 M hydrochloric acid, and absorption was recorded at 450 nm (VersaMax plate reader; Molecular Devices). Sialidase measured in the presence of preimmune serum was considered to have 100% activity.
Analysis of virus shedding by qRT-PCR.
Oropharyngeal and cloacal swabs from infected chickens were sampled daily for a period of 7 days, suspended in 2 ml of MEM, and stored at −70°C. Total RNA was extracted from the samples using the NucleoSpin 96 Virus kit (Macherey-Nagel AG, Düren, Germany). For detection of viral RNA, a quantitative RT-PCR based on the amplification of the conserved viral RNA segment 7 was performed in triplicate, employing enhanced green fluorescent protein (eGFP) as an internal control (54, 55).
Serological tests.
For titration of anti-HAH5, anti-HAH7, and anti-NAN1 antibodies, commercially available competitive enzyme-linked immunosorbent assays (ELISAs) were used according to the manufacturer's protocols (ID-Vet, Montpellier, France). Antibody titers were calculated as the reciprocal value of the highest immune serum dilution passing the threshold value defined by the manufacturer.
HI tests were performed with V-bottom 96-well microplates. Viruses were diluted to obtain a suspension with 4 hemagglutination units, and 25 μl of the diluted virus was incubated for 1 h at 4°C with 25 μl of VCNA-treated serially diluted immune sera. Subsequently, 50 μl of chicken erythrocyte suspension (0.5% in PBS) was added to each well and incubated for 1 h at 4°C. HI units were expressed as the reciprocal value of the highest serum dilution causing complete inhibition of hemagglutination.
Serum levels of α1-acid glycoprotein (α1-AGP) were determined as previously described (56), using a commercially available immunodiffusion test (Cardiotech Services, Inc., Spring Lake, NJ, USA).
Statistical analysis.
Mean values and standard deviations were calculated where indicated. Statistical analysis was performed using the Student t test, with P values of <0.05 considered significant.
RESULTS
Expression of functionally active neuraminidase.
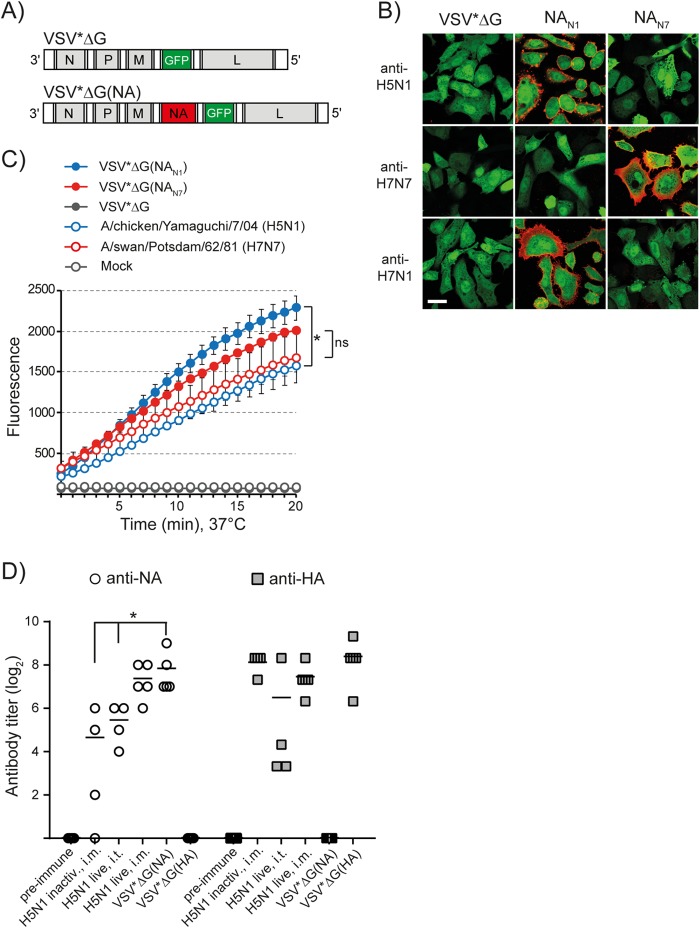
For expression of influenza virus antigens, a modified VSV vector lacking the glycoprotein G gene was used (51). Influenza virus NA antigens derived from either A/chicken/Yamaguchi/7/04 (H5N1), A/swine/Belzig/2/01 (H1N1), or A/swan/Potsdam/62/81 (H7N7) were expressed from the fourth transcription unit of the viral RNA genome (Fig. 1A). The recombinant viruses were propagated on helper cells providing the VSV G glycoprotein in trans (44). Since they are unable to propagate in nonhelper cells beyond a single round of infection, we refer to these viral vectors as VRPs. To study expression of recombinant antigens in vitro, Vero cells were infected with recombinant VRPs using an MOI of 3 FFU/cell. Analysis of the infected cells by indirect immunofluorescence and incubation of the cells with the synthetic sialidase substrate MU-NANA revealed that the recombinant NA antigens were expressed at the cell surface (Fig. 1B) in enzymatically active form (Fig. 1C). When cells were infected with A/swan/Potsdam/62/81 (H7N7) using the same MOI, sialidase activity was at as high a level as in VSV*ΔG(NAN7)-infected cells. In contrast, cells infected with A/chicken/Yamaguchi/7/04 (H5N1) showed significantly lower sialidase activity than VSV*ΔG(NAN1)-infected cells.
FIG 1.
Expression of functionally active NA antigen using propagation-incompetent virus replicon particles. (A) Genome maps of recombinant VSV vectors. VSV*ΔG contains five transcription units encoding the nucleoprotein N, the phosphoprotein P, the matrix protein M, eGFP, and the large RNA polymerase protein L. Note that VSV*ΔG lacks the gene encoding the VSV envelope glycoprotein G. VSV*ΔG(NA) expresses influenza virus NA antigen from the fourth gene position, while GFP is expressed from an additional transcription unit downstream of NA. (B) Immunofluorescence analysis of Vero cells infected with either VSV*ΔG, VSV*ΔG(NAN1), or VSV*ΔG(NAN7). At 6 h p.i., the cells were incubated at 4°C with the indicated immune sera, washed with PBS, and fixed with formalin. Antibodies bound to cell surface NA were visualized with anti-chicken IgY serum conjugated with Alexa Fluor 546 (red fluorescence). Expression of GFP is indicated by green fluorescence. The scale bar represents 24 μm. (C) At 12 h p.i. with the indicated replicon particles, Vero cells were incubated with the sialidase substrate MU-NANA for 20 min at 37°C. Fluorescence was recorded at 355 nm for excitation and 460 nm for emission. The asterisk denotes significantly different sialidase activities (P < 0.05); ns, nonsignificantly different. The error bars indicate standard deviations. (D) Quantification of serum antibody levels. SPF chickens (n = 4/5) were immunized with either inactivated or live A/duck/Hokkaido/Vac-1/04 (H5N1) via the indicated vaccination routes. In addition, chickens were immunized i.m. with VRPs expressing either the NA antigen of A/chicken/Yamaguchi/7/04 (H5N1), the HA antigen of A/duck/Hokkaido/Vac-1/04 (H5N1), or GFP as a control (VSV*ΔG). With the exception of animals receiving live virus via the intratracheal route, all chickens were immunized a second time 3 weeks after the first immunization. Immune sera were collected 3 weeks after the boost and titrated using competitive N1 and H5 ELISAs. The scatter plots show individual and mean antibody titers. The asterisk indicates significantly different values (P < 0.05).
I.m. immunization of SPF chickens with recombinant VRPs encoding the NA antigen of A/chicken/Yamaguchi/7/04 (H5N1) resulted in the induction of serum anti-NAN1 antibodies with a mean titer of 230, as determined by competitive ELISA (Fig. 1D). For comparison, vaccination (i.m.) with inactivated H5N1 virus resulted in a significantly lower anti-NAN1 titer (mean titer, 25) even though an anti-HAH5 titer of 280 was induced by the vaccine. When chickens were infected via the intratracheal route with live H5N1 virus vaccine, serum antibody titers of 44 (for anti-NAN1) and 90 (for anti-HAH5) were detected. Interestingly, anti-NAN1 antibody titers were significantly higher if live H5N1 virus was applied via the intramuscular route (mean titer, 166). In conclusion, these findings indicate that the expression of native NA antigens by recombinant VRPs triggers a robust humoral immune response. The endpoint titers of pooled immune sera are listed in Table 1.
TABLE 1.
Antibody endpoint titers in sera of vaccinated animals
| Vaccinea | Immunization route | Antibody endpoint titerb |
||
|---|---|---|---|---|
| Anti-NAN1 | Anti-HAH5 | Anti-HAH7 | ||
| Control (preimmune serum) | <2 | <5 | <10 | |
| A/duck/Hokkaido/Vac-1/04 (H5N1), inactivated | I.m. | 16 | 160 | <10 |
| A/duck/Hokkaido/Vac-1/04 (H5N1), live | I.t. | 32 | 80 | <10 |
| A/duck/Hokkaido/Vac-1/04 (H5N1), live | I.m. | 128 | 160 | <10 |
| A/turkey/Italy/3675/99 (H7N1), live | I.t. | 32 | <5 | 160 |
| VSV*ΔG(NAN1) A/chicken/Yamaguchi/7/04 (H5N1) | I.m. | 256 | <5 | ND |
| VSV*ΔG(NAN1) A/swine/Belzig/2/01 (H1N1) | I.m. | 512 | ND | ND |
| VSV*ΔG(HAH5) A/chicken/Yamaguchi/7/04 (H5N1) | I.m. | <2 | 160 | ND |
| VSV*ΔG(HAH5) A/duck/Hokkaido/Vac-1/04 (H5N1) | I.m. | <2 | 320 | ND |
| VSV*ΔG(HAH7) A/turkey/Italy/3675/99 (H7N1) | I.m. | ND | <5 | 160 |
All immune sera were produced in SPF chickens, except serum directed to the NA antigen of A/swine/Belzig/2/01 (H1N1), which was raised in SPF pigs. Animal groups (n = 4/5) were immunized with the vaccines listed. The origins of antigens encoded by recombinant VRPs are indicated.
The immune sera from each vaccination group were pooled and titrated using competitive ELISAs for the specific detection of antibodies to NAN1, HAH5, and HAH7. Antibody endpoint titers were calculated as the reciprocal value of the highest antibody dilution that produced a signal lower than the defined threshold value. ND, not determined.
NA antibodies inhibit hemagglutination.
HA-specific antibodies are known to inhibit influenza virus-mediated hemagglutination by interfering with the receptor-binding activity of HA. However, previous work suggested that NA-specific antibodies may also have HI activity (57). We therefore produced monospecific immune sera directed to different HA and NA antigens by taking advantage of recombinant VRPs and analyzed their capacities to inhibit hemagglutination by avian, porcine, and human influenza viruses. Immune sera raised against avian HAH1, HAH5, and HAH7 and porcine HAH1 showed strong subtype-specific HI activity, in particular against the homologous viruses (Table 2). Immune serum directed to NAN1 of A/chicken/Yamaguchi/7/04 (H5N1) revealed HI titers similar to those of antibodies raised against the HA of the virus. The anti-NAN1 serum also inhibited hemagglutination by A/duck/Italy/1447/05 (H1N1) and A/turkey/Italy/3675/99 (H7N1), i.e., avian influenza viruses of the same NA subtype but different HA subtypes. In addition, the antiserum reacted with A/swine/Belzig/2/01 (H1N1) and A/Puerto Rico/8/34 (H1N1), although HI titers were lower than those observed with AIV. A similar HI pattern was found with serum raised against the NA of A/swine/Belzig/2/01 (H1N1). Interestingly, this serum showed higher HI activity against A/duck/Italy/1447/05 (H1N1) and A/chicken/Yamaguchi/7/04 (H5N1) than against A/swine/Belzig/2/01 (H1N1), suggesting that the avian viruses were more susceptible to inhibition by NA-specific antibodies. Antiserum directed to NAN7 inhibited hemagglutination by A/swan/Potsdam/62/81 (H7N7) and A/teal/Italy/1398/06 (H10N7), whereas viruses of the N1 subtype were not affected. These findings demonstrate that the immune sera reacted in an NA subtype-specific manner, showing extensive cross-reactivity with other NA molecules of the same subtype.
TABLE 2.
Analysis of immune sera for hemagglutination inhibition activity
| Virus | HI titer using immune serum raised againsta: |
||||||
|---|---|---|---|---|---|---|---|
| NAN1 Yamaguchi/7/04 | NAN1 Belzig/2/01 | NAN7 Potsdam/62/81 | HAH1 Italy/1447/05 | HAH1 Belzig/2/01 | HAH5 Yamaguchi/7/04 | HAH7 Italy/3675/99 | |
| A/chicken/Yamaguchi/7/04 (H5N1) | 512 | 512 | <16 | <16 | <16 | 512 | <16 |
| A/duck/Italy/1447/05 (H1N1) | 512 | 256 | <16 | 512 | 64 | <16 | <16 |
| A/turkey/Italy/3675/99 (H7N1) | 256 | 64 | <16 | <16 | <16 | <16 | 512 |
| A/swine/Belzig/2/01 (H1N1) | 128 | 128 | <16 | <16 | 256 | <16 | <16 |
| A/PuertoRico/8/34 (H1N1) | 64 | 128 | <16 | <16 | <16 | <16 | <16 |
| A/WSN/33 (H1N1) | <16 | <16 | <16 | <16 | <16 | <16 | <16 |
| A/swan/Potsdam/62/81 (H7N7) | <16 | <16 | 512 | <16 | <16 | <16 | 512 |
| A/teal/Italy/1398/06 (H10N7) | <16 | <16 | 256 | <16 | <16 | <16 | <16 |
All sera were produced in SPF chickens, except sera directed to HA and NA of A/swine/Belzig/2/01 (H1N1), which were from SPF pigs. The animals were immunized twice (i.m) with VRPs expressing HA or NA of either A/chicken/Yamaguchi/7/04 (H5N1), A/swine/Belzig/2/01 (H1N1), A/swan/Potsdam/62/81 (H7N7), A/duck/Italy/1447/05 (H1N1), or A/turkey/Italy/3675/99 (H7N1). Immune sera were treated with V. cholerae sialidase and tested for hemagglutination inhibition activity using chicken erythrocytes and the virus strains listed. HI titers obtained with homotypic viruses are underlined. HI titers ≥16 are depicted in bold.
NA antibodies interfere with infection of MDCK cells.
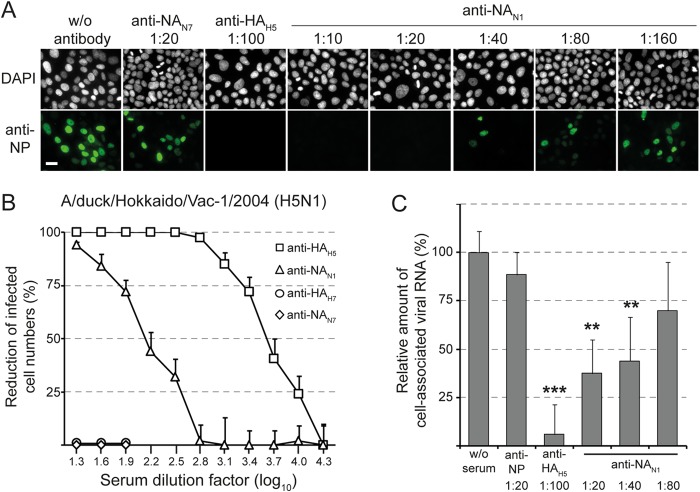
The observation that NA-specific antibodies exhibited HI activity suggested that these antibodies may also interfere with the binding of influenza virus to cell surface sialoglycoconjugates. To address this hypothesis, MDCK cells were inoculated for 1 h at 4°C with A/chicken/Yamaguchi/7/04 (H5N1) in the presence or absence of immune sera raised against either NA or HA of the virus. Each of the two immune sera showed an HI titer of 512 against A/chicken/Yamaguchi/7/04 (H5N1) (Table 2). After inoculation with the virus, the cells were washed, incubated for 5 h at 37°C in the absence of immune serum, and analyzed for the presence of NP antigen by indirect immunofluorescence (Fig. 2A). When virus binding was performed in the absence of any immune serum, NP antigen was subsequently detected in the cell nucleus, indicating that the early phase of virus entry, including adsorption, internalization, and fusion, had taken place. In contrast, virus entry was efficiently blocked in the presence of anti-NAN1 serum (1:20). This inhibitory effect declined with decreasing anti-NAN1 concentrations. Anti-NAN1 serum also inhibited infection of MDCK cells by the LPAIV A/duck/Hokkaido/Vac-01/04 (H5N1), whereas anti-NAN7 and anti-HAH7 immune sera had no effect (Fig. 2B). In addition, anti-NAN1 and anti-NAN7 sera showed inhibitory activity against A/duck/Italy/1447/05 (H1N1) and A/swan/Potsdam/62/81 (H7N7), respectively (data not shown).
FIG 2.
Anti-NA antibodies interfere with influenza virus infection. (A) A/chicken/Yamaguchi/7/04 (H5N1) was incubated with MDCK cell monolayers (0.5 TCID50/cell) for 1 h at 4°C in the presence or absence of the indicated immune sera. The cells were washed, incubated for 5 h at 37°C, fixed with formalin, and permeabilized with Triton X-100. The influenza virus NP antigen was detected by indirect immunofluorescence. The nuclei were stained with DAPI. The scale bar indicates 20 μm. (B) MDCK cells were infected with A/duck/Hokkaido/Vac-1/04 (H5N1) (MOI = 0.0025) in the presence of immune serum directed to HAH5 or NAN1 of A/chicken/Yamaguchi/7/04 (H5N1). Immune sera directed to HAH7 of A/turkey/Italy/3675/99 (H7N1) and NAN7 of A/swan/Potsdam/62/81 (H7N7) were used as controls. The cells were washed, maintained for 24 h in the absence of antibodies, and fixed. Infected cells were visualized by immunostaining with a monoclonal antibody directed to the influenza virus NP antigen. The number of infected cells was calculated as a percentage of the cells infected in the presence of nonimmune serum. The plaque reduction test was run in quadruplicate. Mean values and standard deviations are shown. (C) A/chicken/Yamaguchi/7/04 (H5N1) was incubated with MDCK cell monolayers (0.0025 TCID50/cell) for 1 h at 4°C in the presence or absence of the indicated immune sera. The cells were washed, total RNA was extracted, and viral RNA segment 7 was detected by quantitative RT-PCR. Virus bound to cells in the absence of immune serum was used as a reference (100% virus binding). The mean values and standard deviations of three independent experiments, each run in duplicate, are shown. The asterisks denote significantly different threshold cycle (CT) values compared to virus incubation in the absence of serum (**, P < 0.005; ***, P < 0.0005).
To investigate whether virus entry was affected due to inhibition of virus binding to cell surface receptors or whether the antibodies interfered with subsequent steps in virus entry, A/chicken/Yamaguchi/7/04 (H5N1) virus was adsorbed to MDCK cells at 4°C in the absence or presence of immune sera. The cells were subsequently washed, and virus still associated with the cells was detected by quantitative RT-PCR. Immune serum directed to the NAN1 antigen of A/chicken/Yamaguchi/7/04 (H5N1) significantly reduced virus binding when used at dilutions of 1:20 or 1:40, but not at higher dilutions (1:80 and higher) (Fig. 2C and data not shown). In contrast, immune serum raised against the HAH5 antigen of A/chicken/Yamaguchi/7/04 (H5N1) still abolished virus attachment if used at a dilution of 1:100 (Fig. 2C).
NA antibodies inhibit sialidase activity of influenza virions.
The abilities of NA-specific antibodies to inhibit NA sialidase activity were tested using immobilized glycophorin A, which is the major sialoglycoprotein of erythrocytes containing multiple O-linked oligosaccharides. Desialylation of this mucin-type glycoprotein after incubation with whole influenza virions was detected with biotinylated PNA as a probe. This lectin binds specifically to the carbohydrate sequence Gal-β(1-3)-GalNAc (T antigen), which is exposed in O-linked oligosaccharides following the removal of terminal sialic acid residues (2, 58, 59). The sialidase activity of A/swan/Potsdam/62/81 (H7N7) was efficiently inhibited by submicromolar concentrations of oseltamivir carboxylate, a synthetic neuraminic acid analog (Fig. 3A). Likewise, immune serum directed to NAN7, but not preimmune serum, inhibited the NAN7 sialidase with high potency. Moreover, anti-NAN1 serum revealed low inhibitory activity if used at high concentration. Similar experiments with A/duck/Hokkaido/Vac-1/04 (H5N1) demonstrated that sialidase activity was efficiently inhibited by anti-NAN1 immune serum, while anti-NAN7 showed some cross-inhibitory activity (Fig. 3B). Surprisingly, the sialidases of both viruses were affected to some extent by anti-HA immune sera if they matched the HA subtype of the virus used (Fig. 3A and B). Taken together, these results demonstrate that immune sera directed to NA are powerful inhibitors of influenza virus sialidase.
FIG 3.
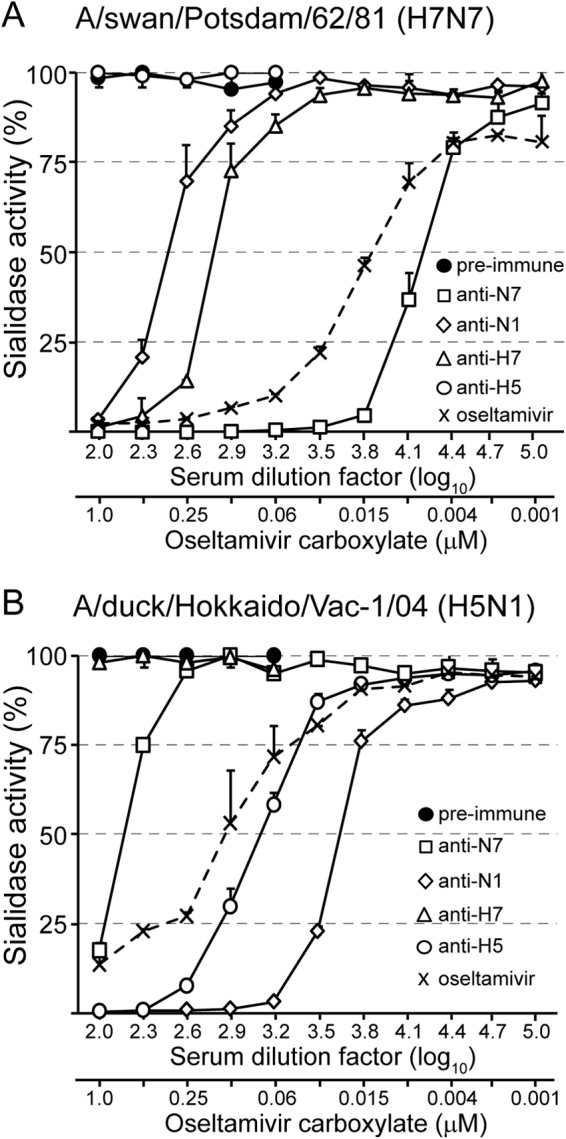
Anti-NA antibodies inhibit the sialidase of influenza virions. (A) A/swan/Potsdam/62/81 (H7N7) was mixed with serially diluted preimmune serum or immune serum raised against either NAN7 of A/swan/Potsdam/62/81 (H7N7), NAN1 of A/chicken/Yamaguchi/7/04 (H5N1), HAH7 of A/turkey/Italy/3675/99 (H7N1), or HAH5 of A/chicken/Yamaguchi/7/04 (H5N1). In parallel, virus was mixed with oseltamivir carboxylate at the indicated concentrations. The mixtures were incubated for 18 h at 37°C with immobilized glycophorin A. Desialylation of glycophorin A was detected by subsequent incubations with biotin-labeled peanut agglutinin, streptavidin-peroxidase conjugate, and TMB peroxidase substrate. (B) A/duck/Hokkaido/Vac-1/04 (H5N1) was mixed with the indicated immune sera or oseltamivir carboxylate prior to incubation with immobilized glycophorin A. Experiments were performed twice, each time with serum quadruplicates. The mean values and standard deviations of a representative experiment are shown.
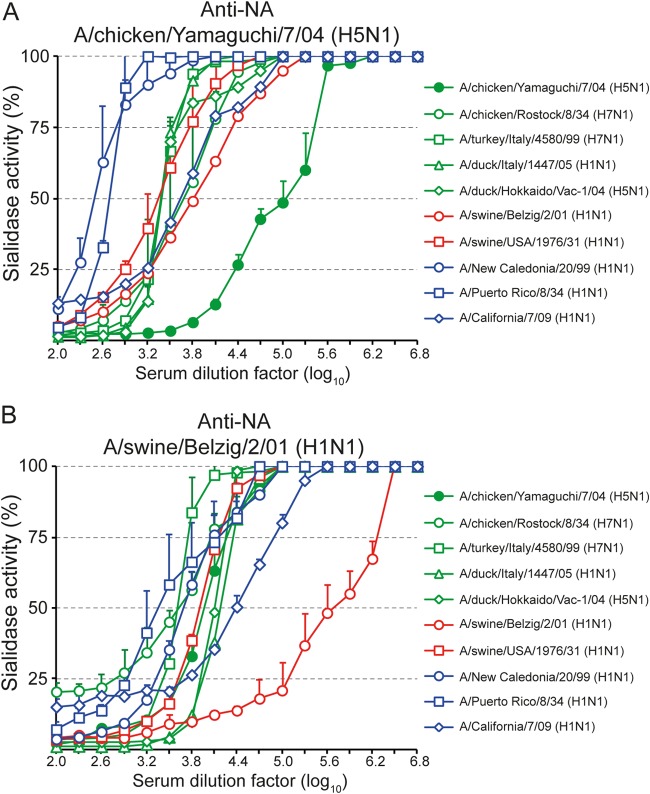
The monospecific immune sera were also analyzed with respect to their inhibitory activities against the sialidases of different human, porcine, and avian influenza viruses. Immune serum raised against the NA of A/chicken/Yamaguchi/7/04 (H5N1) reacted with all N1 viruses tested, including H5N1, H7N1, and H1N1 viruses (Fig. 4A). However, the serum dilutions leading to 50% inhibition of sialidase activity (NI50) differed greatly depending on the virus used. While the homotypic NA was inhibited most efficiently (NI50, about 105), the sialidases of the human influenza viruses A/Puerto Rico/8/33 (H1N1) and A/New Caledonia/20/99 (H1N1) were inhibited to a much lesser extent (NI50s, 500 and 300, respectively). The NI50 titers for all other viruses, including the 1934 isolate A/chicken/Rostock/8/34 (H7N1), were in the middle range, with NI50 values between 2,400 and 6,400. Similar sialidase inhibition tests were also performed with immune serum raised against the NA of A/swine/Belzig/2/01 (H1N1) (Fig. 4B). As expected, this immune serum inhibited the homotypic sialidase most efficiently (NI50, 4 × 105), while the sialidases of A/chicken/Yamaguchi/8/04 (H5N1), A/duck/Italy/1447/05 (H1N1), A/swine/USA/1976/31 (H1N1), and A/New Caledonia/20/99 (H1N1) required higher concentrations of immune serum to cause 50% inhibition of sialidase activity. The sialidases of A/Puerto Rico/8/33 (H1N1) and A/chicken/Rostock/8/34 (H7N1) were inhibited with the least efficacy (NI50s, 2,400 and 4,000, respectively).
FIG 4.
Anti-NA antibodies cross-react with NAN1s of various influenza viruses. Immune sera raised against the NA of either A/chicken/Yamaguchi/7/04 (H5N1) (A) or A/swine/Belzig/2/01 (H1N1) (B) were tested for the ability to inhibit the sialidases of the indicated viruses using immobilized glycophorin A as the substrate. The mean values and standard deviations of quadruplicate measurements are shown. The test was performed at least two times, and a representative experiment is depicted.
NA antibodies inhibit virus spread in vitro.
A major function of NA is to mediate the release of progeny virus at the plasma membranes of infected cells by removing sialic acid residues from viral and cellular glycoproteins (9). Consequently, synthetic neuraminidase inhibitors have been shown to suppress influenza virus dissemination (60). To test whether NA antibodies would also interfere with virus spread, MDCK cells were infected with A/chicken/Yamaguchi/7/04 (H5N1) at an MOI of 0.0025 TCID50/cell. This HPAIV was chosen because it does not depend on exogenous trypsin for propagation and therefore can be grown in the presence of serum. After inoculation with the virus for 1 h, the cells were washed and further cultured in the presence of specific immune sera. At 24 h postinfection, the cells were fixed and stained with a monoclonal antibody specifically recognizing the influenza virus NP antigen (Fig. 5A). The homotypic anti-NAN1 and anti-HAH5 immune sera reduced virus spread in a dose-dependent manner, with anti-HAH5 serum being more efficient than anti-NAN1 serum. The synthetic neuraminidase inhibitor oseltamivir carboxylate also reduced the infected area of the cell monolayer if used in the submicromolar range.
FIG 5.
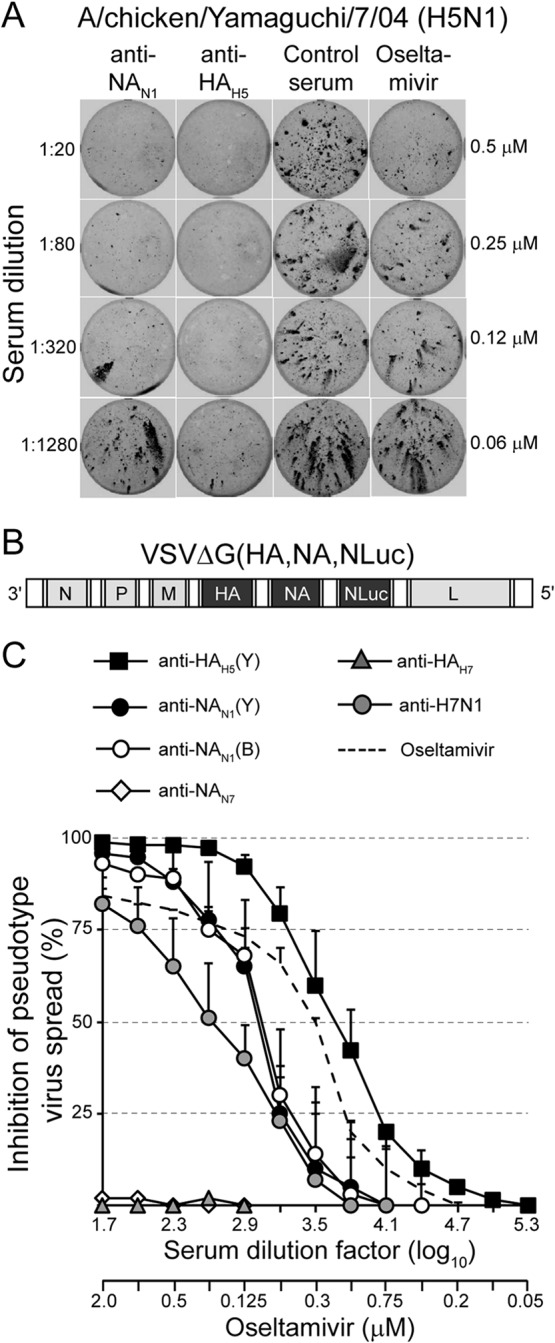
Anti-NA antibodies inhibit virus spread in vitro. (A) MDCK monolayers were infected with the highly pathogenic avian influenza virus A/chicken/Yamaguchi/7/04 (H5N1) (MOI = 0.0025). The cells were washed and maintained for 18 h at 37°C in the presence of either anti-NAN1, anti-HAH5, nonimmune serum, or oseltamivir carboxylate. The serum dilutions and oseltamivir carboxylate concentrations used are indicated on the left and right sides of the graph, respectively. The cells were fixed, permeabilized, and stained using a monoclonal antibody directed to influenza virus NP antigen and a peroxidase-labeled anti-mouse IgG serum. (B) Genome map of recombinant VSV pseudotype vector expressing HA and NA antigens of A/chicken/Yamaguchi/7/04 (H5N1) and a secreted luciferase (NLuc) from O. gracilirostris. The vector lacks the VSV glycoprotein G gene. (C) Inhibition of pseudotype virus spread. MDCK cells were infected with the propagation-competent HA/NA pseudotype virus (MOI = 0.005) and incubated in the presence of the indicated concentrations of either immune sera or oseltamivir carboxylate. At 48 h p.i., the luciferase activity released into the cell culture supernatant was determined. The luciferase expression relative to that of cells that were incubated without immune serum is shown. Mean values and standard deviations of three infection experiments are shown.
In order to allow quantification of anti-NA-mediated inhibition of virus spread, we took advantage of a propagation-competent pseudotype virus expressing the HA and NA glycoproteins of highly pathogenic A/chicken/Yamaguchi/7/04 (H5N1) (52). The virus was further modified to express a secreted luciferase reporter protein (61) instead of the previously used green fluorescent protein (GFP) reporter (Fig. 5B). MDCK cells were infected with VSVΔG(HA,NA,NLuc) at 0.0025 PFU/cell and subsequently incubated for 48 h in the presence of serially diluted immune sera. The NLuc reporter protein secreted by infected cells into the cell culture supernatant was detected with a luminescent luciferase substrate. As expected, immune serum raised against the HA antigen of A/chicken/Yamaguchi/7/04 (H5N1) inhibited the spread of pseudotype virus efficiently, while anti-HAH7 serum did not show any inhibitory activity (Fig. 5C). Immune sera raised against NA of A/chicken/Yamaguchi/7/04 (H5N1) and A/swine/Belzig/2/01 (H1N1) also inhibited virus spread, but not as efficiently as anti-HAH5 serum. Furthermore, reconvalescent sera from chickens experimentally infected with low-pathogenic A/turkey/Italy/3575/99 (H7N1) demonstrated some inhibitory activity. These results indicate that NA-specific antibodies are able to inhibit influenza virus dissemination in cell culture in a concentration-dependent manner. Interestingly, the antibody concentrations required to achieve 50% reduction of virus spread were higher than those required for 50% inhibition of sialidase activity (compare Fig. 4).
Vaccination with NA antigen affects virus shedding in vivo.
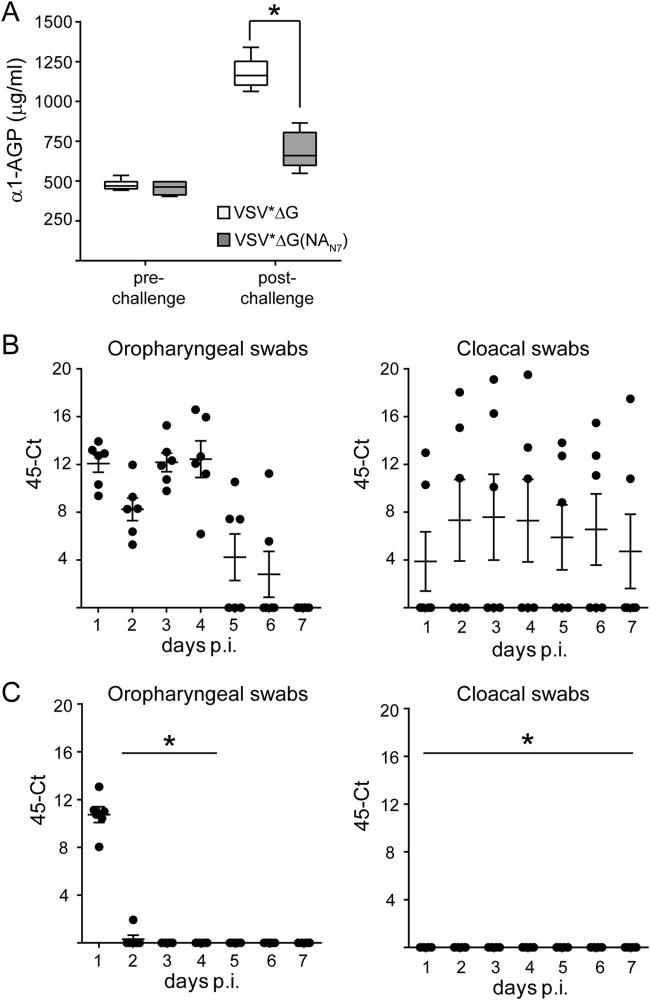
LPAIVs normally do not cause disease in chickens. However, LPAIV may be shed from the respiratory and/or gastrointestinal tract of the host, resulting in undetected virus dissemination. To elucidate whether vaccination with recombinant VRPs would reduce LPAIV shedding, chickens were immunized twice via the intramuscular route with VRPs encoding the NA antigen of A/swan/Potsdam/62/81 (H7N7). All six animals in the group produced NA-specific antibodies that inhibited hemagglutination by A/swan/Potsdam/62/81 (H7N7) (mean HI titer, 554). In contrast, VSV*ΔG-immunized control animals did not develop antibodies with HI activity (data not shown). The vaccinated chickens were infected via the intratracheal route with 107 TCID50 of LPAIV A/swan/Potsdam/62/81 (H7N7). The virus was previously found to be efficiently shed from infected chickens (unpublished results). The infection with A/swan/Potsdam/62/81 (H7N7) did not cause any obvious clinical symptoms in chickens but led to increased levels of the acute-phase serum protein α1-AGP (Fig. 6A, VSV*ΔG group). In contrast, chickens immunized with VSV*ΔG(NAN7) showed significantly lower levels of serum α1-AGP.
FIG 6.
Vaccination with NA-recombinant VRP reduces virus shedding from infected chickens and inflammation. SPF chickens (n = 6/group) were immunized (i.m.) twice with either VSV*ΔG or VSV*ΔG(NAN7) using 108 FFU per dose. Three weeks after the second immunization, the chickens were infected via the intratracheal route with 107 TCID50 of A/swan/Potsdam/62/81 (H7N7). (A) Detection of α1-AGP 2 days after infection. Box-and-whisker plots indicate maximum, minimum, and median values. The asterisk denotes significantly different α1-AGP levels between the two animal groups. (B and C) Detection of challenge virus in oropharyngeal- and cloacal-swab samples collected from chickens vaccinated with VSV*ΔG (B) or VSV*ΔG(NAN7) (C). Swab samples were taken from the animals at daily intervals for a period of 7 days. RNA was extracted from the samples and analyzed for the presence of viral RNA segment 7 by quantitative RT-PCR. Maximum 45 threshold cycles minus the threshold cycles detected are shown (45-Ct). Mean values and standard deviations are indicated. The asterisk denotes significantly different viral RNA loads in swabs taken from VSV*ΔG(NAN7)-vaccinated animals compared to control animals.
In order to monitor virus shedding from infected chickens, oropharyngeal and cloacal swabs were sampled every day and analyzed for the presence of viral RNA by quantitative RT-PCR. All animals in the control group shed virus from the oropharyngeal site for at least 4 days, and thereafter it dropped (Fig. 6B). Three control animals also shed virus from the cloacal site, which lasted for at least 7 days. In contrast, A/swan/Potsdam/62/81 (H7N7) was shed from the respiratory tracts of VSV*ΔG(NAN7)-vaccinated chickens only on day 1 postinfection and not later on. Cloacal shedding was even completely eliminated in this group. These findings indicate that immunization with recombinant NA antigen significantly reduced virus shedding from LPAIV-infected chickens.
DISCUSSION
In the present study, it was shown that infection of cells with recombinant RNA VRPs resulted in expression of enzymatically active NA antigen. Furthermore, intramuscular immunization of animals with these VRPs induced high titers of NA-specific serum antibodies with inhibitory activity against influenza viruses. The expression of NA in the native conformation is believed to be crucial for the induction of anti-NA antibodies with inhibitory activity (62). As only the native tetrameric NA is enzymatically active (63, 64), sialidase activity is regarded as an indicator of NA immunogenicity (62). In contrast to vaccination with VRPs, intramuscular immunization of chickens with inactivated influenza virus resulted in significantly lower titers of anti-NA antibodies, although relatively high anti-HA titers were induced. This is likely due to the lower abundance of NA than of the HA antigen in the viral envelope (26) and the competition between the two antigens (28, 31). In addition, inactivation of the virus and formulation with adjuvant may have had a negative effect on NA stability and immunogenicity (26). The idea that the amount of correctly folded NA antigen might be critical for the induction of antibodies is also supported by the observation that intramuscular immunization with live influenza virus resulted in levels of anti-NA titers similar to those from intramuscular immunization with VSV*ΔG(NA). In conclusion, propagation-incompetent VRPs represent a safe and powerful vector vaccine for the induction of antibodies to NA.
NA-specific serum antibodies were able to inhibit influenza viruses in at least two different ways. First, NA-specific immune sera inhibited sialidase activity of influenza virions in a concentration-dependent and subtype-specific manner. Antibody-mediated inhibition of sialidase was observed with glycophorin A as the substrate but was not detected with the small synthetic substrate MU-NANA, suggesting that antibodies are able to hinder the access of large but not small substrates to the catalytic domain in the globular head of NA (65). Monospecific immune sera directed to NA of either A/chicken/Yamaguchi/7/04 (H5N1) or A/swine/Belzig/2/01 (H1N1) exhibited very broad reactivity and inhibited all N1 viruses tested, although with differing efficacies depending on the virus strain used. For example, immune serum directed to NA of A/chicken/Yamaguchi/7/04 (H5N1) inhibited the sialidase of the virus very efficiently (50% inhibition at a serum dilution of 1:105). In contrast, a 250-fold-higher antibody concentration was required to cause 50% inhibition of the sialidases of A/Puerto Rico/8/33 (H1N1) and A/New Caledonia/20/99. These two neuraminidases were the ones most distantly related to the NA of A/chicken/Yamaguchi/7/04 (H5N1), as indicated by phylogenetic analysis (Fig. 7). The cross-reactivity of immune sera directed to avian and porcine NAN1 with different avian, porcine, and human influenza viruses led us to speculate that antibodies directed to human influenza virus NAN1 would also react with other human influenza viruses of the N1 lineage. However, this hypothesis still needs to be experimentally proven.
FIG 7.
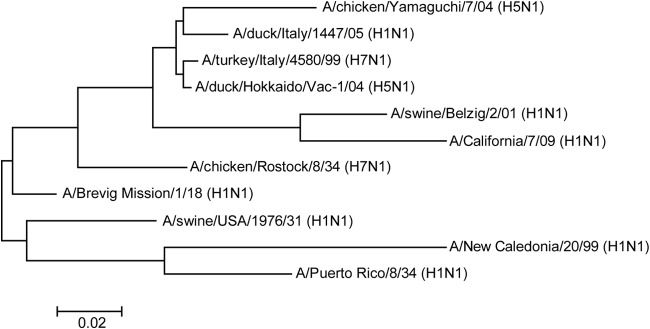
Phylogenetic analysis of influenza virus N1 neuraminidases by ClustalW alignment (78) and subsequent neighbor-joining tree construction by MEGA 4 software (79). The GenBank accession numbers for the NA protein sequences used are as follows: BAD89307 for A/chicken/Yamaguchi/7/04 (H5N1), ACS93996 for A/duck/Italy/1447/05 (H1N1), ACZ45212 for A/turkey/Italy/4580/99 (H7N1), BAE94701 for A/duck/Hokkaido/Vac-1/04 (H5N1), ACQ63272 for A/California/07/09 (H1N1), CAA36475 for A/chicken/Rostock/8/34 (H7N1), AAF77036 for A/Brevig Mission/1/18 (H1N1), ACV52104 for A/swine/USA/1976/31 (H1N1), ACF41881 for A/New Caledonia/20/99 (H1N1), and NP_040981 for A/Puerto Rico/8/34 (H1N1). The Global Initiative on Sharing All Influenza Data (GISAID) accession number for the NA sequence of A/swine/Belzig/2/01 (H1N1) is EPI302494.The scale bar indicates the number of nucleotide substitutions per site.
Sialidase inhibition experiments with heterosubtypic anti-NA sera showed that anti-NAN7 serum had some inhibitory activity against N1 sialidase and anti-NAN1 serum had low inhibitory activity against N7 sialidase, despite the classification of these neuraminidases into different phylogenetic groups (8). This heterosubtype inhibitory activity might be mediated by antibodies that bind to a highly conserved epitope found in all 9 classical NA subtypes (66). However, future studies must show which epitopes are actually recognized by cross-inhibiting antibodies. It will also be interesting to explore whether conformation-dependent or linear NA epitopes are recognized by these antibodies.
A surprising finding for us was that monospecific anti-HA immune sera were able to inhibit the sialidase activity of influenza virions, albeit not as efficiently as NA antibodies. This phenomenon has been previously noted in the WHO Manual on Animal Influenza Diagnosis and Surveillance (67). It can be speculated that anti-HA antibodies that are bound to the virion sterically interfere with the interaction between NA and its substrate.
A second intriguing property of monospecific anti-NA sera was their ability to inhibit hemagglutination by AIV as efficiently as HA antibodies. The NA-specific antibodies reacted in an NA subtype-specific manner independently of the HA subtype given. This phenomenon has been noted previously (57) and is possibly due to antibodies that sterically interfere with the HA-mediated binding of influenza viruses to erythrocytes. In contrast to anti-avian HAH1 serum, which did not react with porcine and human H1 viruses, immune serum directed to avian NAN1 showed HI activity against avian, as well as porcine and human, N1 viruses. Conversely, avian NAN1 was also recognized by immune serum directed to porcine NAN1. These findings suggest that there are common “neutralizing” epitopes present in the NAN1s of different influenza A virus lineages.
Antiserum directed to NAN1 of A/swine/Belzig/2/01 (H1N1) showed stronger HI activity against A/duck/Italy/1447/05 (H1N1) than against the homologous porcine virus. The reason for this phenomenon might be the presence of a second sialic acid-binding site in the globular head of avian NA that has hemadsorption activity but is enzymatically silent (68, 69). Antibodies directed to this domain may be particularly active in terms of hemagglutination inhibition. In contrast to NAs of avian influenza viruses, NAs of human influenza viruses normally lack obvious hemadsorption activity (69). However, a certain level of sialic acid-binding activity may still be associated with the NAs of mammalian influenza A viruses. Recently, it was found that a single mutation (D151G) in the N2 neuraminidase of human H3N2 viruses enabled the glycoprotein to avidly bind to sialic acid-containing receptors (70). Likewise, N1 neuraminidase containing the single amino acid substitution G147R was reported to completely coopt the receptor-binding function normally executed by HA (71).
The HI activity of anti-NA antibodies led us to hypothesize that these antibodies would also be able to interfere with the binding of influenza viruses to cellular sialoglycoproteins. This hypothesis was supported by the finding that anti-NA antibodies significantly affected infection of MDCK cells. However, anti-NA antibodies were less efficient than anti-HA antibodies in preventing infection, suggesting that the binding of influenza viruses to epithelial cells is less sensitive to inhibition by anti-NA serum than agglutination of erythrocytes. When the amount of cell-associated influenza virus was quantified by quantitative RT-PCR, it turned out that anti-NA immune serum reduced virus binding to the cell surface by about 60%. However, immunofluorescence analysis of internalized NP antigen suggested that virus entry was completely inhibited at the same antibody concentration. It is therefore possible that antibodies to NA interfere not only with virus binding to the cell surface, but also with subsequent steps of virus entry, such as membrane fusion or uncoating. Interestingly, sialidase activity of influenza A virus in the endocytic pathway has been shown to enhance viral replication (72). A role of sialidase in virus entry has also been suggested by the finding that treatment of human influenza A viruses with a synthetic neuraminidase inhibitor reduced infection of well-differentiated human respiratory epithelial cells (10). The exact mechanism behind this phenomenon is not known, but it was hypothesized that NA sialidase activity may be important for the inactivation of mucins and other secreted sialoglycoproteins that compete with membrane-bound glycoproteins for binding of influenza virus (73). Sialidase activity may also help viruses to surf along the cell surface until appropriate receptors are found that guarantee efficient virus uptake by receptor-mediated endocytosis (74).
Sialidase activity is believed to be important for influenza A virus release and spread (9). Interestingly, we observed that the effective antibody concentration required to inhibit virus spread was higher than the antibody concentration needed to inhibit sialidase activity. For example, the antibody titer leading to 50% inhibition of sialidase activity of A/chicken/Yamaguchi/7/04 (H5N1) was about 105, whereas the titer for 50% inhibition of pseudotype virus spread was about 103. This difference may reflect the different experimental settings. In the sialidase inhibition assay, the amount of sialidase was fixed, whereas the virus spread assay made use of infected cells continuously supplying new NA. On the other hand, it is possible that minute amounts of sialidase activity are already sufficient to guarantee efficient virus spread. In particular, viruses with a low receptor-binding affinity may rely on sialidase to a lesser extent (75). The ability of anti-NA antibodies to inhibit virus entry has certainly contributed to the inhibition of virus spread, but it is not known what the relative level of inhibition actually was.
Immunization of chickens with NA recombinant VRPs affected shedding of LPAIV significantly, suggesting that anti-NA antibodies may also inhibit virus dissemination in vivo. The animals were immunized via the intramuscular route, which is also the route of choice for application of inactivated virus vaccines. Parenteral vaccination normally results in induction of serum antibodies and not in local mucosal immunity. Nevertheless, there was limited shedding of H5N1 challenge virus from the respiratory and intestinal tracts when chickens were immunized with an inactivated influenza virus vaccine (76). It is believed that serum IgY can be transported across epithelia lining the respiratory and gastrointestinal tracts by transcytosis (77). Most likely, the higher serum antibody levels are, the more antibodies may reach the mucosa in this way. The viral RNA detected in oropharyngeal swabs on day 1 after infection is believed to represent input virus. Virus shedding from the cloacal site was not detected, suggesting that anti-NA antibodies efficiently blocked virus dissemination from the respiratory to the gastrointestinal tract and/or efficiently suppressed virus propagation in the gastrointestinal tract.
Conventional inactivated influenza virus vaccines primarily rely on the immune response directed to HA but do not sufficiently induce antibodies to NA. The NA antigen has been largely neglected in past vaccine development because it is underrepresented in virus particles and is more labile than the HA antigen (23). In this study, we demonstrated that immunization with recombinant VRPs driving expression of native NA antigen resulted in high titers of NA-specific antibodies showing potent inhibitory activity against influenza A viruses. Moreover, these antibodies showed inhibitory activity against highly diverse influenza A viruses of the same NA subtype irrespective of the HA subtype. Thus, a broader immune response to influenza viruses might be achieved if the NA antigen could be successfully incorporated into a new generation of vaccines.
ACKNOWLEDGMENTS
We thank Daniel Brechbühl for helping with the animal trials and Markus Mader for running the quantitative RT-PCR. We are grateful to Timm Harder and Jürgen Stech (FLI, Riems), William Dundon (Legnaro, Italy), Yoshihiro Sakoda (Sapporo, Japan), Gabriele Neumann (Madison, WI, USA), and Veronika von Messling (Paul-Ehrlich-Institut, Langen, Germany) for providing influenza viruses and plasmids.
This study was funded by the Swiss National Science Foundation (project number 310030_129669), the Federal Food Safety and Veterinary Office (project number 1.10.11), and the European Union FP7 project PREDEMICS (grant agreement 278433).
REFERENCES
- 1.Krammer F, Palese P. 2013. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3:521–530. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lotan R, Skutelsky E, Danon D, Sharon N. 1975. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea). J Biol Chem 250:8518–8523. [PubMed] [Google Scholar]
- 3.Xu X, Cox NJ, Bender CA, Regnery HL, Shaw MW. 1996. Genetic variation in neuraminidase genes of influenza A (H3N2) viruses. Virology 224:175–183. doi: 10.1006/viro.1996.0519. [DOI] [PubMed] [Google Scholar]
- 4.Luther P, Bergmann KC, Oxford JS. 1984. An investigation of antigenic drift of neuraminidases of influenza A (H1N1) viruses. J Hyg 92:223–229. doi: 10.1017/S002217240006424X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilbourne ED, Johansson BE, Grajower B. 1990. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc Natl Acad Sci U S A 87:786–790. doi: 10.1073/pnas.87.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, Eichelberger MC. 2011. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci U S A 108:20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Air GM. 2012. Influenza neuraminidase. Influenza Other Respir Viruses 6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palese P, Tobita K, Ueda M, Compans RW. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 10.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol 78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson BE, Cox MM. 2011. Influenza viral neuraminidase: the forgotten antigen. Expert Rev Vaccines 10:1683–1695. doi: 10.1586/erv.11.130. [DOI] [PubMed] [Google Scholar]
- 12.Marcelin G, Sandbulte MR, Webby RJ. 2012. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev Med Virol 22:267–279. doi: 10.1002/rmv.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulman JL, Khakpour M, Kilbourne ED. 1968. Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. J Virol 2:778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilbourne ED, Laver WG, Schulman JL, Webster RG. 1968. Antiviral activity of antiserum specific for an influenza virus neuraminidase. J Virol 2:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. 1974. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J Infect Dis 129:411–420. doi: 10.1093/infdis/129.4.411. [DOI] [PubMed] [Google Scholar]
- 16.Rott R, Becht H, Orlich M. 1974. The significance of influenza virus neuraminidase in immunity. J Gen Virol 22:35–41. doi: 10.1099/0022-1317-22-1-35. [DOI] [PubMed] [Google Scholar]
- 17.Johansson BE, Bucher DJ, Kilbourne ED. 1989. Purified influenza virus hemagglutinin and neuraminidase are equivalent in stimulation of antibody response but induce contrasting types of immunity to infection. J Virol 63:1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson BE, Grajower B, Kilbourne ED. 1993. Infection-permissive immunization with influenza virus neuraminidase prevents weight loss in infected mice. Vaccine 11:1037–1039. doi: 10.1016/0264-410X(93)90130-P. [DOI] [PubMed] [Google Scholar]
- 19.Sandbulte MR, Jimenez GS, Boon AC, Smith LR, Treanor JJ, Webby RJ. 2007. Cross-reactive neuraminidase antibodies afford partial protection against H5N1 in mice and are present in unexposed humans. PLoS Med 4:e59. doi: 10.1371/journal.pmed.0040059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster RG, Reay PA, Laver WG. 1988. Protection against lethal influenza with neuraminidase. Virology 164:230–237. doi: 10.1016/0042-6822(88)90640-X. [DOI] [PubMed] [Google Scholar]
- 21.Eichelberger MC, Wan H. 2015. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol 386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- 22.Rockman S, Brown LE, Barr IG, Gilbertson B, Lowther S, Kachurin A, Kachurina O, Klippel J, Bodle J, Pearse M, Middleton D. 2013. Neuraminidase-inhibiting antibody is a correlate of cross-protection against lethal H5N1 influenza virus in ferrets immunized with seasonal influenza vaccine. J Virol 87:3053–3061. doi: 10.1128/JVI.02434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wohlbold TJ, Krammer F. 2014. In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses 6:2465–2494. doi: 10.3390/v6062465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams MS. 1993. Single-radial-immunodiffusion as an in vitro potency assay for human inactivated viral vaccines. Vet Microbiol 37:253–262. doi: 10.1016/0378-1135(93)90027-5. [DOI] [PubMed] [Google Scholar]
- 25.Hardy S, Eichelberger M, Griffiths E, Weir JP, Wood D, Alfonso C. 2011. Confronting the next pandemic—workshop on lessons learned from potency testing of pandemic (H1N1) 2009 influenza vaccines and considerations for future potency tests, Ottawa, Canada, July 27–29, 2010. Influenza Other Respir Viruses 5:438–442. doi: 10.1111/j.1750-2659.2011.00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murti KG, Webster RG. 1986. Distribution of hemagglutinin and neuraminidase on influenza virions as revealed by immunoelectron microscopy. Virology 149:36–43. doi: 10.1016/0042-6822(86)90084-X. [DOI] [PubMed] [Google Scholar]
- 27.Kendal AP, Bozeman FM, Ennis FA. 1980. Further studies of the neuraminidase content of inactivated influenza vaccines and the neuraminidase antibody responses after vaccination of immunologically primed and unprimed populations. Infect Immun 29:966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilbourne ED, Cerini CP, Khan MW, Mitchell JW Jr, Ogra PL. 1987. Immunologic response to the influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin. I. Studies in human vaccinees. J Immunol 138:3010–3013. [PubMed] [Google Scholar]
- 29.Johansson BE, Moran TM, Bona CA, Kilbourne ED. 1987. Immunologic response to influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin. III. Reduced generation of neuraminidase-specific helper T cells in hemagglutinin-primed mice. J Immunol 139:2015–2019. [PubMed] [Google Scholar]
- 30.Johansson BE, Moran TM, Bona CA, Popple SW, Kilbourne ED. 1987. Immunologic response to influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin. II. Sequential infection of mice simulates human experience. J Immunol 139:2010–2014. [PubMed] [Google Scholar]
- 31.Johansson BE, Kilbourne ED. 1993. Dissociation of influenza virus hemagglutinin and neuraminidase eliminates their intravirionic antigenic competition. J Virol 67:5721–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Kadowaki S, Hagiwara Y, Yoshikawa T, Matsuo K, Kurata T, Tamura S. 2000. Cross-protection against a lethal influenza virus infection by DNA vaccine to neuraminidase. Vaccine 18:3214–3222. doi: 10.1016/S0264-410X(00)00149-3. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z, Yoshikawa T, Kadowaki S, Hagiwara Y, Matsuo K, Asanuma H, Aizawa C, Kurata T, Tamura S. 1999. Protection and antibody responses in different strains of mouse immunized with plasmid DNAs encoding influenza virus haemagglutinin, neuraminidase and nucleoprotein. J Gen Virol 80:2559–2564. [DOI] [PubMed] [Google Scholar]
- 34.Qiu M, Fang F, Chen Y, Wang H, Chen Q, Chang H, Wang F, Wang H, Zhang R, Chen Z. 2006. Protection against avian influenza H9N2 virus challenge by immunization with hemagglutinin- or neuraminidase-expressing DNA in BALB/c mice. Biochem Biophys Res Commun 343:1124–1131. doi: 10.1016/j.bbrc.2006.03.088. [DOI] [PubMed] [Google Scholar]
- 35.DiNapoli JM, Nayak B, Yang L, Finneyfrock BW, Cook A, Andersen H, Torres-Velez F, Murphy BR, Samal SK, Collins PL, Bukreyev A. 2010. Newcastle disease virus-vectored vaccines expressing the hemagglutinin or neuraminidase protein of H5N1 highly pathogenic avian influenza virus protect against virus challenge in monkeys. J Virol 84:1489–1503. doi: 10.1128/JVI.01946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Easterbrook JD, Schwartzman LM, Gao J, Kash JC, Morens DM, Couzens L, Wan H, Eichelberger MC, Taubenberger JK. 2012. Protection against a lethal H5N1 influenza challenge by intranasal immunization with virus-like particles containing 2009 pandemic H1N1 neuraminidase in mice. Virology 432:39–44. doi: 10.1016/j.virol.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CY, Yeh YC, Chan JT, Yang YC, Yang JR, Liu MT, Wu HS, Hsiao PW. 2012. A VLP vaccine induces broad-spectrum cross-protective antibody immunity against H5N1 and H1N1 subtypes of influenza A virus. PLoS One 7:e42363. doi: 10.1371/journal.pone.0042363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deroo T, Jou WM, Fiers W. 1996. Recombinant neuraminidase vaccine protects against lethal influenza. Vaccine 14:561–569. doi: 10.1016/0264-410X(95)00157-V. [DOI] [PubMed] [Google Scholar]
- 39.Bosch BJ, Bodewes R, de Vries RP, Kreijtz JH, Bartelink W, van Amerongen G, Rimmelzwaan GF, de Haan CA, Osterhaus AD, Rottier PJ. 2010. Recombinant soluble, multimeric HA and NA exhibit distinctive types of protection against pandemic swine-origin 2009 A(H1N1) influenza virus infection in ferrets. J Virol 84:10366–10374. doi: 10.1128/JVI.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavlova SP, Veits J, Keil GM, Mettenleiter TC, Fuchs W. 2009. Protection of chickens against H5N1 highly pathogenic avian influenza virus infection by live vaccination with infectious laryngotracheitis virus recombinants expressing H5 hemagglutinin and N1 neuraminidase. Vaccine 27:773–785. doi: 10.1016/j.vaccine.2008.11.033. [DOI] [PubMed] [Google Scholar]
- 41.Nayak B, Kumar S, DiNapoli JM, Paldurai A, Perez DR, Collins PL, Samal SK. 2010. Contributions of the avian influenza virus HA, NA, and M2 surface proteins to the induction of neutralizing antibodies and protective immunity. J Virol 84:2408–2420. doi: 10.1128/JVI.02135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramp K, Veits J, Deckers D, Rudolf M, Grund C, Mettenleiter TC, Romer-Oberdorfer A. 2011. Coexpression of avian influenza virus H5 and N1 by recombinant Newcastle disease virus and the impact on immune response in chickens. Avian Dis 55:413–421. doi: 10.1637/9652-011111-Reg.1. [DOI] [PubMed] [Google Scholar]
- 43.Sylte MJ, Hubby B, Suarez DL. 2007. Influenza neuraminidase antibodies provide partial protection for chickens against high pathogenic avian influenza infection. Vaccine 25:3763–3772. doi: 10.1016/j.vaccine.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Hanika A, Larisch B, Steinmann E, Schwegmann-Wessels C, Herrler G, Zimmer G. 2005. Use of influenza C virus glycoprotein HEF for generation of vesicular stomatitis virus pseudotypes. J Gen Virol 86:1455–1465. doi: 10.1099/vir.0.80788-0. [DOI] [PubMed] [Google Scholar]
- 45.Sutter G, Ohlmann M, Erfle V. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett 371:9–12. doi: 10.1016/0014-5793(95)00843-X. [DOI] [PubMed] [Google Scholar]
- 46.Halbherr SJ, Brostoff T, Tippenhauer M, Locher S, Berger Rentsch M, Zimmer G. 2013. Vaccination with recombinant RNA replicon particles protects chickens from H5N1 highly pathogenic avian influenza virus. PLoS One 8:e66059. doi: 10.1371/journal.pone.0066059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manzoor R, Sakoda Y, Nomura N, Tsuda Y, Ozaki H, Okamatsu M, Kida H. 2009. PB2 protein of a highly pathogenic avian influenza virus strain A/chicken/Yamaguchi/7/2004 (H5N1) determines its replication potential in pigs. J Virol 83:1572–1578. doi: 10.1128/JVI.01879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soda K, Sakoda Y, Isoda N, Kajihara M, Haraguchi Y, Shibuya H, Yoshida H, Sasaki T, Sakamoto R, Saijo K, Hagiwara J, Kida H. 2008. Development of vaccine strains of H5 and H7 influenza viruses. Jpn J Vet Res 55:93–98 http://hdl.handle.net/2115/32369. [PubMed] [Google Scholar]
- 49.Ocana-Macchi M, Bel M, Guzylack-Piriou L, Ruggli N, Liniger M, McCullough KC, Sakoda Y, Isoda N, Matrosovich M, Summerfield A. 2009. Hemagglutinin-dependent tropism of H5N1 avian influenza virus for human endothelial cells. J Virol 83:12947–12955. doi: 10.1128/JVI.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brownie C, Statt J, Bauman P, Buczynski G, Skjolaas K, Lee D, Hotta J, Roth NJ. 2011. Estimating viral titres in solutions with low viral loads. Biologicals 39:224–230. doi: 10.1016/j.biologicals.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 51.Kalhoro NH, Veits J, Rautenschlein S, Zimmer G. 2009. A recombinant vesicular stomatitis virus replicon vaccine protects chickens from highly pathogenic avian influenza virus (H7N1). Vaccine 27:1174–1183. doi: 10.1016/j.vaccine.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 52.Zimmer G, Locher S, Berger Rentsch M, Halbherr SJ. 2014. Pseudotyping of vesicular stomatitis virus with the envelope glycoproteins of highly pathogenic avian influenza viruses. J Gen Virol 95:1634–1639. doi: 10.1099/vir.0.065201-0. [DOI] [PubMed] [Google Scholar]
- 53.Berger Rentsch M, Zimmer G. 2011. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One 6:e25858. doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hofmann MA, Renzullo S, Baumer A. 2008. Phylogenetic characterization of H5N1 highly pathogenic avian influenza viruses isolated in Switzerland in 2006. Virus Genes 37:407–413. doi: 10.1007/s11262-008-0285-2. [DOI] [PubMed] [Google Scholar]
- 55.Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sylte MJ, Suarez DL. 2012. Vaccination and acute phase mediator production in chickens challenged with low pathogenic avian influenza virus; novel markers for vaccine efficacy? Vaccine 30:3097–3105. doi: 10.1016/j.vaccine.2012.02.055. [DOI] [PubMed] [Google Scholar]
- 57.Webster RG, Brown LE, Laver WG. 1984. Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology 135:30–42. doi: 10.1016/0042-6822(84)90114-4. [DOI] [PubMed] [Google Scholar]
- 58.Maupin KA, Liden D, Haab BB. 2012. The fine specificity of mannose-binding and galactose-binding lectins revealed using outlier motif analysis of glycan array data. Glycobiology 22:160–169. doi: 10.1093/glycob/cwr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogerieux F, Belaise M, Terzidis-Trabelsi H, Greffard A, Pilatte Y, Lambre CR. 1993. Determination of the sialic acid linkage specificity of sialidases using lectins in a solid phase assay. Anal Biochem 211:200–204. doi: 10.1006/abio.1993.1257. [DOI] [PubMed] [Google Scholar]
- 60.von Itzstein M, Thomson R. 2009. Anti-influenza drugs: the development of sialidase inhibitors. Handb Exp Pharmacol 189:111–154. doi: 10.1007/978-3-540-79086-0_5. [DOI] [PubMed] [Google Scholar]
- 61.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. 2012. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sultana I, Yang K, Getie-Kebtie M, Couzens L, Markoff L, Alterman M, Eichelberger MC. 2014. Stability of neuraminidase in inactivated influenza vaccines. Vaccine 32:2225–2230. doi: 10.1016/j.vaccine.2014.01.078. [DOI] [PubMed] [Google Scholar]
- 63.Bucher DJ, Kilbourne ED. 1972. A 2 (N2) neuraminidase of the X-7 influenza virus recombinant: determination of molecular size and subunit composition of the active unit. J Virol 10:60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.von Grafenstein S, Wallnoefer HG, Kirchmair J, Fuchs JE, Huber RG, Schmidtke M, Sauerbrei A, Rollinger JM, Liedl KR. 2015. Interface dynamics explain assembly dependency of influenza neuraminidase catalytic activity. J Biomol Struct Dyn 33:104–120. doi: 10.1080/07391102.2013.855142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colman PM, Varghese JN, Laver WG. 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 66.Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, Wang J, Smith D, Cyr T, Farnsworth A, He R, Hurt AC, Brown EG, Li X. 2013. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res 100:567–574. doi: 10.1016/j.antiviral.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 67.World Health Organization. 2002. WHO manual on animal influenza diagnosis and surveillance. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf. [Google Scholar]
- 68.Hausmann J, Kretzschmar E, Garten W, Klenk HD. 1995. N1 neuraminidase of influenza virus A/FPV/Rostock/34 has haemadsorbing activity. J Gen Virol 76:1719–1728. [DOI] [PubMed] [Google Scholar]
- 69.Uhlendorff J, Matrosovich T, Klenk HD, Matrosovich M. 2009. Functional significance of the hemadsorption activity of influenza virus neuraminidase and its alteration in pandemic viruses. Arch Virol 154:945–957. doi: 10.1007/s00705-009-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. 2012. Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic acid receptors. J Virol 86:13371–13383. doi: 10.1128/JVI.01426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hooper KA, Bloom JD. 2013. A mutant influenza virus that uses an n1 neuraminidase as the receptor-binding protein. J Virol 87:12531–12540. doi: 10.1128/JVI.01889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suzuki T, Takahashi T, Guo CT, Hidari KI, Miyamoto D, Goto H, Kawaoka Y, Suzuki Y. 2005. Sialidase activity of influenza A virus in an endocytic pathway enhances viral replication. J Virol 79:11705–11715. doi: 10.1128/JVI.79.18.11705-11715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gerlach T, Kuhling L, Uhlendorff J, Laukemper V, Matrosovich T, Czudai-Matwich V, Schwalm F, Klenk HD, Matrosovich M. 2012. Characterization of the neuraminidase of the H1N1/09 pandemic influenza virus. Vaccine 30:7348–7352. doi: 10.1016/j.vaccine.2012.09.078. [DOI] [PubMed] [Google Scholar]
- 74.Ohuchi M, Asaoka N, Sakai T, Ohuchi R. 2006. Roles of neuraminidase in the initial stage of influenza virus infection. Microbes Infect 8:1287–1293. doi: 10.1016/j.micinf.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 75.Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 76.Swayne DE, Lee CW, Spackman E. 2006. Inactivated North American and European H5N2 avian influenza virus vaccines protect chickens from Asian H5N1 high pathogenicity avian influenza virus. Avian Pathol 35:141–146. doi: 10.1080/03079450600597956. [DOI] [PubMed] [Google Scholar]
- 77.Ewert DL, Barger BO, Eidson CS. 1979. Local antibody response in chickens: analysis of antibody synthesis to Newcastle disease virus by solid-phase radioimmunoassay and immunofluorescence with class-specific antibody for chicken immunoglobulins. Infect Immun 24:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]