ABSTRACT
Since its emergence, Schmallenberg virus (SBV), a novel insect-transmitted orthobunyavirus which predominantly infects ruminants, has caused a large epidemic in European livestock. Newly developed inactivated vaccines are available, but highly efficacious and safe live vaccines are still not available. Here, the properties of novel recombinant SBV mutants lacking the nonstructural protein NSs (rSBVΔNSs) or NSm (rSBVΔNSm) or both of these proteins (rSBVΔNSs/ΔNSm) were tested in vitro and in vivo in type I interferon receptor knockout mice (IFNAR−/−) and in a vaccination/challenge trial in cattle. As for other bunyaviruses, both nonstructural proteins of SBV are not essential for viral growth in vitro. In interferon-defective BHK-21 cells, rSBVΔNSs and rSBVΔNSm replicated to levels comparable to that of the parental rSBV; the double mutant virus, however, showed a mild growth defect, resulting in lower final virus titers. Additionally, both mutants with an NSs deletion induced high levels of interferon and showed a marked growth defect in interferon-competent sheep SFT-R cells. Nevertheless, in IFNAR−/− mice, all mutants were virulent, with the highest mortality rate for rSBVΔNSs and a reduced virulence for the NSm-deleted virus. In cattle, SBV lacking NSm caused viremia and seroconversion comparable to those caused by the wild-type virus, while the NSs and the combined NSs/NSm deletion mutant induced no detectable virus replication or clinical disease after immunization. Furthermore, three out of four cattle immunized once with the NSs deletion mutant and all animals vaccinated with the virus lacking both nonstructural proteins were fully protected against a challenge infection. Therefore, the double deletion mutant will provide the basis for further developments of safe and efficacious modified live SBV vaccines which could be also a model for other viruses of the Simbu serogroup and related orthobunyaviruses.
IMPORTANCE SBV induces only mild clinical signs in adult ruminants but causes severe fetal malformation and, thereby, can have an important impact on animal welfare and production. As SBV is an insect-transmitted pathogen, vaccination will be one of the most important aspects of disease control. Here, mutant viruses lacking one or two proteins that essentially contribute to viral pathogenicity were tested as modified live vaccines in cattle. It could be demonstrated that a novel recombinant double deletion mutant is a safe and efficacious vaccine candidate. This is the first description of a putative modified live vaccine for the complete genus Orthobunyavirus, and in addition, such a vaccine type has never been tested in cattle for any virus of the entire family Bunyaviridae. Therefore, the described vaccine also represents the first model for a broad range of related viruses and is of high importance to the field.
INTRODUCTION
Schmallenberg virus (SBV), which belongs to the Simbu serogroup within the genus Orthobunyavirus, family Bunyaviridae, was detected for the first time in Germany in late 2011 (1). Since it was first recognized, the virus spread rapidly across Europe (2, 3). SBV mainly infects ruminants and occasionally causes fever, diarrhea, reduction in milk yield, and after infection of pregnant animals during a critical phase of gestation, abortions, stillbirth, and severe congenital malformations (1, 4). Apart from domestic ruminants, the SBV genome or specific antibodies could also be detected in wild ruminants, New World camelids, and a dog (5–7), and adult type I interferon receptor knockout mice (IFNAR−/−) are susceptible to an experimental SBV infection (8). However, a zoonotic potential of SBV is very unlikely (9, 10). In accordance with other orthobunyaviruses, which are frequently isolated from Culicoides midges or mosquitoes, SBV is transmitted by insect vectors; viral genome was detected in various field-collected Culicoides species (11, 12).
As in other members of the genus Orthobunyavirus, the SBV genome comprises three negative-stranded single-stranded RNA (ssRNA) segments. The small segment (S) encodes the nucleocapsid protein N and in an alternative overlapping reading frame for the nonstructural protein NSs. The M segment encodes a precursor protein, which is posttranslationally processed to two glycoproteins (Gn and Gc) and to the nonstructural protein NSm (13). The large segment (L) encodes the RNA-dependent RNA polymerase (RdRp). Genomes are encapsidated by the N proteins in the form of ribonucleoprotein (RNP) complexes. The N protein is the most abundant protein in virus particles and infected cells and, therefore, the main target in serological and molecular SBV diagnostics (14, 15). Like the NSs protein of the prototype orthobunyavirus Bunyamwera virus (BUNV), La Crosse virus (LACV), or Akabane virus (AKAV), NSs of SBV was also found to be nonessential for virus viability in mammalian and insect cells (16–20). However, in infected mammalian cells, the NSs protein counteracts the shutoff of host cell protein synthesis and the induction of interferon (IFN) similar to BUNV (21), but it is also involved in other functions like regulation of translation, apoptosis, and viral polymerase activity (19, 22–24). In both in vitro and in vivo mouse experiments, NSs deletion mutants displayed an attenuated phenotype in IFN-competent cells and animals, but not in systems lacking the IFN receptor (16, 25). Besides its function in mammalian hosts, the importance of bunyaviral NSs had also been demonstrated for insect hosts (25, 26).
The function of the nonstructural protein NSm of orthobunyaviruses has not been elucidated in detail until now. It is a small transmembrane protein which is colocalized with the two viral glycoproteins Gn and Gc in the Golgi complex and is probably a scaffold protein involved in virus assembly and morphogenesis. In these processes, the N-terminal part of BUNV NSm is essential, while the C terminus is dispensable (27). However, for Rift Valley fever virus (RVFV), a mosquito-transmitted phlebovirus (another genus within the family Bunyaviridae), lacking NSm was highly immunogenic, but it was only slightly attenuated and retained the ability to induce lethal hepatic and neurological disease in rats (28).
NSs/NSm double deletion RVFV mutants combine the characteristics of the single NSs and NSm mutants. These double deletion mutants replicated efficiently in cell culture and, therefore, have been evaluated regarding their suitability as marker vaccines. The double mutants are highly attenuated, confer protection from viremia in rats and sheep, and allowed differentiation between infected and vaccinated animals (29).
Shortly after the emergence of SBV in Europe, efforts started to develop vaccines to prevent a further spread of the disease. The cross-protection of an inactivated trivalent vaccine against the Simbu serogroup viruses AKAV and Aino virus (AINOV) and the reovirus Chuzan virus was tested, but this vaccine was unable to prevent an experimental SBV infection (30). However, newly developed SBV-specific vaccine candidates based on inactivated virus formulations protected animals from an SBV infection (31). For the United Kingdom market, the Veterinary Medicines Directorate (VMD) granted provisional marketing authorization for an inactivated vaccine against SBV (32). Although inactivated vaccines are safe and useful tools to circumvent the spread of newly emerging disease, live attenuated vaccines are often more efficient in relation to both the onset of immunity and duration of immunity. However, they bear the safety risk of contamination with virulent strains or revertance to virulence by reversion of attenuating nucleotide mutations (33–35). Therefore, genetically attenuated vaccines that contain nonrevertible mutations or genomic deletions within viral proteins could provide a new approach for the development of novel vaccines.
Facilitated by the establishment of a reverse genetics system for the generation of recombinant SBV mutants lacking NSs, NSm, and NSs/NSm in combination were tested in mice but also in cattle, one of the major target species of SBV, regarding their suitability to prevent a subsequent experimental SBV infection. Our results demonstrate that both the NSs mutant and NSs/NSm double mutant are promising candidates for the development of safe and effective SBV vaccines.
MATERIALS AND METHODS
Cells and viruses.
The following cells were obtained from the Collection of Cell Lines in Veterinary Medicine (CCLV) at the Federal Research Institute of Animal Health, Insel Riems, Germany: BHK-21 (RIE164 [CCLV]), BSR-T7/5 (RIE583 [CCLV]) (36), SFT-R (RIE043 [CCLV]), Vero-76, (RIE0228 [CCLV]), KC (RIE1062 [CCLV]) (37), MDBK-t2 (RIE1268 [CCLV]) (38), and SK6-MxLuc (RIE1272 [CCLV]) (39). Cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% BVDV-free fetal calf serum (FCS) or in Schneiders insect medium.
The wild-type SBV (wtSBV) isolate BH80/11 was originally isolated from blood from an infected cow on KC cells (1) and passaged in BHK-21 cells, KC cells, and again BHK-21 cells. The recombinant SBV (rSBV) and the deletion mutants rSBVΔNSs, rSBVΔNSm, and rSBVΔNSs/ΔNSm were generated by reverse genetics. The viruses were grown and titrated on BHK-21 cells, and virus titers were calculated as 50% tissue culture infective dose per ml (TCID50/ml). Plaque phenotypes of the viruses were investigated by a standard plaque assay in SFT-R cells under an overlay comprising minimum essential medium (MEM) supplemented with 2% FCS and 0.75% methylcellulose (Carl Roth) and an incubation at 37°C for 6 days. Infected cells were fixed with 4% formaldehyde and stained with 0.1% crystal violet in 13% formaldehyde.
Plasmids.
All plasmids were generated by fusion PCR, a modified restriction-free cloning method (40) and confirmed by DNA sequencing. Plasmid DNA was purified by using Qiagen Plasmid Mini Kits or Midi Kits (Qiagen). For cloning purposes, PCR was carried out with Phusion DNA polymerase (Finnzymes) and SBV-specific primers (Biomers) according to the published sequences of the SBV genome segments (GenBank accession numbers HE649912, HE649913, and HE649914). The terminal sequences of the genome segments were determined by 5′ and 3′ rapid amplification of cDNA ends (RACE) systems (Invitrogen) according to the manufacturer's instructions. Total RNA was extracted from SBV-infected cells using TRIzol reagent (Invitrogen). For cDNA synthesis, gene-specific primers and the Transcriptor high-fidelity cDNA synthesis kit (Roche) were used according to the manufacturer's instructions. PCR fragments amplified with gene-specific primers and inserted into pT7riboSM2 (41), kindly provided by F. Weber (Philipps University Marburg), served as the template to generate the full-length antigenomic T7 polymerase-driven constructs pT7ribo_SBV_S, pT7ribo_SBV_L, and pT7ribo_SBV_M. While pT7ribo_SBV_S was generated from one PCR fragment, the pT7ribo_SBV_L and pT7ribo_SBV_M plasmids were constituted from five overlapping PCR fragments (segment L) and four overlapping PCR fragments (segment M), respectively. For construction of the NSs knockout mutant pT7ribo_SBVΔNSs, three point mutations were introduced in two cloning steps into pT7ribo_SBV_S (T→G at nucleotide [nt] positions 58, 73, and 103) by fusion PCR. These mutations altered all three NSs initiation codons. To generate plasmid pT7ribo_SBVΔNSm, nucleotides 1233 to 1352 within the SBV M segment were deleted by using primers Mut_dNSm and M_2231R, resulting in a deletion of 60 amino acids (aa) within NSm. Further information about primer sequences and cloning procedures of all described constructs is available on request.
Generation of recombinant viruses from cDNAs.
To recover recombinant SBV, a three-plasmid rescue system in analogy to a protocol for the rescue of BUNV (42) was applied. Semiconfluent layers of BSR-T7/5 cells were grown in six-well plates. Each well was transfected with 3 μg each of pT7ribo_SBV_L, pT7ribo_SBV_M or pT7ribo_SBVΔNSm, or pT7ribo_SBV_S or pT7ribo_SBVΔNSs, using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's protocol. Supernatants of transfected cells were harvested on day 5 posttransfection (p.t.). In order to screen for the presence of recombinant virus, BHK-21 cells grown in six-well plates were inoculated with 500 μl supernatant at 37°C and incubated further with 1 ml fresh DMEM. The presence of recombinant virus was then confirmed either by the appearance of a cytopathic effect (CPE) or immunofluorescence (IF) analysis. Virus stocks were generated from the third passage on BHK-21 cells.
Virus titration and growth kinetics.
In vitro growth kinetic experiments were performed using BHK-21 cells or SFT-R cells. Cells were inoculated with wtSBV or the recombinant viruses rSBV, rSBVΔNSm, rSBVΔNSs, and rSBVΔNSs/ΔNSm with a multiplicity of infection (MOI) of 0.1. Supernatants were collected at 0, 8, 24, 48, and 72 h postinfection (p.i.). Titers were calculated by counting CPE-positive wells of BHK-21 cells and displayed as 50% tissue culture infective dose per ml.
Electron microscopy.
Vero monolayer cells (RIE0228, Vero-76) were infected at an MOI of 0.5 with wild-type and mutant viruses and fixed at 24 h postinfection for 60 min with 2.5% glutaraldehyde buffered in 0.1 M Na cacodylate, pH 7.2 (300 mM osmol; Merck). The cells were then scraped off the plate, pelleted by low-speed centrifugation, and embedded in low-melting-point (LMP) agarose (Biozym). Small pieces were postfixed in 1.0% aqueous OsO4 (Polysciences Europe) and stained en bloc with uranyl acetate. After stepwise dehydration in ethanol, the cells were cleared in propylene oxide, embedded in glycid ether 100 (Serva) and polymerized at 59°C for 4 days. Ultrathin sections of embedded material, counterstained with uranyl acetate and lead salts, were examined with an electron microscope (FEI Tecnai G2 Spirit microscope).
Immunofluorescence staining.
SBV-infected cells were fixed with 80% acetone for 15 min on ice. For immunofluorescence (IF) staining, monoclonal antibodies (MAbs) specific for SBV N or Gc proteins, kindly provided by Emiliana Brocchi (IZSLER, Brescia, Italy) were used. Finally, an Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes) was added as a secondary antibody.
Western blotting.
Western blots were performed from total cell lysates of SBV-infected BHK-21 cells after freeze-thawing 24 h p.i. The proteins were separated by SDS-PAGE under nonreducing conditions and transferred onto nitrocellulose membranes (Bio-Rad). SBV was detected using MAbs against SBV N or SBV Gc, diluted 1:40 in Tris-buffered saline with 0.1% Tween (TBS-T) for 1 h. A horseradish peroxidase-conjugated anti-rabbit antibody (Dianova) (1:20,000 in TBS-T) was used as a secondary antibody.
IFN bioassays.
Two reporter gene assays specific for type I interferon (IFN) were carried out, an assay using luciferase as the genetic reporter and an Mx/CAT (chloramphenicol acetyltransferase) reporter gene assay (38). The first IFN reporter gene assay was carried out in SK6-MxLuc cells, porcine kidney cells expressing firefly luciferase (Luc). Briefly, a total of 1 × 105 SFT-R cells were inoculated with the viruses indicated in Fig. 4 at an MOI of 0.1. Two hours p.i., cell culture supernatants were discarded, cells were washed twice with phosphate-buffered saline (PBS), and 1.0 ml of culture medium was added. Supernatants were collected at 48 p.i. and UV light treated for 3 min to inactivate the virus present in the samples. Twofold serial dilutions of the UV-inactivated supernatants were applied to SK6-MxLuc cells and incubated for 24 h at 37°C. Supernatants of mock-infected SFT-R cells were used as negative controls. The measurement of the firefly luciferase activity (ovine alpha/beta interferon [IFN-α/β]) was carried out by using the Bright-Glo luciferase assay system (Promega).
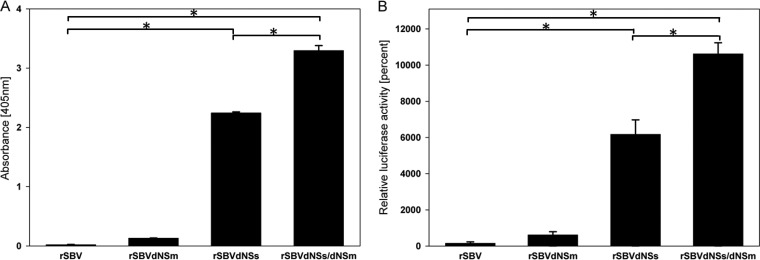
FIG 4.
IFN induction by the different recombinant viruses was measured with two IFN bioassays relying on the Mx promoter either with Mx/CAT (A), or with luciferase (B) as the respective reporter. SFT-R cells were inoculated with the indicated viruses at an MOI of 0.1. Supernatants were collected at 48 p.i., UV light treated, and applied to reporter cells. Supernatants of mock-infected SFT-R cells were used as negative controls. Statistically significant differences (P < 0.01) are indicated by an asterisk and bar.
For the Mx/CAT bioassay (Fray et al. [38]), UV-inactivated supernatants from virus-infected SFT-R cells were applied to MDBK-t2 cells in 2-fold serial dilutions and incubated for 24 h at 35°C. CAT expression was determined using the CAT enzyme-linked immunosorbent assay kit (Roche) according to the manufacturer's instructions. Finally, the absorbance of the samples was determined at 405 nm using an enzyme-linked immunosorbent assay (ELISA) reader (Tecan Infinite F200 PRO reader).
Differences between groups were examined by a paired t test, and P values of <0.01 were considered significant.
Infection of type I interferon receptor knockout mice.
In a pretrial, 36 adult IFNAR−/− mice on a C57BL/6 genetic background were divided into six groups with 6 animals in each group, mice of both sexes were distributed evenly among the groups. Mice were obtained from the specific-pathogen-free breeding unit of the Friedrich-Loeffler-Institut (Insel Riems, Germany) or were kindly provided by Ulrich Kalinke (Twincore, Hannover, Germany). wtSBV was 10-fold diluted in PBS from 106 down to 101 TCID50/ml, and 100 μl of each dilution step was subcutaneously inoculated into mice of one group. Three uninfected mice were kept as controls. All animals were weighed and examined for clinical symptoms daily, and mice showing severe symptoms (ataxia, apathy) were euthanized immediately. All remaining mice were euthanized after 21 days. When infected with 105 TCID50/ml, 50% of the inoculated mice died, which was the lowest survival rate of all groups, so in further experiments with infectious recombinant SBV, this viral titer was used.
Forty-eight IFNAR−/− mice were assigned to four groups with 12 animals in each group and inoculated subcutaneously in the scruff of the neck with one of the following viruses: rSBV for group A, rSBVΔNSm for group B, rSBVΔNSs for group C, and rSBVΔNSs/ΔNSm for group D. Three other mice were kept as controls. All mice were weighed and examined daily by veterinarians. Two mice of each group were euthanized 3 days after inoculation, and samples of brain, lungs, liver, spleen, and small intestine were taken and tested by an SBV S-segment-specific real-time PCR (14). An identical organ panel was tested after premature death or at the end of the study after 21 days. Sera taken on day 21 were 1/10 diluted and tested by a commercially available ELISA (ID Screen Schmallenberg virus competition; IDvet) which detects antibodies against the N protein of SBV (15) using the recommended cutoff of 40% relative optical density compared to the optical density of the positive control.
Immunization and infection of cattle.
Twelve SBV-naive, male Holstein-Friesian calves were assigned to three groups and subcutaneously inoculated with 106 TCID50/ml of either rSBVΔNSs, rSBVΔNSm, or rSBVΔNSs/ΔNSm. Two animals were infected with wtSBV, and two animals were infected with rSBV. Two animals were inoculated with infectious cattle serum (43), and after 21 days, cattle of all groups and five additional control mice were injected with infectious serum (43). During the entire study, rectal body temperatures of cattle were measured daily, and the animals were examined for clinical signs by veterinarians. Serum samples were taken daily from cattle during the first 10 days after immunization/infection and challenge infection, respectively, and analyzed by real-time PCR (14). Additionally, serum samples were taken weekly and analyzed by a commercially available SBV antibody ELISA (ID Screen Schmallenberg virus competition; IDvet) and a standard microneutralization assay as described previously (44). The mice were euthanized after 22 days, and their sera were tested by ELISA as well.
All experimental protocols were reviewed by a state ethics commission and have been approved by the competent authority (State Office for Agriculture, Food Safety and Fisheries of Mecklenburg-Vorpommern, Rostock, Germany, reference LALLF M-V TSD/7221.3-1.1-004/12).
RESULTS
Construction of the rescue plasmids.
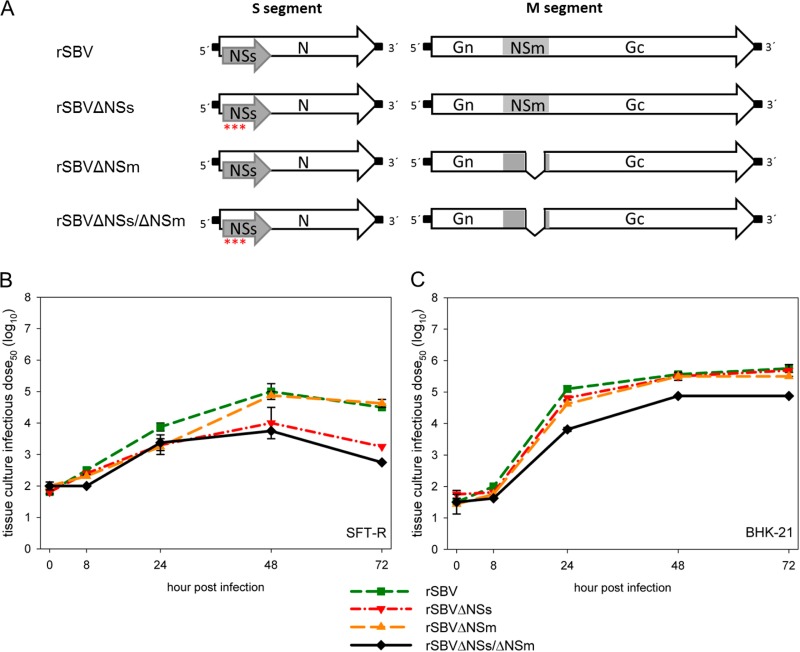
The initial SBV isolate BH80/11, described by Hoffmann et al. (1) was used as the basis of a plasmid-based genetic system for the rescue of rSBV. The three rescue plasmids containing the complete copy of a genomic segment (pT7ribo_SBV_S, pT7ribo_SBV_M, and pT7ribo_SBV_L) downstream of a T7 promoter were constructed within the pT7riboSM2 vector (41) by fusion PCR. The plasmid pT7ribo_SBV_S was also used to generate the plasmid pT7ribo_SBV_ΔNSs by mutation of three potential NSs translation initiation codons in order to abrogate the expression of NSs by the recombinant virus rSBVΔNSs. In addition, an M-segment-derived rescue plasmid pT7ribo_SBV_ΔNSm with a partial deletion within the NSm-encoding region was constructed for the rescue of rSBVΔNSm expressing a partially deleted NSm protein (60 amino acids). Both mutant plasmids were also used for the generation of the double mutant virus rSBVΔNSs/ΔNSm (Fig. 1A).
FIG 1.
(A) Overview of the genome structure of the cDNA constructs of the S and M segments of the full-length recombinant SBV (rSBV) and the deletion mutants rSBVΔNSs, rSBVΔNSm, and rSBVΔNSs/ΔNSm. Asterisks indicate mutations within the three potential translation start codons of the NSs-encoding sequence that abrogate the expression of the NSs protein. The introduced deletion within the NSm-encoding sequence is indicated by a serrated line. (B and C) Growth kinetics of recombinant viruses rSBV, rSBVΔNSs, rSBVΔNSm, and rSBVΔNSs/ΔNSm in SFT-R cells (B) and BHK-21 cells (C). The cells were infected with a multiplicity of infection (MOI) of 0.1; supernatants were collected at the times indicated in the figure, and titers were measured by endpoint titration in BHK-21 cells.
Recovery and in vitro characterization of rSBV.
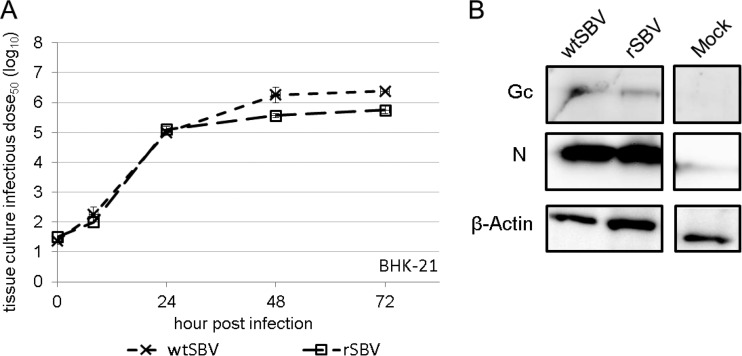
For the recovery of rSBV of the different plasmids, pT7ribo_SBV_S, pT7ribo_SBV_M, and pT7ribo_SBV_L were transfected into BSR-T7/5 cells constitutively expressing the T7 RNA polymerase (36). Three days p.t., rSBV could be recovered from transfection supernatants of BSR-T7/5 cells. rSBV could be propagated in several mammalian and insect cell lines such as Vero, BHK-21, SFT-R, and KC cells. In standard plaque assays using SFT-R cells, rSBV produced a phenotype similar compared to that produced by wtSBV (data not shown). Virus stocks were generated after passaging rSBV three times on BHK-21 cells, resulting in final titers of about 107.5 TCID50/ml, and complete genome sequencing of the recombinant virus stock revealed that no mutations had arisen. In rSBV-infected and wtSBV-infected Vero cells, no significant differences in expression of the nucleoprotein N and the glycoprotein Gc could be detected by IF staining using N- and Gc-specific monoclonal antibodies, and a typical CPE could be observed (data not shown). Growth kinetics studies of rSBV and wtSBV were performed in BHK-21 cells and indicated a similar growth of both viruses with final titers of at least 105.5 TCID50/ml at 72 h p.i. (Fig. 2A).
FIG 2.
In vitro characterization of wild-type SBV (wtSBV) and recombinant SBV (rSBV). (A) Growth curves of wtSBV and rSBV. BHK-21 cells were infected at an MOI of 0.1 Supernatants were collected at the indicated times postinfection, and titers were calculated by counting CPE-positive wells and displayed as 50% tissue culture infective dose per ml (TCID50/ml). (B) Western blot analysis of BHK-21 cells infected with wtSBV and rSBV by using SBV N- and Gc-specific monoclonal antibodies. Mock-infected BHK-21 cells were used as negative controls. For control purposes, lysates were also tested for β-actin expression.
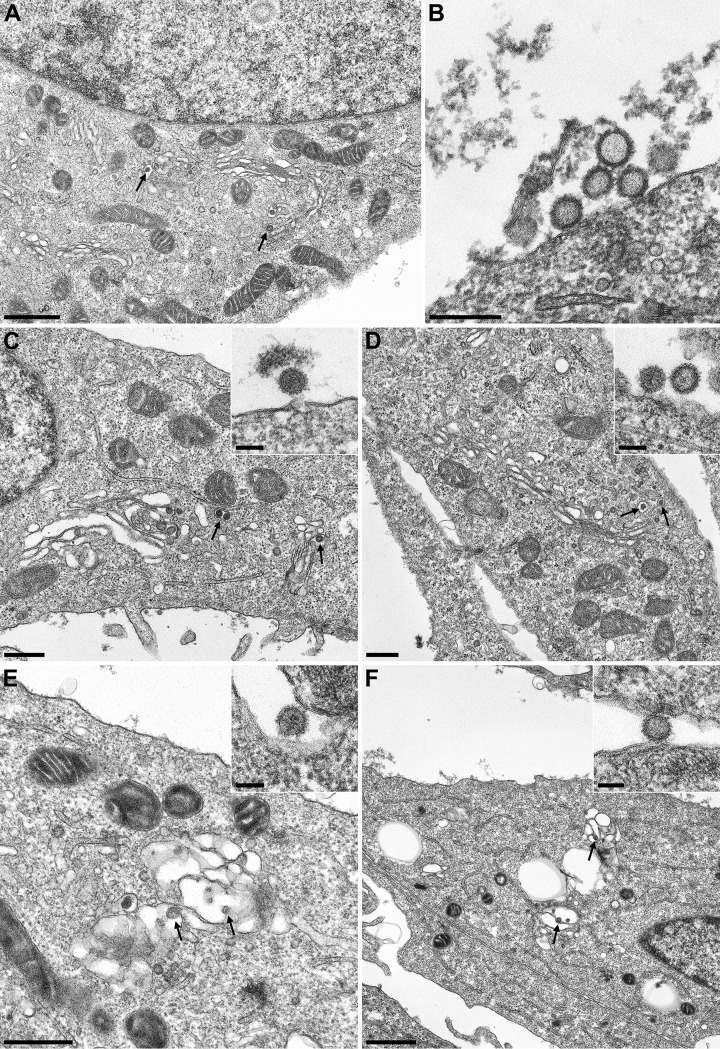
Electron microscopic (EM) analysis of the morphogenesis of the indicated viruses demonstrated virus replication in the cytoplasm. All mature virus particles were found inside Golgi cisterna or extracellularly located, indicating that in comparison to wild-type or recombinant virus, virus assembly was not influenced by mutations of nonstructural proteins (Fig. 3A to C).
FIG 3.
Electron microscopy analysis of wtSBV (A and B), rSBV (C), rSBVΔNSm (D), rSBVΔNSs (E), and rSBVΔNSs/ΔNSm (F) in Vero-76 cells 24 h p.i. Bars, 1 μm (A and F), 250 nm (B), 500 nm (C to E), and 100 nm (insets). Maturing bunyaviruses inside Golgi cisterna are indicated by black arrows.
In addition, Western blot analysis of rSBV resulted in detection of the N and Gc proteins that were indistinguishable in size from the proteins of wtSBV (Fig. 2B). Neither SBV NSs nor NSm proteins could be detected in wtSBV or rSBV by IF staining and Western blot analysis using polyclonal SBV-specific antisera (data not shown), and unfortunately, SBV-specific monoclonal antibodies for the detection of NSs and NSm have not been available until now.
Generation and in vitro characterization of rSBV with mutated NSs and NSm proteins.
For the directed attenuation of rSBV, plasmids containing mutations of the NSs gene (pT7ribo_SBV_ΔNSs) or a partial gene deletion of the NSm genomic region (pT7ribo_SBV_ΔNSm) were generated. To avoid the expression of the NSs protein, the pT7ribo_SBV_ΔNSs plasmid was generated by introduction of three silent mutations within the N-protein-encoding region, which resulted in the mutation of three NSs translation start codons. Plasmid pT7ribo_SBV_ΔNSm was constructed by deletion of nt 1233 to 1352 (60 aa) within the NSm-encoding region of plasmid pT7ribo_SBV_M. Mutant recombinant viruses could be rescued as described above by transfection of pT7ribo_SBV_L in combination with either pT7ribo_SBV_ΔNSs and pT7ribo_SBV_M (rSBVΔNSs) or with pT7ribo_SBV_S and pT7ribo_SBV_ΔNSm (rSBVΔNSm) or with pT7ribo_SBV_ΔNSs and pT7ribo_SBV_ΔNSm (rSBVΔNSs/ΔNSm) (Fig. 1A). Virus stocks were generated from the third passage on BHK-21 cells, and final titers of 107.625 TCID50/ml (rSBVΔNSs), 107.5 TCID50/ml (rSBVΔNSm), and 106.5 TCID50/ml (rSBVΔNSs/ΔNSm) could be determined. The identity of the mutant viruses was confirmed by sequencing. Sequence analysis also showed that no mutations had arisen in any virus stock compared with the original input cDNA plasmid used for virus rescue (data not shown).
In order to investigate whether the NSs knockout, the deletion within the NSm region, or the combination of both mutations resulted in a growth defect, comparative one-step growth kinetics studies of rSBV, rSBVΔNSs, rSBVΔNSm, and rSBVΔNSs/ΔNSm were conducted in IFN-defective BHK-21 and IFN-competent SFT-R cells. In BHK-21 cells, rSBVΔNSs and SBVΔNSm replicated efficiently to similar final virus titers (105.75 TCID50/ml for rSBV, 105.68 TCID50/ml for rSBVΔNSs, and 105.5 TCID50/ml for rSBVΔNSm). The mutant lacking both nonstructural proteins, NSs and NSm (rSBVΔNSs/ΔNSm), showed a reduced replication efficacy with an approximately 10-fold-lower final titer of 104.875 TCID50/ml (Fig. 1C). In interferon-competent SFT-R cells, all recombinant viruses replicated at a level lower than that in BHK-21 cells. However, final titers of 104.6 TCID50/ml were observed for rSBV and rSBVΔNSm, and even markedly lower virus titers were seen for rSBVΔNSs/ΔNSm (102.75 TCID50/ml) and rSBVΔNSs (103.25 TCID50/ml) (Fig. 1B). Additionally, there was no significant difference in plaque morphology between the genetically intact rSBV and the NSm deletion mutant rSBVΔNSm in SFT-R cells, whereas no plaque formation could be detected in cells infected with the NSs deletion mutant rSBVΔNSs and the double deletion mutant rSBVΔNSs/ΔNSm within 6 days p.i. (data not shown). EM analysis of morphogenesis revealed that the deletion of NSs and NSm had no influence on the virus production and virus morphology (Fig. 3D to F).
Interferon induction.
A characteristic feature of the NSs protein of orthobunyaviruses is an effective IFN antagonism to circumvent the innate defense in infected cells. However, the role of NSm in this process had not been elucidated until now. Therefore, we investigated IFN induction by the recombinant mutants lacking either NSs or NSm or both nonstructural proteins in comparison to the recombinant backbone rSBV and performed two established IFN bioassays relying on the Mx promoter, one with luciferase and one with CAT as the respective reporter. Although the IFN reporter gene assay specifically detecting firefly luciferase activity is sensitive to bovine IFN-α/β, it can also be used to detect ovine IFN. Almost no luciferase activity could be determined in the culture media of cells infected with rSBV at 48 h p.i. Interestingly, the mutant with a partial NSm deletion (rSBVΔNSm) induced low, but visible reporter signaling, indicating that very small amounts of IFN were present in those samples (Fig. 4B).
In contrast, supernatants collected from cells, which were infected with the mutant lacking NSs (rSBVΔNSs) or the rSBVΔNSs/ΔNSm double mutant revealed very high levels of IFN at 48 h p.i. At this time point, in supernatants of rSBVΔNSs-infected cells, a 6,000-fold increase of luciferase expression compared to supernatants of rSBV-infected cells could be measured with the reporter assay (P < 0.01). Surprisingly, in supernatants of cells infected with the double mutant rSBVΔNSs/ΔNSm, the expression of luciferase was significantly higher than in supernatants of rSBVΔNSs-infected cells, demonstrating not only an additive, but a potentiating effect caused by the NSm deletion in the double mutant. Very similar results could be obtained by using the Mx/CAT reporter gene assay. Figure 4A illustrates the level of CAT induction in correlation to the IFN-suppressing activity of the appropriate viruses at 48 h p.i. Cells infected with rSBV did not induce any significant CAT expression, while the SBV-NSs protein-expressing NSm deletion mutant rSBVΔNSm did, but at a very low level. In contrast, cells infected with rSBVΔNSs or rSBVΔNSs/ΔNSm showed very high CAT signals due to IFN induction with a significantly higher value for rSBVΔNSs/ΔNSm at 48 h p.i.
Infection of type I interferon receptor knockout mice with recombinant SBV mutants.
In order to evaluate the virulence and growth of the novel recombinant SBV in an interferon type 1-independent model system, IFNAR−/− mice were inoculated with the different virus mutants.
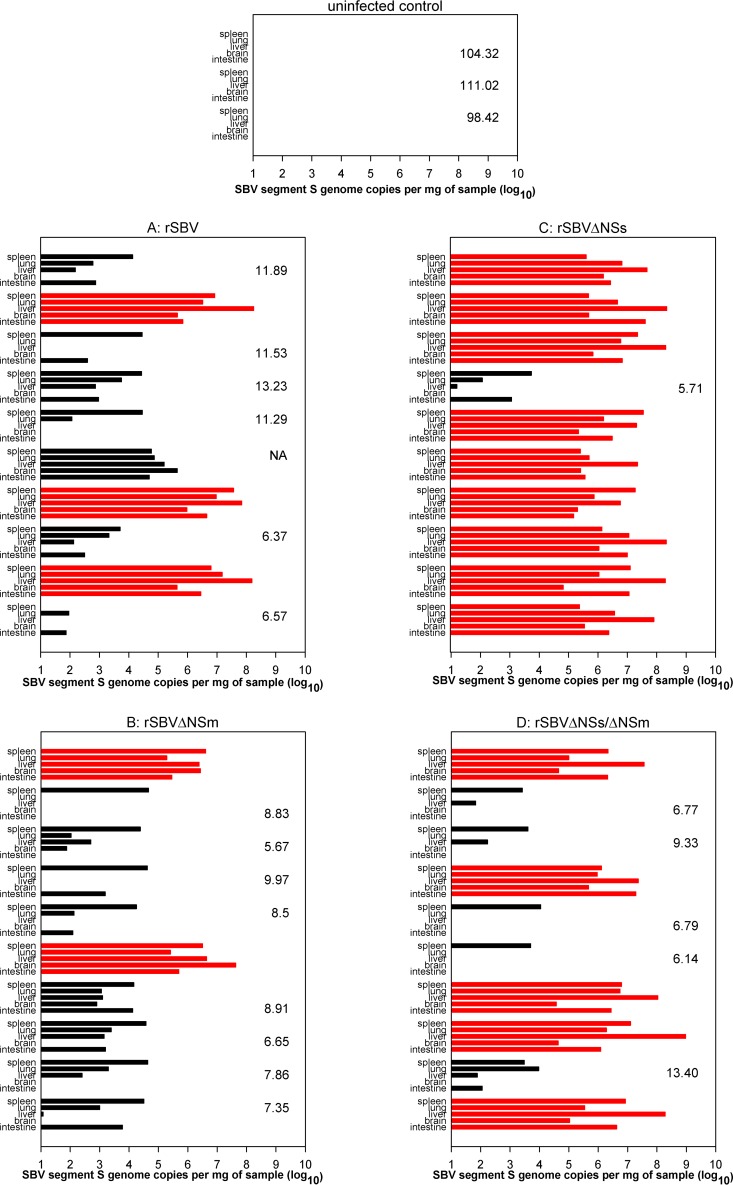
In organ samples from each mouse euthanized on day 3 after inoculation, the SBV genome was present (data not shown). From day two onwards, mice of all groups except the uninfected control animals started to lose weight. Until day 8 after infection, 6 out of 10 mice inoculated with rSBV, 2 out of 10 animals infected with rSBVΔNSm, 9 out of 10 mice that received rSBVΔNSs, and 5 out of 10 mice injected with rSBVΔNSs/ΔNSm were found dead or had to be humanely killed (Fig. 5). Samples taken from deceased mice were highly positive in the SBV-specific real-time PCR (Fig. 6). In all surviving mice of groups A to D, the SBV genome was detectable in at least one organ sample at the end of the study (21 days after infection; Fig. 6), while every organ sample taken from the control mice tested negative. SBV-specific antibodies were present in every surviving mouse, but not in uninfected control animals (Fig. 6).
FIG 5.
Mortality curve of type I interferon receptor-deficient mice infected with either rSBV, rSBVΔNSm, rSBVΔNSs, or rSBVΔNSs/ΔNSm.
FIG 6.
Real-time RT-PCR and SBV antibody ELISA results of type I interferon receptor-deficient mice. The PCR results for the panel of organs from an individual mouse are shown. The red bars indicate animals that died spontaneously or had to be euthanized prematurely. The number to the right of PCR data of a mouse or in the right portion of the graph represents the ELISA result (sample optical density [OD] relative to negative-control OD; values above 40% were considered negative) in sera obtained from surviving mice at the end of the study. NA, not available.
Antibody response of cattle after inoculation with the recombinant SBV mutants.
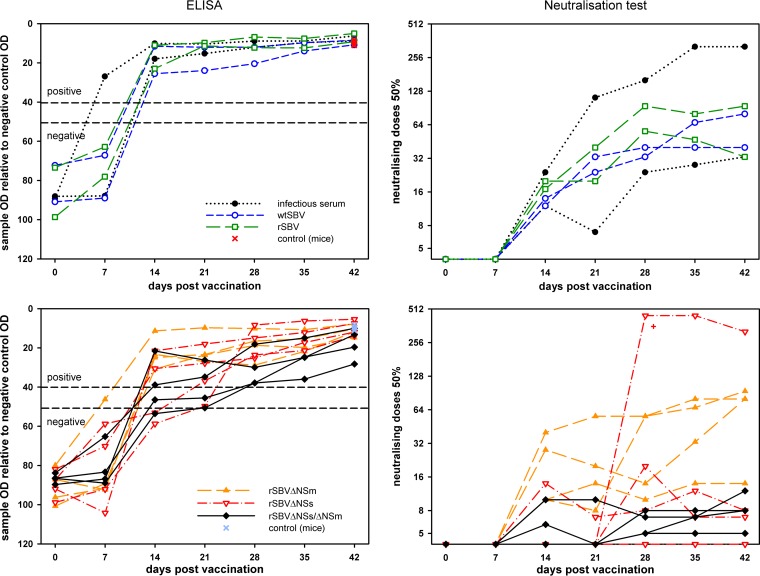
On the day of the inoculation with infectious cattle serum, wtSBV or rSBV, all cattle were negative in both serological assays (Fig. 7).
FIG 7.
Results of an SBV antibody ELISA and a neutralization assay of blood samples taken from cattle after inoculation with recombinant SBV mutants. IFNAR−/− mice were used as unvaccinated challenge controls. The cutoff values of the antibody ELISA are marked by dashed lines. The results of the animal that are shown labeled with a red plus sign (bottom right panel) correspond to the real-time PCR results of the identical animal shown in Fig. 8. OD, optical density.
The first antibodies were detectable by an SBV-specific ELISA 1 week after infection in one animal inoculated with infectious serum and 2 weeks after infection in all cattle injected with wtSBV, rSBV, rSBVΔNSm, or infectious serum and in two animals inoculated with rSBVΔNSs or rSBVΔNSs/ΔNSm (Fig. 7). Neutralizing antibodies could be detected from 2 weeks after inoculation onwards in all animals inoculated with wtSBV, rSBV, rSBVΔNSm, or infectious serum and in one animal each of group rSBVΔNSs and rSBVΔNSs/ΔNSm. One cow inoculated with rSBVΔNSs/ΔNSm was positive 2 weeks after immunization (titer 1/6) and negative 1 week later (Fig. 7).
Three weeks after the challenge infection, SBV-specific antibodies were detectable in samples from all cattle and from the control mice by ELISA, and all but one cow (rSBVΔNSs) had a positive result by the neutralization test (Fig. 7).
Clinical observations and RNA detection in cattle.
None of the cattle showed any relevant clinical signs during the entire study.
In all animals inoculated with wtSBV, rSBV, or infectious serum and in three out of four cattle injected with rSBVΔNSm, viral RNA was detectable for at least two consecutive days after infection. The remaining animal of group rSBVΔNSm and all cattle inoculated with rSBVΔNSs and rSBVΔNSs/ΔNSm tested negative by the sensitive real-time RT-PCR analysis. The mean SBV genome copy number values for all groups are depicted in Fig. 8.
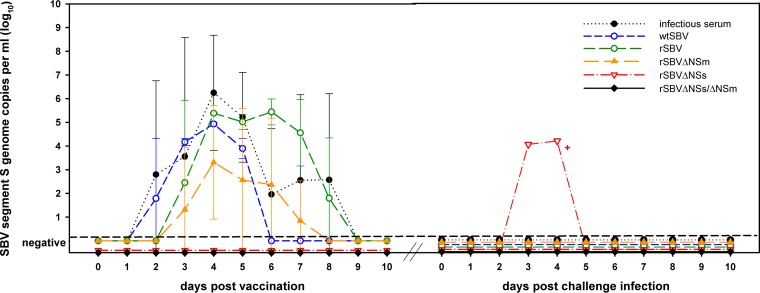
FIG 8.
Real-time RT-PCR results of cattle after a single-shot immunization and subsequent challenge infection. The results of the animal that are marked by a red plus sign correspond to the results of the serum neutralization test of the same animal shown in Fig. 7.
After challenge infection with our standardized infectious bovine serum, all but one cow (previously inoculated with rSBVΔNSs) tested negative by real-time PCR; the remaining animal was PCR positive on days 3 and 4 after infection (Fig. 8). Every SBV-naive control mouse injected with the identical challenge material as the vaccinated cattle seroconverted (Fig. 7), proving the infectious nature of the challenge material used.
In summary, a single-shot immunization with recombinant SBV mutants resulted in complete protection from SBV challenge infection.
DISCUSSION
Since its emergence in 2011, SBV has caused a large epidemic in European livestock (3), and it is used more and more as a model virus for studying orthobunyaviruses.
For the reduction of virus circulation and to prevent clinical disease, especially the induction of fetal malformation and premature birth or stillbirth, vaccines based on inactivated-virus formulations were developed in a short time frame in analogy to the related AKAV. Most SBV-specific vaccine candidates were able to elicit protective immune responses in cattle and sheep (31). However, high-titer preparations, the use of adjuvants, and repeated inoculations of conventional vaccines are required to induce a protective immune response (35). Compared to those inactivated vaccines, live attenuated vaccines are in most cases easier to produce and more efficacious. The first available modified live vaccines were based on randomly introduced mutations via serial passages in cell culture or in susceptible animals or on in vitro passage in the presence of chemical mutagens (45–47), but reversion to wild-type and pathogenic forms cannot be excluded for this type of vaccines (35, 47), and differentiation between infected and vaccinated animals is not possible in most cases (45). The complete or partial deletion of genes, nonessential for replication or induction of immunity, reduces the risk of revertance to virulence and could enable us to distinguish vaccinated from field-infected animals (marker strategy) when used in combination with a corresponding test system (47). Generally, reversion to virulence becomes very unlikely when two or more independent attenuating deletions are introduced, while viruses lacking a single protein may show residual virulence or revertance to virulence, e.g., by compensating mutations (35).
Therefore, live vaccine candidates either lacking parts of the sequence encoding NSm or expression of the NSs protein, which is a major virulence factor of orthobunyaviruses, or a recombinant SBV with a combination of both genome alterations were generated. An SBV double deletion mutant virus should have significant safety advantages as has been demonstrated here for the first time, e.g., by the lack of viremia in inoculated cattle, which is especially important for viruses transmitted by insect vectors.
The SBV NSs protein mediates multiple functions in infected cells, including IFN antagonism (17, 18) and inhibition of the cellular protein synthesis and antiviral response by direct proteasomal degradation of the RPB1 subunit of the RNA polymerase II similar to LACV (48), thus playing a critical role in mammalian host pathogenesis. NSs-deleted BUNV and SBV replicated at comparable levels in established cell lines lacking a functional IFN system (BHK-21, CPT-Tert) but displayed a smaller plaque size and reached lower final titers than wild-type (wt) viruses did (16). In contrast to that, but in agreement with previous results (23), the virus replication of SBV NSs deletion mutants was clearly reduced in cells with a functional IFN system (SFT-R) compared to wtSBV. The involvement of NSs in the innate immune response was further demonstrated in the present study by both IFN bioassays, the lack of viremia in interferon-competent adult cattle infected with the NSs deletion mutant, and an unaltered virulence of that virus when infecting mice lacking a functional IFN-α/β receptor (49). Apart from the mammalian host, NSs could also play a role in the insects (mosquitos or midges) involved in virus transmission. In vitro experiments with BUNV demonstrated the importance of NSs for the infection of mosquitoes. However, for the infection of insect cell lines with BUNV or LACV, NSs was nonessential (25, 26). Accordingly, the SBV NSs deletion mutant generated in this study did not show a reduced growth rate compared to rSBV in KC cells (data not shown).
Owing to its recent discovery, little is known about the role of SBV's NSm protein; for other bunyaviruses, however, it was shown that NSm could also be important for virulence in mammalian and insect hosts (29, 50–53). Because NSm-coding strategies of phleboviruses differ from those of the orthobunyaviruses, viable RVFV mutants completely lacking the NSm-coding region could be successfully rescued. For orthobunyaviruses such as BUNV, viable mutants were generated by partial deletion of the central domains (27). The N-terminal region of BUNV NSm is required for virus assembly, while the C-terminal hydrophobic domain probably has a function as an internal signal sequence for the Gc glycoprotein (27). Our rSBVΔNSm mutant also displays a large deletion within the central region of the M-segment-encoded nonstructural protein. Though not definitely proven because NSm-specific antibodies were not available, it is very unlikely that a virus with such a large deletion in a protein-coding sequence displays a fully functional form of that protein.
In agreement with other bunyaviruses such as BUNV or RVFV, both nonstructural proteins of SBV, NSs and NSm, are not required for viral growth in vitro, as has been demonstrated by the replication of both mutant viruses in IFN-defective BHK-21 cells at a level comparable to that of the parental rSBV. Furthermore, similar to RVFV (28), NSm of SBV seems to be not necessary for replication in interferon-competent mammalian hosts, since the rSBVΔNSm virus with the partial deletion induced viremia lasting several days in cattle, one of the major target species of SBV. Consequently, despite the prevention of disease after a challenge infection with an SBV field strain 3 weeks after immunization, SBVΔNSm alone is not an adequate vaccine candidate due to the risk of transmission of the mutant virus by insect vectors. In contrast, both viruses containing an NSs deletion (rSBVΔNSs and rSBVΔNSs/ΔNSm) caused no detectable viremia or any adverse side effects in immunized cattle, but in most cases, they induced a measurable antibody response after a single-shot vaccination. The duration of immunity, however, needs to be further evaluated.
In addition, the double deletion mutant completely prevented viremia in all cattle after a subsequent challenge infection with virulent wild-type SBV, making it a safe candidate vaccine. Furthermore, rSBVΔNSs/ΔNSm could enable in the absence of both nonstructural proteins the differentiation between vaccinated and field-infected animals if a companion serological test system is established in the future.
Until now, modified live vaccine candidates lacking both nonstructural proteins in combination were not available for any orthobunyavirus, but they have been described for the phlebovirus RVFV and were proven to be safe and protective (29, 50). In contrast to that RVFV NSs/NSm double deletion mutant which grew to high titers in Vero E6 cells, the SBV double mutant virus rSBVΔNSs/ΔNSm had a visible growth defect (ca. 1 log10 unit) in BHK-21 cells compared to the parental rSBV, and both single mutant viruses (rSBVΔNSs and rSBVΔNSm) and lower final virus titers were obtained. In mice lacking a functional IFN-α/β receptor, the injection of both single as well as the double deletion mutants led to infection, as has been demonstrated by the presence of viral genome and/or the induction of an antibody response. The infection with SBV, however, varied between the groups and was generally not as lethal as described before (18), but the results match those of previous infection studies with this mouse line (8), which is presumably dependent on the age of the mice as described for the Simbu serogroup virus AKAV as well (54), route of inoculation, or applied viral dose. Furthermore, virus dosages that are too high may activate other defense mechanisms, even when an IFN-α/β receptor is lacking, which might lead to higher survival rates as seen in the present study in the IFNAR−/− mouse group inoculated with 106 TCID50/ml. However, the remarkably high mortality rate for mice infected with rSBVΔNSs might imply additional effects of NSs apart from its interaction with the interferon system of the host, which needs to be evaluated further in future studies.
In contrast to IFN-deficient systems, substantial differences between the mutant viruses were observed in systems that have an intact IFN system. As an example, in SFT-R cells, both mutants containing an NSs deletion (rSBVΔNSs and rSBVΔNSs/ΔNSm) were attenuated and showed a marked growth defect compared to the parental rSBV, and no plaque formation could be observed.
Since rSBVΔNSm and rSBV showed similar growth properties in SFT-R cells and indistinguishable plaques, only NSs seems to interfere with the host IFN response, while SBV NSm should be dispensable in this process. Indeed, both mutants lacking NSs induced high levels of IFN in SFT-R cells compared to the genetically intact rSBV. Interestingly, IFN expression in rSBVΔNSs/ΔNSm-infected cells was nearly twice as high as in supernatants of rSBVΔNSs-infected cells. This presumably potentiating effect caused by the additional NSm deletion in the double mutant did not correlate with the small amounts of IFN that could be detected in supernatants of rSBVΔNSm-infected SFT-R cells. The reasons for that observation need to be investigated further.
Not only in in vitro systems with an intact IFN system but also in IFN-competent cattle, substantial differences could be observed for the mutant viruses. In contrast to the unmodified virus and the NSm deletion mutant, both viruses lacking NSs (rSBVΔNSs and rSBVΔNSs/ΔNSm) were completely attenuated, and no RNAemia was observed in the natural host, confirming the safety of these vaccine candidates in terms of potential transmission by insect vectors. Despite the reduction in viral virulence, the mutant viruses maintained their immunogenicity, which is one of the major challenges in the development of modified live vaccines (35); in most immunized cattle, a measurable antibody response was elicited. The antibody titers, however, were low, as has been observed after two applications of some chemically inactivated whole-virus SBV-specific prototype vaccines (31), and nearly no increase was noticed after challenge infection. The only exception was one animal out of four immunized with rSBVΔNSs and not protected in full from challenge with virulent virus. In agreement with genetics-derived RVFV vaccine candidates, only rSBV lacking NSs and NSm in combination completely prevented viremia after challenge infection (29, 50). However, since the most important pathogenic effect of SBV is the induction of fetal malformation and stillbirth or premature birth, the safety and efficacy of this newly designed SBV-specific vaccine candidate need to be evaluated in further studies in pregnant ruminants as well.
To conclude, SBVs lacking both nonstructural proteins in combination have been demonstrated to be innocuous and efficient when cattle were challenged 3 weeks after vaccination, and this potentially enables the differentiation between vaccinated and field-infected animals once a companion serological test system is established for NSs or NSm. Therefore, the double deletion mutant will provide a basis for further developments of highly efficient modified live SBV vaccines which are also a model for related orthobunyaviruses.
ACKNOWLEDGMENTS
This work was financially supported by Boehringer Ingelheim Vetmedica GmbH.
We are grateful to Friedemann Weber, Philipps University Marburg, for kindly providing the plasmid vector pT7riboSM2, Martin D. Fray, Pirbright Institute, Compton, United Kingdom, for kindly providing MDBK-t2 cells, and Nicolas Ruggli, Institute of Virology and Immunoprophylaxis, Mittelhäusern, for making the SK6-MxLuc cells available. We thank Gabriela Adam, Anja Landmesser, and Petra Meyer for outstanding technical assistance and Mandy Jörn for photographic help. Dedicated animal care was provided by the staff of the biosafety level 3 (BSL-3) facility of the Friedrich-Loeffler-Institut.
REFERENCES
- 1.Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, Schirrmeier H, Eschbaumer M, Goller KV, Wernike K, Fischer M, Breithaupt A, Mettenleiter TC, Beer M. 2012. Novel orthobunyavirus in cattle, Europe, 2011. Emerg Infect Dis 18:469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer M, Conraths FJ, van der Poel WH. 2013. ‘Schmallenberg virus’ – a novel orthobunyavirus emerging in Europe. Epidemiol Infect 141:1–8. doi: 10.1017/S0950268812002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wernike K, Conraths F, Zanella G, Granzow H, Gache K, Schirrmeier H, Valas S, Staubach C, Marianneau P, Kraatz F, Höreth-Böntgen D, Reimann I, Zientara S, Beer M. 2014. Schmallenberg virus—two years of experiences. Prev Vet Med 116:423–434. doi: 10.1016/j.prevetmed.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 4.Conraths FJ, Peters M, Beer M. 2013. Schmallenberg virus, a novel orthobunyavirus infection in ruminants in Europe: potential global impact and preventive measures. N Z Vet J 61:63–67. doi: 10.1080/00480169.2012.738403. [DOI] [PubMed] [Google Scholar]
- 5.Jack C, Anstaett O, Adams J, Noad R, Brownlie J, Mertens P. 2012. Evidence of seroconversion to SBV in camelids. Vet Rec 170:603. doi: 10.1136/vr.e3939. [DOI] [PubMed] [Google Scholar]
- 6.Wensman JJ, Blomqvist G, Hjort M, Holst BS. 2013. Presence of antibodies to Schmallenberg virus in a dog in Sweden. J Clin Microbiol 51:2802–2803. doi: 10.1128/JCM.00877-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linden A, Desmecht D, Volpe R, Wirtgen M, Gregoire F, Pirson J, Paternostre J, Kleijnen D, Schirrmeier H, Beer M, Garigliany MM. 2012. Epizootic spread of Schmallenberg virus among wild cervids, Belgium, fall 2011. Emerg Infect Dis 18:2006–2008. doi: 10.3201/eid1812.121067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wernike K, Breithaupt A, Keller M, Hoffmann B, Beer M, Eschbaumer M. 2012. Schmallenberg virus infection of adult type I interferon receptor knock-out mice. PLoS One 7:e40380. doi: 10.1371/journal.pone.0040380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ducomble T, Wilking H, Stark K, Takla A, Askar M, Schaade L, Nitsche A, Kurth A. 2012. Lack of evidence for Schmallenberg virus infection in highly exposed persons, Germany, 2012. Emerg Infect Dis 18:1333–1335. doi: 10.3201/eid1808.120533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reusken C, van den Wijngaard C, van Beek P, Beer M, Bouwstra R, Godeke GJ, Isken L, van den Kerkhof H, van Pelt W, van der Poel W, Reimerink J, Schielen P, Schmidt-Chanasit J, Vellema P, de Vries A, Wouters I, Koopmans M. 2012. Lack of evidence for zoonotic transmission of Schmallenberg virus. Emerg Infect Dis 18:1746–1754. doi: 10.3201/eid1811.120650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen LD, Kristensen B, Kirkeby C, Rasmussen TB, Belsham GJ, Bodker R, Bøtner A. 2012. Culicoids as vectors of Schmallenberg virus. Emerg Infect Dis 18:1204–1206. doi: 10.3201/eid1807.120385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elbers AR, Meiswinkel R, van Weezep E, Kooi EA, van der Poel WH. 24July2013. Schmallenberg virus in Culicoides biting midges in the Netherlands in 2012. Transbound Emerg Dis doi: 10.1111/tbed.12128. [DOI] [PubMed] [Google Scholar]
- 13.Walter CT, Barr JN. 2011. Recent advances in the molecular and cellular biology of bunyaviruses. J Gen Virol 92:2467–2484. doi: 10.1099/vir.0.035105-0. [DOI] [PubMed] [Google Scholar]
- 14.Bilk S, Schulze C, Fischer M, Beer M, Hlinak A, Hoffmann B. 2012. Organ distribution of Schmallenberg virus RNA in malformed newborns. Vet Microbiol 159:236–238. doi: 10.1016/j.vetmic.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 15.Breard E, Lara E, Comtet L, Viarouge C, Doceul V, Desprat A, Vitour D, Pozzi N, Cay AB, De Regge N, Pourquier P, Schirrmeier H, Hoffmann B, Beer M, Sailleau C, Zientara S. 2013. Validation of a commercially available indirect Elisa using a nucleocapsid recombinant protein for detection of Schmallenberg virus antibodies. PLoS One 8:e53446. doi: 10.1371/journal.pone.0053446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc Natl Acad Sci U S A 98:664–669. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott RM, Blakqori G, van Knippenberg IC, Koudriakova E, Li P, McLees A, Shi X, Szemiel AM. 2013. Establishment of a reverse genetics system for Schmallenberg virus, a newly emerged orthobunyavirus in Europe. J Gen Virol 94:851–859. doi: 10.1099/vir.0.049981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varela M, Schnettler E, Caporale M, Murgia C, Barry G, McFarlane M, McGregor E, Piras IM, Shaw A, Lamm C, Janowicz A, Beer M, Glass M, Herder V, Hahn K, Baumgartner W, Kohl A, Palmarini M. 2013. Schmallenberg virus pathogenesis, tropism and interaction with the innate immune system of the host. PLoS Pathog 9:e1003133. doi: 10.1371/journal.ppat.1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blakqori G, Weber F. 2005. Efficient cDNA-based rescue of La Crosse bunyaviruses expressing or lacking the nonstructural protein NSs. J Virol 79:10420–10428. doi: 10.1128/JVI.79.16.10420-10428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa Y, Sugiura K, Kato K, Tohya Y, Akashi H. 2007. Rescue of Akabane virus (family Bunyaviridae) entirely from cloned cDNAs by using RNA polymerase I. J Gen Virol 88:3385–3390. doi: 10.1099/vir.0.83173-0. [DOI] [PubMed] [Google Scholar]
- 21.Weber F, Bridgen A, Fazakerley JK, Streitenfeld H, Kessler N, Randall RE, Elliott RM. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J Virol 76:7949–7955. doi: 10.1128/JVI.76.16.7949-7955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Knippenberg I, Fragkoudis R, Elliott RM. 2013. The transient nature of Bunyamwera orthobunyavirus NSs protein expression: effects of increased stability of NSs protein on virus replication. PLoS One 8:e64137. doi: 10.1371/journal.pone.0064137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barry G, Varela M, Ratinier M, Blomstrom AL, Caporale M, Seehusen F, Hahn K, Schnettler E, Baumgartner W, Kohl A, Palmarini M. 2014. The NSs protein of Schmallenberg virus counteracts the antiviral response of the cell by inhibiting its transcriptional machinery. J Gen Virol 95:1640–1646. doi: 10.1099/vir.0.065425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohl A, Clayton RF, Weber F, Bridgen A, Randall RE, Elliott RM. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J Virol 77:7999–8008. doi: 10.1128/JVI.77.14.7999-8008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J Virol 81:4991–4999. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szemiel AM, Failloux AB, Elliott RM. 2012. Role of Bunyamwera orthobunyavirus NSs protein in infection of mosquito cells. PLoS Negl Trop Dis 6:e1823. doi: 10.1371/journal.pntd.0001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi X, Kohl A, Leonard VH, Li P, McLees A, Elliott RM. 2006. Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis. J Virol 80:8089–8099. doi: 10.1128/JVI.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird BH, Albarino CG, Nichol ST. 2007. Rift Valley fever virus lacking NSm proteins retains high virulence in vivo and may provide a model of human delayed onset neurologic disease. Virology 362:10–15. doi: 10.1016/j.virol.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 29.Bird BH, Maartens LH, Campbell S, Erasmus BJ, Erickson BR, Dodd KA, Spiropoulou CF, Cannon D, Drew CP, Knust B, McElroy AK, Khristova ML, Albarino CG, Nichol ST. 2011. Rift Valley fever virus vaccine lacking the NSs and NSm genes is safe, nonteratogenic, and confers protection from viremia, pyrexia, and abortion following challenge in adult and pregnant sheep. J Virol 85:12901–12909. doi: 10.1128/JVI.06046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hechinger S, Wernike K, Beer M. 2013. Evaluating the protective efficacy of a trivalent vaccine containing Akabane virus, Aino virus and Chuzan virus against Schmallenberg virus infection. Vet Res 44:114. doi: 10.1186/1297-9716-44-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wernike K, Nikolin VM, Hechinger S, Hoffmann B, Beer M. 2013. Inactivated Schmallenberg virus prototype vaccines. Vaccine 31:3558–3563. doi: 10.1016/j.vaccine.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 32.British Veterinary Association. 2013. VMD authorises SBV vaccine for use in the UK. Vet Rec 172:543. doi: 10.1136/vr.f3336.23709393 [DOI] [Google Scholar]
- 33.Luongo C, Winter CC, Collins PL, Buchholz UJ. 2012. Increased genetic and phenotypic stability of a promising live-attenuated respiratory syncytial virus vaccine candidate by reverse genetics. J Virol 86:10792–10804. doi: 10.1128/JVI.01227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins SR, Yoder HW Jr. 1986. Reversion to virulence of chicken-passaged infectious bronchitis vaccine virus. Avian Dis 30:221–223. doi: 10.2307/1590639. [DOI] [PubMed] [Google Scholar]
- 35.Shams H. 2005. Recent developments in veterinary vaccinology. Vet J 170:289–299. doi: 10.1016/j.tvjl.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wechsler SJ, McHolland LE, Wilson WC. 1991. A RNA virus in cells from Culicoides variipennis. J Invertebr Pathol 57:200–205. doi: 10.1016/0022-2011(91)90117-9. [DOI] [PubMed] [Google Scholar]
- 38.Fray MD, Mann GE, Charleston B. 2001. Validation of an Mx/CAT reporter gene assay for the quantification of bovine type-I interferon. J Immunol Methods 249:235–244. doi: 10.1016/S0022-1759(00)00359-8. [DOI] [PubMed] [Google Scholar]
- 39.Ocana-Macchi M, Bel M, Guzylack-Piriou L, Ruggli N, Liniger M, McCullough KC, Sakoda Y, Isoda N, Matrosovich M, Summerfield A. 2009. Hemagglutinin-dependent tropism of H5N1 avian influenza virus for human endothelial cells. J Virol 83:12947–12955. doi: 10.1128/JVI.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geiser M, Cebe R, Drewello D, Schmitz R. 2001. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques 31:88–90, 92. [DOI] [PubMed] [Google Scholar]
- 41.Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Muhlberger E, Mirazimi A, Weber F. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowen AC, Noonan C, McLees A, Elliott RM. 2004. Efficient bunyavirus rescue from cloned cDNA. Virology 330:493–500. doi: 10.1016/j.virol.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Wernike K, Eschbaumer M, Breithaupt A, Hoffmann B, Beer M. 2012. Schmallenberg virus challenge models in cattle: infectious serum or culture-grown virus? Vet Res 43:84. doi: 10.1186/1297-9716-43-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wernike K, Eschbaumer M, Schirrmeier H, Blohm U, Breithaupt A, Hoffmann B, Beer M. 2013. Oral exposure, reinfection and cellular immunity to Schmallenberg virus in cattle. Vet Microbiol 165:155–159. doi: 10.1016/j.vetmic.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 45.Vannie P, Capua I, Le Potier MF, Mackay DK, Muylkens B, Parida S, Paton DJ, Thiry E. 2007. Marker vaccines and the impact of their use on diagnosis and prophylactic measures. Rev Sci Tech 26:351–372. [PubMed] [Google Scholar]
- 46.Plotkin SA. 2009. Vaccines: the fourth century. Clin Vaccine Immunol 16:1709–1719. doi: 10.1128/CVI.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henderson LM. 2005. Overview of marker vaccine and differential diagnostic test technology. Biologicals 33:203–209. doi: 10.1016/j.biologicals.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 48.Verbruggen P, Ruf M, Blakqori G, Overby AK, Heidemann M, Eick D, Weber F. 2011. Interferon antagonist NSs of La Crosse virus triggers a DNA damage response-like degradation of transcribing RNA polymerase II. J Biol Chem 286:3681–3692. doi: 10.1074/jbc.M110.154799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 50.Bird BH, Albarino CG, Hartman AL, Erickson BR, Ksiazek TG, Nichol ST. 2008. Rift valley fever virus lacking the NSs and NSm genes is highly attenuated, confers protective immunity from virulent virus challenge, and allows for differential identification of infected and vaccinated animals. J Virol 82:2681–2691. doi: 10.1128/JVI.02501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crabtree MB, Kent Crockett RJ, Bird BH, Nichol ST, Erickson BR, Biggerstaff BJ, Horiuchi K, Miller BR. 2012. Infection and transmission of Rift Valley fever viruses lacking the NSs and/or NSm genes in mosquitoes: potential role for NSm in mosquito infection. PLoS Negl Trop Dis 6:e1639. doi: 10.1371/journal.pntd.0001639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morrill JC, Laughlin RC, Lokugamage N, Wu J, Pugh R, Kanani P, Adams LG, Makino S, Peters CJ. 2013. Immunogenicity of a recombinant Rift Valley fever MP-12-NSm deletion vaccine candidate in calves. Vaccine 31:4988–4994. doi: 10.1016/j.vaccine.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weingartl HM, Nfon CK, Zhang S, Marszal P, Wilson WC, Morrill JC, Bettinger GE, Peters CJ. 2014. Efficacy of a recombinant Rift Valley fever virus MP-12 with NSm deletion as a vaccine candidate in sheep. Vaccine 32:2345–2349. doi: 10.1016/j.vaccine.2013.12.064. [DOI] [PubMed] [Google Scholar]
- 54.Kurogi H, Inaba Y, Takahashi E, Sato K, Akashi H, Satoda K, Omori T. 1978. Pathogenicity of different strains of Akabane virus for mice. Natl Inst Anim Health Q 18:1–7. [PubMed] [Google Scholar]