ABSTRACT
Adenovirus vectors are widely used as vaccine candidates for a variety of pathogens, including HIV-1. To date, human and chimpanzee adenoviruses have been explored in detail as vaccine vectors. The phylogeny of human and chimpanzee adenoviruses is overlapping, and preexisting humoral and cellular immunity to both are exhibited in human populations worldwide. More distantly related adenoviruses may therefore offer advantages as vaccine vectors. Here we describe the primary isolation and vectorization of three novel adenoviruses from rhesus monkeys. The seroprevalence of these novel rhesus monkey adenovirus vectors was extremely low in sub-Saharan Africa human populations, and these vectors proved to have immunogenicity comparable to that of human and chimpanzee adenovirus vaccine vectors in mice. These rhesus monkey adenoviruses phylogenetically clustered with the poorly described adenovirus species G and robustly stimulated innate immune responses. These novel adenoviruses represent a new class of candidate vaccine vectors.
IMPORTANCE Although there have been substantial efforts in the development of vaccine vectors from human and chimpanzee adenoviruses, far less is known about rhesus monkey adenoviruses. In this report, we describe the isolation and vectorization of three novel rhesus monkey adenoviruses. These vectors exhibit virologic and immunologic characteristics that make them attractive as potential candidate vaccine vectors for both HIV-1 and other pathogens.
INTRODUCTION
Recombinant adenoviruses (Ads) are currently being explored as candidate vaccine vectors for multiple pathogens (1–6), as a result of their safety profile, manufacturability, and ability to induce broad and strong immune responses (7–16). Multiple human and chimpanzee adenovirus vectors have been developed to date (8, 9, 11–13). The majority of these adenovirus vectors are from species B, C, D, and E. Adenovirus vectors from avian, bovine, and other species have also been constructed, but their different genomic structures may necessitate the development of a novel manufacturing platform for clinical development (17, 18). Old World monkey adenoviruses have been hypothesized to be distinct from both human and chimpanzee adenoviruses and may offer unique advantages, such as the ability to more efficiently bypass preexisting immunity to human adenoviruses (12, 19–24), while maintaining the genomic structure and growth properties of human adenoviruses.
We isolated simian adenoviruses from fecal samples from rhesus monkeys (Macaca mulatta) that were positive for adenoviruses by metagenomics sequencing (25). We performed whole-genome sequencing of these novel adenoviruses and determined their phylogeny in comparison with that of other human and simian adenoviruses. The basic genetic structure of these novel rhesus monkey adenoviruses proved similar to that of human adenoviruses (7, 16, 26). We vectorized three rhesus monkey adenoviruses and then assessed humans in sub-Saharan Africa for seroprevalence of these adenoviruses and assessed these vectors for immunogenicity in mice. In addition, we assessed receptor usage, innate immune stimulation, and adaptive immune responses.
MATERIALS AND METHODS
Isolation and vectorization of novel Old World monkey adenoviruses.
Stool samples from rhesus monkeys (Macaca mulatta) were resuspended in unsupplemented Dulbecco modified Eagle medium (DMEM) and filtered through a 0.45-μm-pore-size filter. Human E1-complementing cells were infected with the filtrates, incubated at 37°C with 10% CO2, and monitored for cytopathic effects (CPEs). The crude lysates were subjected to two rounds of plaque purification, and adenovirus cultures were expanded and purified by cesium chloride density gradient centrifugation as previously described (16). At each step, the lysates were tested for the presence of adenovirus using primers that were defined by metagenomics sequencing (25). The growth of purified wild-type rhesus monkey adenoviruses was assessed on human complementing and noncomplementing cell lines, including the HEK293, Per55K, and A549 cell lines (data not shown). Viral DNA was extracted by standard phenol-chloroform extraction, and whole-genome shotgun sequencing was done at GE Seqwright (Houston, TX).
Vectorization of wild-type adenoviruses was performed essentially as previously described (7). Briefly, the left region of the adenovirus genome was cloned into a pBR322 bacterial backbone. The E1 gene was deleted and replaced with a transgene expression cassette consisting of a cytomegalovirus (CMV) promoter, a multiple-cloning site, and a simian virus 40 (SV40) polyadenylation signal. The remainder of the adenovirus genome was cloned into a pWE15 cosmid backbone from which the E3 gene was deleted. Virus was grown in E1-complementing cell lines and purified by CsCl density gradient centrifugation.
Phylogeny and recombination.
For phylogenetic analysis, we used the software FastTree (27) to infer a maximum likelihood (ML) phylogenetic tree using a data set of 115 full genome sequences for adenoviruses infecting human and nonhuman primate hosts. We used the general time-reversible (GTR) model (which was inferred by maximizing likelihood) and a gamma distribution for variable mutation rates with 20 categories. Bootstrap support was calculated for phylogenetic splits using 10,000 bootstrap replicates. The unrooted tree was rooted using midpoint rooting.
The sequences were aligned using the default settings of the web server MAFFT (http://mafft.cbrc.jp/alignment/server/) (28).
The following adenovirus types were included in the phylogenetic analysis (GenBank accession numbers are given in parentheses): for human adenoviruses, adenovirus type 1 (Ad1; AC_000017), Ad2 (AC_000007), Ad3 (AY599834), Ad4 (AY594253), Ad5 (AC_000008), Ad6 (FJ349096), Ad7 (AY594255), Ad8 (AB448769), Ad9 (NC_010956), Ad10 (JN226746), Ad11 (AY163756), Ad12 (NC_001460), Ad13 (JN226747), Ad14 (AY803294), Ad15 (JN226748), Ad16 (AY601636), Ad17 (AC_000006), Ad18 (GU191019), Ad19 (JQ326209), Ad20 (JN226749), Ad21 (AY601633), Ad22 (FJ619037), Ad23 (JN226750), Ad24 (JN226751), Ad25 (JN226752), Ad26 (EF153474), Ad27 (JN226753), Ad28 (FJ824826), Ad29 (JN226754), Ad30 (JN226755), Ad31 (AM749299), Ad32 (JN226756), Ad33 (JN226758), Ad34 (AY737797), Ad35 (AY271307), Ad36 (GQ384080), Ad37 (DQ900900), Ad38 (JN226759), Ad39 (JN226760), Ad40 (NC_001454), Ad41 (DQ315364), Ad42 (JN226761), Ad43 (JN226762), Ad44 (JN226763), Ad45 (JN226764), Ad46 (AY875648), Ad47 (JN226757), Ad48 (EF153473), Ad49 (DQ393829), Ad50 (AY737798), Ad51 (JN226765), Ad52 (DQ923122), Ad53 (AB605245), Ad54 (NC_012959), Ad55 (FJ643676), Ad56 (HM770721), Ad57 (HQ003817), Ad58 (HQ883276), Ad60 (JN162672), Ad61 (JF964962), Ad62 (JN162671), Ad63 (JN935766), Ad64 (JQ326206), Ad65 (AP012285), Ad66 (JN860676), Ad67 (AP012302), and Ad68 (JN860678) and for simian adenoviruses, simian adenovirus type 1 (sAd1; AY771780), sAd3 (AY598782), sAd6 (JQ776547), sAd7 (DQ792570), sAd18 (NC_022266), sAd20 (HQ605912), sAd21 (AC_000010), sAd22 (AY530876), sAd23 (AY530877), sAd24 (AY530878), sAd25 (AC_000011), sAd26 (FJ025923), sAd27.1 (FJ025909), sAd28.1 (FJ025914), sAd29 (FJ025916), sAd30 (FJ025920), sAd31.1 (FJ025906), sAd32 (FJ025911), sAd34 (FJ025905), sAd35.1 (FJ025912), sAd36 (FJ025917), sAd37.1 (FJ025921), sAd38 (FJ025922), sAd39 (FJ025924), sAd40.1 (FJ025907), sAd41.1 (FJ025913), sAd42.1 (FJ025903), sAd43 (FJ025900), sAd44 (FJ025899), sAd45 (FJ025901), sAd46 (FJ025930), sAd47 (FJ025929), sAd48 (HQ241818), sAd49 (HQ241819), sAd50 (HQ241820), simian adenovirus type A1139 (JN880448), simian adenovirus type A1163 (JN880449), simian adenovirus type A1327 (JN880455), simian adenovirus type A1335 (JN880456), simian adenovirus type A1296 (JN880453), simian adenovirus type A1312 (JN880454), simian adenovirus type A1173 (JN880450), simian adenovirus type A1285 (JN880452), simian adenovirus type A1258 (JN880451), baboon Ad1 (KC693021), baboon Ad2 (KC693022), and chimpanzee adenovirus type Y25 (NC_017825. Novel sequences for rhesus monkey adenovirus type Ad51 (RhAd51), RhAd52, and RhAd53 were used in this study.
We investigated potential across-species recombination events for the novel vectors using the program RIP (29) at the Los Alamos National Laboratory (LANL) HIV database (http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html). We constructed consensus full-length genomes for each species, human adenovirus species A to G, and simian adenovirus species A, using the sequences from the data set mentioned above and the Consensus Maker tool at the LANL HIV database (http://www.hiv.lanl.gov/content/sequence/CONSENSUS/consensus.html). For species G, the consensus sequence was constructed using the Ad52, sAd1, and sAd7 sequences. Using RIP, the similarity of the sequences of novel vectors was compared to the consensus sequence of each species in sliding windows of 1 kb, with the significance threshold for best similarity being set to 0.01.
Seroprevalence.
The seroprevalence of the novel rhesus monkey adenovirus vectors in 80 South African and 64 Rwandan serum samples as well as in 108 naive rhesus monkey serum samples was tested using a luciferase-based neutralization assay, as previously described (30). Human samples were obtained with informed consent, and studies were approved by the Beth Israel Deaconess Medical Center Institutional Review Board (IRB). Briefly, serum was serially diluted in a 96-well plate, with the exception that the last column served as maximum infectivity. Virus was added, which was followed by addition of A549 cells. The plates were incubated for 24 h before the medium was removed and 100 μl phosphate-buffered saline (PBS) and 100 μl Steady-Glo substrate (Promega) were added to the wells. Luminescence was read on a Victor 3 multilabel counter (PerkinElmer, Waltham, MA). The seroprevalence titer was determined to be the dilution of serum where 90% of the virus was neutralized in the presence of serum.
Transgene expression.
Transgene expression of purified virus was determined by Western blotting. A549 cells were infected with 1 × 109 viral particles (vp) of purified virus encoding a human immunodeficiency virus type 1 (HIV-1) envelope (Env) antigen and incubated at 37°C in 10% CO2. After a 48-h incubation period, the cells were washed with 1× PBS and harvested on ice in a lysis buffer containing a protease inhibitor tablet (Roche). Samples were centrifuged and frozen at −20°C. For the Western blotting assay, sample reducing agent was added to the samples and the mixture was heated to 96°C for 10 min, after which it was run on a 10% SDS-polyacrylamide reducing gel (NuPAGE; Invitrogen). Proteins were transferred from the gel to a polyvinylidene difluoride (PVDF) membrane using an iBlot system (Life Technologies, Grand Island, NY). Monoclonal antibody (MAb) 5F3 (Polymun, Austria) was used as the primary antibody. Horseradish peroxidase (HRP)-conjugated anti-human IgG (Jackson ImmunoResearch, West Grove, PA) was used as the secondary antibody, and then chemiluminescence detection, using the ECL Plus substrate (GE Healthcare), was performed. Light-sensitive films were exposed to the membranes and developed and fixed using a Kodak X-OMAT 2000A processor.
Immunogenicity in mice.
To determine the ability of these novel rhesus monkey adenovirus vectors to induce humoral immune responses, BALB/c mice (n = 4) were immunized intramuscularly with 1 × 109 or 1 × 108 vp of vectors expressing HIV-1 459C Env gp140 at day 0. Vectors expressing HIV-1 Env were utilized in this experiment, as Env is considered an important antibody target. Serum was taken at day 0 preimmunization and at day 28 postimmunization. Purified 459C gp140 was used as a positive control and formulated in 15% (vol/vol) Emulsigen oil-in-water emulsion (MVP Laboratories) and 50 μg CpG (Midland Reagent Company) as adjuvants. The titers of antibodies to heterologous HIV-1 MosI or C97ZA012 Env gp140 or to homologous 459C Env gp140 in these sera were assayed by enzyme-linked immunosorbent assay (ELISA), in which the proteins were developed with SureBlue tetramethylbenzidine microwell peroxidase substrate (KPL). Log10 values were plotted using GraphPad Prism (version 6) software.
To assess the cellular immunogenicity of these novel rhesus monkey adenovirus vectors, C57BL/6 mice (n = 8) were immunized with 109, 108, or 107 vp of vectors expressing simian immunodeficiency virus (SIV) mac239 Gag. SIV Gag-specific CD8+ T lymphocytes were assessed at weekly intervals by major histocompatibility complex class I-restricted Db/AL11 tetramer staining as described previously (31). Further assessment was done using gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISPOT) assays with splenocytes from spleens harvested at day 28. Splenocytes were isolated and stimulated in vitro with a SIV mac239 Gag peptide pool, the CD8+ T-lymphocyte epitopes AL11 (AAVKNWMTQTL) and KV9 (KSLYNTVCV), and the CD4+ T-lymphocyte epitope DD13 (DRFYKSLRAEQTD), as described previously (32). Results reflect those from at least two separate experiments. All animal studies were approved by the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee (IACUC).
Receptor binding.
Receptor binding was tested in the A549 human carcinoma cell line (CCL-185; American Type Culture Collection [ATCC]). Cells were maintained in DMEM supplemented with 10% fetal bovine serum (Seradigm, UT, USA). Cells were infected with enhanced green fluorescent protein (eGFP)-expressing vectors in the absence or presence of various concentrations of anti-CAR antibodies (antibodies E1-1 and 3C100; Santa Cruz) or anti-CD46 antibodies (antibodies M177 [Hycult Biotech] and MEM-258 [Abnova]). After 24 h incubation at 37°C, cells were harvested in 10% CO2 (TryplE Select; Invitrogen), washed with magnetically activated cell sorting buffer, and collected by centrifugation. The cells were resuspended in 2% formaldehyde and subjected to flow cytometry. FlowJo (version 8) software (Tree Star, Inc.) was used to gate eGFP-positive cells, and counts were normalized to 100% transduction in the absence of any antibody.
Simian Ad cytokine elicitation.
Stimulation of innate cytokines by the novel rhesus monkey adenoviruses was assessed by Luminex assays (Milliplex nonhuman primate premix 23-plex), as previously described (33, 34). Fresh rhesus monkey peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density gradient centrifugation. Rhesus monkey PBMCs rather than human PBMCs were utilized in this experiment to assess the natural biology of these viruses. Cells (1 × 106) were stimulated with adenovirus vectors at a multiplicity of infection (MOI) of 1,000. At 24 h postinfection, the supernatants were harvested and analyzed for cytokine and chemokine levels by Luminex assays according to the manufacturer's protocol.
Nucleotide sequence accession numbers.
The sequences of RhAd51, RhAd52, and RhAd53 were submitted to GenBank and can be found under accession numbers KM591901, KM591902, and KM591903, respectively.
RESULTS
Isolation of rhesus monkey adenoviruses.
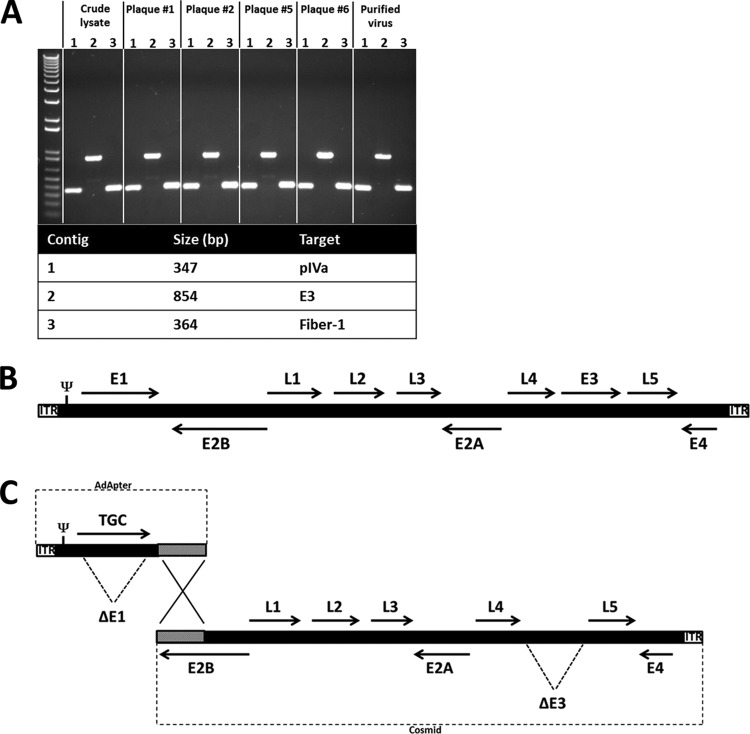
Metagenomics sequencing was used to define the enteric virome of SIV-infected rhesus monkeys, as previously reported (25). In the 25 rhesus monkeys studied, 19 possible novel adenoviruses were detected. We isolated three novel rhesus monkey adenoviruses, RhAd51, RhAd52, and RhAd53, from stool samples that tested positive for adenovirus by two rounds of plaque purification. Primers for screening of the viral cultures were designed from the metagenomics sequence reads, and PCR results were assessed by gel electrophoresis, as shown in Fig. 1A. Plaques that tested positive for adenovirus by PCR were amplified, and virus was purified by CsCl density gradient purification as previously described (7). In addition, we propagated several additional simian adenoviruses obtained from ATCC, including sAd16, sAd19, chimpanzee Ad24 (ChAd24; also known as sAd24 or chimpanzee AdC7), and sAd33. Whole-genome sequencing of the novel rhesus monkey adenoviruses as well as the adenoviruses obtained from ATCC revealed a genetic structure similar to that of other known adenoviruses, as shown in Fig. 1B (7, 14, 16, 26).
FIG 1.
Adenovirus identification and genomic structure. (A) PCR-amplified regions in the first crude lysate after infection of cells with filtered stool samples (crude lysate), 4 separate plaques after the second round of plaque purification, and CsCl density gradient-purified virus. Listed are the targeted regions in the adenovirus genome and their expected band sizes. First lane, a 1-kb-plus DNA ladder. (B) The layout of the early (E) and late (L) genes of the novel simian adenoviruses is similar to that of known human adenoviruses. (C) The AdApter plasmid comprises the left end of the adenovirus genome in which E1 is replaced with a transgene cassette (TGC). The cosmid contains the remaining right end of the adenovirus genome in which E3 is deleted. Ψ, virus packaging signal.
Vectorization of rhesus monkey adenoviruses.
Adenovirus vectors with an E1/E3 deletion were produced as previously described (7, 16, 26) and as shown in Fig. 1C. The left end of the adenovirus from the left inverted terminal repeat (lITR) through pIX was placed into an AdApter plasmid in which E1 was deleted and was replaced by a transgene cassette containing a CMV promoter, a multiple-cloning site, and an SV40 poly(A) tail. The remainder of the adenovirus genome from pIX through the right inverted terminal repeat (rITR) was placed into a cosmid in which the E3 region was deleted. The adapter plasmid and the cosmid have approximately 2.5 kb of overlap that facilitates homologous recombination during transfection in an E1-complementing cell line. Table 1 shows the source and growth characteristics of these simian adenoviruses. The novel rhesus monkey adenovirus vectors were viable and grew to high titers, with ratios ranging from 3 to 30 vp/PFU, similar to those seen with human adenovirus vectors (data not shown), suggesting that these vectors might be suitable for large-scale production in the standard cell lines that are currently being utilized to manufacture human adenovirus vaccine vectors (1, 35). We also constructed vectors that expressed various transgenes, including HIV/SIV Gag, Pol, Env, luciferase, and eGFP (data not shown), suggesting the generalizability of these vector platforms.
TABLE 1.
Adenovirus subtypesa
| Subtype | Sourceb | Virus growthc |
|---|---|---|
| ChAd23 (VR-592) | ATCC | No growth |
| ChAd24 (VR-593) | ATCC | Growth |
| sAd16 (VR-941) | ATCC | No growth |
| sAd19 (VR-275) | ATCC | No growth |
| sAd33 (VR-206) | ATCC | Slow growth |
| RhAd51 | Novel | Growth |
| RhAd52 | Novel | Growth |
| RhAd53 | Novel | Growth |
A vector from which E1 and E3 were deleted was used for cloning.
Indicated is whether the virus was obtained from our metagenomics screen (Novel) or from ATCC.
Growth of the replication-incompetent virus was determined after transfection into an E1-complementing cell line.
Phylogeny of rhesus monkey adenoviruses.
We inferred a maximum likelihood phylogenetic tree using full-length genome sequences for the novel vectors and 115 reported human and simian adenoviruses, which included adenoviruses that were classified into species A to G, as well as unclassified viruses, as shown in Fig. 2. Whereas previous typing has primarily been done using hemagglutination or sequence alignment of only hexon, fiber, or other regions, we used whole-genome alignments to determine placement in the phylogenetic tree. The three novel rhesus monkey adenoviruses all clustered with the poorly studied species G, which includes human Ad52, sAd1, sAd7, and sAd11. The rhesus monkey adenoviruses similarly clustered with species G if we compared hexon or fiber sequences alone (data not shown).
FIG 2.
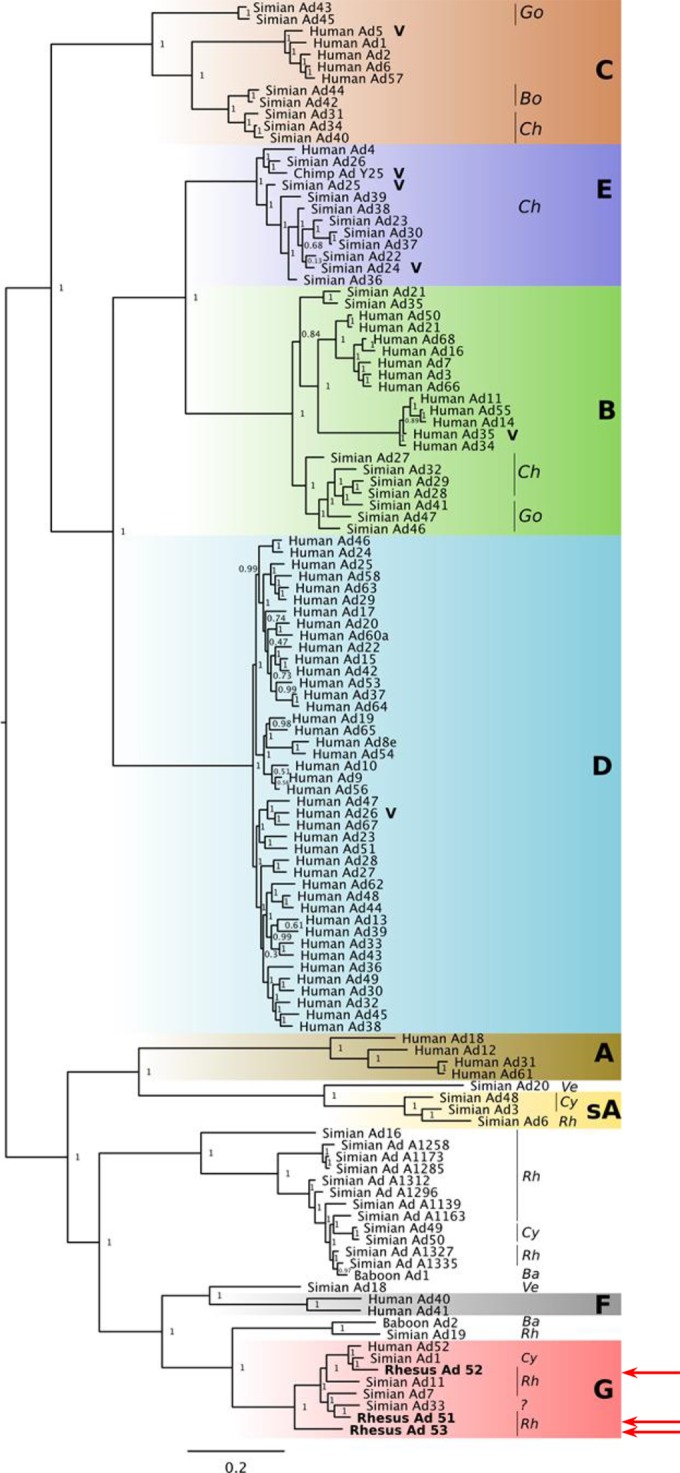
Maximum likelihood phylogenetic tree of adenovirus genomes. Full-length genomes from classified (species A to G) and unclassified human and simian adenoviruses, in addition to the novel vectors, were used to infer an ML phylogenetic tree. The different species are highlighted by colored rectangles (sA, simian species A), and in the case of simian vectors, the host species are noted (Rh, rhesus macaque; Cy, cynomolgus macaque; Go, gorilla; Ve, vervet monkey; Ba, baboon; Bo, bonobo; Ch, chimpanzee). The novel vectors were closest to species G adenoviruses (red arrows). Other vaccine candidates are highlighted with a V.
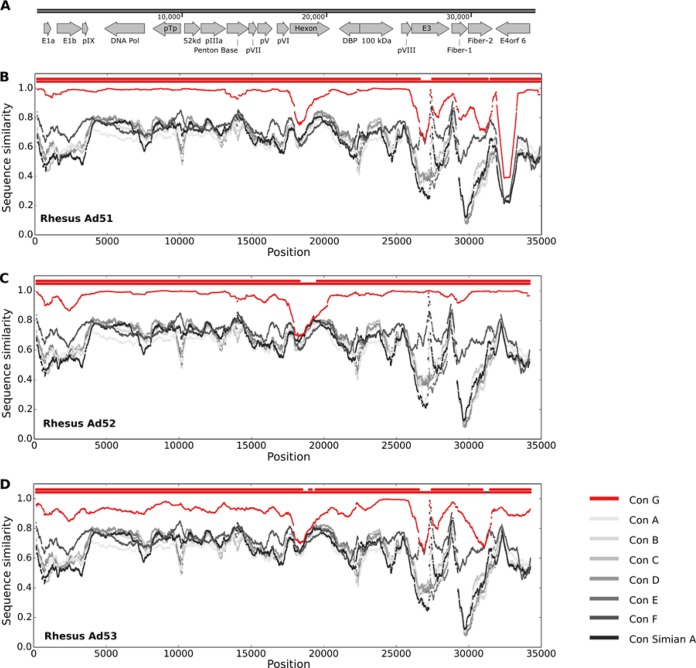
Given that recombination can occur among adenoviruses (36–39), we wanted to test whether these novel vectors were species G throughout their complete genomes. The similarity of the sequences of the novel vectors to the consensus sequence for each species, obtained using sliding windows of 1 kb along the genome, is shown in Fig. 3. Throughout their genomes, the sequence of each of the novel vectors was most closely related to species G sequences. The similarity to species G adenoviruses was higher than that to the unclassified adenoviruses as well (data not shown).
FIG 3.
Novel vectors are most similar to species G throughout their genomes. (A) Gene map for the novel vector RhAd52. For simplicity, some genes are not shown. (B to D) The similarity of the sequences of the novel vectors to the consensus from each of species A to G is shown in sliding windows of 1 kb along the genome. The similarity score is shown as a fraction and is plotted at the midpoint of each sliding window. Each subplot has 2 horizontal lines of dots on the top. The lower line shows the color of the best matching consensus sequence for a sliding window. The consensus species G sequence is shown in red. The top line shows a dot only when the best match is significant, at a P value of <0.01. Con, consensus.
Studies of seroprevalence in sub-Saharan African human sera and rhesus monkey sera.
The seroprevalence of the novel rhesus monkey adenoviruses was determined by luciferase-based neutralization assays as previously described (30). A set of 80 South African and 64 Rwandan human serum samples was tested for the seroprevalence of the viruses, as shown in Fig. 4A. As previously reported (10, 19, 40), human Ad5 neutralizing antibody titers were >200 in 67.4% of individuals in sub-Saharan Africa and >1,000 in 43.1%. Neutralizing antibody titers to human Ad26 were lower, with titers being >200 in 27.1% of individuals in sub-Saharan Africa and >1,000 in 6.3% of individuals in sub-Saharan Africa. For the chimpanzee virus ChAd24, antibody titers were generally low (<200), but 45% of individuals were still seropositive. In contrast, the seroprevalence of the novel rhesus monkey adenoviruses was extremely low in these populations, with less than 10 to 15% being seropositive and nearly all having low titers of <200. The seroprevalence of these rhesus monkey adenovirus vectors was also lower than the published seroprevalence of other human, chimpanzee, and simian Ad vectors in sub-Saharan African human populations (19, 21, 23). We next determined the seroprevalence in 108 naive rhesus monkey serum samples, as shown in Fig. 4B. As expected, monkeys showed little to no seroprevalence of the human and chimpanzee adenoviruses, but the rhesus monkey adenoviruses demonstrated intermediate neutralizing antibody titers, in particular, for RhAd52.
FIG 4.
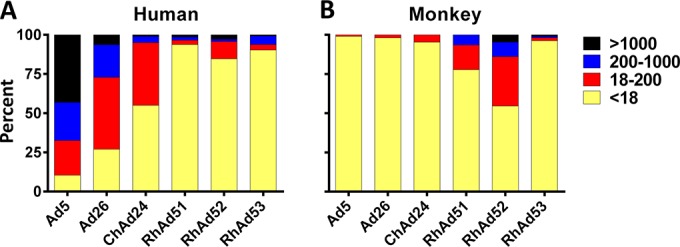
Seroprevalence of antibodies to the novel rhesus monkey adenoviruses. The seroprevalence of the novel rhesus monkey adenoviruses compared to that of human Ad5 and Ad26 and ChAd24 in 80 South African and 64 Rwandan human serum samples (A) and in 108 naive rhesus monkey serum samples (B) was determined. Undetectable and low titers are shown in yellow and red, respectively; medium and high titers are shown in blue and black, respectively.
Transgene expression and immunogenicity in mice.
To determine if the novel vectors would express the encoded transgenes, A549 cells were transduced at an MOI of 4,000 with Ad26, ChAd24, RhAd51, RhAd52, or RhAd53 expressing HIV-1 459C Env gp140. Transgene expression in cell lysates was assessed utilizing the MAb 5F3, as shown in Fig. 5. All novel rhesus monkey adenovirus vectors expressed the full-length transgene at levels comparable to those at which the Ad26 vector expressed the same transgene.
FIG 5.
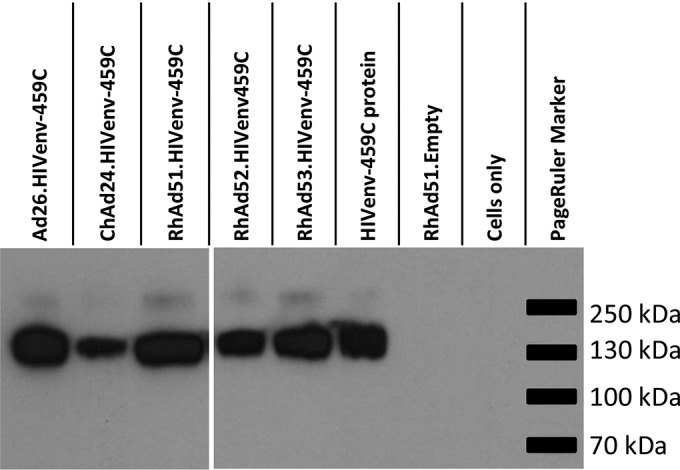
Transgene expression by novel rhesus monkey adenovirus vectors. A Western blot of lysates of cells collected 48 h postinfection with HIV-1 Env-expressing adenoviruses is shown. MAb 5F3 was used as the primary antibody, and purified HIV-1 Env protein was used as the positive control.
To assess Env-specific humoral immune responses, BALB/c mice were immunized (n = 4) with a single injection of 1 × 109 or 1 × 108 vp of adenovirus vectors expressing HIV-1 459C Env gp140. Sera from day 0 preimmunization and day 28 postimmunization were evaluated by ELISA. As shown in Fig. 6, all vectors induced binding antibodies to heterologous HIV-1 C97ZA012 and mosaic Env gp140 and to the homologous HIV-1 459C Env gp140. The titers were comparable to those induced by immunization with purified HIV-1 459C Env gp140 with CpG and Emulsigen adjuvant for all vectors except that carrying RhAd51, which induced antibody titers lower than those induced by the other vectors.
FIG 6.
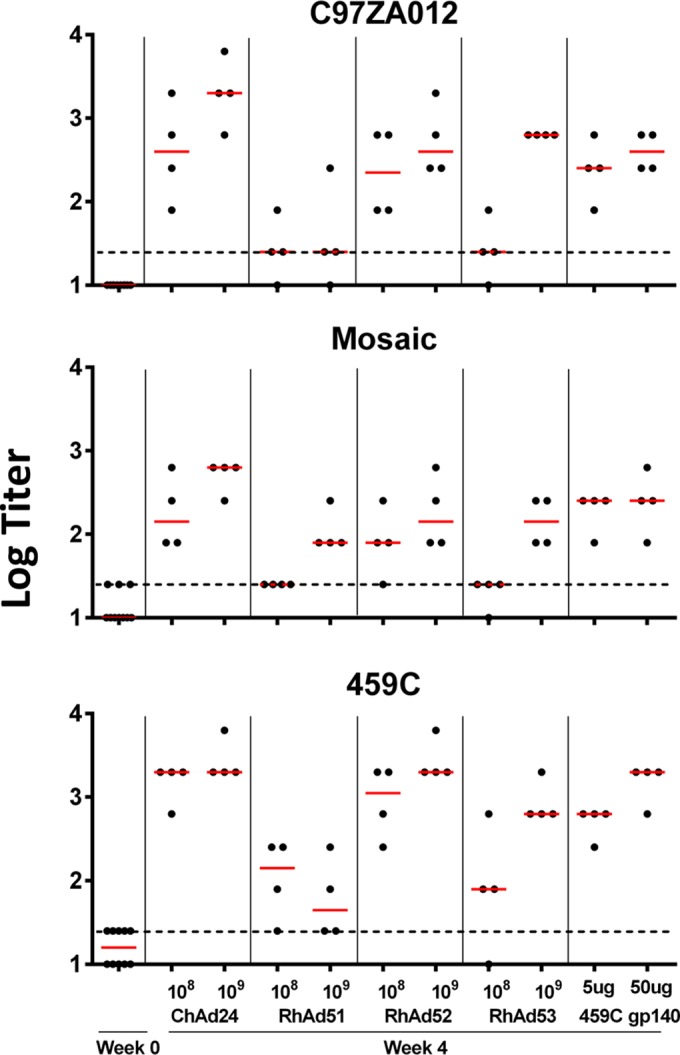
Humoral immune responses in mice. BALB/c mice (n = 4) received a single immunization with 108 or 109 vp of Ad vectors expressing HIV-1 459C Env gp140 at day 0. Immunization with purified protein was used as the positive control. The titers of antibodies to heterologous HIV-1 C97ZA012 and Mosaic Env gp140 as well as to homologous 459C Env gp140 in serum were determined by ELISA on day 0 and day 28. Red lines, medians; dotted lines, titer cutoff 2 times the average background titer.
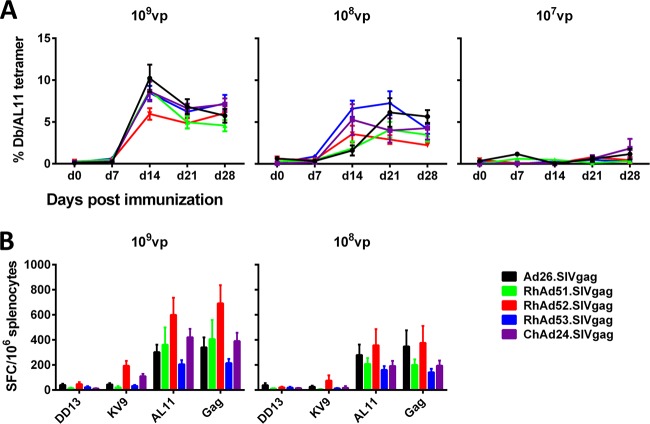
To assess Gag-specific cellular immune responses, C57BL/6 mice (n = 8) were immunized with a single injection of 1 × 109, 1 × 108, or 1 × 107 vp of ChAd24, RhAd51, RhAd52, RhAd53, or human Ad26 expressing SIV mac239 Gag. Blood was obtained at weekly intervals to determine SIV Gag-specific CD8+ T-lymphocyte responses by Db/AL11 tetramer binding assays as previously described (31). As seen in Fig. 7A, all the rhesus monkey vectors induced CD8+ T-cell responses comparable to those induced by Ad26 and ChAd24. With a dose of 1 × 107 vp, cellular immune responses were low. These results were confirmed by IFN-γ ELISPOT assays with splenocytes harvested at day 28, as shown in Fig. 7B. All novel rhesus monkey vectors induced comparable cellular immune responses at both 1 × 109 and 1 × 108 vp following stimulation with a Gag peptide pool, the CD8+ T-lymphocyte epitopes AL11 and KV9, and the CD4+ T-lymphocyte epitope DD13.
FIG 7.
Cellular immune responses in mice. (A) A single immunization of C57BL/6 mice with 1 × 109, 1 × 108, or 1 × 107 vp of vectors expressing SIV Gag at day 0. Gag-specific CD8+ T cells were detected in serum by Db/AL11 tetramer binding assays at weekly intervals. (B) Spleens were harvested at day 28 and evaluated by IFN-γ ELISPOT assays after stimulation with the DDR13, KV9, and AL11 epitopes as well as a whole SIV Gag peptide pool. SFC, number of spot-forming cells.
Receptor studies.
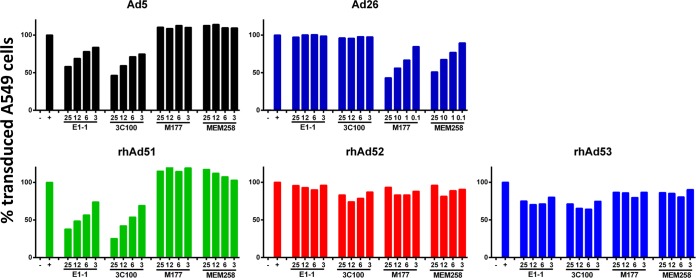
We next assessed whether the novel rhesus monkey Ads utilized the well-described CAR or CD46 cellular receptors that are used by Ad5 and Ad26, respectively (41, 42). A549 cells were infected with human Ad5, human Ad26, RhAd51, RhAd52, or RhAd53 expressing eGFP and analyzed by flow cytometry. To evaluate receptor usage, cells were preincubated with various concentrations of anti-CAR (E1-1 and 3C100) and anti-CD46 (M177 and MEM-258) monoclonal antibodies (41). As shown in Fig. 8, Ad5 was inhibited by anti-CAR MAbs but not by anti-CD46 MAbs, as expected (41). RhAd51 was also inhibited by anti-CAR MAbs, suggesting that RhAd51 can utilize CAR for viral entry. Ad26 was inhibited by anti-CD46 but not anti-CAR MAbs, as we have previously reported (41). RhAd52 and RhAd53 were not inhibited by either anti-CAR or anti-CD46 MAbs under the experimental conditions tested, suggesting that these viruses may utilize a different, as yet unidentified receptor for cell entry.
FIG 8.
Receptor binding assays. A549 cells were infected with eGFP-labeled adenovirus vectors alone or after 1 h preincubation with anti-CAR (E1-1, 3C100) or anti-CD46 (M177, MEM-258) antibodies. Cells were washed, harvested, fixed after a 48-h incubation period, and analyzed by flow cytometry. Antibody concentrations are in μg/ml. The value for transduced cells in the absence of antibody was normalized to 100%.
Innate immune cytokine profiles.
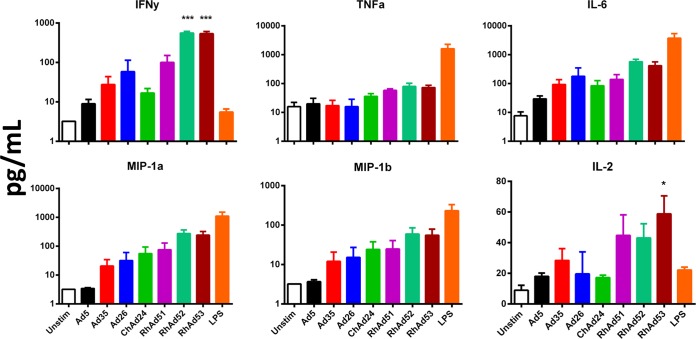
To assess innate immune cytokine profiles, rhesus monkey PBMCs were isolated and stimulated at an MOI of 1,000 vp/cell of adenovirus vectors for 24 h, after which the supernatants were analyzed by Luminex assays, as previously described (33). As shown in Fig. 9, RhAd52 and RhAd53 induced significantly higher IFN-γ responses than human Ad5, human Ad26, human Ad35, and ChAd24. RhAd53 also induced significantly higher interleukin-2 (IL-2) responses than all other vectors. In addition, trends toward higher levels of multiple other cytokines were also observed, including tumor necrosis factor alpha (TNF-α), IL-6, macrophage inflammatory protein 1α (MIP-1α), and MIP-1β. The chemokine and cytokine responses induced by RhAd51 were consistently lower than those induced by RhAd52 and RhAd53 for all chemokines and cytokines tested. These data suggest that the novel rhesus monkey adenoviruses induced distinct innate immune profiles compared with those induced by the human and chimpanzee adenoviruses that were tested.
FIG 9.
Cytokine and chemokine responses elicited in rhesus monkey PBMCs. PBMCs (n = 3) were stimulated with 1 × 103 vp/cell of various adenovirus vectors and incubated for 24 h. Cytokine and chemokine responses were measured by Luminex assays. Lipopolysaccharide at 1 ng/ml was used as a positive control. Statistical analysis was done with one-way analysis of variance between all groups. ***, P ≤ 0.0001; *, P < 0.03.
DISCUSSION
In this study, we isolated three novel rhesus monkey adenoviruses by metagenomics sequencing of stool samples followed by virus expansion and whole-genome sequencing. These rhesus monkey adenoviruses proved to be most closely related to adenovirus species G, and their genetic structure resembled that of other human or chimpanzee adenoviruses. We then vectorized these novel adenoviruses, and the resultant vectors exhibited growth characteristics similar to those of human adenovirus vectors in standard human complementing and noncomplementing cell lines. Furthermore, they stably expressed multiple transgenes and proved highly immunogenic in mice. These data suggest that these rhesus monkey adenoviruses may prove useful as a novel class of vaccine vectors.
A major research focus over the past several years has been the development of novel adenovirus vaccine vectors, apart from Ad5, that exhibit a lower seroprevalence in humans in the developing world. Most efforts to date have focused on alternative serotypes of human adenovirus vectors and chimpanzee adenovirus vectors, both of which are currently being evaluated in clinical trials (1, 13, 43, 44). However, these vectors still show a degree of seroprevalence in the developing world, and human and chimpanzee adenoviruses exhibit interdigitating phylogeny. Bovine and avian adenovirus vectors have also been produced and are more distantly related to human and chimpanzee adenovirus vectors, but the clinical development of these vectors may be slowed by the potential need to develop novel production cell lines (17, 18). In contrast, the novel rhesus monkey adenoviruses that we describe here show the unique combination of extremely low seroprevalence in sub-Saharan Africa and efficient growth in typical human complementing cell lines, which provide both theoretical and practical advantages.
The novel rhesus monkey adenovirus vectors expressed transgenes and induced humoral and cellular immune responses in mice comparable to the immunogenicity observed with human adenovirus vectors. However, their biology was substantially different, as RhAd52 and RhAd53 did not utilize CAR or CD46 for cell entry. Moreover, the cytokine and chemokine profiles in cells transduced with the RhAd vectors were markedly different from those in cells transduced with Ad5, although the impact of these differential innate immune profiles remains unclear.
Evaluations of a wide variety of adenovirus vectors in preclinical models and phase I clinical trials for multiple pathogens have shown promising results, although HIV-1 vaccine efficacy trials using human Ad5 in humans have failed (45, 46). We have reported partial protection against repeated challenges with the difficult-to-neutralize simian-human immunodeficiency virus (SHIV) SF162P3 by the use of human Ad26 vectors in combination with modified vaccinia Ankara (MVA) vectors expressing HIV-1 antigens in rhesus monkeys (3). In a comparative study in mice, the ChAd3 vector was shown to be more immunogenic than Ad28, sAd11, ChAd63, sAd16, and Ad35 vectors in terms of the magnitude, quality, phenotype, and protective capacity of CD8+ T-cell responses (23). In addition, a ChAd3 hepatitis C virus vaccine vector in a phase I clinical trial has been shown to induce strong T-cell responses with a good safety profile (44). ChAd3 is also currently being explored as a vaccine vector in a clinical trial for Ebola virus disease (www.ClinicalTrials.gov registration no. NCT02231866). Moreover, a single immunization with ChAd68 induced long-lasting protection against rabies virus by the induction of neutralizing antibodies in nonhuman primates (47). The Ad vector PanAd3, which was isolated from bonobos and which was engineered to express an influenza A virus matrix 1 and nucleoprotein fusion antigen, provided significant protection against a lethal dose of a highly pathogenic challenge influenza virus in mice (48). The novel rhesus monkey adenovirus vectors described in the present study represent a new class of vaccine vectors with unique advantages and distinct biology.
In conclusion, we have produced and characterized a set of novel rhesus monkey adenovirus vaccine vectors which phylogenetically cluster as adenovirus species G and which have not previously been investigated as vaccine candidates. The immunogenicity of these novel vectors is comparable to that of other highly immunogenic human and chimpanzee adenovirus vectors. These novel vectors will likely not be subject to preexisting immunity in human populations and are biologically different from existing human and chimpanzee adenoviruses. Further studies are therefore warranted to examine their potential as vaccine candidates.
ACKNOWLEDGMENTS
We thank Z. Cama, F. Ball, C. D. Bleckwehl, M. Kirilova, N. Paulino, M. Boyd, R. A. Larocca, N. M. Provine, F. Stephens, and R. Hamel for generous advice, assistance, and reagents and the National Institutes of Health Tetramer Core Facility (Emory University, GA) for providing the Db/AL11 monomers.
We acknowledge support from grants OD011170, AI078526, AI084794, and AI096040; Bill and Melinda Gates Foundation grant OPP1040741; and the Ragon Institute of MGH, MIT, and Harvard.
We declare no financial conflicts of interest.
REFERENCES
- 1.Baden LR, Walsh SR, Seaman MS, Tucker RP, Krause KH, Patel A, Johnson JA, Kleinjan J, Yanosick KE, Perry J, Zablowsky E, Abbink P, Peter L, Iampietro MJ, Cheung A, Pau MG, Weijtens M, Goudsmit J, Swann E, Wolff M, Loblein H, Dolin R, Barouch DH. 2013. First-in-human evaluation of the safety and immunogenicity of a recombinant adenovirus serotype 26 HIV-1 Env vaccine (IPCAVD 001). J Infect Dis 207:240–247. doi: 10.1093/infdis/jis670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, SanMiguel A, Seaman MS, Ferrari G, Forthal DN, Ourmanov I, Hirsch VM, Carville A, Mansfield KG, Stablein D, Pau MG, Schuitemaker H, Sadoff JC, Billings EA, Rao M, Robb ML, Kim JH, Marovich MA, Goudsmit J, Michael NL. 2012. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, Liu J, Abbink P, Maxfield LF, Seaman MS, Dugast AS, Alter G, Ferguson M, Li W, Earl PL, Moss B, Giorgi EE, Szinger JJ, Eller LA, Billings EA, Rao M, Tovanabutra S, Sanders-Buell E, Weijtens M, Pau MG, Schuitemaker H, Robb ML, Kim J H, Korber BT, Michael NL. 2013. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell 155:531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosseini SY, Sabahi F, Moazzeni SM, Modarressi MH, Saberi Firoozi M, Ravanshad M. 2012. Construction and preparation of three recombinant adenoviruses expressing truncated NS3 and core genes of hepatitis C virus for vaccine purposes. Hepat Mon 12:e6130. doi: 10.5812/hepatmon.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roshorm Y, Cottingham MG, Potash MJ, Volsky DJ, Hanke T. 2012. T cells induced by recombinant chimpanzee adenovirus alone and in prime-boost regimens decrease chimeric EcoHIV/NDK challenge virus load. Eur J Immunol 42:3243–3255. doi: 10.1002/eji.201242624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smaill F, Jeyanathan M, Smieja M, Medina MF, Thanthrige-Don N, Zganiacz A, Yin C, Heriazon A, Damjanovic D, Puri L, Hamid J, Xie F, Foley R, Bramson J, Gauldie J, Xing Z. 2013. A human type 5 adenovirus-based tuberculosis vaccine induces robust T cell responses in humans despite preexisting anti-adenovirus immunity. Sci Transl Med 5:205ra134. doi: 10.1126/scitranslmed.3006843. [DOI] [PubMed] [Google Scholar]
- 7.Abbink P, Lemckert AA, Ewald BA, Lynch DM, Denholtz M, Smits S, Holterman L, Damen I, Vogels R, Thorner AR, O'Brien KL, Carville A, Mansfield KG, Goudsmit J, Havenga MJ, Barouch DH. 2007. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J Virol 81:4654–4663. doi: 10.1128/JVI.02696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belousova N, Mikheeva G, Xiong C, Soghomonian S, Young D, Le Roux L, Naff K, Bidaut L, Wei W, Li C, Gelovani J, Krasnykh V. 2010. Development of a targeted gene vector platform based on simian adenovirus serotype 24. J Virol 84:10087–10101. doi: 10.1128/JVI.02425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capone S, D'Alise AM, Ammendola V, Colloca S, Cortese R, Nicosia A, Folgori A. 2013. Development of chimpanzee adenoviruses as vaccine vectors: challenges and successes emerging from clinical trials. Expert Rev Vaccines 12:379–393. doi: 10.1586/erv.13.15. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Xiang ZQ, Li Y, Kurupati RK, Jia B, Bian A, Zhou DM, Hutnick N, Yuan S, Gray C, Serwanga J, Auma B, Kaleebu P, Zhou X, Betts MR, Ertl HC. 2010. Adenovirus-based vaccines: comparison of vectors from three species of adenoviridae. J Virol 84:10522–10532. doi: 10.1128/JVI.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colloca S, Barnes E, Folgori A, Ammendola V, Capone S, Cirillo A, Siani L, Naddeo M, Grazioli F, Esposito ML, Ambrosio M, Sparacino A, Bartiromo M, Meola A, Smith K, Kurioka A, O'Hara GA, Ewer KJ, Anagnostou N, Bliss C, Hill AV, Traboni C, Klenerman P, Cortese R, Nicosia A. 2012. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci Transl Med 4:115ra112. doi: 10.1126/scitranslmed.3002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dicks MD, Spencer AJ, Edwards NJ, Wadell G, Bojang K, Gilbert SC, Hill AV, Cottingham MG. 2012. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One 7:e40385. doi: 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Hara GA, Duncan CJ, Ewer KJ, Collins KA, Elias SC, Halstead FD, Goodman AL, Edwards NJ, Reyes-Sandoval A, Bird P, Rowland R, Sheehy SH, Poulton ID, Hutchings C, Todryk S, Andrews L, Folgori A, Berrie E, Moyle S, Nicosia A, Colloca S, Cortese R, Siani L, Lawrie AM, Gilbert SC, Hill AV. 2012. Clinical assessment of a recombinant simian adenovirus ChAd63: a potent new vaccine vector. J Infect Dis 205:772–781. doi: 10.1093/infdis/jir850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy S, Medina-Jaszek A, Wilson MJ, Sandhu A, Calcedo R, Lin J, Wilson JM. 2011. Creation of a panel of vectors based on ape adenovirus isolates. J Gene Med 13:17–25. doi: 10.1002/jgm.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatsis N, Tesema L, Robinson ER, Giles-Davis W, McCoy K, Gao GP, Wilson JM, Ertl HC. 2006. Chimpanzee-origin adenovirus vectors as vaccine carriers. Gene Ther 13:421–429. doi: 10.1038/sj.gt.3302675. [DOI] [PubMed] [Google Scholar]
- 16.Vogels R, Zuijdgeest D, van Rijnsoever R, Hartkoorn E, Damen I, de Bethune MP, Kostense S, Penders G, Helmus N, Koudstaal W, Cecchini M, Wetterwald A, Sprangers M, Lemckert A, Ophorst O, Koel B, van Meerendonk M, Quax P, Panitti L, Grimbergen J, Bout A, Goudsmit J, Havenga M. 2003. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J Virol 77:8263–8271. doi: 10.1128/JVI.77.15.8263-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy PS, Idamakanti N, Chen Y, Whale T, Babiuk LA, Mehtali M, Tikoo SK. 1999. Replication-defective bovine adenovirus type 3 as an expression vector. J Virol 73:9137–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tutykhina IL, Shmarov MM, Logunov D, Grabko VI, Sevast'ianova GA, Naroditskii BS, Gintsburg AL. 2008. Construction of the vector based on the CELO avian adenovirus genome providing enhanced expression of secreted alkaline phosphatase gene in a non-permissive system in vitro and in vivo. Mol Gen Mikrobiol Virusol 2008:26–30 (In Russian.) doi: 10.3103/S089141680804006X. [DOI] [PubMed] [Google Scholar]
- 19.Barouch DH, Kik SV, Weverling GJ, Dilan R, King SL, Maxfield LF, Clark S, Ng'ang'a D, Brandariz KL, Abbink P, Sinangil F, de Bruyn G, Gray GE, Roux S, Bekker LG, Dilraj A, Kibuuka H, Robb ML, Michael NL, Anzala O, Amornkul PN, Gilmour J, Hural J, Buchbinder SP, Seaman MS, Dolin R, Baden LR, Carville A, Mansfield KG, Pau MG, Goudsmit J. 2011. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine 29:5203–5209. doi: 10.1016/j.vaccine.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bukh I, Calcedo R, Roy S, Carnathan DG, Grant R, Qin Q, Boyd S, Ratcliffe SJ, Veeder CL, Bellamy SL, Betts MR, Wilson JM. 2014. Increased mucosal CD4+ T cell activation in rhesus macaques following vaccination with an adenoviral vector. J Virol 88:8468–8478. doi: 10.1128/JVI.03850-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ersching J, Hernandez MI, Cezarotto FS, Ferreira JD, Martins AB, Switzer WM, Xiang Z, Ertl HC, Zanetti CR, Pinto AR. 2010. Neutralizing antibodies to human and simian adenoviruses in humans and New-World monkeys. Virology 407:1–6. doi: 10.1016/j.virol.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 22.Jian L, Zhao Q, Zhang S, Huang W, Xiong Y, Zhou X, Jia B. 2014. The prevalence of neutralising antibodies to chimpanzee adenovirus type 6 and type 7 in healthy adult volunteers, patients with chronic hepatitis B and patients with primary hepatocellular carcinoma in China. Arch Virol 159:465–470. doi: 10.1007/s00705-013-1828-y. [DOI] [PubMed] [Google Scholar]
- 23.Quinn KM, Da Costa A, Yamamoto A, Berry D, Lindsay RW, Darrah PA, Wang L, Cheng C, Kong WP, Gall JG, Nicosia A, Folgori A, Colloca S, Cortese R, Gostick E, Price DA, Gomez CE, Esteban M, Wyatt LS, Moss B, Morgan C, Roederer M, Bailer RT, Nabel GJ, Koup RA, Seder RA. 2013. Comparative analysis of the magnitude, quality, phenotype, and protective capacity of simian immunodeficiency virus gag-specific CD8+ T cells following human-, simian-, and chimpanzee-derived recombinant adenoviral vector immunization. J Immunol 190:2720–2735. doi: 10.4049/jimmunol.1202861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, Keough M, Sandhu A, Wang Q, Medina-Jaszek CA, Plotkin JB, Wilson JM. 2009. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog 5:e1000503. doi: 10.1371/journal.ppat.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, Stanley K, Kramer J, Macri SC, Permar SR, Schmitz JE, Mansfield K, Brenchley JM, Veazey RS, Stappenbeck TS, Wang D, Barouch DH, Virgin HW. 2012. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemckert AA, Grimbergen J, Smits S, Hartkoorn E, Holterman L, Berkhout B, Barouch DH, Vogels R, Quax P, Goudsmit J, Havenga MJ. 2006. Generation of a novel replication-incompetent adenoviral vector derived from human adenovirus type 49: manufacture on PER.C6 cells, tropism and immunogenicity. J Gen Virol 87:2891–2899. doi: 10.1099/vir.0.82079-0. [DOI] [PubMed] [Google Scholar]
- 27.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siepel AC, Halpern AL, Macken C, Korber BT. 1995. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses 11:1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 30.Sprangers MC, Lakhai W, Koudstaal W, Verhoeven M, Koel BF, Vogels R, Goudsmit J, Havenga MJ, Kostense S. 2003. Quantifying adenovirus-neutralizing antibodies by luciferase transgene detection: addressing preexisting immunity to vaccine and gene therapy vectors. J Clin Microbiol 41:5046–5052. doi: 10.1128/JCM.41.11.5046-5052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barouch DH, Pau MG, Custers JH, Koudstaal W, Kostense S, Havenga MJ, Truitt DM, Sumida SM, Kishko MG, Arthur JC, Korioth-Schmitz B, Newberg MH, Gorgone DA, Lifton MA, Panicali DL, Nabel GJ, Letvin NL, Goudsmit J. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J Immunol 172:6290–6297. doi: 10.4049/jimmunol.172.10.6290. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Ewald BA, Lynch DM, Nanda A, Sumida SM, Barouch DH. 2006. Modulation of DNA vaccine-elicited CD8+ T-lymphocyte epitope immunodominance hierarchies. J Virol 80:11991–11997. doi: 10.1128/JVI.01348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teigler JE, Iampietro MJ, Barouch DH. 2012. Vaccination with adenovirus serotypes 35, 26, and 48 elicits higher levels of innate cytokine responses than adenovirus serotype 5 in rhesus monkeys. J Virol 86:9590–9598. doi: 10.1128/JVI.00740-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teigler JE, Phogat S, Franchini G, Hirsch VM, Michael NL, Barouch DH. 2014. The canarypox virus vector ALVAC induces distinct cytokine responses compared to the vaccinia virus-based vectors MVA and NYVAC in rhesus monkeys. J Virol 88:1809–1814. doi: 10.1128/JVI.02386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barouch DH, Liu J, Peter L, Abbink P, Iampietro MJ, Cheung A, Alter G, Chung A, Dugast AS, Frahm N, McElrath MJ, Wenschuh H, Reimer U, Seaman MS, Pau MG, Weijtens M, Goudsmit J, Walsh SR, Dolin R, Baden LR. 2013. Characterization of humoral and cellular immune responses elicited by a recombinant adenovirus serotype 26 HIV-1 Env vaccine in healthy adults (IPCAVD 001). J Infect Dis 207:248–256. doi: 10.1093/infdis/jis671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dehghan S, Seto J, Liu EB, Walsh MP, Dyer DW, Chodosh J, Seto D. 2013. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 443:197–207. doi: 10.1016/j.virol.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujimoto T, Yamane S, Ogawa T, Hanaoka N, Ogura A, Hotta C, Niwa T, Chiba Y, Gonzalez G, Aoki K, Koyanagi KO, Watanabe H. 2014. A novel complex recombinant form of type 48-related human adenovirus species D isolated in Japan. Jpn J Infect Dis 67:282–287. doi: 10.7883/yoken.67.282. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez G, Koyanagi KO, Aoki K, Kitaichi N, Ohno S, Kaneko H, Ishida S, Watanabe H. 2014. Intertypic modular exchanges of genomic segments by homologous recombination at universally conserved segments in human adenovirus species D. Gene 547:10–17. doi: 10.1016/j.gene.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 39.Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, Yousuf MA, Betensky RA, Jones MS, Dyer DW, Seto D, Chodosh J. 2013. Molecular evolution of human adenoviruses. Sci Rep 3:1812. doi: 10.1038/srep01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mast TC, Kierstead L, Gupta SB, Nikas AA, Kallas EG, Novitsky V, Mbewe B, Pitisuttithum P, Schechter M, Vardas E, Wolfe ND, Aste-Amezaga M, Casimiro DR, Coplan P, Straus WL, Shiver JW. 2010. International epidemiology of human pre-existing adenovirus (Ad) type-5, type-6, type-26 and type-36 neutralizing antibodies: correlates of high Ad5 titers and implications for potential HIV vaccine trials. Vaccine 28:950–957. doi: 10.1016/j.vaccine.2009.10.145. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Rhee EG, Masek-Hammerman K, Teigler JE, Abbink P, Barouch DH. 2012. Adenovirus serotype 26 utilizes CD46 as a primary cellular receptor and only transiently activates T lymphocytes following vaccination of rhesus monkeys. J Virol 86:10862–10865. doi: 10.1128/JVI.00928-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Bergelson JM. 2005. Adenovirus receptors. J Virol 79:12125–12131. doi: 10.1128/JVI.79.19.12125-12131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baden LR, Liu J, Li H, Johnson JA, Walsh SR, Kleinjan JA, Engelson BA, Peter L, Abbink P, Milner DA Jr, Golden KL, Viani KL, Stachler MD, Chen BJ, Pau MG, Weijten M, Carey BR, Miller CA, Swann EM, Wolff M, Loblein H, Seaman MS, Dolin R, Barouch DH. 26August2014. Induction of HIV-1-specific mucosal immune responses following intramuscular recombinant adenovirus serotype 26 HIV-1 vaccination of humans. J Infect Dis. doi: 10.1093/infdis/jiu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barnes E, Folgori A, Capone S, Swadling L, Aston S, Kurioka A, Meyer J, Huddart R, Smith K, Townsend R, Brown A, Antrobus R, Ammendola V, Naddeo M, O'Hara G, Willberg C, Harrison A, Grazioli F, Esposito ML, Siani L, Traboni C, Oo Y, Adams D, Hill A, Colloca S, Nicosia A, Cortese R, Klenerman P. 2012. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci Transl Med 4:115ra111. doi: 10.1126/scitranslmed.3003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, Koblin BA, Buchbinder SP, Keefer MC, Tomaras GD, Frahm N, Hural J, Anude C, Graham BS, Enama ME, Adams E, DeJesus E, Novak RM, Frank I, Bentley C, Ramirez S, Fu R, Koup RA, Mascola JR, Nabel GJ, Montefiori DC, Kublin J, McElrath MJ, Corey L, Gilbert PB. 2013. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med 369:2083–2092. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang ZQ, Greenberg L, Ertl HC, Rupprecht CE. 2014. Protection of non-human primates against rabies with an adenovirus recombinant vaccine. Virology 450-451:243–249. doi: 10.1016/j.virol.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vitelli A, Quirion MR, Lo CY, Misplon JA, Grabowska AK, Pierantoni A, Ammendola V, Price GE, Soboleski MR, Cortese R, Colloca S, Nicosia A, Epstein SL. 2013. Vaccination to conserved influenza antigens in mice using a novel simian adenovirus vector, PanAd3, derived from the bonobo Pan paniscus. PLoS One 8:e55435. doi: 10.1371/journal.pone.0055435. [DOI] [PMC free article] [PubMed] [Google Scholar]