ABSTRACT
Enterovirus 71 (EV71) is responsible for seasonal outbreaks of hand, foot, and mouth disease in the Asia-Pacific region. The virus has the capability to cause severe disease and death, especially in young children. Although several vaccines are currently in clinical trials, no vaccines or therapeutics have been approved for use. Previous structural studies have revealed that two antigenically distinct capsid forms are produced in EV71-infected cells: an expanded empty capsid, sometimes called a procapsid, and the infectious virus. Specifically, an immunodominant epitope of EV71 that maps to the virus canyon is structurally different in the procapsid and virus. This structure-function study shows that the procapsid can sequester antibodies, thus enhancing EV71 infection in vitro. The results presented here suggest that, due to conformational differences between the EV71 procapsid and virus, the presence of the procapsid in natural virus infections should be considered in the future design of vaccines or therapeutics.
IMPORTANCE In a picornavirus infection, both an infectious and a noninfectious empty capsid, sometimes referred to as a procapsid, are produced. It was novel to discover that the procapsid form of EV71 was expanded and antigenically distinct from the infectious virus. Previously, it had been supposed that this empty capsid was an off-pathway dead end or at best served for storage of pentameric subunits, which was later shown to be unlikely. It remains unexplained why picornaviruses evolutionarily conserve the wasteful production of so much noninfectious capsid. Here, we demonstrate that the EV71 procapsid has different antigenic properties than the infectious virus. Thus, the procapsid has the capacity to sequester neutralizing antibody and protect the virus, promoting or restoring a successful infection in vitro. This important observation should be considered in the future design and development of vaccines and therapeutics.
INTRODUCTION
Enterovirus 71 (EV71) is a human pathogen of the family Picornaviridae, a group of nonenveloped, single-stranded, plus-sense RNA viruses, many of which share common structural features in their icosahedrally symmetric capsids (1–4). These characteristics include a raised “mesa” at each 5-fold vertex that is surrounded by a depression called the “canyon.” Beneath the canyon of infectious capsids is a hydrophobic “pocket,” likely filled with a lipid moiety called the “pocket factor.” For most picornaviruses, a specific host cellular receptor binds into the canyon and dislodges the pocket factor, which triggers the transition to an altered state, or “A-particle.” A-particles subsequently release their genomes into the host cytoplasm to initiate an infection, leaving behind an empty capsid, or 80S particle (5).
Several proteins have been demonstrated to serve as host receptors for EV71. Mutational evidence indicates that scavenger receptor B2 (SCARB2) may serve as the canyon-binding receptor for EV71 (6). P-selectin glycoprotein ligand 1 (PSGL-1) binds at the 5-fold vertex and enhances EV71 infection of cells that express low levels of SCARB2 (7, 8). The binding sites for sialylated glycans, annexin II, heparin sulfate, and extracellular vimentin are unknown (9–14). After receptor-mediated cell entry, the virus replicates in the cytoplasm. A successful infection produces two icosahedral capsid forms: an empty noninfectious capsid, sometimes termed the procapsid (15), and an infectious virus with an encapsidated genome (16). Cryo-electron microscopy (cryo-EM) reconstructions and X-ray crystallography structures are available for both naturally occurring capsid forms and highlight important differences between the particles (4, 17–19). The EV71 procapsid is composed of 60 copies each of the structural proteins VP0, VP1, and VP3 and has an increased diameter compared to the infectious virus. Although the procapsid is produced in amounts equal to those of the infectious virus, it is not known if the procapsid serves as an assembly intermediate into which the genome is packaged or if it exists as a dead-end by-product of the capsid morphogenesis process (20). After autocatalytic cleavage of VP0 to form VP2 and VP4, the infectious virus is composed of 60 copies each of VP1 to VP4. These proteins enclose the viral genome within an icosahedral particle that is smaller and more compact than the procapsid. Protein rearrangements must occur to accommodate the different sizes of the procapsid and the virus capsid, resulting in antigenically distinct particles (4).
Currently, EV71 poses a worldwide health threat, as it is a causative agent of seasonal epidemics of hand, foot, and mouth disease (HFMD) in the Asia-Pacific region and can be detected globally (21). EV71 infections can progress through distinct stages of increasing clinical severity: stage one, HFMD/herpangina; stage two, central nervous system involvement, often presenting as meningitis or brainstem encephalitis; stage three, cardiopulmonary failure; and stage four, death (22). Most often, EV71 infection is asymptomatic or does not advance past stage one of clinical presentation. The majority of cases that progress to severe disease occur in young children. Follow-up studies have demonstrated that survivors of these severe infections can suffer long-term complications, such as reduced cognitive function and delayed development (23, 24). There are currently no effective treatments or vaccines to combat EV71 infection, and ongoing research efforts have been complicated by the diversity of the capsid sequence.
EV71 is classified in the species enterovirus A, one of nine species (A to J) within the genus Enterovirus of the family Picornaviridae. This virus circulates as one serotype and is divided into three genogroups (A, B, and C) that are further separated into 11 genotypes (A, B1 to B5, and C1 to C5). Classification of EV71 using this system is based on the VP1 sequence, which is the most variable, surface-exposed, and immunogenic structural protein. Several recent studies have identified regions of the EV71 capsid that can elicit strong neutralizing antibodies in mice by using a successive peptide-screening method (25, 26). These immunogens, named SP55 and SP70, are located in the VP1 protein and span amino acids 163 to 177 and 208 to 222, respectively. The epitope corresponding to SP70 is of particular interest, as its sequence is conserved throughout the 11 genotypes, and it is readily accessible on the capsid exterior. Mice inoculated with purified infectious capsids generate antibodies to the SP70 epitope (27). This epitope maps to the VP1 “GH” loop at the edge of the canyon. Li et al. generated a monoclonal antibody (MAb) (22A12) against the SP70 peptide that efficiently neutralized virus infection of RD cells in culture (28). However, the mechanism of neutralization for antibody 22A12 is unknown.
Our current study examines the neutralization capacity and mechanism of monoclonal antibody 22A12 against EV71 infection. We show that MAb 22A12 has stronger binding to the procapsid form of EV71 than to the infectious virus. The cryo-EM reconstruction of fragments of antibody (Fab) 22A12 in complex with EV71 procapsid suggests that MAb 22A12 has the ability to block receptor binding and subsequent cellular entry. However, the failure of Fab to neutralize EV71 infection indicates that the predominant mechanism of neutralization conferred by MAb 22A12 is likely cross-linking of viral capsids. Evolutionary conservation of the production of the picornavirus procapsid has yet to be explained. The work presented here shows that the procapsid can act in vitro as an antibody “sink,” effectively sequestering antibody, which consequently allows virus infection.
MATERIALS AND METHODS
Virus propagation and purification.
EV71 strain 1095/Shiga (genogroup C2) was propagated and purified as described previously (29). Briefly, confluent HeLa cell (Hafenstein laboratory cell collection) monolayers were infected (multiplicity of infection, 0.1), incubated at 37°C for 24 h, and lysed by three freeze-thaw cycles. After removal of cellular debris, the virus was precipitated overnight with 8% polyethylene glycol 8K and 0.5 M NaCl and purified by ultracentrifugation. The EV71 procapsid and infectious virus formed distinct bands in the gradient, as reported previously (4, 29, 30). The virus concentration was determined by absorbance at 280 nm for procapsid and 260 nm for infectious virus.
Antibody fragment preparation.
Purified murine monoclonal antibody 22A12 raised against the SP70 peptide of EV71 was obtained from SydLabs (Maiden, MA). Fab were generated using the Pierce Fab Micro Preparation kit (Thermo Scientific). In brief, MAb 22A12 at 1 mg/ml was incubated with immobilized papain for 5 h at 37°C with gentle rocking. Fab molecules were separated from Fc fragments and undigested Fab with a protein A column, the buffer was changed to phosphate-buffered saline (PBS), and the concentration was assessed by absorbance at a wavelength of 280 nm.
Complex formation and negative-stain transmission electron microscopy (TEM).
Purified EV71 procapsid or infectious virus (0.1-mg/ml concentration) was incubated with excess Fab 22A12 (4 mg/ml) at a ratio of two Fab per virus binding site on ice for 15 min. Three microliters of sample was applied to a freshly glow-discharged continuous carbon-coated copper EM grid and negatively stained with 3 μl of uranyl formate. The grids were visualized with the JEOL 1400 transmission electron microscope housed in the imaging facility at The Pennsylvania State University College of Medicine.
BLItz binding assay.
For the biolayer interferometry (BLItz) binding assay, 0.3 μl of 1 mM EZ-Link Sulfo-NHS-LC-LC-Biotin (Themo Scientific) was added to 2 ml of EV71 procapsid (0.1 mg/ml in PBS) to achieve a molecular-coupling ratio of 10:1. After incubation at room temperature for 30 min, the preparations were placed in kinetics buffer (PBS, 0.1% bovine serum albumin [BSA], 0.02% Tween 20) and concentrated to 0.5 mg/ml using 100-kDa molecular-mass cutoff centricons (Millipore) to remove any unreacted biotin. Procapsid was loaded onto a streptavidin (SA) biosensor for 4 min. The immobilized procapsid was allowed to associate with Fab 22A12 (0.7 mg/ml in kinetics buffer) for 2 min. The sensor was then placed in kinetics buffer to allow dissociation of Fab for 2 min. The association of Fab 22A12 with an unloaded SA biosensor was used to assess and minimize the nonspecific binding of Fab molecules. Procapsid bound to an SA biosensor was allowed to associate with kinetics buffer alone (no Fab 22A12) to serve as a loading control. The same protocol was performed with infectious virus. Five concentrations of Fab 22A12, increasing from 0.04 to 0.7 mg/ml, were tested independently with both immobilized procapsid and infectious virus. Only the highest concentration of Fab was detected by infectious virus in four independent replicates, and these data was presented (see Fig. 6). Only biosensors with the same amount of loading signal (2 nM) for procapsid and infectious virus were used for Fab 22A12 association.
FIG 6.
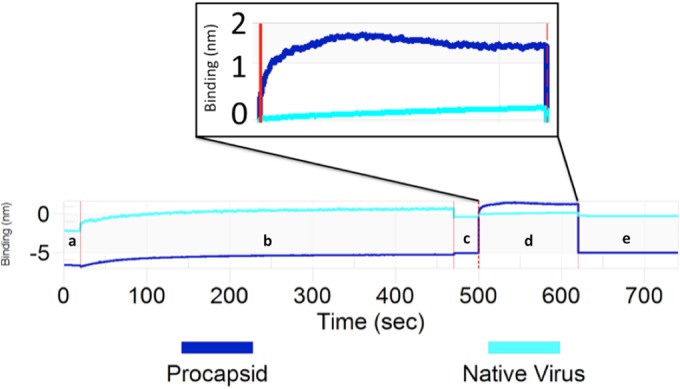
BLItz was used to determine the relative Fab 22A12 binding capability of the EV71 procapsid compared to that of infectious virus. After an initial baseline reading in running buffer (a), biotinylated EV71 procapsid or infectious virus was immobilized on a streptavidin sensor (b). The increase in line slope from a to c indicates 2 nM loading for both. A second baseline was recorded (c) before the sensor was inserted into the solution of purified Fab 22A12. The Fab and sensor association step (d) (2 minutes) is followed by a subsequent dissociation step (e) (2 minutes). The vertical red lines indicate transition from one experimental step to the next. The EV71 procapsid (dark blue) associates readily with Fab 22A12; however, EV71 infectious virus (light blue) binds Fab 22A12 at low levels. The association step has been aligned and magnified in the inset to highlight the differences in the binding signal intensity.
Cryo-EM data collection and processing.
Aliquots of EV71 procapsid or infectious virus incubated with Fab 22A12 were vitrified for cryo-EM data collection using an FEI Vitrobot Mark III freezing robot (FEI, Hillsboro, OR). A sample was applied to freshly glow-discharged holey carbon Quantifoil EM grids, blotted, and plunge frozen in a 60:40 mixture of liquid ethane and propane cooled in a bath of liquid nitrogen. Data were collected using an FEI Tecnai TF-20 transmission electron microscope equipped with a field emission gun operating at an accelerating voltage of 200 kV. Images were captured at ×50,000 magnification on Kodak SO-163 film (Kodak, Rochester, NY). A Nikon Super Coolscan 9000 scanner was used with a sampling rate of 6.35 μm/pixel to digitize the micrographs, creating a final pixel size of 1.27 Å at the sample. The preliminary reconstruction generated from EV71 infectious virus and Fab 22A12 had poor Fab occupancy and was not further processed (31). The EV71 procapsid-Fab complexes (with noticeable occupancy of Fab 22A12) were selected using the interactive boxing function of EMAN2 (32). Linearization, normalization, apodization, contrast transfer function correction, and three-dimensional (3-D) reconstruction were carried out using AUTO3DEM (31). The resolution was determined by comparing half data set maps using Fourier shell correlation at a cutoff of 0.5 (see Fig. 2b).
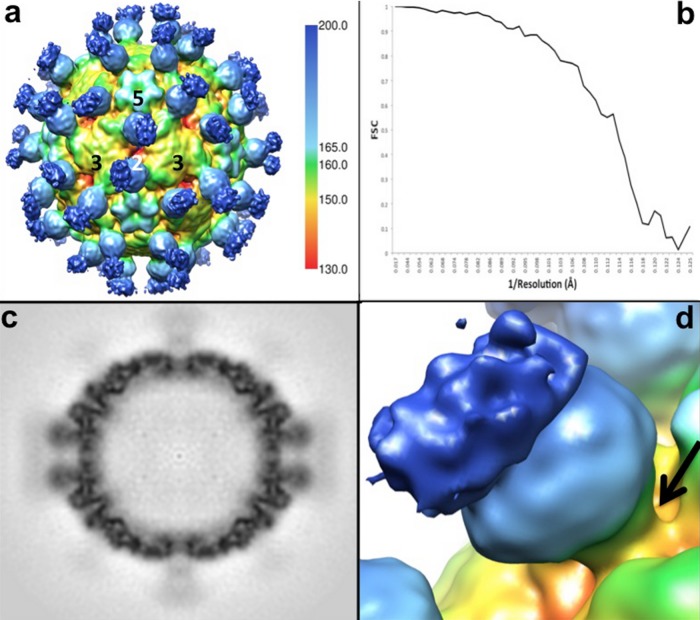
FIG 2.
The EV71 procapsid binds Fab 22A12 at the canyon. (a) The surface-rendered cryo-EM density map of Fab 22A12 in complex with EV71 procapsid is displayed at a contour level of 1 σ above background, radially colored as depicted by the scale bar, and labeled to indicate icosahedral symmetry axes. (b) The Fourier shell correlation (FSC) for the cryo-EM reconstruction falls below 0.5 at a resolution of 8.8 Å. (c) The central section of the cryo-EM density shown in panel a shows Fab density less than that of the capsid, indicating that Fab was not bound to all 60 binding sites. The blurred density of the constant region suggests flexibility of the hinge region of the Fab. (d) Closeup view of one Fab molecule binding at the canyon (arrow) surrounding the icosahedral 5-fold mesa of the EV71 procapsid. The constant region of the Fab (dark blue) has less density than the variable region (light blue), likely due to flexibility of the Fab hinge region.
Absolute pixel size assessment and difference map calculation.
The EV71-1095 procapsid crystal structure (Protein Data Bank [PDB] ID 4GMP) was used to calculate a density map with similar quality and resolution of the procapsid-Fab complex (17). This calculated map was then scaled and compared to the cryo-EM reconstruction to determine the absolute pixel size of 1.27 Å/pixel (33). The difference map was made by subtracting the calculated procapsid density from the procapsid-Fab 22A12 complex density map (34).
Fitting analysis.
The procapsid crystal structure (PDB ID 4GMP) (17) was fitted and refined into the cryo-EM density map using the CoLoRes package (34). The crystal structures of a murine Fab and its variable domain alone (PDB ID 3GK8) (35) were fitted into the Fab difference map. Icosahedral symmetry operators were applied to the best fits of 4GMP and 3GK8 as assessed by correlation coefficients, and this psuedoatomic model was refined to minimize clashes (34). Contacts between the EV71 procapsid and Fab were determined using CCP4 (36) with a 7-Å distance between atoms to identify any possible interactions. After splicing the ordered structure of VP1 amino acid residues 210 to 220 from the virus into the fitted procapsid, potential contact residues were identified 4 Å from the fitted Fab structure. The radial Fab density at the connection point between Fab and procapsid (radii, 155 to 160 Å) was projected onto the roadmap of the procapsid using RIVEM (37). The distance between the most C-terminal C-α atoms of two adjacent Fab molecules in the pseudoatomic model was measured in Chimera (33).
Antibody neutralization assay.
Minimally purified cell lysate was used as virus inoculum. Infected HeLa cells were harvested, frozen at −80°C, and thawed at room temperature for three cycles. Centrifugation at 4,500 × g for 5 min was used to pellet cellular debris. The clarified virus inoculum was aliquoted and stored at −80°C. HeLa cells (100 μl) at a density of 7 × 104 cells/ml were seeded into each well of a 96-well plate and incubated at 37°C for 24 h. The cells were rinsed with PBS and then incubated at 37°C with 50 μl of EV71 inoculum (3.2 × 104 PFU/ml) that had been preincubated at 37°C for 1 h with 2-fold dilutions of MAb 22A12 (starting concentration, 1 mg/ml) or Fab 22A12 (starting concentration, 0.5 mg/ml) in Dulbecco's modified Eagle's medium (DMEM). The amounts of inoculum and virus required to induce cytopathic effects (CPE) in HeLa cells in 48 h were assessed independently for normalization. After 1 h, 50 μl of DMEM supplemented with 5% fetal bovine serum (FBS) was added to each well, and the infection was allowed to proceed for 48 h at 37°C. The cells were then rinsed with PBS, fixed with 4% formaldehyde for 5 min, and stained with 0.1% crystal violet in 20% ethanol for 30 min. Wells with no blue staining were scored as positive for CPE. Each assay included PBS-treated cells as a positive control and cells infected with EV71 inoculum as negative controls. The same protocol was used for purified infectious virus (1.0 ×10−6 mg/ml). Each experiment contained 5 replicates and was repeated two times independently. The figures were generated in Excel.
Procapsid competition assay.
HeLa cells (100 μl) at a density of 7 × 104 cells/ml were seeded into each well of a 96-well plate and incubated at 37°C for 24 h. The cells were rinsed with PBS and incubated with 50 μl of a mixture of purified EV71 infectious virus (10 × 10−5 mg/ml), MAb 22A12 (0.0625 mg/ml), and a 2-fold dilution series of purified EV71 procapsid (starting concentration, 1 mg/ml) in DMEM that had been preincubated for 1 h at 37°C. The inoculated cells were incubated at 37°C for 1 h, 50 μl of DMEM supplemented with 5% FBS was added to each well, and the incubation was allowed to proceed for 48 h at 37°C. The cells were rinsed with PBS, fixed with 4% formaldehyde for 5 min, and stained with 0.1% crystal violet in 20% ethanol for 30 min. CPE was evaluated as described above. PBS-treated cells and cells incubated with purified infectious EV71 and MAb 22A12, procapsid, and infectious virus alone served as controls. Native virus treated with 1:2, 1:4, and 1:8 dilutions of mock lysate (HeLa cells lysed by three freeze-thaw cycles and centrifugation at 4,500 rpm for 5 min) was used to determine the effect of cellular debris on virus infectivity. Additionally, native virus incubated with procapsid in the above ratios without MAb served as a control for the effect of procapsid in the absence of antibody. Each experiment contained 5 replicates and was repeated two times independently.
Accession numbers.
The EV71 procapsid-Fab 22A12 cryo-EM density map was deposited in the EMDB (EMD-6200). The fitted EV71 procapsid crystal structure and murine Fab molecules were deposited in the PDB (3J91 and 3J93, respectively).
RESULTS
Fab 22A12 decorates the EV71 procapsid, but not the infectious virus.
Incubation of purified EV71 infectious virus with excess Fab 22A12 resulted in only a few Fab binding to each capsid. This low occupancy visualized in the cryoEM data did not yield sufficient density of Fab for interpretation in the 3-D image reconstruction (data not shown). However, a small percentage of empty capsids, which are either procapsid or 80S, visible in the micrographs were decorated with Fab molecules (Fig. 1a). Therefore, purified EV71 procapsids were incubated with Fab 22A12. Negative-stain TEM verified that the Fab bound efficiently to the procapsids (data not shown), and the sample was subsequently vitrified and used for cryo-EM data collection (Fig. 1b).
FIG 1.
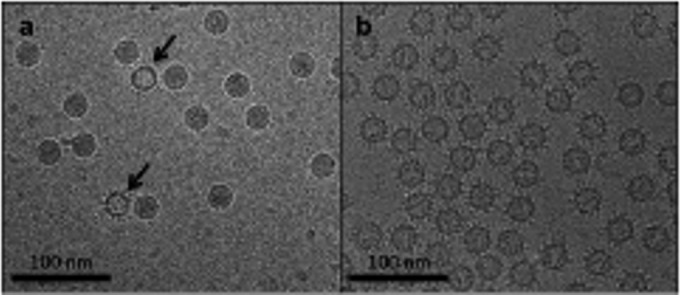
EV71 infectious virus (a) and procapsid (b) incubated with excess Fab 22A12. (a) Few Fab 22A12 molecules are bound to infectious virus, whereas empty capsids are clearly decorated with Fab (arrows). (b) Fab 22A12 binding to EV71 procapsid is visually evident.
Fab 22A12 binds EV71 procapsid at the canyon in the region of the VP1 GH loop.
The EV71 procapsid-Fab 22A12 complex was reconstructed to 8.8-Å resolution (Fig. 2a and b). The central section of the resulting cryo-EM density map showed that the Fab variable-domain density was weaker overall than the capsid shell. These densities suggest that Fab 22A12 does not bind all 60 of the available binding sites on the surface of the icosahedron (Fig. 2c). The Fab density extends radially from the EV71 canyon region (Fig. 2d) with very weak outer density corresponding to the Fab constant domain (Fig. 2c and d). The flexible hinge of Fab 22A12 likely allows movement of the constant domain, resulting in the weak cryo-EM density. The location of Fab 22A12 binding is similar to that of the receptor binding of other picornaviruses (38–41) that require interaction with a host molecule in the virus canyon to initiate entry. Thus, the Fab binding suggested that antibody 22A12 might act to block receptor binding or mimic a canyon-binding receptor.
Fitting of the EV71 procapsid crystal structure (PDB ID 4GMP) (17) yielded a correlation coefficient (cc) of 0.87, indicating that Fab binding induced no detectable conformational changes. The procapsid density was subtracted from the complex map (see Materials and Methods) to generate a difference map (Fig. 3a), into which a murine Fab crystal structure (PDB ID 3GK8) (35) was fitted (cc = 0.79). Due to the low density of the constant domain, the Fab variable domain was fitted alone and yielded a higher correlation (cc = 0.87). The independently fitted Fab and variable-domain structures were similar (root mean square deviation [RMSD] = 1.88 Å), suggesting that the weak constant-domain density did interfere with fitting analyses (Fig. 3b). Therefore, the variable domain alone was used for refinement of the fitted crystal structures to produce a pseudoatomic model (see Materials and Methods). The interface between Fab and procapsid mapped to the VP1 GH loop, which is disordered in the EV71 procapsid crystal structure (Fig. 3b). Thus, a pseudoatomic model could not be used to identify all contacts, and an alternative approach was used to map the antibody footprint. At the binding interface of the Fab, a radial section of the cryo-EM complex map density was projected onto the surface of the EV71 procapsid to identify the potential binding footprint (37). The Fab 22A12 density was represented as contour lines on the roadmap of the EV71 procapsid surface (Fig. 4) (42). This analysis showed that the Fab 22A12 footprint extended into the shallow EV71 canyon (Fig. 4), suggesting that the Fab may block the canyon.
FIG 3.
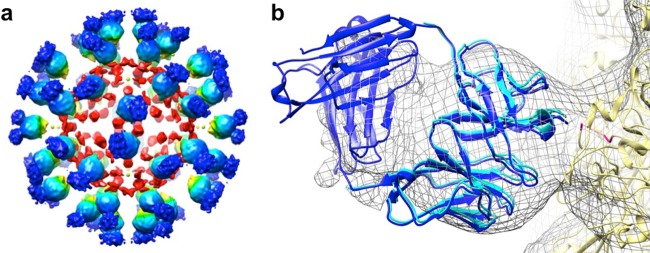
(a) A calculated procapsid map was subtracted from the EV71 procapsid-Fab 22A12 complex to generate a difference map, which is shown surface rendered at 1 σ, indicating strong Fab density (blue) and interior noise (red). (b) The crystal structures of the EV71 procapsid (yellow; PDB ID 4GMP), a murine Fab molecule (dark blue; PDB ID 3GK8), and a murine Fab variable domain (light blue) were fitted into the cryo-EM density map (gray mesh). The fitted variable domain superimposes onto the fitted Fab with an RMSD of 1.88 Å. The pseudoatomic model shows that the disordered GH loop of VP1 (magenta) is at the interface between the procapsid and Fab.
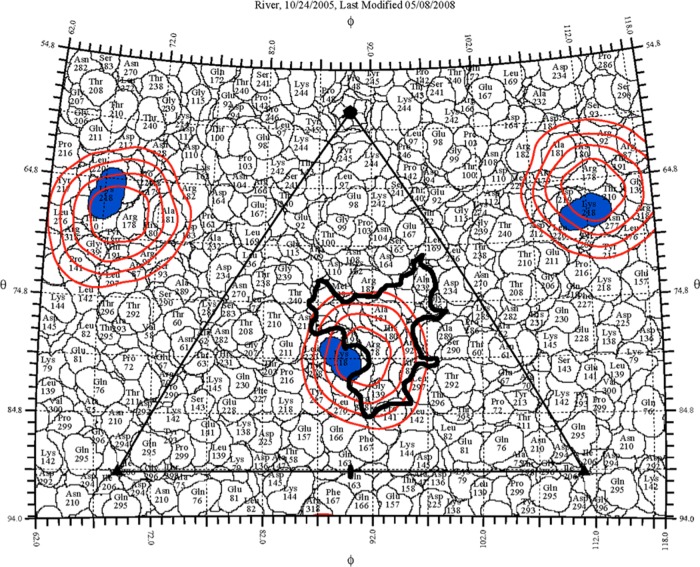
FIG 4.
The EV71 procapsid surface is shown as a stereographic projection in which the polar angles θ and ϕ represent latitude and longitude, respectively (37), with the procapsid topology represented as a quilt of amino acids (42). The icosahedral asymmetric unit of the virus is indicated by the triangular boundary. Due to the disordered VP1 GH loop, only one of the three identified contacts (blue) is shown. The radial density of the complex map corresponding to the interface between Fab and virus (radii, 155 to 160 Å) was projected onto the procapsid surface, and the red contours are shown overlapping a region of the canyon (thick black outline).
The epitope corresponding to the SP70 peptide is structurally different between procapsid and infectious virus.
The SP70 peptide spans VP1 amino acids 208 to 222. Amino acid residues 211 to 217 of the GH loop are disordered in the procapsid but form a structured loop in the infectious virus (Fig. 5). To identify the Fab 22A12 footprint, amino acid residues 208 to 220 of VP1 from the infectious virus crystal structure (PDB ID 3VBS) (4) were superimposed on the fitted EV71 procapsid. This hybrid model was used to identify contact residues 215, 217, and 218 of the VP1 protein, which are all part of the SP70 peptide. Alanine substitution experiments have shown previously that these residues, as well as amino acid 219, are essential for binding of MAb 22A12 to EV71 capsids (43). The different structures of the GH loop in the procapsid and infectious virus suggest altered affinity and/or accessibility of the epitope to the 22A12 antibody.
FIG 5.
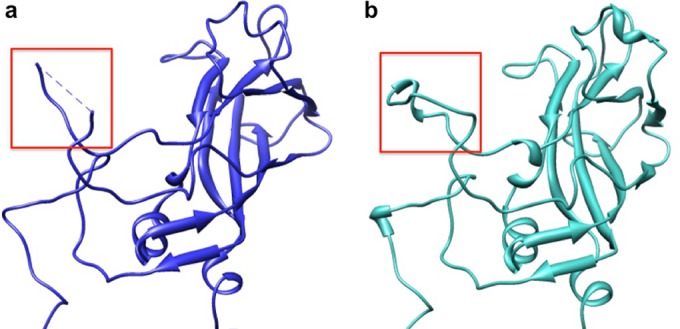
Location and structure of the VP1 SP70 peptide. Ribbon diagrams depicting the VP1 protein in the EV71 procapsid (PDB ID 4GMP) (a) and EV71 infectious virus (PDB ID 3VBS) (b) reveal conformational differences. Amino acids 211 to 218 of the SP70 peptide (red boxes) are flexible and disordered in the procapsid (a) but form an ordered structural loop in the infectious virus (b).
Procapsid interacts with Fab 22A12 more strongly than infectious virus in a binding assay.
BLItz was used to quantify the association of EV71 procapsid or infectious virus with Fab 22A12 (44). Biotinylated procapsid or infectious virus was immobilized on a streptavidin sensor before association and dissociation with Fab 22A12 (see Materials and Methods). EV71 procapsid associated with Fab 22A12 in a dose-dependent manner. However, the association of Fab 22A12 with EV71 infectious virus was detectable only at the highest concentration tested (see Materials and Methods). These BLItz data showed that Fab 22A12 readily associated with immobilized procapsid at a detectable signal corresponding to ∼2 nM. However, infectious virus binding was poor, with an association signal of less than 1 nM in each of the independent experiments (44) (Fig. 6). Thus, procapsid binds Fab 22A12 more effectively than infectious virus.
MAb 22A12 cannot bind capsids bivalently.
Although Fab 22A12 binds infectious virus only weakly, MAb 22A12 is nonetheless effective at neutralizing infectivity. The procapsid-Fab complex structure can be used to distinguish between different modes of binding virions, including cross-linking different capsids and binding bivalently to the same capsid (45, 46). The distance between the two closest adjacently bound Fab molecules on the EV71 procapsid surface was measured as 64 Å (see Materials and Methods), demonstrating that an intact antibody could not bind bivalently. However, the infectious virus is 4% smaller in diameter than the procapsid. Therefore, the refined pseudoatomic structure of a Fab and protomer was superimposed on a infectious capsid protomer (PDB ID 3VBS) (4) to generate a simulated virus-Fab complex. In this structure, the distance between the closest symmetry-related Fabs (61 Å) was still too great to allow bivalent binding of an antibody (45, 46). These results suggest that avidity is not required for MAb binding.
MAb 22A12, and not Fab 22A12, neutralizes purified EV71 infectious virus.
To investigate the neutralization ability of MAb 22A12 and its Fab, microneutralization assays were performed. Increasing concentrations of MAb 22A12 (or Fab) were incubated with purified EV71 infectious virus at 37°C for 1 h before infecting HeLa cell monolayers; 48 h postinfection, the neutralization capacity was determined by the survival of HeLa cells. The relatively low affinity for virus binding does not prevent MAb 22A12 from effectively neutralizing purified virions at a low concentration of MAb (7.8 × 10−3 mg/ml) (Fig. 7). However, after 48 h of infection, all the cells incubated with EV71-Fab 22A12 were no longer viable (data not shown). Thus, Fab does not have the capacity to neutralize virus. This result demonstrates that the mechanism of neutralization depends on the presence of antigen binding fragments and/or the Fc region of the antibody. One possible mechanism of neutralization may be cross-linking of virus capsids.
FIG 7.
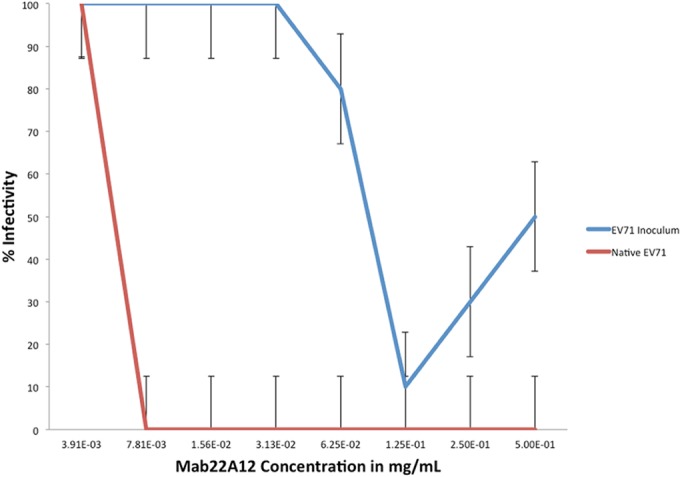
As the amount of MAb increases, infectivity decreases, and there are significant differences in the neutralization of inoculum and purified infectious virus. EV71 infectious virus preincubated with MAb 22A12 (red line) was incapable of infecting HeLa cells at all but one tested concentration of antibody, indicating efficient neutralization of purified virus. However, the EV71 inoculum (a mixture of infectious virus and procapsid) preincubated with the same amount of MAb 22A12 (blue line) readily infected cells, causing severe CPE and death. The data were collected from two independent experiments that contained five replicates each. The error bars denote standard errors.
MAb 22A12 does not effectively neutralize the virus inoculum, which is a mixture of procapsid and infectious virus.
Clarified cell lysate (inoculum) is often used for laboratory assessment of virus infectivity (see Materials and Methods). This inoculum includes a mixture of procapsid and infectious virus, along with minor amounts of cellular debris, and represents an in vivo infection. The ability of MAb 22A12 to neutralize EV71 inoculum was tested. The inoculum was preincubated with MAb 22A12 before infecting HeLa cells, and cell survival was recorded after 48 h, as described above. MAb 22A12 poorly neutralized the virus inoculum, with only two antibody concentrations (0.5 and 0.25 mg/ml) preventing CPE (Fig. 7). This loss of 22A12 neutralization ability indicates that a component of the inoculum prevents antibody from neutralizing infectious virus.
EV71 procapsid protects infectious virus from antibody neutralization.
The putative role of procapsid in preventing antibody neutralization of the inoculum was investigated by a microneutralization competition assay. Under all conditions, the amount of neutralizing MAb 22A12 used was greater than that required to neutralize the purified infectious virus. This excess neutralizing ratio of MAb to infectious virus was held constant throughout the experiment. Purified procapsid was added at increasing concentrations until the number of procapsids exceeded the number of infectious viruses. Thus, the effect of the presence of procapsid on virus infectivity was assessed and quantified directly. When the ratio of the number of procapsids to that of infectious viruses reached 1:4 (procapsid/virus), the infectivity of EV71 was restored to 50%. At ratios where the number of procapsids exceeded the number of viruses, infectivity was nearly 100%, even though neutralizing antibody was present (Fig. 8). The experiment was repeated without MAb 22A12; incubating purified virus and procapsid alone had no effect on virus infectivity, revealing that procapsid does not enhance the infectivity of virus (data not shown). This result indicates that the procapsid rescues EV71 infection in vitro by preferentially binding neutralizing antibody, as suggested by our initial microscopy experiment (Fig. 1a).
FIG 8.
EV71 procapsid inhibits neutralization of infectious virus by MAb 22A12. Purified EV71 infectious virus (1 × 10−5 mg/ml) preincubated with MAb 22A12 (6.25 × 10−2 mg/ml) was incapable of infecting HeLa cells. As increasing amounts of purified procapisd were added to the mixture of virus and antibody, the infectivity level of infectious virus increased, causing severe CPE and death of HeLa cells. The x axis indicates the number of procapsids present per infectious virus. The data were collected from two independent experiments that contained five replicates each. The error bars denote standard errors.
DISCUSSION
Here, we present a structure-function investigation of the EV71 neutralization mechanism of MAb 22A12. The known epitope, VP1 amino acids 208 to 222, includes the GH loop, which is disordered in the procapsid but has secondary structure in the infectious virus (Fig. 5). These structural differences in the binding interface likely dictate the ability of antibody to bind the procapsid better than infectious virus. MAb 22A12 was raised against a linear peptide (28), supporting the conclusion that the flexible procapsid epitope is a better binding site than the structured epitope presented by the infectious virus.
Previously published structural studies of fragments of neutralizing antibodies bound to picornavirus capsids have identified multiple mechanisms of neutralization (45–49), which include cross-linking, capsid stabilization, blocking receptor binding, inducing genome release, and marking capsids for clearance by host immune systems. Our 8.8-Å-resolution reconstruction of the Fab 22A12-procapsid complex reveals Fab bound to the canyon region (Fig. 2 and 4), suggesting that the Fab might mimic the host cell receptor. Electron micrographs of infectious virus incubated with Fab 22A12 showed no loss of RNA, and the 3-D reconstruction revealed no changes to the capsid (data not shown). Thus, Fab 22A12 does not act as a canyon-binding receptor to trigger A-particle formation. Alternatively, antibodies bound at the canyons of other picornaviruses have been shown to neutralize by capsid stabilization and/or blocking receptor binding (50, 51). However, the low affinity of Fab 22A12 argues against the ability of antibody to stabilize capsids or outcompete the receptor effectively.
The failure of antibody 22A12 to neutralize as Fab, even at high concentrations (0.5 mg/ml), suggests that the Fc region mediates the antibody's neutralization mechanism. Two possibilities are bivalent binding and cross-linking of capsids, both of which require complete antibody molecules with two Fab domains (45, 46). However, the distance between adjacently bound Fab 22A12 molecules demonstrates that MAb 22A12 could not bind bivalently to a single procapsid or infectious virus. Thus, it is likely that antibody 22A12 neutralizes EV71 infections predominantly by cross-linking capsids, although the ability to block receptor binding may play a minor role.
Despite the low binding affinity of MAb 22A12 for infectious virus, in vitro infection is efficiently neutralized at the low molar ratio of one antibody molecule per virus binding site (Fig. 7). However, significantly more antibody (12 antibody molecules per binding site) is required to neutralize virus when small amounts of purified EV71 procapsid (one procapsid to every 16 infectious viruses) are added to the incubation (Fig. 8). Even this large excess of MAb is insufficient for neutralization when the procapsid/virus ratio increases to one procapsid for every four infectious viruses. This competition for antibody binding demonstrates that the presence of procapsid is sufficient to rescue infectivity.
Picornaviruses produce both procapsid and infectious virus during the course of an infection. A function for the production of large amounts of procapsids has yet to be determined. For EV71, the procapsid is radially expanded compared to the infectious virus and is antigenically distinct. Here, we propose that the EV71 procapsid has the capacity to advance the infectivity of infectious virus. The EV71 procapsid contains a more favorable MAb 22A12 binding site than the virus, suggesting the procapsid binds and sequesters antibody to rescue the infectivity of EV71 (Fig. 9). Although MAb 22A12 was raised against the SP70 peptide, it is important to note that mice inoculated with purified virus capsids also raise antibodies against the MAb 22A12 epitope (27). This is the first time a function has been demonstrated for a picornavirus procapsid and may explain the evolutionary conservation of procapsid assembly during infection.
FIG 9.
The antigenically different EV71 procapsid (yellow) binds efficiently and sequesters neutralizing MAb 22A12 (blue) in vitro. This preferential binding minimizes the amount of unbound MAb, which protects infectious virus (red), allowing successful binding to susceptible cells to initiate an infection. This result suggests a model for a potential function of procapsid to enhance infectious virus infectivity because of altered antigenicity between the two capsid forms.
Our findings also indicate a need for a revised strategy for the design and evaluation of vaccines and therapeutics targeting EV71 infection. Using inoculum, procapsid, or peptide fragments does have the capability of producing neutralizing antibodies (25, 48, 52–54). However, consideration of both antigenic forms of the EV71 capsid is critical. An increased understanding of the antigenicity of procapsid and virus will build a comprehensive model for the natural infection and will be a better indicator of the in vivo success of potential vaccines and therapeutics.
ACKNOWLEDGMENTS
This project is funded, in part, under a grant from the Pennsylvania Department of Health using Tobacco CURE funds. The work was made possible by an NIH SIG 1S10RR031780-01A1 award to S.H.
We thank the Microscopy Imaging Shared Core Facility at the Pennsylvania State University College of Medicine.
REFERENCES
- 1.Hogle JM, Chow M, Filman DJ. 1985. Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 2.Muckelbauer JK, Kremer M, Minor I, Tong L, Zlotnick A, Johnson JE, Rossmann MG. 1995. Structure determination of coxsackievirus B3 to 3.5 A resolution. Acta Crystallogr D Biol Crystallogr 51:871–887. doi: 10.1107/S0907444995002253. [DOI] [PubMed] [Google Scholar]
- 3.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol 19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lonberg-Holm K, Gosser LB, Kauer JC. 1975. Early alteration of poliovirus in infected cells and its specific inhibition. J Gen Virol 27:329–342. doi: 10.1099/0022-1317-27-3-329. [DOI] [PubMed] [Google Scholar]
- 6.Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, Koike S. 2009. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat Med 15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura Y, Lee H, Hafenstein S, Kataoka C, Wakita T, Bergelson JM, Shimizu H. 2013. Enterovirus 71 binding to PSGL-1 on leukocytes: VP1-145 acts as a molecular switch to control receptor interaction. PLoS Pathog 9:e1003511. doi: 10.1371/journal.ppat.1003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura Y, Shimojima M, Tano Y, Miyamura T, Wakita T, Shimizu H. 2009. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat Med 15:794–797. doi: 10.1038/nm.1961. [DOI] [PubMed] [Google Scholar]
- 9.Du N, Cong H, Tian H, Zhang H, Zhang W, Song L, Tien P. 2014. Cell surface vimentin is an attachment receptor for enterovirus 71. J Virol 88:5816–5833. doi: 10.1128/JVI.03826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. 2013. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J Virol 87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su PY, Liu YT, Chang HY, Huang SW, Wang YF, Yu CK, Wang JR, Chang CF. 2012. Cell surface sialylation affects binding of enterovirus 71 to rhabdomyosarcoma and neuroblastoma cells. BMC Microbiol 12:162. doi: 10.1186/1471-2180-12-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan CW, Poh CL, Sam I-C, Chan YF. 2013. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J Virol 87:611–620. doi: 10.1128/JVI.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang B, Chuang H, Yang KD. 2009. Sialylated glycans as receptor and inhibitor of enterovirus 71 infection to DLD-1 intestinal cells. Virol J 6:141. doi: 10.1186/1743-422X-6-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S-L, Chou Y-T, Wu C-N, Ho M-S. 2011. Annexin II binds to capsid protein VP1 of enterovirus 71 and enhances viral infectivity. J Virol 85:11809–11820. doi: 10.1128/JVI.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips BA, Fennell R. 1973. Polypeptide composition of poliovirions, naturally occurring empty capsids, and 14S precursor particles. J Virol 12:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson MF, Baltimore D. 1968. Morphogenesis of poliovirus. I. Association of the viral RNA with coat protein. J Mol Biol 33:369–378. [DOI] [PubMed] [Google Scholar]
- 17.Cifuente JO, Lee H, Yoder JD, Shingler KL, Carnegie MS, Yoder JL, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2013. Structures of the procapsid and mature virion of enterovirus 71 strain 1095. J Virol 87:7637–7645. doi: 10.1128/JVI.03519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyu K, Ding J, Han J-F, Zhang Y, Wu X-Y, He Y-L, Qin C-F, Chen R. 2014. Human enterovirus 71 uncoating captured at atomic resolution. J Virol 88:3114–3126. doi: 10.1128/JVI.03029-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. 2012. Crystal structure of human enterovirus 71. Science 336:1274. doi: 10.1126/science.1218713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Wang JC-Y, Taylor MW, Zlotnick A. 2012. In vitro assembly of an empty picornavirus capsid follows a dodecahedral path. J Virol 86:13062–13069. doi: 10.1128/JVI.01033-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMinn PC. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol Rev 26:91–107. doi: 10.1111/j.1574-6976.2002.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 22.Alexander JP, Baden L, Pallansch MA, Anderson LJ. 1994. Enterovirus 71 infections and neurologic disease—United States, 1977–1991. J Infect Dis 169:905–908. doi: 10.1093/infdis/169.4.905. [DOI] [PubMed] [Google Scholar]
- 23.Chan LG, Parashar UD, Lye MS, Ong FG, Zaki SR, Alexander JP, Ho KK, Han LL, Pallansch MA, Suleiman AB, Jegathesan M, Anderson LJ. 2000. Deaths of children during an outbreak of hand, foot, and mouth disease in Sarawak, Malaysia: clinical and pathological characteristics of the disease. For the Outbreak Study Group. Clin Infect Dis 31:678–683. doi: 10.1086/314032. [DOI] [PubMed] [Google Scholar]
- 24.Lee M-S, Chang L-Y. 2010. Development of enterovirus 71 vaccines. Expert Rev Vaccines 9:149–156. doi: 10.1586/erv.09.152. [DOI] [PubMed] [Google Scholar]
- 25.Foo DGW, Alonso S, Chow VTK, Poh CL. 2007. Passive protection against lethal enterovirus 71 infection in newborn mice by neutralizing antibodies elicited by a synthetic peptide. Microbes Infect 9:1299–1306. doi: 10.1016/j.micinf.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Foo DGW, Alonso S, Phoon MC, Ramachandran NP, Chow VTK, Poh CL. 2007. Identification of neutralizing linear epitopes from the VP1 capsid protein of enterovirus 71 using synthetic peptides. Virus Res 125:61–68. doi: 10.1016/j.virusres.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Chang H-W, Liu C-C, Lin M-H, Ho H-M, Yang Y-T, Chow Y-H, Chong P, Sia C. 2011. Generation of murine monoclonal antibodies which cross-neutralize human enterovirus genogroup B isolates. J Virol Methods 173:189–195. doi: 10.1016/j.jviromet.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Mao C, Ma S, Wang X, Sun Z, Yi Y, Guo M, Shen X, Sun L, Bi S. 2009. Generation of neutralizing monoclonal antibodies against enterovirus 71 using synthetic peptides. Biochem Biophys Res Commun 390:1126–1128. doi: 10.1016/j.bbrc.2009.09.103. [DOI] [PubMed] [Google Scholar]
- 29.Shingler KL, Yoder JL, Carnegie MS, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2013. The enterovirus 71 A-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS Pathog 9:e1003240. doi: 10.1371/journal.ppat.1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C-C, Guo M-S, Lin FH-Y, Hsiao K-N, Chang KH-W, Chou A-H, Wang Y-C, Chen Y-C, Yang C-S, Chong PC-S. 2011. Purification and characterization of enterovirus 71 viral particles produced from Vero cells grown in a serum-free microcarrier bioreactor system. PLoS One 6:e20005. doi: 10.1371/journal.pone.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol 157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Chacón P, Wriggers W. 2002. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol 317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 35.Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG. 2009. Structural comparison of different antibodies interacting with parvovirus capsids. J Virol 83:5556–5566. doi: 10.1128/JVI.02532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao C, Rossmann MG. 2007. Interpretation of electron density with stereographic roadmap projections. J Struct Biol 158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolatkar PR, Bella J, Olson NH, Bator CM, Baker TS, Rossmann MG. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J 18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Organtini LJ, Makhov AM, Conway JF, Hafenstein S, Carson SD. 2014. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J Virol 88:5755–5765. doi: 10.1128/JVI.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao C, Bator-Kelly CM, Rieder E, Chipman PR, Craig A, Kuhn RJ, Wimmer E, Rossmann MG. 2005. The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure 13:1019–1033. doi: 10.1016/j.str.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 41.Zhang P, Mueller S, Morais MC, Bator CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG. 2008. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc Natl Acad Sci U S A 105:18284–18289. doi: 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossmann MG, Palmenberg AC. 1988. Conservation of the putative receptor attachment site in picornaviruses. Virology 164:373–382. doi: 10.1016/0042-6822(88)90550-8. [DOI] [PubMed] [Google Scholar]
- 43.Liu C-C, Chou A-H, Lien S-P, Lin H-Y, Liu S-J, Chang J-Y, Guo M-S, Chow Y-H, Yang W-S, Chang KH-W, Sia C, Chong P. 2011. Identification and characterization of a cross-neutralization epitope of enterovirus 71. Vaccine 29:4362–4372. doi: 10.1016/j.vaccine.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 44.Concepcion J, Witte K, Wartchow C, Choo S, Yao D, Persson H, Wei J, Li P, Heidecker B, Ma W, Varma R, Zhao L-S, Perillat D, Carricato G, Recknor M, Du K, Ho H, Ellis T, Gamez J, Howes M, Phi-Wilson J, Lockard S, Zuk R, Tan H. 2009. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb Chem High Throughput Screen 12:791–800. doi: 10.2174/138620709789104915. [DOI] [PubMed] [Google Scholar]
- 45.Hewat EA, Blaas D. 1996. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J 15:1515–1523. [PMC free article] [PubMed] [Google Scholar]
- 46.Hewat EA, Marlovits TC, Blaas D. 1998. Structure of a neutralizing antibody bound monovalently to human rhinovirus 2. J Virol 72:4396–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H, Cifuente JO, Ashley RE, Conway JF, Makhov AM, Tano Y, Shimizu H, Nishimura Y, Hafenstein S. 2013. A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with Fab from neutralizing antibody. J Virol 87:11363–11370. doi: 10.1128/JVI.01926-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plevka P, Lim P-Y, Perera R, Cardosa J, Suksatu A, Kuhn RJ, Rossmann MG. 2014. Neutralizing antibodies can initiate genome release from human enterovirus 71. Proc Natl Acad Sci U S A 111:2134–2139. doi: 10.1073/pnas.1320624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith TJ, Olson NH, Cheng RH, Liu H, Chase ES, Lee WM, Leippe DM, Mosser AG, Rueckert RR, Baker TS. 1993. Structure of human rhinovirus complexed with Fab fragments from a neutralizing antibody. J Virol 67:1148–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schotte L, Strauss M, Thys B, Halewyck H, Filman DJ, Bostina M, Hogle JM, Rombaut B. 2014. Mechanism of action and capsid-stabilizing properties of VHHs with an in vitro antipolioviral activity. J Virol 88:4403–4413. doi: 10.1128/JVI.03402-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Smith TJ, Lee WM, Mosser AG, Rueckert RR, Olson NH, Cheng RH, Baker TS. 1994. Structure determination of an Fab fragment that neutralizes human rhinovirus 14 and analysis of the Fab-virus complex. J Mol Biol 240:127–137. doi: 10.1006/jmbi.1994.1427. [DOI] [PubMed] [Google Scholar]
- 52.Gong M, Zhu H, Zhou J, Yang C, Feng J, Huang X, Ji G, Xu H, Zhu P. 2014. Cryo-electron microscopy study of insect cell-expressed enterovirus 71 and coxsackievirus a16 virus-like particles provides a structural basis for vaccine development. J Virol 88:6444–6452. doi: 10.1128/JVI.00200-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ku Z, Ye X, Huang X, Cai Y, Liu Q, Li Y, Su Z, Huang Z. 2013. Neutralizing antibodies induced by recombinant virus-like particles of enterovirus 71 genotype C4 inhibit infection at pre- and post-attachment steps. PLoS One 8:e57601. doi: 10.1371/journal.pone.0057601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye X, Ku Z, Liu Q, Wang X, Shi J, Zhang Y, Kong L, Cong Y, Huang Z. 2014. Chimeric virus-like particle vaccines displaying conserved enterovirus 71 epitopes elicit protective neutralizing antibodies in mice through divergent mechanisms. J Virol 88:72–81. doi: 10.1128/JVI.01848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]