Abstract
Chondrocyte differentiation in the growth plate is an important process for the longitudinal growth of endochondral bones. Sox9 and Runx2 are the most often-studied transcriptional regulators of the chondrocyte differentiation process, but the importance of additional factors is also becoming apparent. Mafs are a subfamily of the basic ZIP (bZIP) transcription factor superfamily, which act as key regulators of tissue-specific gene expression and terminal differentiation in many tissues. There is increasing evidence that c-Maf and its splicing variant Lc-Maf play a role in chondrocyte differentiation in a temporal-spatial manner. This review summarizes the functions of c-Maf in chondrocyte differentiation and discusses the possible role of c-Maf in osteoarthritis progression.
Keywords: signaling molecules, chondrocytes, chondrogenesis, mesenchymal stem cells, articular cartilage
Introduction
Chondrogenesis is the earliest phase of skeletal development, involving mesenchymal cell recruitment and migration, condensation of progenitors, and chondrocyte differentiation and maturation. These processes lead to the formation of cartilage and bone during endochondral ossification.1-3 Mutations or deregulation of these processes lead to skeletal malformation, skeletal malfunction, and/or susceptibility to injury.4
Each step of the chondrocytic differentiation process is characterized by specific morphological change and gene expression profiles. Several cytokines, growth factors, extracellular matrix (ECM) components, and transcription factors have important roles in these processes. Two groups of transcription factors have been identified that control the key steps of chondrocyte differentiation: the Sox and Runx families.
The Sox trio of transcription factors (Sox9/Sox5/Sox6) has important regulatory functions in the early stages of chondrogenesis. Sox5 and Sox6 are coexpressed with Sox9 in pre-chondrocytes and proliferating chondrocytes, and the three Sox proteins cooperate with each other to directly activate type II collagen (Col2a1) and aggrecan in vitro.5,6 Sox9-null mutant cells in mouse embryo chimera are excluded from mesenchymal condensations and fail to express Col2a1 and other chondrocyte-specific markers (Col9a2, Col11a2, and aggrecan), indicating that Sox9 is required for mesenchymal condensation and subsequent cartilage formation.7 Sox5 and Sox6 have essential but somewhat redundant roles in promoting chondrogenesis, and double-null mice fetuses die at embryonic day 16.5 (E16.5) from heart failure with severe chondrodysplasia.8
A key transcription factor controlling pre-hypertrophic and hypertrophic chondrocyte differentiation is Runx2. Runx2 is required for osteoblast formation but also necessary for normal chondrocyte maturation. Runx2 is mainly expressed when chondrocytes are about to become pre-hypertrophic, and it remains expressed through hypertrophy and terminal differentiation.9 Runx2-null mice have abnormal hypertrophic chondrocyte differentiation in addition to failure of bone formation.10,11 However, there is failure of terminal differentiation only in the humerus and femur and delayed terminal differentiation in other long bones, including tibia, fibula, radius, and ulna.10,12 These data would suggest that additional transcription factors play a role during terminal differentiation.2
The transcription factor c-Maf is specifically expressed in late hypertrophic and terminal chondrocytes,13,14 making it a candidate for controlling terminal differentiation. In addition, the expression of c-Maf and Lc-Maf, an mRNA splicing variant of c-Maf, partially overlaps with the expression of both Sox9 and Runx2.15
c-Maf Is a bZIP Transcription Factor
The founding member of Maf family, v-Maf (musculoaponeurotic fibrosarcoma), was originally identified as a retroviral oncoprotein, and then cellular counterparts, c-Mafs, were cloned from a number of vertebrate genomes.16 The Maf family is currently subdivided into two groups, small Maf (MafF, MafG, MafK, MafT, and MafS) and large Maf (c-Maf, MafA, MafB, and Nrl [neural retina leucine zipper]) proteins based on their domain structure and function (Table 1). A transcript variant of c-Maf, Lc-Maf, also has been reported15 (Fig. 1). The large Maf proteins contain a conserved N-terminal transactivation domain, whereas the small Maf proteins lack this transactivation domain.17 Based on the strong sequence conservation, the members of the Maf family are classified under the bZIP transcription factor superfamily.
Table 1.
Binding Partners of Mafs
| Mafs | Binding Partners | References |
|---|---|---|
| Large Mafs | ||
| c-Maf | c-Maf | Civil et al., 200270 |
| MafB | Hale et al., 200049 | |
| Nrl | Kerppola and Curran, 199420 | |
| Jun | Kerppola and Curran, 199420 | |
| Fos | Kerppola and Curran, 199420 | |
| TBP | Friedman et al., 200432 | |
| CBP/p300 | Chen et al., 200233 | |
| MafA | MafA | Benkhelifa et al., 199837 |
| c-Jun | Benkhelifa et al., 199837 | |
| TBP | Friedman et al., 200432 | |
| MafB | c-Maf | Civil et al., 200270 |
| MafB | Kataoka et al., 199417 | |
| Fos | Kataoka et al., 199417 | |
| Nrl | c-Maf | Kerppola and Curran, 199420 |
| Jun | Kerppola and Curran, 199420 | |
| TBP | Friedman et al., 200432 | |
| Small Mafs | ||
| MafF | MafF | Kimura et al., 200771 |
| MafG | MafG | Kimura et al., 200771 |
| MafK | MafK | Kataoka et al., 199421 |
| MafF/MafK | Fos | Kataoka et al., 199572 |
| MafG/MafK | p45NF-E2 | Toki et al., 199773 |
| MafF | Nrf1 | Johnsen et al., 199674 |
| MafG | Nrf2 | Kimura et al., 200771 |
Homo- and heterodimers form between Mafs and other bZIP family proteins. Mafs also bind to general transcription factors and coactivators.
Figure 1.
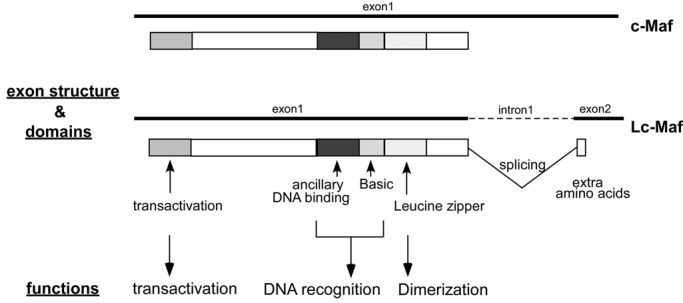
Schematic representation of c-Maf genomic structure and predicted functional domains. c-Maf and its RNA splice variant Lc-Maf (long form c-Maf) contain the same 5′ untranslated region. c-Maf is transcribed from a single exon, and through mRNA splicing, Lc-Maf has a frame shift that alters the last amino acid of c-Maf Exon 1 and codes for an additional 10 amino acids (in mouse) or 30 amino acids (in human). The mechanism of splicing is unknown. Intron/exon sizes are not drawn to scale.
The bZIP transcription factor superfamily consists of 53 different proteins in humans18 and has been subdivided into several subgroups, including the hallmark AP-1 complex (c-Fos/c-Jun), CREB/ATF, CNC, C/EBP, PAR, and Maf families.19 bZIP transcription factors are activated by a variety of signal transduction pathways to mediate critical aspects of cellular differentiation.19 The bZIP structural motif is a long bipartite α-helix of 60 to 80 amino acids in length and comprises a basic region and a leucine zipper. The N-terminal basic region contains an 18–amino acid basic peptide responsible for specific DNA recognition, whereas the C-terminal leucine zipper domain mediates homo- and heterodimerization with other bZIP proteins.18
The Maf family members are unique among the bZIP superfamily in that they contain a highly conserved ancillary DNA binding domain N-terminal to the typical basic domain (Fig. 1), which increases the number of specific DNA sequences that can be bound.20 Through this extended DNA binding domain, Maf proteins bind to relatively long consensus DNA sequences termed Maf-recognition elements (MAREs).21 Early in vitro selection studies identified two 13– and 14–base pair palindromic sequences, TGCTGACTCAGCA (T-MARE) and TGCTGACGTCAGCA (C-MARE), as the optimal binding sites for c-Maf.21 The T-MARE contains a phorbol 12-O-tetradecanoate (TPA)–Response Element (TRE: TGACTCA), and the C-MARE contains a cyclic AMP-responsive element (CRE: TGACGTCA) in the middle region, respectively. However, in vitro studies have shown that some base mismatches from the consensus sequences are allowed in Maf binding to the MARE and that Maf homodimers also bind efficiently to MARE half-palindromic sites when they are preceded by an AT-rich sequence.22 Naturally occurring c-Maf binding sites in IL-4, crystallin, and other genes are not intact palindromic sequences, as the full-consensus sequences are present only in the 5′ half-region.23 Therefore, although there is a degree of specificity in the Maf binding to the target DNA sequences, there is also considerable flexibility.
c-Maf in Chondrocytes
In the developing mouse embryo, c-Maf is strongly expressed in the lens fiber cells,24 and studies have focused on c-Maf’s function in lens development. c-Maf expression in chondrocytes was first reported in rat.13 Expression of c-Maf and MafB was detected at the RNA level in limb and rib cartilage as well as eye lens and spinal cord of the E15 rat embryo. The analysis of postnatal rat femur epiphysis in the same study showed that c-Maf is expressed strongly in the hypertrophic chondrocytes but low in immature proliferating chondrocytes. A detailed localization study in mouse embryonic cartilage showed that c-Maf is expressed mostly in late hypertrophic and terminally differentiated chondrocytes with a maximal expression at E15.5-E16.5 and at low levels in the perichondrium and in the primary spongiosa.14 On the other hand, Lc-Maf is expressed in E10.5-E12.5 mesenchymal condensations.15 At later stages, Lc-Maf expression is detected only in the perichondrium, with no expression in differentiated chondrocytes.15 Although the differential expression patterns of c-Maf and Lc-Maf during chondrogenesis were described, the functional differences between c-Maf and Lc-Maf are still unknown. Thus, the regulation mechanisms that control temporal-spatial expression of the different c-Maf isoforms need to be further investigated.
The prominent phenotype of the c-Maf-null mice is defective lens fiber cell differentiation.24-26 Direct transactivation of lens-specific crystallin genes by c-Maf has been related to this phenotype. Although c-Maf (+/−) heterozygous mice have normal lenses, naturally occurring heterozygous point mutations in c-Maf cause mild cataracts in both mice and humans.27,28 These mutations in the basic domain may have a dominant negative effect on wild-type c-Maf protein. Whether these mutations are related to abnormal cartilage formation remains unknown. It is interesting to note that lens and cartilage share similar gene expression profiles, especially genes involved in ECM such as collagens, other noncollagenous ECM proteins, and matrix metalloproteinases (MMPs).29,30 Hence, similar regulation mechanisms for gene expression may occur in both tissues.
The role of c-Maf in cartilage was examined in endochondral bone development in mice lacking c-Maf.14 c-Maf-null mice have abnormal terminal differentiation of hypertrophic chondrocytes with the most dramatic abnormality at E15.5 to E16.5, which corresponds to the time points of the highest c-Maf expression in chondrocytes.14 Terminal differentiation of the hypertrophic chondrocytes is delayed, and the domain of late hypertrophic chondrocytes is expanded in both the E16.5 to E18.5 embryos and 4-week postnatal c-Maf-null mice. The expanded hypertrophic zone is associated with delayed vascular invasion and reduced ossification. This suggests that c-Maf facilitates both the initiation and the completion of terminal differentiation in chondrocytes.14
Interaction of Maf Proteins with Other Transcription Factors
The Maf proteins dimerize with themselves and with other bZIP proteins through the leucine zipper domain. They can also interact with several non-bZIP factors.15,31-33 The Maf binding partners that have been identified so far are summarized in Table 1, and the various types of c-Maf binding interactions are illustrated diagrammatically in Figure 2. There is specificity in Maf dimerization, and the different binding interactions affect the transactivation function of the Maf proteins. However, the circumstances that determine the binding of Maf to its partners are not fully understood. The small Maf proteins, which do not dimerize with large Mafs, generally repress transcription when they form homodimers. In contrast, when they form heterodimers with other members of bZip proteins, they can activate or repress transcription of genes regulating many aspects of cellular differentiation.34,35 A heterodimeric partner of small Maf, the transcription factor nuclear factor E2 p45-related factor-2 (Nrf2), displays decreased expression during chondrocyte differentiation and inhibits numerous aspects of chondrocyte differentiation in vitro.36 This suggests the possibility that small Maf/Nrf2 heterodimers may be involved in the negative regulation of chondrocyte differentiation, although no expression data for small Mafs are available in chondrocytes. Large Mafs efficiently form homodimers both in vitro and in vivo. They also have the ability to form heterodimers with AP-1 family members in vitro, although heterodimers have not yet been detected in vivo.21 In vitro, c-Maf and Nrl can form heterodimers with c-Fos and c-Jun.20 MafA can heterodimerize with c-Jun but not with c-Fos, whereas MafB can form heterodimers with c-Fos but not with c-Jun.37 Considering that Maf proteins are expressed in a wide variety of cells and tissues, it has been suggested that mixing and matching of dimeric partners might create specific complexes within cells to affect target gene expression and cell differentiation.38
Figure 2.
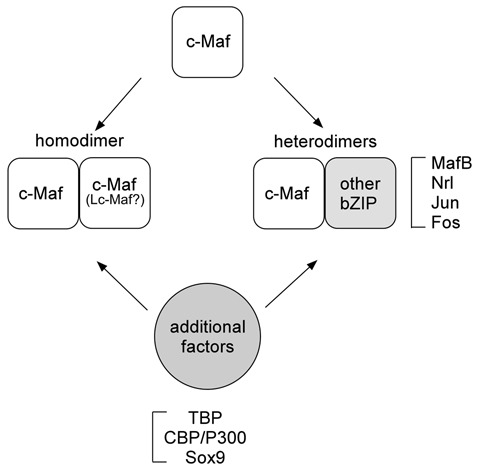
Different types of c-Maf binding interactions. c-Maf can form homodimers as well as heterodimers with other bZIP transcription factors. In addition, several factors, including general transcription factors (TBP), cofactors (CBP), and specific transcription factors (Sox9), interact with c-Maf.
Mafs can also physically and/or functionally interact with several non-bZIP proteins, including general transcription factors, common coactivators, and specific transcription factors (Fig. 2). Nrl directly interacts with TATA binding protein (TBP) in vitro, and TBP is co-immunoprecipitated with the Nrl-, c-Maf- and MafA-containing complexes in vivo.32 c-Maf recruits the CREB binding protein (CBP) and p300 to the promoters of crystallin genes, resulting in the synergistic activation of the genes.33 This synergistic activation requires histone acetyltransfease activity, indicating that chromatin remodeling is involved in c-Maf transactivation. Synergy in transcriptional activation by c-Maf and other specific transcription factors was also observed. c-Maf acts synergistically with Sox1, Sox2, and Sox3, which are important regulators of lens development, to activate the γF-crystallin gene.39 Similarly, Lc-Maf acts synergistically with Sox9 to activate the Col2a1 and type XXVII collagen (Col27a1) gene transcription in chondrocytes.15,40
c-Maf Target Genes and Pathways in Chondrocytes
Although Maf was first identified as a retroviral oncoprotein, the physiological functions of Maf family proteins include regulatory roles in a wide variety of cell- and tissue-specific gene expression and in cell differentiation during development.41 The target genes of Maf transcription factors in tissues are well summarized (see review by Kataoka41). This section focuses on c-Maf targets in chondrocytes, summarizing various lines of evidence that c-Maf regulates genes related to chondrocyte differentiation and the progression of osteoarthritis. This growing list of genes includes matrix metalloproteinase-13 (MMP-13), connective tissue growth factor (CTGF), Col2a1, and Col27a1 (Fig. 3).
Figure 3.
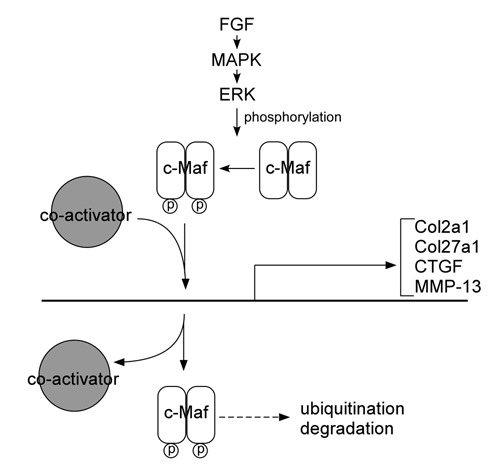
Schematics of c-Maf target genes in chondrocytes and possible pathways regulating c-Maf regulation. FGF/MAPK/ERK pathway may activate and affect the stability of c-Maf, based on homology with interactions affecting MafA. Some interactions are proposed based on homology of the phosphorylation/ubiquitination sites between MafA and c-Maf. These interactions still need to be verified for c-Maf, as clarified in the text.
MMP-13, along with other matrix metalloproteinases, affects hypertrophic differentiation and vascular invasion through matrix proteolysis.42 MMP-13 is induced in terminally differentiated chondrocytes in the developing growth plate and is highly upregulated in osteoarthritic cartilage. MMP-13 expression in the hypertrophic chondrocytes region was decreased specifically in the c-Maf-null mice embryo,14 suggesting a regulatory role for c-Maf in MMP-13 expression. This is supported by a study of c-Maf activity on the MMP-13 proximal promoter.43 However, there is the possibility that c-Maf indirectly regulates MMP-13 expression through the activation or repression of other factors, and further molecular and biological examination will be required.
CTGF is involved in chondrocyte differentiation and angiogenesis during skeletal development.44 CTGF and c-Maf are co-localized in hypertrophic chondrocytes, and interestingly, c-Maf-null and CTGF-null mice share common phenotypes, including expanded hypertrophic zones in the E16.5 embryo and reduced ossification of the tibia.14,44 CTGF is expressed by proliferating and hypertrophic chondrocytes in the mouse growth plate,45 and it was identified as a c-Maf-responsive gene in chondrocytes.40 In vitro studies using reporter transfection assays have shown that c-Maf and Lc-Maf activate the CTGF promoter, and chromatin immunoprecipitation assays demonstrated a direct interaction between the CTGF promoter and Maf.
Col2a1 was also upregulated by Lc-Maf via direct interactions with the intronic enhancer.15 Lc-Maf interacts with Sox9 to synergistically activate Col2a1 transcription in vitro. Lc-Maf is coexpressed with Sox9 and Col2a1 in the precartilaginous mesenchymal condensations during mouse embryogenesis, suggesting a role for Lc-Maf in early chondrocyte differentiation. The same study identified an Lc-Maf binding site adjacent to the well-known Sox9 binding site, although this site does not contain the typical MARE consensus sequence. The timing of in vivo expression of the Sox proteins and Lc-Maf during chondrogenesis led to the hypothesis that Lc-Maf cooperates early with Sox9 to activate target genes, including Col2a1, during mesenchymal condensation and that Sox9/Sox5/Sox6 function at later steps of chondrocyte differentiation.15
Col27a1 was recently identified as a target gene of Lc-Maf.31 Col27a1 is most abundant in proliferating and pre-hypertrophic chondrocytes during endochondral ossification.46 The enhancers of Col27a1 contain a Sox9-responsive element similar to that of Col2a1, and Lc-Maf and Sox9 synergistically activate the expression of both Col27a1 and Col2a1.15 Further evidence of the physiological significance of this activation comes from the co-localization of Lc-Maf and Col27a1 in growth plate, with maximal expression of both genes occurring in the proliferating and pre-hypertrophic chondrocytes.31
c-Maf may be involved in chondrocyte apoptosis either in the normal bone formation process or in the progression of pathologic conditions. The targets of c-Maf in apoptosis pathways have been identified in nonchondrocyte cells and appear to be dependent on the specific cell context. It has been shown that c-Maf interacts with the transcription factor c-Myb to downregulate Bcl-2 expression and increase apoptosis in peripheral CD4 cells47 and that c-Maf increases apoptosis by direct transactivation of caspase-6 in peripheral CD8 cells.48 Apoptosis through a p53-dependent pathway may also be relevant in both normal chondrocyte maturation and pathologic conditions. The promoter of p53 contains MARE consensus sequences, and overexpression of c-Maf induces p53-dependent cell death in primary rat embryo fibroblast cells.49 The p53-null embryonic mice have defects in the skeletal growth, abnormal chondrocyte maturation, and abnormal chondrocyte apoptosis, suggesting that p53 plays a role in normal chondrocyte development.50 Increased expression of p53 was reported in rheumatoid arthritis.51,52
The target genes regulated by c-Maf and Lc-Maf, as well as the phenotypes of c-Maf-null mice, generate the general types of abnormalities commonly seen with factors that affect endochondral ossification or chondrocytic differentiation. A causative role for c-Maf will require more hard evidence from direct experimentation.
Regulation of c-Maf
The Maf proteins control multiple biological processes, and they are synthesized at varying levels and in distinct spatial-temporal patterns; therefore, it is important to determine how they are regulated. There must be regulation mechanisms that control the levels of Maf members or splicing variants either on mRNA and/or protein levels. Several upstream regulators of Maf have been identified in the nonchondrocyte cells, and one can speculate that similar Maf regulation exists in chondrocytes (Fig. 3).
The fibroblast growth factor (FGF) pathway is a candidate for regulation of c-Maf expression. FGF signaling plays an important role in lens fiber differentiation, which is severely disrupted in c-Maf-null mice.25 Deletion of three FGF receptors (Fgfr1-3) demonstrated that the expression of FGFR is required for normal lens development, and FGFR deletion is involved in reduced expression of c-Maf and its target genes, crystallins, in the lens.53 The regulation of Maf by FGF signaling is at the posttranslational level. MAPK pathway is activated upon interaction of FGFR and ligand, and the FGF/MAPK cascade leads to the activation of ERK1/2 kinases, which can phosphorylate MafA.54 Two conserved serine residues (Ser14 and Ser65) located in the Maf transactivation domains are phosphorylated by ERK. These phosphorylations affect the stability of Maf and are required for Maf-dependent transcriptional activation.
Phosphorylation of Maf family proteins is a tightly controlled regulation mechanism. The transforming activity of MafA is controlled by glycogen synthase kinase–3 (GSK-3)–dependent phosphorylations, at conserved serine/threonine residues that are also present on c-Maf.55 This phosphorylation resulted in the recruitment of coactivators to increase MafA-dependent transcription, and once released from the coactivators, the phosphorylation induced MafA ubiquitination and degradation. Similarly, glucocorticoids can regulate c-Maf ubiquitination and proteosomal degradation to negatively regulate c-Maf-dependent transcription, although the phosphorylation status of c-Maf was not examined.56 Whether these pathways regulate c-Maf in chondrocytes or during chondrogenic differentiation remains to be determined experimentally.
Osteoarthritis and Terminal Chondrocyte Differentiation
Osteoarthritis (OA) is a degenerative joint disease characterized by erosion of the articular cartilage, hypertrophy of bone at the margins (i.e., osteophytes), subchondral sclerosis, and a range of biochemical and morphological alterations of the chondrocytes. Whereas endochondral ossification involves successive steps of chondrocyte maturation resulting in terminal hypertrophy and mineralization, normal articular chondrocytes are constrained from further differentiation, as evidenced by a lack of hypertrophic marker expression.57 However, during the progression of OA, the articular chondrocytes behave analogously to late hypertrophic and terminally differentiating growth plate chondrocytes with respect to their gene expression and cellular function. Increased expressions of hypertrophic chondrocyte markers and osteoblast markers occur in OA, such as type X collagen, osteopontin, osteocalcin, and alkaline phosphatase.58-60 Moreover, OA chondrocytes actively produce cartilage-degrading enzymes, including MMP-13, which is normally expressed only in the late hypertrophic zone,61-63 and cell apoptosis is also frequently observed in OA cartilage.64,65 Thus, it has been suggested that OA may involve the functional loss of mechanisms that block articular chondrocyte maturation.66 In this context, understanding the mechanisms that control chondrocyte hypertrophy is important for normal skeletal development and growth, as well as cartilage degeneration during the pathogenesis of OA.
Three studies have suggested a possible involvement of Mafs in the pathogenesis of OA.43,67,68 Microarray analysis of a rat model of OA demonstrated an upregulation of MafB expression in degenerating cartilage.68 It is noteworthy that the expression patterns of c-Maf and MafB in rat embryo cartilage are virtually identical.13 Expression of c-Maf was upregulated in OA cartilage compared to normal cartilage by real-time PCR and in situ hybridization.43,67 Because it has been shown that cells in the chondrocyte clusters in OA have increased expression in both cartilage matrix proteins and matrix-degrading proteases,69 it is notable that c-Maf mRNA was detected in proliferating chondrocyte clusters in OA cartilage, whereas almost no c-Maf-positive cells were found in normal cartilage.67
Concluding Remarks
The Maf family proteins form a unique subclass of bZIP transcription factors. Since the identification of v-Maf, many aspects of the physiological roles of Maf proteins have been elucidated, and currently Mafs are widely accepted as regulators of tissue-specific gene expression and cell differentiation. Studies of the roles of Maf proteins in chondrocyte differentiation and cartilage are under way, as is the identification of genes directly regulated by c-Maf. The identification of c-Maf binding partners and additional target genes during chondrogenesis and OA will provide insight into molecular functions of c-Maf. Furthermore, studies to integrate upstream differentiation signal and modulation of c-Maf activity at specific differentiation steps will be important to understanding chondrocyte terminal differentiation and OA pathogenesis.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interests with respect to the authorship and/or publication of this article.
Funding: The authors received no financial support for the research and/or authorship of this article.
References
- 1. DeLise AM, Fischer L, Tuan RS. Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage. 2000;8:309-34. [DOI] [PubMed] [Google Scholar]
- 2. Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97:33-44. [DOI] [PubMed] [Google Scholar]
- 3. Lefebvre V, Smits P. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res C Embryo Today. 2005;75:200-12. [DOI] [PubMed] [Google Scholar]
- 4. Shum L, Nuckolls G. The life cycle of chondrocytes in the developing skeleton. Arthritis Res. 2002;4:94-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. Embo J. 1998;17:5718-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han Y, Lefebvre V. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol. 2008;28:4999-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85-9. [DOI] [PubMed] [Google Scholar]
- 8. Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277-90. [DOI] [PubMed] [Google Scholar]
- 9. Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, et al. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999;214:279-90. [DOI] [PubMed] [Google Scholar]
- 11. Kim IS, Otto F, Zabel B, Mundlos S. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev. 1999;80:159-70. [DOI] [PubMed] [Google Scholar]
- 12. Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15:467-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sakai M, Imaki J, Yoshida K, Ogata A, Matsushima-Hibaya Y, Kuboki Y, et al. Rat maf related genes: specific expression in chondrocytes, lens and spinal cord. Oncogene. 1997;14:745-50. [DOI] [PubMed] [Google Scholar]
- 14. MacLean HE, Kim JI, Glimcher MJ, Wang J, Kronenberg HM, Glimcher LH. Absence of transcription factor c-maf causes abnormal terminal differentiation of hypertrophic chondrocytes during endochondral bone development. Dev Biol. 2003;262:51-63. [DOI] [PubMed] [Google Scholar]
- 15. Huang W, Lu N, Eberspaecher H, De Crombrugghe B. A new long form of c-Maf cooperates with Sox9 to activate the type II collagen gene. J Biol Chem. 2002;277:50668-75. [DOI] [PubMed] [Google Scholar]
- 16. Nishizawa M, Kataoka K, Goto N, Fujiwara KT, Kawai S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci U S A. 1989;86:7711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kataoka K, Fujiwara KT, Noda M, Nishizawa M. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol Cell Biol. 1994;14:7581-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22:6321-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y, Cvekl A. Large Maf transcription factors: cousins of AP-1 proteins and important regulators of cellular differentiation. Einstein J Biol Med. 2007;23:2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kerppola TK, Curran T. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene. 1994;9:3149-58. [PubMed] [Google Scholar]
- 21. Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol. 1994;14:700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshida T, Ohkumo T, Ishibashi S, Yasuda K. The 5’-AT-rich half-site of Maf recognition element: a functional target for bZIP transcription factor Maf. Nucleic Acids Res. 2005;33:3465-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973-83. [DOI] [PubMed] [Google Scholar]
- 24. Kawauchi S, Takahashi S, Nakajima O, Ogino H, Morita M, Nishizawa M, et al. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem. 1999;274:19254-60. [DOI] [PubMed] [Google Scholar]
- 25. Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci U S A. 1999;96:3781-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307-17. [DOI] [PubMed] [Google Scholar]
- 27. Lyon MF, Jamieson RV, Perveen R, Glenister PH, Griffiths R, Boyd Y, et al. A dominant mutation within the DNA-binding domain of the bZIP transcription factor Maf causes murine cataract and results in selective alteration in DNA binding. Hum Mol Genet. 2003;12:585-94. [PubMed] [Google Scholar]
- 28. Jamieson RV, Munier F, Balmer A, Farrar N, Perveen R, Black GC. Pulverulent cataract with variably associated microcornea and iris coloboma in a MAF mutation family. Br J Ophthalmol. 2003;87:411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ihanamaki T, Pelliniemi LJ, Vuorio E. Collagens and collagen-related matrix components in the human and mouse eye. Prog Retin Eye Res. 2004;23:403-34. [DOI] [PubMed] [Google Scholar]
- 30. Tsonis PA, Goetinck PF. Expression of cartilage-matrix genes and localization of their translation products in the embryonic chick eye. Exp Eye Res. 1988;46:753-64. [DOI] [PubMed] [Google Scholar]
- 31. Mayo JL, Holden DN, Barrow JR, Bridgewater LC. The transcription factor Lc-Maf participates in Col27a1 regulation during chondrocyte maturation. Exp Cell Res. 2009;315:2293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Friedman JS, Khanna H, Swain PK, Denicola R, Cheng H, Mitton KP, et al. The minimal transactivation domain of the basic motif-leucine zipper transcription factor NRL interacts with TATA-binding protein. J Biol Chem. 2004;279:47233-41. [DOI] [PubMed] [Google Scholar]
- 33. Chen Q, Dowhan DH, Liang D, Moore DD, Overbeek PA. CREB-binding protein/p300 co-activation of crystallin gene expression. J Biol Chem. 2002;277:24081-9. [DOI] [PubMed] [Google Scholar]
- 34. Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1-12. [DOI] [PubMed] [Google Scholar]
- 35. Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal. 2006;8:107-18. [DOI] [PubMed] [Google Scholar]
- 36. Hinoi E, Takarada T, Fujimori S, Wang L, Iemata M, Uno K, et al. Nuclear factor E2 p45-related factor 2 negatively regulates chondrogenesis. Bone. 2007;40:337-44. [DOI] [PubMed] [Google Scholar]
- 37. Benkhelifa S, Provot S, Lecoq O, Pouponnot C, Calothy G, Felder-Schmittbuhl MP. mafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene. 1998;17:247-54. [DOI] [PubMed] [Google Scholar]
- 38. Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437-41. [DOI] [PubMed] [Google Scholar]
- 39. Rajaram N, Kerppola TK. Synergistic transcription activation by Maf and Sox and their subnuclear localization are disrupted by a mutation in Maf that causes cataract. Mol Cell Biol. 2004;24:5694-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Omoteyama K, Ikeda H, Imaki J, Sakai M. Activation of connective tissue growth factor gene by the c-Maf and Lc-Maf transcription factors. Biochem Biophys Res Commun. 2006;339:1089-97. [DOI] [PubMed] [Google Scholar]
- 41. Kataoka K. Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J Biochem. 2007;141:775-81. [DOI] [PubMed] [Google Scholar]
- 42. Wu CW, Tchetina EV, Mwale F, Hasty K, Pidoux I, Reiner A, et al. Proteolysis involving matrix metalloproteinase 13 (collagenase-3) is required for chondrocyte differentiation that is associated with matrix mineralization. J Bone Miner Res. 2002;17:639-51. [DOI] [PubMed] [Google Scholar]
- 43. Li T, Xiao J, Wu Z, Qiu G, Ding Y. Transcriptional activation of human MMP-13 gene expression by c-Maf in osteoarthritic chondrocyte. Connect Tissue Res. 2010;51:48-54. [DOI] [PubMed] [Google Scholar]
- 44. Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, et al. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development. 2003;130:2779-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nishida T, Kubota S, Fukunaga T, Kondo S, Yosimichi G, Nakanishi T, et al. CTGF/Hcs24, hypertrophic chondrocyte-specific gene product, interacts with perlecan in regulating the proliferation and differentiation of chondrocytes. J Cell Physiol. 2003;196:265-75. [DOI] [PubMed] [Google Scholar]
- 46. Hjorten R, Hansen U, Underwood RA, Telfer HE, Fernandes RJ, Krakow D, et al. Type XXVII collagen at the transition of cartilage to bone during skeletogenesis. Bone. 2007;41:535-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peng S, Lalani S, Leavenworth JW, Ho IC, Pauza ME. c-Maf interacts with c-Myb to down-regulate Bcl-2 expression and increase apoptosis in peripheral CD4 cells. Eur J Immunol. 2007;37:2868-80. [DOI] [PubMed] [Google Scholar]
- 48. Peng S, Wu H, Mo YY, Watabe K, Pauza ME. c-Maf increases apoptosis in peripheral CD8 cells by transactivating Caspase 6. Immunology. 2009;127:267-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hale TK, Myers C, Maitra R, Kolzau T, Nishizawa M, Braithwaite AW. Maf transcriptionally activates the mouse p53 promoter and causes a p53-dependent cell death. J Biol Chem. 2000;275:17991-9. [DOI] [PubMed] [Google Scholar]
- 50. Ohyama K, Chung CH, Chen E, Gibson CW, Misof K, Fratzl P, et al. p53 influences mice skeletal development. J Craniofac Genet Dev Biol. 1997;17:161-71. [PubMed] [Google Scholar]
- 51. Sun Y, Cheung HS. p53, proto-oncogene and rheumatoid arthritis. Semin Arthritis Rheum. 2002;31:299-310. [DOI] [PubMed] [Google Scholar]
- 52. Firestein GS, Nguyen K, Aupperle KR, Yeo M, Boyle DL, Zvaifler NJ. Apoptosis in rheumatoid arthritis: p53 overexpression in rheumatoid arthritis synovium. Am J Pathol. 1996;149:2143-51. [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao H, Yang T, Madakashira BP, Thiels CA, Bechtle CA, Garcia CM, et al. Fibroblast growth factor receptor signaling is essential for lens fiber cell differentiation. Dev Biol. 2008;318:276-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Benkhelifa S, Provot S, Nabais E, Eychene A, Calothy G, Felder-Schmittbuhl MP. Phosphorylation of MafA is essential for its transcriptional and biological properties. Mol Cell Biol. 2001;21:4441-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rocques N, Abou Zeid N, Sii-Felice K, Lecoin L, Felder-Schmittbuhl MP, Eychene A, et al. GSK-3-mediated phosphorylation enhances Maf-transforming activity. Mol Cell. 2007;28:584-97. [DOI] [PubMed] [Google Scholar]
- 56. Mao X, Stewart AK, Hurren R, Datti A, Zhu X, Zhu Y, et al. A chemical biology screen identifies glucocorticoids that regulate c-maf expression by increasing its proteasomal degradation through up-regulation of ubiquitin. Blood. 2007;110:4047-54. [DOI] [PubMed] [Google Scholar]
- 57. Sandell LJ, Aigner T. Articular cartilage and changes in arthritis. An introduction: cell biology of osteoarthritis. Arthritis Res. 2001;3:107-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. von der Mark K, Kirsch T, Nerlich A, Kuss A, Weseloh G, Gluckert K, et al. Type X collagen synthesis in human osteoarthritic cartilage: indication of chondrocyte hypertrophy. Arthritis Rheum. 1992;35:806-11. [DOI] [PubMed] [Google Scholar]
- 59. Pullig O, Weseloh G, Gauer S, Swoboda B. Osteopontin is expressed by adult human osteoarthritic chondrocytes: protein and mRNA analysis of normal and osteoarthritic cartilage. Matrix Biol. 2000;19:245-55. [DOI] [PubMed] [Google Scholar]
- 60. Pullig O, Weseloh G, Ronneberger D, Kakonen S, Swoboda B. Chondrocyte differentiation in human osteoarthritis: expression of osteocalcin in normal and osteoarthritic cartilage and bone. Calcif Tissue Int. 2000;67:230-40. [DOI] [PubMed] [Google Scholar]
- 61. Mitchell PG, Magna HA, Reeves LM, Lopresti-Morrow LL, Yocum SA, Rosner PJ, et al. Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase-13 from human osteoarthritic cartilage. J Clin Invest. 1996;97:761-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moldovan F, Pelletier JP, Hambor J, Cloutier JM, Martel-Pelletier J. Collagenase-3 (matrix metalloprotease 13) is preferentially localized in the deep layer of human arthritic cartilage in situ: in vitro mimicking effect by transforming growth factor beta. Arthritis Rheum. 1997;40:1653-61. [DOI] [PubMed] [Google Scholar]
- 63. Shlopov BV, Lie WR, Mainardi CL, Cole AA, Chubinskaya S, Hasty KA. Osteoarthritic lesions: involvement of three different collagenases. Arthritis Rheum. 1997;40:2065-74. [DOI] [PubMed] [Google Scholar]
- 64. Hashimoto S, Ochs RL, Komiya S, Lotz M. Linkage of chondrocyte apoptosis and cartilage degradation in human osteoarthritis. Arthritis Rheum. 1998;41:1632-8. [DOI] [PubMed] [Google Scholar]
- 65. Kim HA, Lee YJ, Seong SC, Choe KW, Song YW. Apoptotic chondrocyte death in human osteoarthritis. J Rheumatol. 2000;27:455-62. [PubMed] [Google Scholar]
- 66. Drissi H, Zuscik M, Rosier R, O’Keefe R. Transcriptional regulation of chondrocyte maturation: potential involvement of transcription factors in OA pathogenesis. Mol Aspects Med. 2005;26:169-79. [DOI] [PubMed] [Google Scholar]
- 67. Li T, Xiao J, Wu Z, Qiu G. Over-expression of c-maf by chondrocytes in osteoarthritis. J Int Med Res. 2009;37:129-35. [DOI] [PubMed] [Google Scholar]
- 68. Solomon LA, Berube NG, Beier F. Transcriptional regulators of chondrocyte hypertrophy. Birth Defects Res C Embryo Today. 2008;84:123-30. [DOI] [PubMed] [Google Scholar]
- 69. Hambach L, Neureiter D, Zeiler G, Kirchner T, Aigner T. Severe disturbance of the distribution and expression of type VI collagen chains in osteoarthritic articular cartilage. Arthritis Rheum. 1998;41:986-96. [DOI] [PubMed] [Google Scholar]
- 70. Civil A, van Genesen ST, Lubsen NH. c-Maf, the gammaD-crystallin Maf-responsive element and growth factor regulation. Nucleic Acids Res. 2002;30:975-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kimura M, Yamamoto T, Zhang J, Itoh K, Kyo M, Kamiya T, et al. Molecular basis distinguishing the DNA binding profile of Nrf2-Maf heterodimer from that of Maf homodimer. J Biol Chem. 2007;282:33681-90. [DOI] [PubMed] [Google Scholar]
- 72. Kataoka K, Igarashi K, Itoh K, Fujiwara KT, Noda M, Yamamoto M, et al. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol Cell Biol. 1995;15:2180-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Toki T, Itoh J, Kitazawa J, Arai K, Hatakeyama K, Akasaka J, et al. Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif. Oncogene. 1997;14:1901-10. [DOI] [PubMed] [Google Scholar]
- 74. Johnsen O, Skammelsrud N, Luna L, Nishizawa M, Prydz H, Kolsto AB. Small Maf proteins interact with the human transcription factor TCF11/Nrf1/LCR-F1. Nucleic Acids Res. 1996;24:4289-97. [DOI] [PMC free article] [PubMed] [Google Scholar]


