Abstract
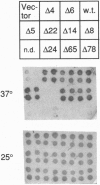
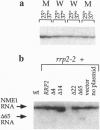
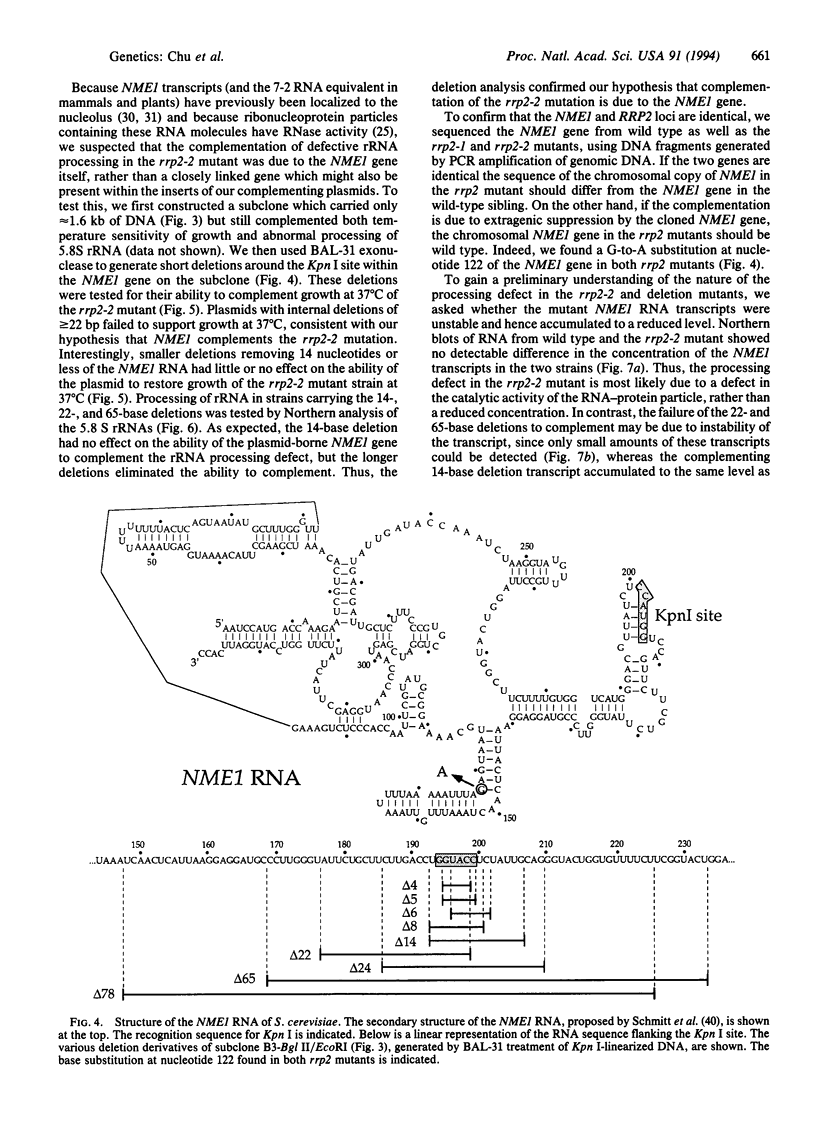
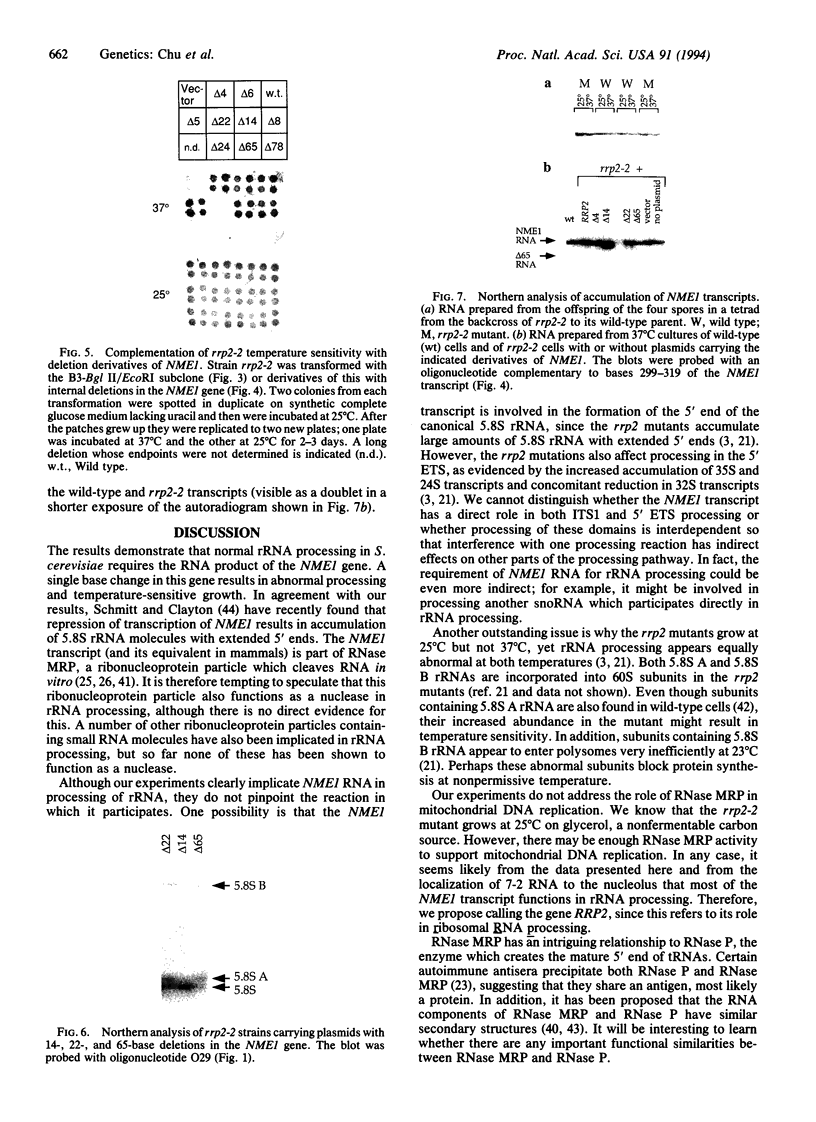
We have isolated clones which complement the temperature sensitivity and abnormal rRNA processing pattern of the rrp2-2 mutant of Saccharomyces cerevisiae we previously described. DNA sequencing and restriction analysis demonstrated that all clones contain the NME1 gene encoding the RNA of the ribonucleprotein particle RNase MRP. Deletion analysis showed that the NME1 gene is responsible for the complementation of the rrp2-2 phenotype. A single base change was identified in the nme1 gene in the rrp2 mutant, confirming that the RRP2 and NME1 genes are identical. Our experiments therefore indicate that RNase MRP, in addition to its previously reported role in formation of RNA primers for mitochondrial DNA replication [Clayton, D. A. (1991) Trends Biochem. Sci. 16, 107-111], is involved in rRNA processing.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992 Apr;11(4):1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. Nuclear gadgets in mitochondrial DNA replication and transcription. Trends Biochem Sci. 1991 Mar;16(3):107–111. doi: 10.1016/0968-0004(91)90043-u. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990 Aug 10;62(3):407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Fournier M. J., Maxwell E. S. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993 Apr;18(4):131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Girard J. P., Lehtonen H., Caizergues-Ferrer M., Amalric F., Tollervey D., Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992 Feb;11(2):673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold H. A., Topper J. N., Clayton D. A., Craft J. The RNA processing enzyme RNase MRP is identical to the Th RNP and related to RNase P. Science. 1989 Sep 22;245(4924):1377–1380. doi: 10.1126/science.2476849. [DOI] [PubMed] [Google Scholar]
- Hughes J. M., Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991 Dec;10(13):4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J. A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990 Mar 23;60(6):897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Kiss T., Filipowicz W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell. 1992 Jul 10;70(1):11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- Kiss T., Marshallsay C., Filipowicz W. 7-2/MRP RNAs in plant and mammalian cells: association with higher order structures in the nucleolus. EMBO J. 1992 Oct;11(10):3737–3746. doi: 10.1002/j.1460-2075.1992.tb05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K., Inouye M. Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J Biol Chem. 1992 Aug 15;267(23):16252–16258. [PubMed] [Google Scholar]
- Kondo K., Kowalski L. R., Inouye M. Cold shock induction of yeast NSR1 protein and its role in pre-rRNA processing. J Biol Chem. 1992 Aug 15;267(23):16259–16265. [PubMed] [Google Scholar]
- Lee W. C., Zabetakis D., Mélèse T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992 Sep;12(9):3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Archer R. H., Zengel J. M. A new rRNA processing mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Jan 25;20(2):295–301. doi: 10.1093/nar/20.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W., Tollervey D., Séraphin B. Small nuclear RNAs in messenger RNA and ribosomal RNA processing. FASEB J. 1993 Jan;7(1):47–53. doi: 10.1096/fasebj.7.1.8422974. [DOI] [PubMed] [Google Scholar]
- Morrissey J. P., Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993 Apr;13(4):2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis B. A., Steitz J. A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993 Jun 18;73(6):1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Stotz A., Scherf C. DNA of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- Raué H. A., Planta R. J. Ribosome biogenesis in yeast. Prog Nucleic Acid Res Mol Biol. 1991;41:89–129. doi: 10.1016/s0079-6603(08)60007-0. [DOI] [PubMed] [Google Scholar]
- Reimer G., Raska I., Scheer U., Tan E. M. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988 May;176(1):117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. Three forms of the 5.8-S ribosomal RNA species in Saccharomyces cerevisiae. Eur J Biochem. 1974 Jan 3;41(1):197–202. doi: 10.1111/j.1432-1033.1974.tb03260.x. [DOI] [PubMed] [Google Scholar]
- Russell I. D., Tollervey D. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J Cell Biol. 1992 Nov;119(4):737–747. doi: 10.1083/jcb.119.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R., Gerbi S. A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990 Jul;9(7):2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Bennett J. L., Dairaghi D. J., Clayton D. A. Secondary structure of RNase MRP RNA as predicted by phylogenetic comparison. FASEB J. 1993 Jan;7(1):208–213. doi: 10.1096/fasebj.7.1.7678563. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Clayton D. A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Dec;13(12):7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. E., Clayton D. A. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev. 1992 Oct;6(10):1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- Shuai K., Warner J. R. A temperature sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res. 1991 Sep 25;19(18):5059–5064. doi: 10.1093/nar/19.18.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl L. L., Clayton D. A. Saccharomyces cerevisiae contains an RNase MRP that cleaves at a conserved mitochondrial RNA sequence implicated in replication priming. Mol Cell Biol. 1992 Jun;12(6):2561–2569. doi: 10.1128/mcb.12.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987 Dec 20;6(13):4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E. C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991 Mar;10(3):573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper J. N., Bennett J. L., Clayton D. A. A role for RNAase MRP in mitochondrial RNA processing. Cell. 1992 Jul 10;70(1):16–20. doi: 10.1016/0092-8674(92)90529-l. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Singh R., Reddy R. Rat nucleolar 7-2 RNA is homologous to mouse mitochondrial RNase mitochondrial RNA-processing RNA. J Biol Chem. 1989 Sep 5;264(25):14835–14839. [PubMed] [Google Scholar]