Abstract
The anaphase–promoting complex/cyclosome (APC/C) is a 22 S ubiquitin ligase complex that initiates chromosome segregation and mitotic exit. We have used biochemical and electron microscopic analyses of Saccharomyces cerevisiae and human APC/C to address how the APC/C subunit Doc1 contributes to recruitment and processive ubiquitylation of APC/C substrates, and to understand how APC/C monomers interact to form a 36 S dimeric form. We show that Doc1 interacts with Cdc27, Cdc16 and Apc1, and is located in vicinity of the cullin–RING module Apc2–Apc11 in the inner cavity of the APC/C. Substrate proteins also bind in the inner cavity, in close proximity to DOC1 and the co–activator CDH1, and induce conformational changes in APC2–APC11. Our results suggest that substrates are recruited to the APC/C by binding to a bipartite substrate receptor composed of a co–activator protein and Doc1.
Sister chromatid separation and exit from mitosis are initiated by a 1.5 MDa ubiquitin ligase (E3) composed of at least 13 subunits, called the anaphase–promoting complex/cyclosome (APC/C)1. The APC/C initiates these events by ubiquitylating substrate proteins such as securin and cyclin B which are subsequently degraded by the 26 S proteasome. Recognition of substrates by the APC/C depends on co–activator proteins, called Cdc20 and Cdh1, which promote substrate recruitment by interacting with both the APC/C and recognition motifs in substrates, called the destruction box (D box) and KEN box. The subsequent ubiquitylation of substrates is mediated by ubiquitin conjugating (E2) enzymes that interact with a RING finger subunit of the APC/C, called Apc11 (ref. 2). Before all chromosomes have been bioriented on the mitotic spindle, the spindle assembly checkpoint (SAC) inhibits the form of the APC/C that is interacting with Cdc20 (APC/CCdc20) by promoting assembly of a mitotic checkpoint complex (MCC), in which Mad2, BubR1 and Bub3 bind to Cdc20 (APC/CMCC)3.
To understand the mechanism of APC/C mediated ubiquitylation reactions structural information will be essential, but so far only crystal structures of the 35 kDa APC/C subunit Doc1/Apc10 (ref. 4,5) and of parts of the tetratricopeptide repeat (TPR) subunits Cdc16, Cdc27 and Apc7 have been solved6-8. Therefore, electron microscopy (EM) has been used to analyze the structure of the APC/C. For human9,10, Xenopus9 and fission yeast APC/C11 this has revealed that the APC/C complex has a roughly triangular shape and is largely composed of two domains, called the “platform” and the “arc lamp”, which together enclose a central cavity (see Fig. 3c below), whereas a model obtained for budding yeast APC/C shows a more globular structure12. Biochemical and EM subunit mapping experiments in different species9-11,13-15 have indicated that the platform domain contains Apc1, Apc4 and Apc5, whereas the arc lamp domain consists of the TPR proteins Cdc16, Cdc23, Cdc27 and, in case of the vertebrate APC/C, presumably also Apc7. Apc2, a member of the cullin protein family, is located between the platform and the “head” of the arc lamp domain, whereas Cdc20 and Cdh1 bind to the arc lamp opposite of Apc2, with Apc2 and co–activators both facing the central cavity.
Figure 3.
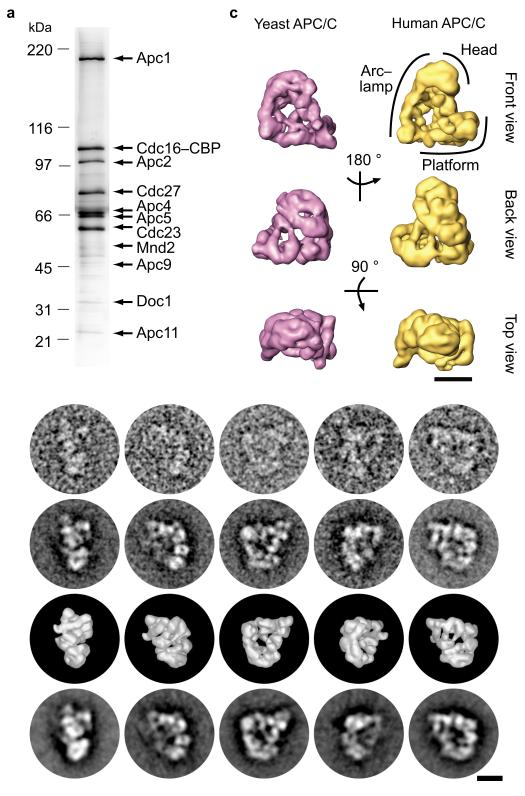
Purification and 3D reconstruction of budding yeast APC/C. (a) SDS-PAGE of APC/C TAP–tag purified via Cdc16. APC/C subunits were identified by their characteristic electrophoretic mobility and mass spectrometry. (b) Top row shows selected EM raw images of yeast APC/C in different orientations. Class averages were obtained by alignment, multivariate statistical analysis and classification of the raw images and are shown in the second row. The third row shows the surface representation of the computed yeast APC/C 3D structure in the corresponding orientations. The fourth row shows re-projections of the yeast APC/C 3D structure in angular directions determined for the views in the third row. (c) Surface representations of yeast and human APC/C 3D structures in three different orientations. Indicated rotations refer to the top structures. The location of APC/C’s head, arc lamp and platform domain is indicated. The size bars indicate a distance of 10 nm.
Because Apc2’s interaction partner Apc11 interacts with E2 enzymes and co–activator proteins help to recruit substrates, it has been speculated that ubiquitylation reactions occur in the inner cavity9,10. However, direct evidence for substrate recruitment to this site is lacking so far. It is also unknown where Doc1/Apc10 is located, a subunit that has been implicated in substrate binding to the APC/C16 and in processive substrate ubiquitylation17. How Doc1 contributes to these processes is unknown, but Doc1 is structurally similar to ligand binding domains in bacterial sialidases and some other enzymes4,5, and related “Doc domains” are also found in other ubiquitin ligases18,19. It is therefore possible that Doc1 uses its putative ligand binding region to interact with APC/C substrates. Consistent with this hypothesis, Doc1’s ligand binding domain is required for its ability to confer processivity to the APC/C20. However, direct evidence for Doc1–substrate interactions has not been obtained.
Here we have used biochemical reconstitution experiments to isolate various forms of budding yeast and human APC/C and have analyzed these complexes by using photo–crosslinking, single particle EM, and 3D reconstruction. Our results show that the structure of the APC/C is conserved from budding yeast to vertebrates, and that Doc1 is located in close vicinity to the cullin–RING module Apc2–Apc11. Structural analysis of APC/CCDH1–substrate complexes revealed that the substrate density is found in the inner cavity, directly intercalated between CDH1 and DOC1. These observations suggest that CDH1 and DOC1 form a bipartite substrate receptor on the APC/C.
RESULTS
Doc1 crosslinks to Cdc27, Cdc16 and Apc1 in yeast APC/C
To understand how Doc1 interacts with the APC/C we generated radiolabeled forms of Doc1 containing a photo–activatable crosslinker at defined sites, allowed these Doc1 proteins to bind to APC/C lacking endogenous Doc1, and used photo–crosslinking followed by SDS polyacrylamide gel electrophoresis (SDS–PAGE) and phosphorimaging to identify crosslink products between Doc1 and APC/C subunits. We used APC/C from budding yeast for these experiments because previous work had shown that APC/C can be purified from yeast strains from which the DOC1 gene has been deleted, and because the processivity defect of the resulting APC/CΔDoc1 complexes can be restored in vitro by addition of recombinant Doc1 (ref. 16,20). Earlier studies used a version of Doc1 containing 283 amino acid residues20,21, whereas the Saccharomyces genome database (SGD; http://www.yeastgenome.org/) reports only 250 amino acids for Doc1 (lacking 38 residues at the N terminus). Because we could only identify the latter form of Doc1 associated with endogenous APC/C (Supplementary Fig. 1a online) we used a cDNA that encodes the 250 amino acid version of Doc1. When this form of Doc1 was synthesized by coupled in vitro transcription–translation in rabbit reticulocyte lysate (RRL) and added to purified APC/CΔDoc1 in the presence of the ubiquitin activating enzyme E1, the E2 enzyme Ubc4, Cdh1 and a fragment of the APC/C substrate Hsl1 (Hsl1667–872), Doc1 restored the ability of APC/CΔDoc1 to generate high molecular weight Hsl1–ubiquitin conjugates (Fig. 1a,b and Supplementary Fig. 1b online), indicating that the short form of Doc1 used here is functional.
Figure 1.
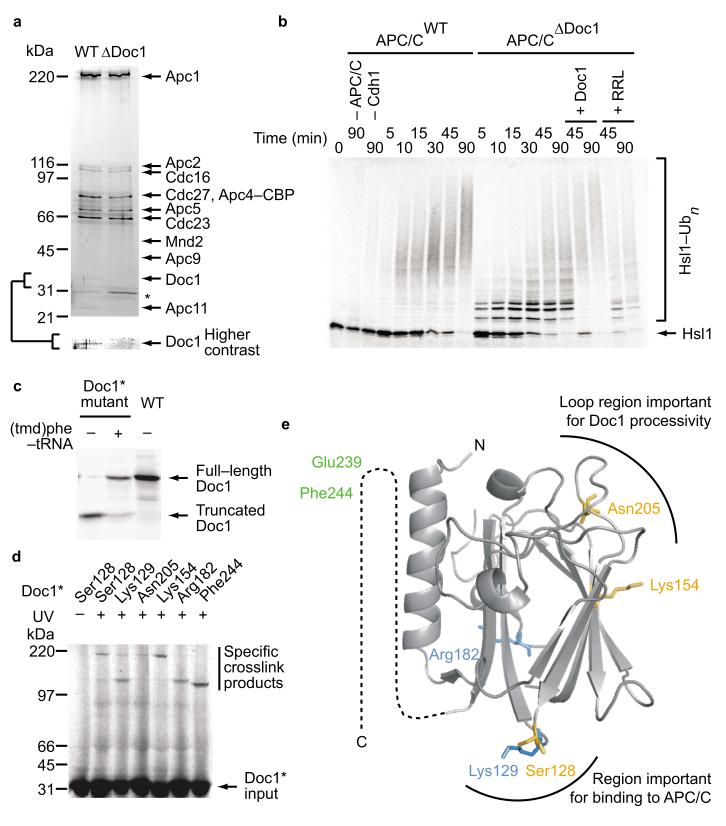
Incorporation of a photo–crosslinker into Doc1 results in crosslink products between Doc1 and APC/C subunits. (a) Composition of wild type APC/C (WT) and APC/C lacking Doc1 (ΔDoc1) affinity–purified via a TAP–tag on Apc4. Apc4-CBP indicates calmodulin binding protein that remains on Apc4 after TEV cleavage. Asterisk marks contaminating band. (b) APC/C ubiquitylation of 35S–methionine–labeled Hsl1667–872 substrate is impaired in the absence of Doc1 compared to the wild type form of APC/C. (c) Crosslinker-incorporation into in vitro translated Doc1. Full–length Doc1 contains the photo–activatable amino acid. (d) Overview of crosslink products obtained with 35S–labeled Doc1amber mutants carrying the photo–crosslinker at 6 different sites. The two consecutive sites Ser128 and Lys129 each created two crosslink products of different sizes. (e) Doc1 crystal structure of S. cerevisiae. Crosslinker incorporation sites are indicated. Predominant crosslinks of APC/C subunits to respective Doc1 sites are color coded (green: Cdc27; blue: Cdc16, orange: Apc1).
To generate forms of Doc1 that contain crosslinkers we introduced “amber” stop codons (TAG) at different sites into the coding sequence of the Doc1 cDNA. We used the resulting mutated cDNAs as templates in in vitro transcription–translation reactions which contained 35S–methionine and 35S–cysteine, and an “amber” suppressor tRNA coupled to the artificial amino acid L–4′–(3–[trifluoromethyl]–3H–diazirine–3yl) phenylalanine (hereafter called (tmd)phe22). Use of the suppresser tRNA allowed translation beyond the “amber” stop codon, resulting in 35S–labeled Doc1 carrying (tmd)phe at the sites specified by the “amber” codon (Fig. 1c). We inserted (tmd)phe at either one of 24 different positions, representing nearly 10% of all amino acid residues in Doc1 (Supplementary Table 1 online). When incubated with APC/CΔDoc1 seven of the 24 Doc1 mutants reproducibly yielded crosslink products with molecular masses of 120 kDa (F224amber, E239amber), 140 kDa (K129amber, R182amber) and 200 kDa (S128amber, K154amber, N205amber; Fig. 1d and Supplementary Table 2 online). Figure 1e shows positions in the Doc1 structure at which (tmd)phe had been inserted in these mutants.
All crosslink products were only obtained in the presence of APC/C (see Fig. 2c below, and data not shown), indicating that these products represented Doc1 that had been crosslinked to APC/C subunits. To determine the identity of these subunits, we fused all APC/C subunits whose molecular mass was consistent with them being part of the Doc1 crosslink products (Apc1, Apc2, Cdc27, Cdc16 and Apc5) to myc6–, myc9– or myc18–tags, which are known to reduce their electrophoretic mobility23. These experiments identified crosslink products between Doc1 and three different APC/C subunits (Fig. 2a–c and Supplementary Table 2 online): Doc1–F244amber yielded crosslink products with Cdc27, consistent with the observation that the IR–tail of Doc1, in which Phe244 (and also Glu239) is located, is required for Cdc27 binding in vitro5. Doc1–S128amber and Doc1–K129amber, which are located in the C–terminal region of Doc1, and Doc1–R182amber, which is located on the back side of Doc1, formed crosslink products with Cdc16. Two Doc1 mutants, Doc1–S128amber and Doc1–K154amber predominantly formed crosslink products with Apc1, but less abundant Apc1 crosslink products were also obtained with Doc1–K129amber and Doc1–N205amber (note that Doc1–S128amber formed predominantly crosslink products with Apc1 but less frequently also with Cdc16, whereas Doc1–K129amber showed the opposite behavior and formed crosslink products preferentially with Cdc16 but to a lesser degree also with Apc1; for possible interpretations of this result see Discussion). Doc1–E239amber was not analyzed because Doc1–E239amber and Doc1–F244amber yielded indistinguishable crosslink products, suggesting that they both interact with the same APC/C subunit (Cdc27). These results suggest that Doc1 interacts with the APC/C by contacting at least three other subunits, Cdc27, Cdc16 and Apc1.
Figure 2.
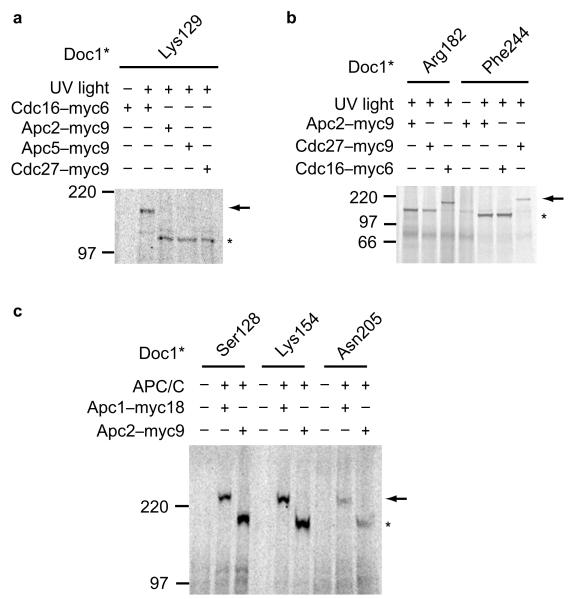
Identification of APC/C subunits that interact with Doc1. (a) Cdc16-myc6 incorporated into the APC/C complex caused a mobility shift of the crosslink product (arrow) when incubated with radiolabeled Doc1 carrying the photo–crosslinker at the position of Lys129, which indicates an interaction between Doc1–Lys129 and Cdc16. (b) Doc1 residue Arg182 contacts Cdc16. Cdc27 interacts with Doc1 via residue Phe244. (c) The large subunit Apc1 contacts the Doc1 protein at residues Ser128, Lys154 and Asn205. Arrows indicate the mobility shift of the crosslink product (asterisk).
Localization of Doc1 in budding yeast and human APC/C
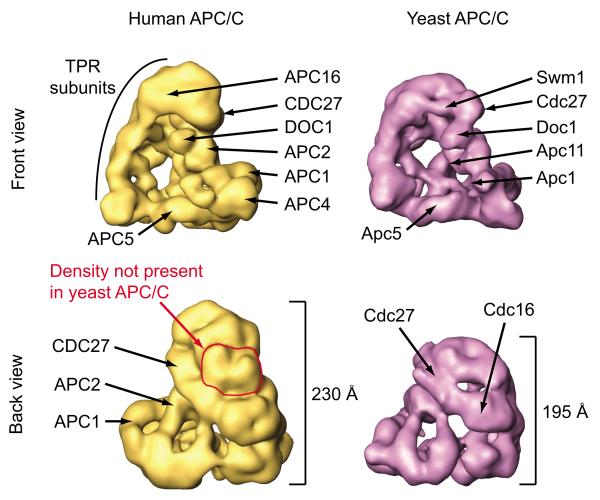
To better understand Doc1’s interactions with other APC/C subunits we identified the position of Doc1 within the structure of APC/C. Because the previously reported 3D model of budding yeast APC/C12 differed substantially from the structures obtained for human and Xenopus APC/C9,10, we first determined a 3D structure of yeast APC/C (Fig. 3a) at ~ 25 Å resolution by cryo–negative staining EM (Fig. 3b,c and Supplementary Table 3 online). This structure revealed that yeast APC/C, like vertebrate APC/C, is composed of a platform and an arc lamp domain which together enclose a central cavity. The dimensions of the platform domain are similar in yeast, Xenopus and human APC/C (135 × 130 Å in yeast APC/C), but the arc lamp domain of yeast APC/C is shorter than the corresponding domain in vertebrate APC/C (195 Å high in yeast versus 230 Å in human APC/C). Otherwise, the structures of yeast and vertebrate APC/C are similar in shape and size, indicating that APC/C’s structure has been largely conserved during evolution.
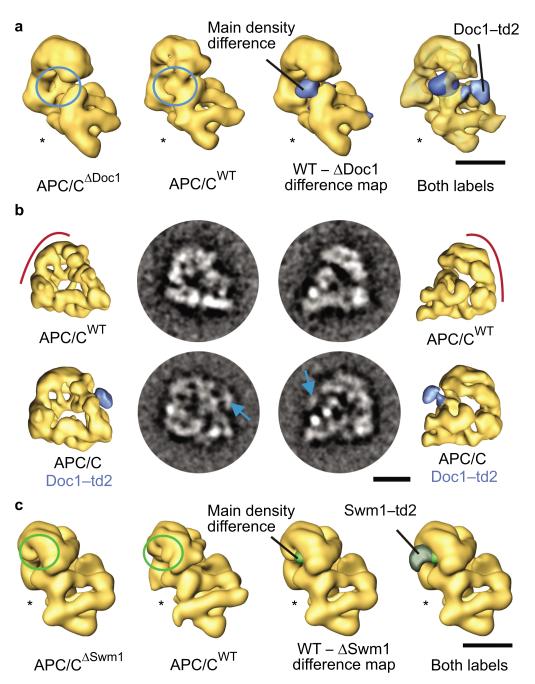
We identified the location of Doc1 within the APC/C by using three different approaches. First, comparison of the 3D structure of wild type yeast APC/C with a structure obtained for APC/CΔDoc1 revealed that a small density located on top of the inner cavity and under the “head” of the arc lamp domain was absent in APC/CΔDoc1 (Fig. 4a). Second, we generated a strain in which Doc1 was fused to a 56 kDa tag called tdimer2 (td2), which is composed of two copies of the red fluorescent protein DsRed24,25 (Supplementary Fig. 1c online). When APC/C containing only this version of Doc1 was analyzed by EM and 3D reconstruction, an extra density was observed next to the density which was absent in the structure of APC/CΔDoc1 (Fig. 4a,b). This observation supports the notion that the density identified by difference mapping between wild type APC/C and APC/CΔDoc1 represents the position of the Doc1 protein. Third, we used antibodies to DOC1/APC10 to map the location of this subunit in human APC/C. We incubated APC/C purified from HeLa cells with DOC1/APC10 antibodies at an APC/C–antibody ratio that led to the formation of trimers in which two APC/C particles were bound by one IgG molecule, enriched these complexes by glycerol density gradient centrifugation, and used negative staining EM to determine the region on APC/C to which the DOC1/APC10 antibodies had been bound (Supplementary Fig. 1d,e online). These experiments indicated that also in the structure of human APC/C DOC1/APC10 is located above the inner cavity and below the “head” of the arc lamp domain.
Figure 4.
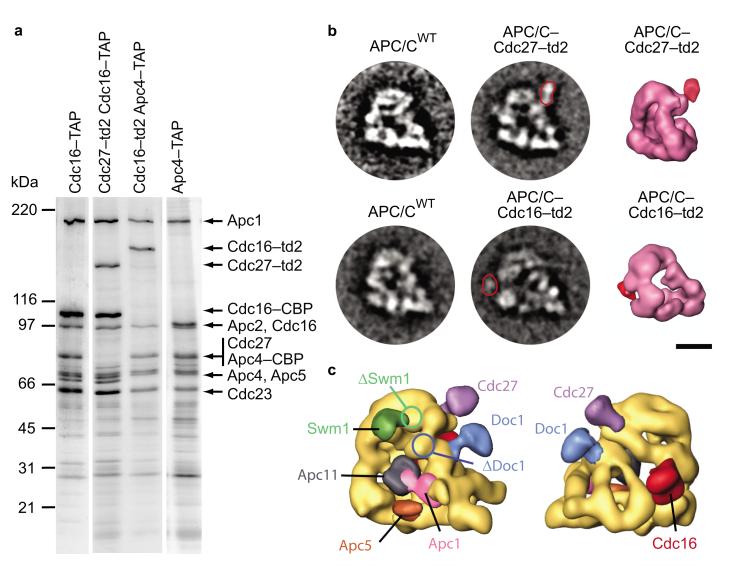
Localization of Doc1 and Swm1 in the yeast APC/C 3D structure by subunit deletion and td2–labeling. (a) 3D models illustrating Doc1 localization. Density not present in APC/CΔDoc1 compared to APC/CWT is labeled as main density difference in the APC/C WT – ΔDoc1 difference map. Subunit deletion and td2–labeling converge at identical locations. (b) Doc1–td2–labeling and EM analysis compared to wild type APC/C. Class averages of wild type and Doc1–td2–labeled APC/C are shown in two different orientations. The additional density caused by the td2–tag is indicated with arrows in the class averages. 3D models computed from the corresponding datasets are depicted in similar orientations. (c) Localization of Swm1 using subunit deletion and td2–labeling. Swm1 is located in the head domain. The size bars illustrate a distance of 10 nm. The asterisks mark the face of APC/C’s central cavity.
Localization of yeast APC/C subunits
To determine if the position of Doc1 within the APC/C structure is consistent with the observation that Doc1 can form crosslink products with Cdc27, Cdc16 and Apc1, we mapped the location of these subunits in the 3D structure of yeast APC/C. We tagged Cdc27 and Cdc16 at their C–termini with td2, and Apc1 with a tag that contained a single copy of DsRed (called “tm” for tmonomer; viable strains expressing Apc1–td2 could not be obtained). We purified APC/C containing Cdc27–td2, Cdc16–td2, or Apc1–tm and obtained 3D structures for these complexes (Fig. 5 and Supplementary Fig. 2 online). These experiments revealed that Cdc27 is located in the “head” of the arc lamp domain and Cdc16 in a central region of the arc lamp domain, next to the putative location of Cdc27 (Fig. 5b,c), i.e. that Cdc27 and Cdc16 are located above and behind Doc1. The tag fused to the C terminus of Apc1 protruded from the platform domain into the inner cavity of APC/C, i.e. was located below Doc1 (Supplementary Fig. 2d online). Although it can not be inferred from these data that Doc1 directly contacts Cdc27, Cdc16 and Apc1, the EM data are consistent with this possibility.
Figure 5.
APC/C subunit localization using td2–labeling. (a) SDS–PAGE analysis of td2–labeled Cdc16 or Cdc27 subunits incorporated into yeast APC/C. (b) Class averages of APC/C-Cdc27-td2 and APC/C-Cdc16-td2 compared to wild type APC/C. Extra densities caused by the td2–label are highlighted in the class averages and marked in red color in the respective 3D models. The size bar equals a distance of 10 nm. (c) Results of all labeling experiments carried out in this study superimposed onto the structure of yeast APC/C.
We also used the techniques developed for localization of Doc1 for identifying the position of other APC/C subunits. In case of Swm1/Apc13, one of the few APC/C subunits that are not essential for viability in yeast14,16,26,27, we purified APC/C from a strain from which the SWM1 gene had been deleted. In the APC/CΔSwm1 structure, a small density was missing on the front side of the “head” of the arc lamp domain, next to where Cdc16 and Cdc27 are located (Fig. 4c and Fig. 5c). A 3D structure of APC/C in which Swm1 had been tagged with td2 revealed an extra globular density right next to the putative Swm1 density identified by difference mapping (Fig. 4c). Swm1 is therefore located in the vicinity of Cdc16 and Cdc27, consistent with the observation that Swm1 stabilizes interactions between these two subunits14.
We attempted to use td2 tagging also for the remaining APC/C subunits but were able to obtain viable yeast strains and APC/C samples suitable for EM only for Apc11 and Apc5. The resulting 3D structures revealed an additional globular domain in the central cavity, close to Doc1, when td2 had been fused to the C terminus of Apc11 (Fig. 5c and Supplementary Fig. 2c,d online), consistent with the previously identified location of APC11’s binding partner, APC2, in human APC/C9,10. C–terminal tagging of Apc5 with td2 revealed an extra density on the left side of the platform domain (Fig. 5c and Supplementary Fig. 2b,d online), also consistent with previous localization of APC5 in human APC/C10.
EM analysis of dimeric forms of yeast APC/C
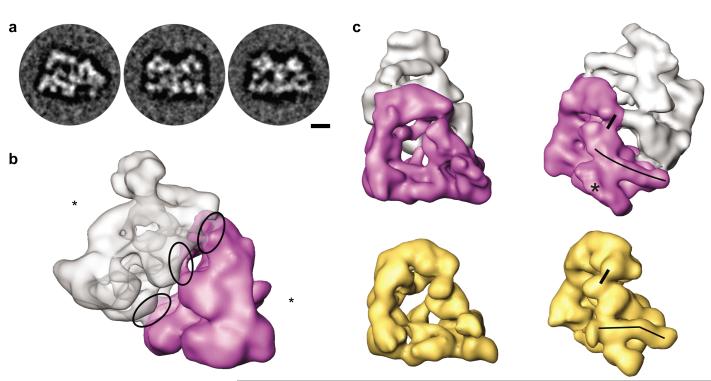
The majority of APC/C observed in our EM specimens was monomeric, but dimeric forms of APC/C were also present (Supplementary Fig. 3a online), confirming previous reports12,16. As shown before23, we found that APC/C sediments corresponding to a sedimentation coefficient of 36 S when yeast extracts were fractionated in the presence of low salt concentrations (50 mM KCl). However, when the same yeast extracts were analyzed in the presence of 400 mM KCl, most APC/C sedimented as a 22 S particle (Supplementary Fig. 3b,c online), as does purified monomeric yeast APC/C28. These observations indicate that many APC/C particles in yeast extracts exist as dimers which dissociate into monomers in the presence of buffers with high ionic strength. Because these findings are consistent with the possibility that yeast APC/C can also exist as a dimer in vivo we also generated a 3D structure of dimeric APC/C at a resolution of ~ 35 Å. Images of these dimers could be sorted into class averages of similar orientations (Fig. 6a), indicating that these dimers represent a homogeneous population of particles in which two APC/C monomers adopt a defined orientation.
Figure 6.
Analysis of APC/C dimers. (a) Class averages of dimeric APC/C with different orientations. Size bar indicates 10 nm. (b) 3D model of dimeric APC/C. The purple APC/C monomer is shown in a side view orientation with the TPR–rich arc lamp domain in the front. The gray APC/C monomer is shown in a bottom view orientation with the bottom of the platform domain in the front. Asterisks mark the face of APC/C’s central cavity. The middle ellipse marks a unique contact lying on the c2 symmetry axis, whereas the other ellipses show contact points that each exist as asymmetric pairs. (c) APC/C dimerization causes conformational rearrangements in the platform domain as well as the head domain compared to monomeric APC/C (yellow). Apc1 has a bent shape in monomeric APC/C and adopts a straight conformation in the dimeric form (illustrated by long black lines). The Apc4 density is better defined in the APC/C monomer compared to the dimer structure (asterisk). The contact between Cdc27 and Doc1 is more pronounced in the monomer (short black lines in the head domain).
The resulting 3D structure revealed that APC/C monomers within the dimer interact via five discrete contact points (three of which can be seen in Fig. 6b, marked by ellipses), mostly on the “back” side of the arc lamp and the platform domains (Fig. 6b,c). One of these points lies on a c2 symmetry axis and is generated by a contact between the same regions of the arc lamp domain where Cdc27 is located, i.e. possibly through a homo–typic interaction between two Cdc27 molecules. The other contacts are not located on the c2 axis and thus occur as asymmetrical pairs of interactions (subunit A of monomer 1 interacts with subunit B of monomer 2 and vice versa). The first pair of contacts is generated by interactions between Apc1 in the platform domain and an unidentified subunit located on the back side of the arc lamp domain, possibly Cdc27, Cdc16 or Cdc26. The second pair of contacts is formed between Cdc16 and an unidentified subunit on the “back” side of the platform, possibly Apc1 or Apc2. During 3D reconstruction we applied two–fold symmetry which prevents the detection of differences in conformation between APC/C monomers within the dimer. However, the conformation of APC/C in the dimer is clearly different from the conformation of monomeric APC/C. Whereas Apc1 has a characteristic bent shape in monomeric APC/C, this subunit adopts a more straight conformation in the dimer, pointing upwards into the direction of Doc1 (the conformational change in Apc1 is indicated by long black lines in Fig. 6c). Smaller conformational changes could also be observed for densities that correspond to Apc4, Cdc27 and Doc1. In the monomer, the density of Apc4 is better defined and the contact between Cdc27 and Doc1 (indicated by short black lines in Fig. 6c) is more pronounced than in the dimer.
Because the dimeric form of yeast APC/C has been reported to ubiquitylate substrates with higher processivity than monomeric APC/C12 we tested if the role of Doc1 in APC/C processivity could be explained by a requirement for Doc1 in APC/C dimerization. However, when we analyzed proteins in whole cell extract from yeast Δdoc1 strains we observed that Cdc16 sedimented corresponding to 36 S, and in EM specimens of APC/CΔDoc1, dimers could still be detected (data not shown), indicating that APC/CΔDoc1 also exists in a dimeric form (see also Supplementary Fig. 3d). Doc1 is therefore not required for APC/C dimerization and must contribute to APC/C processivity through other mechanisms.
Substrate binding to the APC/C occurs between CDH1 and DOC1
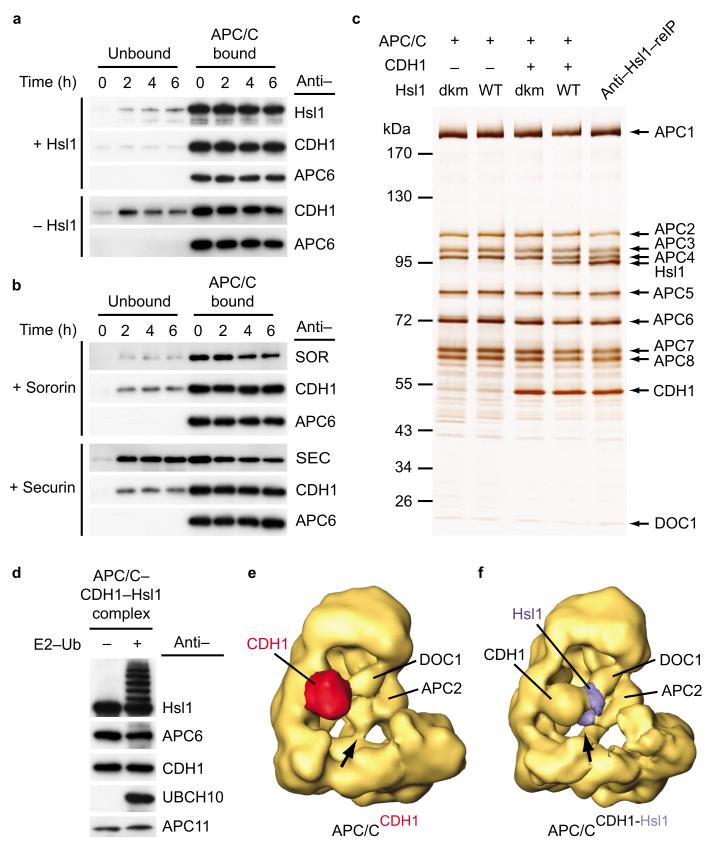
To understand how substrates are recruited to the APC/C and if DOC1 has a direct role in this process we mapped the location of a substrate protein on the APC/C by EM and 3D reconstruction. We used human APC/CCDH1 for this analysis because we were able to generate sufficient amounts of human CDH1 for these experiments (His6–HA–CDH1), but not of yeast co–activator proteins. To evaluate which substrate binds to APC/CCDH1 stably enough to allow EM mapping experiments, we reconstituted and purified APC/CCDH1 bound to either S. pombe Hsl1667–872, human Sororin or human Securin and measured by immunoblotting over a time course of six hours how much substrate remained bound to APC/CCDH1. Many Sororin and Securin molecules dissociated from APC/CCDH1 during the six–hour time course, whereas the majority of Hsl1 remained associated (Fig. 7a,b), consistent with the previous observation that Hsl1 binds to APC/C particularly tightly20,29. These experiments also confirmed that the interaction between APC/C and CDH1 is stabilized by the presence of Hsl1 (ref. 29,30).
Figure 7.
In vitro reconstitution of human APC/C bound to a substrate molecule. (a) Off–rate determination of Hsl1 and/or CDH1. APC CDH1–Hsl1 complexes are compositionally stable for at least six hours. Presence of Hsl1 further increases binding of CDH1 to the APC/C. (b) Reconstituted APC/CCDH1 complexes bound to either Sororin or Securin were subjected to off–rate experiments as described in a. Compared to Hsl1, Sororin and Securin seem to less stably bind to APC/CCDH1 complexes. (c) Human APC/C incubated with wild (WT) type or D box/KEN box mutant (dkm) form of His–Flag–td2–Hsl1667–872 in absence or presence of purified CDH1. Only in the presence of CDH1, Hsl1 containing wild type destruction motifs could be recognized by the APC/C. Re–immunoprecipitation experiments were used to purify stoichiometric APC/CCDH1–Hsl1 complexes. (f) APC/CCDH1–Hsl1 incubated with preformed UBCH10–ubiqtuin complexes results in formation of Hsl1–ubiquitin conjugates. (e) 3D model of human APC/C bound to its co–activator protein CDH1 (red). In the APC/CCDH1 structure, the APC2–APC11 module contacts the platform domain (arrow). (f) 3D model of human APC/CCDH1–Hsl1. The density attributed to the substrate molecule (blue) is intercalated between CDH1 and DOC1. In the APC/CCDH1–Hsl1 structure the APC2/11–platform contact is resolved to form a new connection (blue) to the co–activator protein (arrow).
We therefore performed all subsequent experiments with the Hsl1667–872 fragment. We used a td2–tagged version of Hsl1 for these experiments (His6–Flag–td2–Hsl1667–872), hoping that the td2–domain might help in the subsequent visualization of the substrate in the 3D structure. To ascertain that this protein binds as a bona fide substrate we isolated APC/C on Cdc27–antibody beads and incubated these with wild type or a D box/ KEN box Hsl1 mutant31, in either the absence or presence of CDH1. Unbound CDH1 and Hsl1 were removed by washing the beads, and APC/C was subsequently eluted from the antibody beads by an excess of the antigenic Cdc27 peptide. SDS–PAGE and silver staining confirmed that only the wild type fragment of Hsl1 could efficiently bind to the APC/C, and that this interaction was greatly increased by the presence of CDH1 (Fig. 7c). Furthermore, incubation of APC/CCDH1–Hsl1 complexes with E1, UBCH10 and ubiquitin resulted in the ubiquitylation of Hsl1, further indicating that Hsl1 associated with APC/CCDH1 as a functional substrate (Fig. 7d). Because substrates have to be turned over rapidly in vivo it was surprising to find that substrates bind to the APC/C relatively stably. This observation raises the interesting possibility that in vivo additional factors might promote substrate release from the APC/C.
To obtain complexes composed of APC/C, CDH1 and substrate in a 1:1:1 stoichiometry, we further re–isolated proteins bound to Hsl1 by immunoprecipitation using Flag antibodies, again followed by peptide elution (Fig. 7c, last lane). The resulting APC/CCDH1–Hsl1 complexes were further purified by glycerol density gradient centrifugation using the GraFix method32 and analyzed by cryo–negative staining EM. The difference map calculated for the structures of APC/CCDH1 (ref. 10 and Fig. 7e) and APC/CCDH1–Hsl1 revealed one prominent extra density in substrate bound APC/C (Fig. 7f). This density was intercalated between the densities that we had identified as CDH1 and DOC1. A second extra density was present near the platform domain, but this density disappeared when the threshold for surface rendering was increased, whereas the prominent density between CDH1 and DOC1 remained under these conditions (Supplementary Fig. 4 online). The two densities might represent two domains of His6–Flag–td2–Hsl1667–872 that are connected by a linker region that we were unable to visualize. Because Hsl1 is known to bind to Cdh1 directly in a D box and KEN box dependent manner31 we suspect that the density near CDH1 and DOC1 represents Hsl1667–872, whereas the density near the platform domain may represent the td2–tag whose position might be more flexible and which, therefore, might become invisible at increased surface rendering thresholds.
We also noticed that a contact that normally exists between APC2 and a subunit in the platform domain (indicated by a short arrow in Fig. 7e) is absent in APC/CCDH1–Hsl1 (Fig. 7e,f and Supplementary Fig. 5 online). Instead, in APC/CCDH1–Hsl1 APC2 appears to contact CDH1 directly, in close proximity to where CDH1 contacts the substrate density. These data are consistent with the possibility that the substrate protein forms several contacts on the APC/C, possibly with CDH1 and DOC1, and that substrate binding induces conformational changes in APC2 and/or APC11.
DISCUSSION
Despite being essential for cell division, it remains poorly understood how the APC/C recognizes and ubiquitylates specific substrate proteins, how these processes are controlled in time, and why, in contrast to other ubiquitin ligases, the APC/C is composed of at least 13 different subunits. Because structural information will be important for answering these questions we have used biochemical and electron microscopic approaches to identify how the processivity factor Doc1 and a substrate protein interact with the APC/C. Our results also show for the first time that the structure of the APC/C is largely conserved between budding yeast and vertebrates, and how two APC/C particles interact to form a 36 S dimer.
Although our structural analysis of yeast APC/C revealed many similarities with the structure of human APC/C, there are also important differences. The most notable one is the lack of a density in the arc lamp domain, which results in a reduced height (by 15%) of this domain in yeast APC/C (Fig. 8 and Supplementary Fig. 3e online). Since the arc lamp domain is predominantly composed of TPR subunits it is possible that the density only found in vertebrate APC/C is formed by Apc7, a TPR subunit only identified in higher eukaryotes so far.
Figure 8.
Topological comparison of human and budding yeast APC/C. Human and yeast APC/C models are shown in either front or back view orientation at a resolution of ~ 25 Å. Subunit localization reveals overlapping positions for Cdc27, Doc1, Apc1 and Apc5 in the respective APC/C structures. Overall structural resemblance of human and yeast APC/C 3D models suggest similar positions for most of the subunits, e.g. Apc2, Apc4, Apc11, Cdc16 and Swm1/Apc13. Human APC/C carries a marked extra mass within the TPR–rich arc lamp domain, which might represent a subunit specific for vertebrate APC/C, e.g. Apc7.
A second peculiarity of budding yeast APC/C is its ability to form dimers12,23,33, which is the predominating form in whole cell yeast extracts23 and which, therefore, might also exist in vivo. This notion is further supported by the fact that we could obtain a defined 3D structure of APC/C dimers, which would not be possible if the dimers were formed by unspecific aggregation of monomers. Our 3D structure reveals that two APC/C monomers interact via multiple discrete contacts which involve Cdc27, Cdc16, Apc1 and possibly Apc2. Because these contacts are located on the back side of the arc lamp and the platform domains, the substrate binding sites within the dimer face opposite directions. This observation suggests that each monomer within the APC/C dimer can mediate ubiqutylation reactions independent of its dimerization partner. Understanding the functional relevance of APC/C dimerization, and if such dimers also exist in vivo, will therefore require further investigation.
By combining EM structure determination with the deletion or tagging of different subunits we have identified the position of seven subunits in yeast APC/C. For four of these, we have also mapped the position of their orthologs in human or Xenopus APC/C9,10 (Supplementary Table 4 online). Importantly, for all four of these subunits we found that their positions were corresponding to each other in the yeast and vertebrate 3D structures. This observation, and the finding that the 3D structures of yeast and human APC/C are similar, allows us for the first time to draw an almost complete 3D map of where APC/C subunits are located (Fig. 8). This topographic map is largely consistent with previous biochemical and genetic data13-15 and indicates that the platform domain is composed of Apc1, Apc4 an Apc5, and the arc lamp domain of Cdc27, Apc16, Swm1/Apc13, Cdc16, Cdc26, Cdc23, and in vertebrates also Apc7. Importantly, a third small domain, composed of Apc2, Apc11 and Doc1, is located between the platform and the arc lamp. Because Apc11 is known to recruit E2 enzymes which transfer ubiquitin residues to substrates1, we propose to call this domain the “catalytic core”.
It is well established that Doc1 is required for efficient substrate recruitment to the APC/C and for processive substrate ubiquitylation16,17,20, but how Doc1 performs these functions and how Doc1 itself interacts with the APC/C is poorly understood. In crosslinking experiments we identified Cdc27, Cdc16 and Apc1 as binding partners of Doc1. These results are consistent with our previous observation that Doc1 binds to Cdc27 in vitro5 and with our EM data, which place Doc1 above Apc1, to the right of Cdc16 and below Cdc27 (Fig. 8). Furthermore, it has been previously shown that the Doc1 residues Lys129 and Arg130 are required for efficient binding of Doc1 to the APC/C (ref. 20; note that in this study these residues were referred to as Lys162 and Arg163 because the 283–residue version of Doc1 was used). This observation combined with our finding that Lys129 predominantly crosslinks to Cdc16 and the neighboring residue Ser128 predominantly to Apc1 indicate that the interaction of Doc1 with Cdc16 and Apc1 is required for efficient binding of Doc1 to the APC/C. Because genetic experiments had shown that Apc2 is required for recruitment of Doc1 to yeast APC/C15 it was unexpected that no Doc1–Apc2 crosslink products were obtained, but our EM data reveal that Doc1 is indeed located next to Apc2 and Apc11. It is possible that we were unable to detect Doc1–Apc2 interactions because we did not insert tmd(phe) into Doc1 residues that contact Apc2. Interestingly, we observed that two Doc1 residues (Ser128 and Lys129) could crosslink to two APC/C subunits each, Cdc16 and Apc1 (Supplementary Table 2 online), implying that Ser128 and Lys129 are located directly at the interface of Cdc16 and Apc1, or that Doc1 is able to interact with the APC/C in two different ways. This observation raises the interesting possibility that Doc1 could adopt two different conformations on the APC/C and that this ability might be required for Doc1’s role in processive substrate ubiquitylation.
How might Doc1 confer processivity to APC/C mediated ubiquitylation reactions? Our observation that substrate protein is located directly in between DOC1 and CDH1 strongly implies that DOC1 directly interacts with the substrate, as has previously been shown to be the case for CDH1 (ref. 28). We therefore propose that Doc1 mediates processive substrate ubiquitylation reactions by forming part of a bipartite substrate receptor on the APC/C, composed of Doc1’s ligand binding region4,5,20 and the WD40 propeller domain of the co–activator protein28. This hypothesis is also supported by the previous observation that mutation of the D box in APC/C substrates and deletion of Doc1 reduces substrate ubiquitylation in similar and functionally redundant manners20. It is also interesting to note that the major putative substrate density occupies a position that overlaps with the position of MCC subunits (Supplementary Fig. 5 online), which are believed to inhibit the APC/C as a pseudo-substrate10,34. The overlap between the positions of substrate and MCC proteins provides further support for the hypothesis that MCC inactivates the APC/C by occupying and/or altering its substrate binding site.
Methods
Strains and plasmids
Yeast genetic manipulations were carried out using standard protocols. For untagged Doc1 constructs in pME vectors, pME34 (ref. 20) was used and the original STOP codon of the ORF was kept. To introduce DOC1 including its endogenous promoter into pRS316, the DOC1 ORF plus 154 bases upstream of the translation start and 158 bases downstream were amplified from yeast genomic DNA and cloned into pRS316 via EcoRI sites. Amber (TAG) and ochre (TAA) STOP codons as well as alanine and phenylalanine mutations were generated using the Quikchange method (Stratagene). For tagging of APC/C subunits with tdimer2, pKT146 was used35. Detailed information about N–terminal tagging of Doc1 with tdimer2, as well as a list of yeast strains (Supplementary Table 5 online) used are provided in the Supplementary Material.
APC/C purification
Yeast cells were grown in rich YPD medium (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose). At OD600, cells were harvested by centrifugation and washed with EM lysis buffer (20 mM Hepes–KOH pH 8.0, 200 mM KCl, 10% (v/v) glycerol, 1.5 mM MgCl2). Yeast extracts were prepared by bead beating for crosslinking experiments. For electron microscopy experiments, frozen cells were ground in a mortar grinder (RM100, Retsch) whereas a freezer mill (SPEX Freezer/Mill 6770, 6780) was used for all other applications. APC/C isolation by tandem affinity purification was carried out as described16,36. For each EM experiment cells from 40 liters of yeast culture were lysed in the presence of 0.03% (w/v) octyl–beta–D–glucopyranoside and APC/C was eluted with 3 mM EGTA. 500 μl APC/C eluate were loaded onto a 10 –40% (v/v) glycerol density gradient (20 mM Hepes–KOH pH 7.9, 150 mM NaCl, 2 mM MgCl2, 0.05% (w/v) octyl–β–D–glucopyranoside) containing 0.05–0.2% (v/v) glutaraldehyde32 and centrifuged at 37000 rpm (140601 g) for 14 h at 4°C in a Beckman SW60Ti rotor. APC/C peak fractions were used for subsequent electron microscopy experiments.
Photo–crosslinking
TNT rabbit reticulocyte lysates were used to incorporate photoactivatable crosslinkers into the Doc1 protein. Reactions were supplemented with 1 mM MgOAc and 1.6 μl (tmd)phe–tRNA (see below) per 50 μl reaction. Photo–crosslinking experiments were carried out with freshly prepared yeast cell extracts (50 mM Tris–HCl pH 8, 150 mM Cl, 10% (v/v) glycerol, 0.1% (v/v) NP40). Samples containing photo–crosslinkers were light protected in all steps. APC/C was bound to IgG Sepharose beads by incubating extract and sepharose (1–2 mg protein per 5 μl beads) for 60–90 minutes at 4°C. Beads were washed with buffer used for cell lysis before addition of TNT rabbit reticulocyte lysate containing in vitro translated Doc1–versions carrying a photo–crosslinker at distinct sites. Samples were incubated at room temperature for 20 minutes and subsequently washed 3–4 times with lysis buffer. For photo–crosslinking, samples with lids opened were kept in a thermomixer (1300 rpm, 9°C) and exposed to a black ray long UV–lamp (B–100AP, 100W, UVP) with a wave length of 360 nm at a distance of 10 cm for 5–8 minutes. APC/C was eluted by the addition of SDS–sample buffer.
In vitro reconstitution of APC/C bound to human CDH1 and substrates
Cell extracts were prepared by lysing frozen log–phase HeLa cells in extract buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 10% (v/v) glycerol, 0.05% (v/v) Tween20) using a Dounce homogenizer followed by centrifugation. APC/C was immunoprecipitated (IP) from the soluble fraction by incubation with CDC27 antibodies37 crosslinked to Affi–prep protein A beads (BioRad) for 1h at 4°C. Beads were washed 4 times with at least 20 bead volumes of wash buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 0.05 % (v/v) Tween20) for 3 minutes at 4°C. To in vitro reconstitute co–activator and/or substrate bound APC/C complexes, APC/C–bound CDC27 beads were resuspended in binding buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.05% (v/v) Tween20, 4 mg ml−1 BSA) supplemented with 3.5 μM recombinant human CDH1 and/or 150 nM recombinant wild type or mutant (D box and KEN box mutated) His–Flag–td2–Hsl1667–872, Sororin–td2–Flag–His or Securin–Flag–His. After 1h incubation at 4°C, the excess of recombinant proteins were removed by washing the beads 4 times with at least 20 bead volumes of wash buffer for 3 minutes at 4°C and APC/C complexes were recovered by elution with antigenic peptides. APC/CCDH1–Hsl1 complexes were enriched by re–immunoprecipitation (re–IP) experiments using ANTI–FLAG M2 Agarose (Sigma) and recovered by elution with antigenic peptides. For electron microscopy experiments, APC/C specimens were subjected to GraFix32 to further purify and stabilize the complexes. Human recombinant CDH1 was expressed in Baculovirus–infected Sf9 insect cells. Recombinant His–Flag–td2–Hsl1667–872, Sororin–td2–Flag–His and Securin–Flag–His substrate proteins were expressed and purified from E. coli BL21(DE3) or Rosetta(DE3). The Hsl1667–872 sequence was derived from S. pombe, Sororin and Securin from H. sapiens. To demonstrate activity of in vitro reconstituted APC/CCDH1–Hsl1 complexes, 50 μl eluate was incubated with 5 μl reaction mix containing preloaded UBCH10~monoubiqitin, E1, ATP and DTT for 1h at 4°C.
Off–rate experiments
In vitro reconstituted substrate bound APC/CCDH1 on 25 μl Cdc27–antibody beads were resuspended in 50 μl wash buffer and incubated on a orbital rotator for 1 minute (time point 0), 2, 4 and 6 hours. CDC27–antibody beads were spun down, wash buffer was saved (unbound fraction) for subsequent SDS–PAGE analysis and APC/C complexes were recovered by elution with 50 μl antigenic peptides. APC/C bound and unbound fractions were analyzed by western blotting.
Supplementary Material
Acknowledgements
We are grateful to I. Häcker and M. Madalinski for technical assistance, J. Barrett, J. Brunner, D. Finley, U. Hoya, D. Morgan, K. Nasymth, M. Solomon, W. Zachariae for kindly providing yeast strains and reagents, and J. Barrett, J. Brunner and B. Martoglio for advice on photo–crosslinking. Research in the laboratory of H.S. was supported by grants from the Federal Ministry of Education and Research, Germany, and the Sixth Framework Programme of the European Union via the Integrated Project 3DRepertoire. Research in the laboratory of J.–M.P is supported by Boehringer Ingelheim, the Vienna Spots of Excellence Programme and the Austrian Science Fund.
Footnotes
Database accession numbers
Protein Data Bank Japan
Yeast APC/C monomer: EMD–1820
Yeast APC/C dimer: EMD–1822
Human APC/CCDH1–Hsl1: EMD–1821
References
- 1.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 2.Matyskiela ME, Rodrigo-Brenni MC, Morgan DO. Mechanisms of ubiquitin transfer by the anaphase-promoting complex. J Biol. 2009;8:92. doi: 10.1186/jbiol184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 4.Au SW, Leng X, Harper JW, Barford D. Implications for the ubiquitination reaction of the anaphase-promoting complex from the crystal structure of the Doc1/Apc10 subunit. J Mol Biol. 2002;316:955–68. doi: 10.1006/jmbi.2002.5399. [DOI] [PubMed] [Google Scholar]
- 5.Wendt KS, et al. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat Struct Biol. 2001;8:784–8. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Dye BT, Rajashankar KR, Kurinov I, Schulman BA. Insights into anaphase promoting complex TPR subdomain assembly from a CDC26-APC6 structure. Nat Struct Mol Biol. 2009;16:987–9. doi: 10.1038/nsmb.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, et al. Molecular structure of the N-terminal domain of the APC/C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture. J Mol Biol. 397:1316–28. doi: 10.1016/j.jmb.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 8.Han D, Kim K, Kim Y, Kang Y, Lee JY. Crystal structure of the N-terminal domain of anaphase-promoting complex subunit 7. J Biol Chem. 2009;284:15137–46. doi: 10.1074/jbc.M804887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dube P, et al. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20:867–79. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Herzog F, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–81. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohi MD, et al. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol Cell. 2007;28:871–85. doi: 10.1016/j.molcel.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Passmore LA, et al. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol Cell. 2005;20:855–66. doi: 10.1016/j.molcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol. 2003;13:1459–68. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 14.Schwickart M, et al. Swm1/Apc13 is an evolutionarily conserved subunit of the anaphase-promoting complex stabilizing the association of Cdc16 and Cdc27. Mol Cell Biol. 2004;24:3562–76. doi: 10.1128/MCB.24.8.3562-3576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thornton BR, et al. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–60. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passmore LA, et al. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. Embo J. 2003;22:786–96. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll CW, Morgan DO. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat Cell Biol. 2002;4:880–7. doi: 10.1038/ncb871. [DOI] [PubMed] [Google Scholar]
- 18.Grossberger R, et al. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J Biol Chem. 1999;274:14500–7. doi: 10.1074/jbc.274.20.14500. [DOI] [PubMed] [Google Scholar]
- 19.Gieffers C, Schleiffer A, Peters JM. Cullins and cell cycle control. Protoplasma. 2000;211:20–28. [Google Scholar]
- 20.Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15:11–8. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 21.Hwang LH, Murray AW. A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol Biol Cell. 1997;8:1877–87. doi: 10.1091/mbc.8.10.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunner J. New photolabeling and crosslinking methods. Annu Rev Biochem. 1993;62:483–514. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- 23.Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274:1201–4. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]
- 24.Campbell RE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–82. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacker I, et al. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat Struct Mol Biol. 2008;15:1206–12. doi: 10.1038/nsmb.1506. [DOI] [PubMed] [Google Scholar]
- 26.Hall MC, Torres MP, Schroeder GK, Borchers CH. Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J Biol Chem. 2003;278:16698–705. doi: 10.1074/jbc.M213109200. [DOI] [PubMed] [Google Scholar]
- 27.Yoon HJ, et al. Proteomics analysis identifies new components of the fission and budding yeast anaphase-promoting complexes. Curr Biol. 2002;12:2048–54. doi: 10.1016/s0960-9822(02)01331-3. [DOI] [PubMed] [Google Scholar]
- 28.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18:543–53. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 29.Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell. 2009;34:68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol Cell. 2005;18:533–42. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 31.Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–95. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kastner B, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–5. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 33.Passmore LA, Barford D. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep. 2005;6:873–8. doi: 10.1038/sj.embor.7400482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–67. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
References Methods section
- 35.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–70. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 36.Passmore LA, Barford D, Harper JW. Purification and assay of the budding yeast anaphase-promoting complex. Methods Enzymol. 2005;398:195–219. doi: 10.1016/S0076-6879(05)98017-8. [DOI] [PubMed] [Google Scholar]
- 37.Gieffers C, Dube P, Harris JR, Stark H, Peters JM. Three-dimensional structure of the anaphase-promoting complex. Mol Cell. 2001;7:907–13. doi: 10.1016/s1097-2765(01)00234-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.