Abstract
Hax1 plays an important role in immunodeficiency syndromes and apoptosis. Recently, Chao et al., (Nature, 2008) reported that Hax1 is a Bcl-2-family-related protein required to suppress apoptosis in lymphocytes and neurons via a mechanism that involves association to the rhomboid protease PARL in the mitochondria intermembrane space (IMS). Mechanistically, Hax1/PARL interaction allows the recruitment of the serine protease Omi/HtrA2 and its presentation to PARL, which cleaves it to generate a form of Omi/HtrA2 that may proteolytically eliminate active Bax during mitochondrial outer membrane permeabilization (MOMP). The results of this study imply that the control of cell-type sensitivity to proapoptotic stimuli is governed by the PARL/Hax1 complex in the IMS and, more generally, that Bcl-2-family-related proteins can control MOMP from the inside of the mitochondrion. Further, it defines a novel, antiapoptotic Opa1-independent pathway for PARL. Here we present evidence that Hax1 is not a Bcl-2-family-related protein. Also, that in vivo the activity of Hax1 cannot be mechanistically coupled to PARL because the two proteins are confined in distinct cellular compartments and their interaction in vitro is an artifact. Our results indicate a different function and mechanism of Hax1 in apoptosis and re-opens the question of whether mammalian PARL, in addition to apoptosis, regulates mitochondrial stress response through Omi/HtrA2 processing.
Keywords: Parl, Hax1, Omi, HtrA2, rhomboids, serine protease, mitochondrial stress, mitochondria dynamics, apoptosis, neurodegenerative disease, Parkinson's disease, cancer
Introduction
Mitochondria are key players in central cellular processes, such as ATP production, Ca2+ signaling and apoptosis. Mitochondrial involvement in apoptosis was throughly documented during the last decade. Its two main features include the release of proteins from the mitochondrial intermembrane space (IMS) and the initiation of a programme of dysfunction that includes the loss of the proton electrochemical gradient across the inner mitochondrial membrane (IMM) (1). These two cascades of events appear to be mediated by the crosstalk of several molecular mechanisms that are still not fully characterized (2). Nevertheless, the general consensus is that during apoptosis mitochondria release cytochrome c and other proteins which cooperate to execute programmed cell death (3-5).
The function of mitochondria in the regulation and amplification of the apoptotic cascade is regulated by members of the Bcl-2 protein family (6, 7). These are cytosolic proteins that, under steady state conditions, are mainly peripherally associated to heavy membranes (8), and that share a limited structural similarity with Bcl-2 in the so called Bcl-2 homology (BH) module. Bcl-2-family-related proteins participate in the same process of regulation of apoptosis and are classified as pro- and antiapoptotic, depending on their effect on programmed cell death. Proapoptotic members are further subdivided into ‘multidomain’ ones that share the BH1, BH2, and BH3 modules, and the ‘BH3-only’ proteins that only share the BH3 module. The pro-apoptotic BH3-only proteins “sense” the death stimuli and transduce them to mitochondria, where they activate the ‘multidomain’ pro-apoptotic proteins Bax and Bak, ultimately resulting in mitochondrial outer membrane permeabilization (MOMP) and cytochrome c release. With respect to the antiapoptotic Bcl-2 family members, two models of activity have been proposed and are the subject of intense investigations. In the first model, these proteins are endogenous inhibitors of the multidomain proapoptotic proteins, requiring to be antagonized by ‘BH3-only’ molecules; in the second model, the antiapoptoci Bcl-2 family proteins act by sequestering BH3-only proteins, a possibility that is strongly supported by recent, elegant studies from the Andrews laboratory (9). However, a recent report by Chao and colleagues challenged this view by describing how Hax1, a purported Bcl-2-family-related protein of the mitochondrial IMS, mediates the elimination of active Bax, thereby introducing the concept that antiapoptotic Bcl-2 proteins can antagonize MOMP also from inside the mitochondria via the activation of a proteolytic cascade (10).
To ensure the complete release of cytochrome c, the architecture of the mitochondrial reticulum and the ultrastructure of the organelle changes in the early stages of apoptosis (11-14). A combination of electron tomography and physiological measurements identified a pathway of cristae remodeling characterized by the widening of the narrow tubular junction and by the fusion of individual cristae. These morphological changes support the mobilization of cytochrome c from the cristae to the IMS, and eventually to the cytosol (15). Mechanistically, the mitochondrial rhomboid protease PARL and the dynamin-related GTPase Opa1, two proteins of the IMM, participate in the control of the shape of the cristae and of the diameter of the cristae junctions during apoptosis (16, 17). Under steady state conditions, active Opa1 can prevent the widening of the cristae junctions by forming an oligomer that functions as a molecular staple between the adjacent membranes of the cristae. This high molecular weight complex contains both an IMM-bound form of Opa1 and an IMS-soluble one. Generation of the latter form requires the rhomboid protease PARL, whose ablation is lethal in adult mice owing to a pathology caused by excessive apoptosis in multiple tissues (16, 17). A similar phenotype was recently observed in mice lacking Hax1, which also displayed reduced level of a cleaved form of Omi/HtrA2 (10), a serine protease of the IMS implicated in oxidative stress and apoptosis (18). These observations and the finding that Hax1 could be co-immunoprecipitated with PARL led Chao and colleagues to propose a model where Hax1 presents Omi/HtrA2 to PARL, to generate a form of Omi/HtrA2 that may proteolytically antagonize from the IMS active Bax during MOMP (10), thereby defining a novel antiapoptotic Opa1-independent pathway for PARL (16, 17, 19).
Our present study challenges the conclusions of Chao et al. (10) by showing that Hax1 is not a Bcl-2-family-related protein because it does not contain bona fide BH modules, and that, in vivo, the activity of Hax1 cannot be mechanistically coupled to PARL because the two proteins are confined in different cellular compartments, and their interaction in vitro is an artifact. Hax1 plays a key role in autosomal recessive severe congenital neutropenia (20), a primary immunodeficiency syndrome associated with increased apoptosis in myeloid cells (21); therefore, elucidating the mechanism of Hax1 activity remains an outstanding question that has to be addressed to decipher the molecular pathways that link mitochondrial stress response to apoptosis.
Results
Hax1 lacks BH modules
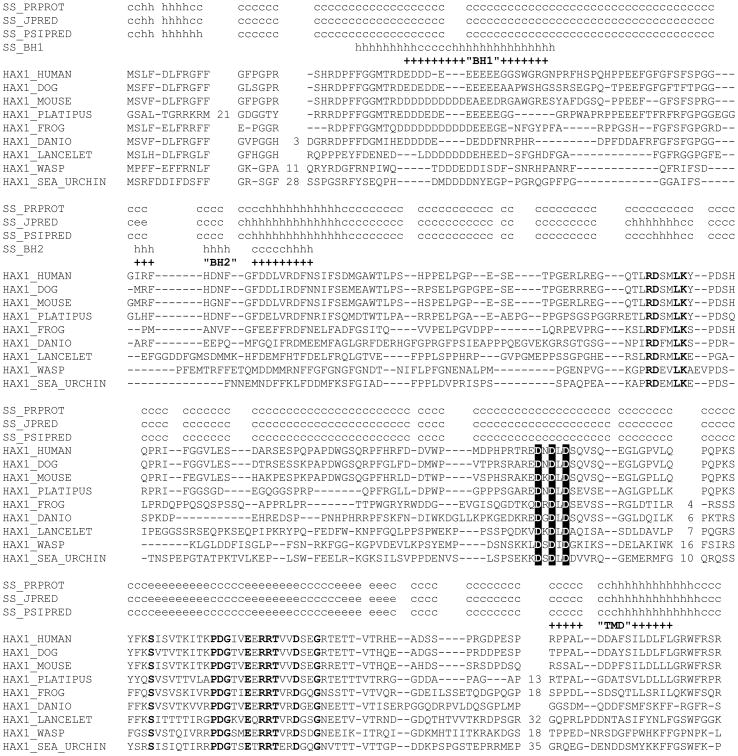
Chao et al. denoted Hax1 a Bcl-2 family protein on the basis of purported structural similarities to Bcl-2 family members including the presence of BH1- and BH2-like domains and a carboxyl transmembrane domain (10). This characterization is based, primarily, on the previous report by Sharp et al. (22). According to these studies, Hax1 contains a BH1 and BH2 module in positions 37-56 and 74-89, respectively, as well as a C-terminal transmembrane domain. However, our sequence analysis and structure prediction do not support the presence of any of these modules in Hax1. Indeed, these purported BH1 and BH2 modules are not recognized by conserved protein domain search, even with the most relaxed threshold. Further, multiple secondary structure predictions show that the regions of Hax1 that were previously aligned with the BH1 and BH2 domains are largely disordered whereas the bona fide BH1 and BH2 modules are stable hairpins formed by hydrophobic α-helices (23, 24). Instead, the corresponding regions of the Hax1 sequence are not well conserved even in closely related animals, such as mammals, and show no sequence conservation at all in more distantly related species (Figure 1), which would be incompatible with the key roles of these regions in the function of Hax1 in apoptosis as a Bcl-2 protein. Our analysis shows instead that Hax1 is an α/β-protein that contains a strongly predicted and relatively well-conserved, in animals, three-strand (β-sheet near the C-terminus (Figure 1), a structural element absent in Bcl-2 proteins. Interestingly, Hax1 also contains a conspicuous pattern of three universally conserved aspartates embedded in a predicted disordered loop, which is suggestive of functionally important metal (possibly, calcium)-binding residues. None of these structural elements are present in any of the Bcl-2 proteins. We conclude, therefore, that Hax1 is not a Bcl-2-family-related protein.
Figure 1. Hax1 does not contain BH1 or BH2 modules or a transmembrane domain.
Multiple alignment of selected Hax1 sequences from diverse animals. The numbers between aligned blocks indicate poorly conserved sequence segments that are not shown. Secondary structure (SS) predicted with three methods is shown above the alignment: “c” indicates random coil (disordered structure), “h” indicates α-helix, and “e” indicates (β-strand (extended conformation). Amino acid residues that are conserved in all aligned sequences are shown in bold type, and the three invariant aspartates that comprise a putative metal-binding site are shown by reverse shading. The positions of purported BH1 and BH2 modules are shown above the respective regions of the alignment using the alignment from (22) for BH1, and arbitrarily centering the alignment on a conserved hydrophobic residue for BH2. “SS_BH” denotes the consensus secondary structure motifs of BH1 and BH2 modules derived from multiple crystal and NMR structures of Bcl-2-family proteins (24). The position of the transmembrane domain (TMD) predicted in (22), but not In our analysis, is indicated in the C-terminal block of the alignment.
Hax1 is not an integral membrane protein
At the C-terminus of Hax1 there is a strongly predicted and conserved C-terminal α-helix that has been purported to constitute the transmembrane domain that anchors Hax1 to both mitochondrial membranes (10). Multiple methods of transmembrane region prediction, as well as visual inspection of Hax1 sequence for long hydrophobic stretches, indicate that Hax1 does not contain such transmembrane domain, either near the C-terminus or anywhere within the protein sequence (Figure 1). To span the lipid bilayer, an α –helix must be composed by a minimum of about 20 amino acids, mainly hydrophobic (25). The purported transmembrane domain of Hax1 consists in an αa-helix of 16 amino acids, 4 of which are charged and one more is polar (RPPALDDAFSILDLFL), which could not form a transmembrane domain capable of anchoring Hax1 within a lipid bilayer. This analysis is consistent with our experimental data from alkaline and high salt extraction of heavy membranes prepared from HeLa cells, which showed that endogenous Hax1 is peripherally associated, but not integrated, to these membranes (Figure 2). We conclude that Hax1 is not an integral membrane protein and suggest that the conserved C-terminal domain might be functionally important in coordinating Hax1 interaction with other proteins.
Figure 2. Hax1 is not an integral membrane protein.
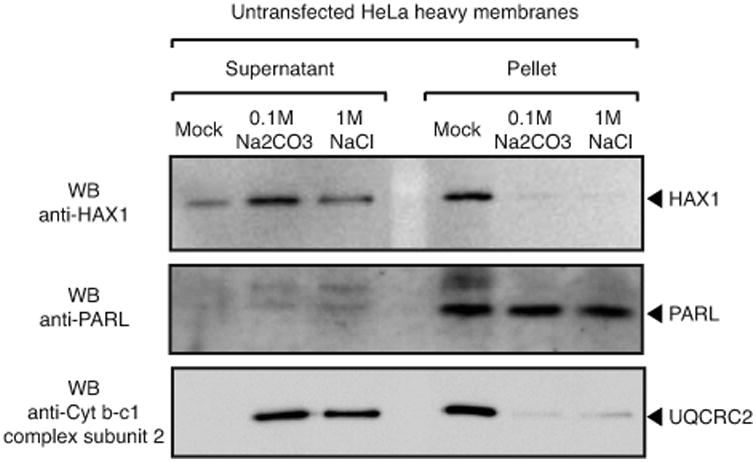
Alkaline extraction of heavy membranes isolated from HeLa cells (200 μg); whereas membrane-associated proteins and proteins associated to membrane-bound proteins (e.g. UQCRC2) are solubilized in the supernatant, integral membrane proteins like PARL are recovered in the membrane pellet. Lack of Hax1 integration in heavy membranes is consistent with our computational analysis, which does not predict any potential transmembrane domain.
Hax1 is not localized inside the mitochondria
Hax1 does not contain cysteine residues required for IMS protein import via the MIA pathway (26), or any predictable mitochondrial import peptide. Further, a recent extensive proteomic study does not list Hax1 as part of the compendium of mitochondrial proteins expressed across 14 mouse tissues (27). Our data show that although heavy membranes contain Hax1, the protein is absent in preparations of highly purified, intact mouse liver mitochondria (Figure 3). Thus, contrary to the claim of Chao et al. that endogenous Hax1 “is localized on the inner and outer mitochondrial membranes and exposed to the intermembrane space (data not shown)” (10), we conclude that the protein might be peripherally associated with mitochondria but not imported inside the organelle.
Figure 3. Hax1 is not detected in mitochondria.
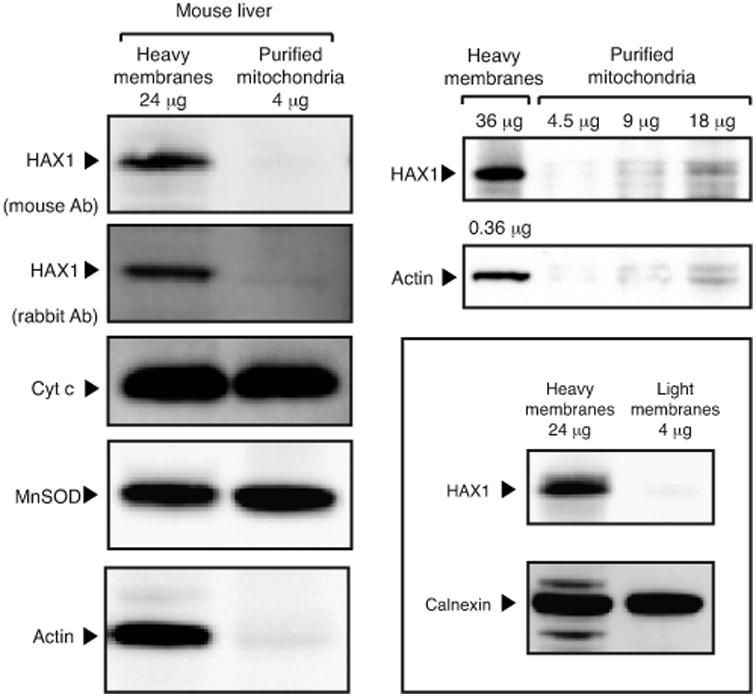
Immunoblot analysis of Percoll-purified fractions of mouse livers. In this preparations, lack of cross-contaminating organelles was assayed by electron microscopy (not shown); inner and outer mitochondria membrane integrity was tested for presence of diffusible proteins of the IMS (cytochrome c) and mitochondrial matrix (MnSOD); purity from membranes associated cytosolic proteins by absence of actin. The left panel shows that, upon normalization for cytohrome c and MnSOD, endogenous Hax1 is detected in heavy membranes but not purified mitochondria, a finding consistent with lack of Hax1 in the human and murine mitochondrial proteome (27). Note that association of Hax1 with heavy membranes, but not with ER-enriched light membranes (framed panel), suggests association of Hax1 to the cytosolic side of the outer mitochondrial membrane.
PARL association to Hax1 is unspecific
PARL is a 7-transmembrane domain-containing IMM protein (28) (29). Topology analysis of this rhomboid protease showed that its N-terminus is exposed to the matrix, and the C-terminus to the IMS (29) (Figure 4a). PARL contains 3 loops exposed to the IMS: one large Loop-A that is functionally dispensable for PARL proteolytic activity (Figure 4b) (29), and two very small loops that are part of the rhomboid domain (28, 30). Chao et al. reported the interaction of Hax1 with PARL in the IMS as being required to mediate PARL cleavage of Omi/HtrA2 (10).
Figure 4. PARL does not have a domain that coordinates interaction to Hax1 in the IMS.
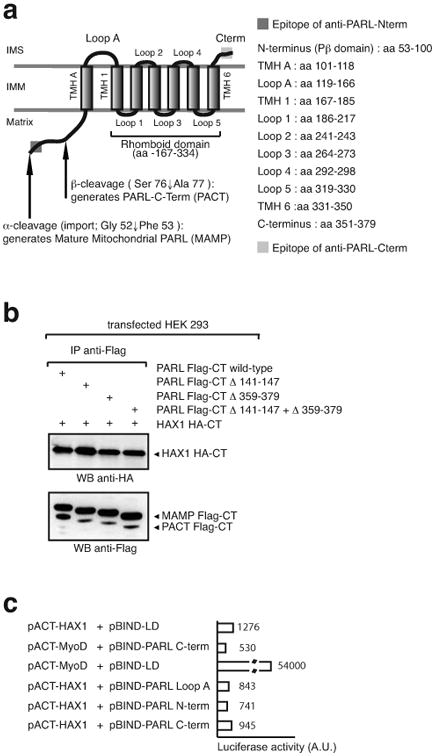
a, schematic representation of the structure and domain composition of PARL (28, 30). b, co-immunoprecipitation of Hax1 with mutant forms of PARL in which the IMS domains (loop-A and C-terminus) have been deleted. Note that PACT formation in these mutants indicate that the proteins are correctly imported and folded in the inner membrane. c, mammalian two-hybrid assays fails to identify a domain in PARL that could mediate interaction to Hax1.
To map the domain of interaction of PARL with Hax1, we deleted the only two domains of PARL that could coordinate the purported PARL/Hax1 association in the IMS, the Loop-A and the C-terminus. Our data showed that, like the wild-type protein, PARL mutants carrying deletion in either or both domains co-immunoprecipitated Hax1 (Figure 4b). Given that neither the Loop-A nor the C-terminus interact with Hax1 in a mammalian two-hybrid system (Figure 4c), it seems most likely that the co-immunoprecipitation of PARL and Hax1 is mediated by non-specific, hydrophobic interactions. To test this possibility, we mixed lysates of cells that were independently transfected with constructs expressing PARL-Flag-CT and Hax1-HA-CT (Figure 5a) and, separately, lysates of heavy membranes isolated from Parl -/- and Hax1 -/- MEFs (Figure 5b). In both cases, PARL/Hax1 association was observed, suggesting that the interaction between these two proteins is an in vitro artifact.
Figure 5. Hax1 binding to PARL is unspecific.
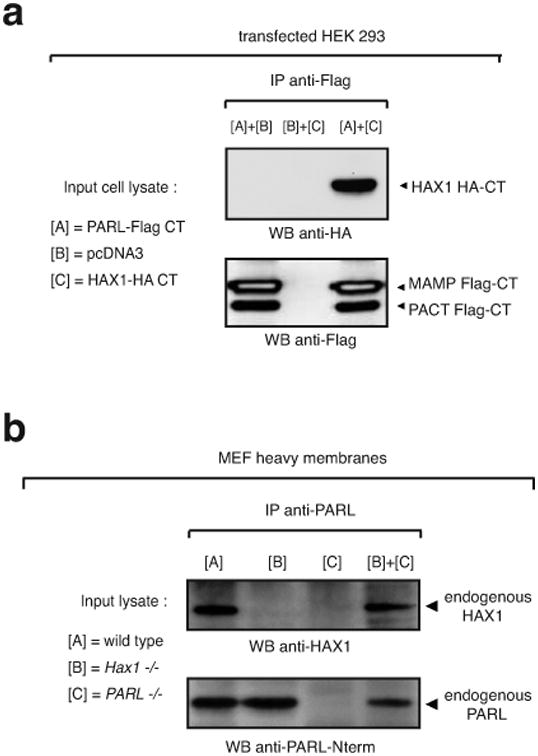
a, in vitro reconstitution of PARL/Hax1 complex by mixing lysates of HEK293 cells transfected with a construct expressing PARL-Flag-CT or Hax-HA-CT. b, in vitro reconstitution of PARL/Hax1 complex by mixing lysates of heavy membranes (150 μg of proteins per genotype used) isolated from MEFs +/+, Hax1 -/-, or Parl -/- .
PARL activity does not require Hax1
Some but not all types of proteases require substrate presentation by an accessory protein; in vitro studies have shown that bacterial rhomboids do not require one (31, 32). Under the model proposed by Chao et al., the PARL-Hax1 interaction allows Hax1 to present Omi/HtrA2 to PARL (10), suggesting a role of Hax1 as a substrate-presenting factor for this rhomboid protease. However, this appears to be an unlikely possibility because loss of Hax1 function only partially reduces the expression of a cleaved form of Omi/HtrA2. Further, genetic ablation of Hax1 compromises neither Opa1 cleavage (10) nor PARL (β-cleavage (Figure 6), an N-terminal processing that requires PARL activity supplied in trans (33). Together, these results indicate that Hax1 is not a PARL substrate-presenting protein, as it has been recently discussed (34), and that PARL rhomboid activity does not require Hax1.
Figure 6. PARL proteolytic activity does not require Hax1.
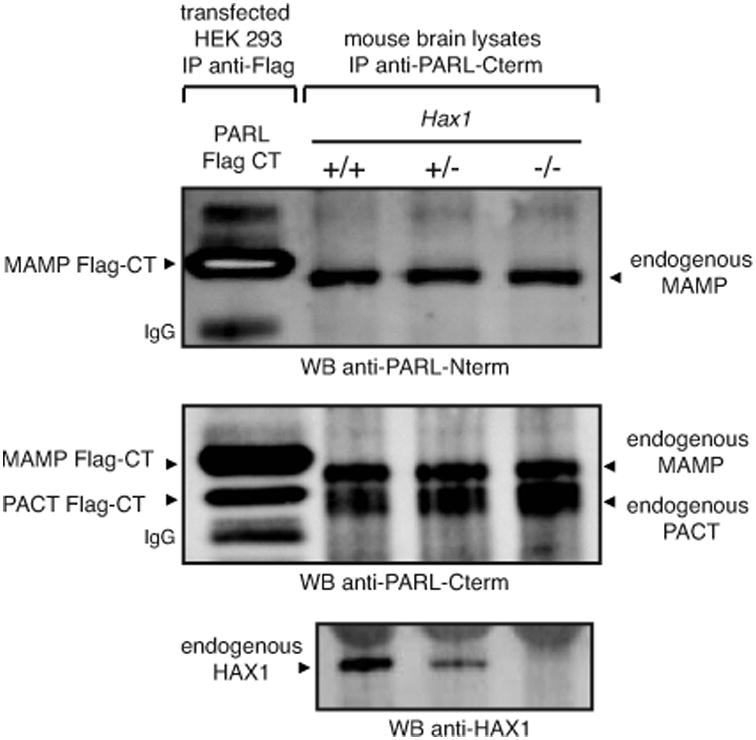
Genetic ablation of Hax1 does not impair the generation of PACT, a shorter form of PARL that requires PARL activity supplied in trans (33).
Discussion
The present study raised from a recent report that addressed the role of Hax1 in mediating the processing of the mitochondrial stress-related Omi/HtrA2 protease, to allow survival of lymphocytes and neurons (10). In this study, Chao and colleagues claimed that cell-type sensitivity to proapoptotic stimuli is governed by the formation of a complex between Hax1, purported Bcl2-family protein, and PARL, a mitochondrial rhomboid protease implicated in apoptosis and mitochondria dynamics regulation (17) (29). Mechanistically, Hax1-PARL association in the IMS was reported to allow the recruitment and presentation of Omi/HtrA2 to PARL, to generate a cleaved, active form of Omi/HtrA2 that, in turn, could proteolytically antagonize MOMP from the IMS. These findings are summarized in the model illustrated in Supplemental Figure 8 of (10).
In order for Hax1 to recruit and present Omi/HtrA2 to PARL, the protein must be localized within the IMS. However, although we observed an association of Hax1 with the heavy membrane fraction, we did not detect Hax1 in Percoll-purified mouse liver mitochondria (Figure 3), a finding consistent with the absence of Hax1 from the mitochondrial proteome (27) as well as with the lack of a predictable mitochondrial targeting peptide or amino acid signature in animal orthologues of this protein (Figure 1 and data not shown). The association of Hax1 with heavy membranes is peripheral as it was removed by alkaline and high salt extraction (Figure 2). Thus, contrary to the claims of Chao and colleagues (10), we found that endogenous Hax1 is neither resident inside the mitochondria nor anchored to the membranes of the organelle. Our findings do not rule out that a minor amount of Hax1 is targeted inside the mitochondria, but indicate that the loss of the heavy membrane- associated form of Hax1 is responsible for the lethal phenotype displayed by Hax1 -/- mice, as well as for the reduced levels of processed Omi/HtrA2 (10).
In this study, we tested the specificity of the interaction between Hax1 and PARL, a highly hydrophobic protein of the IMM (28) (29). Consistent with the data shown by Chao et al., we also observed that Hax1 can be co-immunoprecipitated with PARL, but not vice versa. However, the specificity of this association is dubious at best. Mixing lysates of cells that were independently transfected with constructs expressing PARLFlag-CT and Hax1-HA-CT reconstituted PARL/Hax1 interaction (Figure 5a); similarly, the interaction of endogenous PARL and Hax1 could be recreated by mixing lysates of heavy membranes isolated from Parl -/- and Hax1 -/- MEFs (Figure 5b). Furthermore, we could not identify any domain within PARL that could mediate interaction to Hax1 (Figure 4). Together, these results suggest that Hax1 does not specifically bind PARL and thus it cannot present a substrate to PARL. This conclusion is consistent with the observation that deletion of Hax1 does not fully abolish Omi/HtrA2 processing (10), it is not required for the cleavage of OPA1 (10), and it does not affect PARL (β-cleavage (Figure 6), which is self-regulated (33).
Whether mammalian PARL cleaves Omi/HtrA2, a mitochondrial stress response serine protease (18) (35), remains to be demonstrated, but in the light of the data presented here a direct role of Hax1 in this processing can be ruled out. We further showed by extensive sequence analysis and protein structure prediction that Hax1 is not Bcl2-family protein, thus ruling out the model of Chao et al. in which Hax1 mediates the elimination of active Bax via its BH1 and BH2 modules.
Given the peripheral association of Hax1 to heavy membranes as well as its possible calcium-binding capacity (Figure 1) and its interaction with Phospholamban (36), a sarcoplasmic reticulum protein and a key regulator of Ca2+ homeostasis, the reduced level of processed Omi/HtrA2 observed in Hax1 -/- cells could be explained by defects in calcium signaling and homeostasis (37-44). Thus, our study re-opens the question of whether and how the organelle governs PARL activity to regulate apoptosis and stress, two processes that directly involve mitochondrial dysfunctions in the etiology of cancer (45-47) and several major neurodegenerative disorders, including Parkinson's disease (48, 49).
Materials and Methods
Mitochondria purification
Mitochondria were purified as described (42). Briefly, mouse livers were washed once with PBS, suspended in isolation buffer (200mM sucrose, 1mM EGTA-Tris, and 10mM Tris-MOPS, pH 7.4), and then disrupted by dounce homogenization. The homogenate was spun at 800g for 10 min; the supernatant was recovered and further centrifuged for 10 min at 8,000g. The resulting pellet (mitochondrial fraction) was collected whereas the supernatant was further spun for 30 min at 100,000g. The resulting pellet (light membrane fraction) and supernatant (cytosolic fraction) were spun again at 100,000g to further purify the fractions. The mitochondrial fraction was purified further by centrifuging twice at 8,000g for 10 min. The obtained pellet was purified by centrifugation at 95,000g for 30 min on a 30% Percoll gradient in isolation buffer. The mitochondrial layer was washed free of Percoll and resuspended in isolation buffer. Alternatively, subcellular fractions were obtained by differential centrifugation from mouse liver as described (50). Protein concentration was determined and the indicated amounts of protein were separated by SDS–PAGE and immunoblotted.
Alkaline extraction
Heavy membranes prepared from HeLa cells were diluted to a final concentration of 1 mg/ml in 20 mM HEPES/KOH (pH 7.4). After the addition of an equal volume of freshly prepared 0.2 M sodium carbonate (pH 11.5), samples were incubated for 30 min at 4 °C. The membrane and soluble fractions were separated by ultracentrifugation at 100,000g for 30 min at 4 °C.
Cell culture and transfection
HEK 293T and HeLa cells were purchased from American Type Culture Collection (Manassas, VA). All cell types used in this study were maintained under standard cell culture conditions. Cells were transfected at 40% confluence with FuGENE 6 (Roche). Hax1 -/- MEFs and brains were kindly provided by Dr. J.N. Ihle; Parl -/- MEFs by B. De Strooper.
Cloning and mutagenesis
The pcDNA3 vector was used to express the human PARL and mouse Hax1 protein in mammalian cells. Mutants were obtained by site-directed mutagenesis (Clontech). The identity of every constructs and mutants was confirmed by DNA sequencing.
Computational analysis of protein sequences
Hax1 orthologs from all sequenced animal genomes were detected by searching the Genpept database using the PSI-BLAST program (51), and a multiple alignment of a selected set of diverse sequences was constructed using the T-Coffee program (52). Secondary structure (SS) prediction was performed using the program PSIPRED (53), JPRED (54) and PredictProtein (PRPROT) (55). The search for conserved protein domains was performed using the RPS-BLAST program and the Conserved Domain Database (56, 57). Mitochondrial import prediction was done using the MitoProt II program (58). Transmembrane domains were predicted using PredictProtein (55) as well as TMHMM (59) and TMPred (60).
Co-immunoprecipitations
Transfected HeLa cells were lysed in STEN buffer; heavy membranes in CHAPS buffer. Immunoprecipitations were conducted as described (10) (29).
Antibodies
rabbit anti-PARL (33) (29); rabbit anti-MnSOD (Stressgen; 1:1000); mouse anti-UQCR (clone 13G12, Molecular Probes; 1:3000); mouse anti-HA (Roche); mouse anti-Flag M2 (Sigma); mouse anti-actin (clone C4, Cedarlane; 1:1000); mouse anti-cytochrome c (clone 7H8.2C12, BD Pharmingen; 1:1000); mouse anti-Hax1 (clone-52, BD Biosciences, 1:250); rabbit anti-calnexin (Stressgen; 1:4000). The polyclonal rabbit anti-Hax1 antibody was raised against peptides encompassing amino acid 126-142 and 201-217 of mouse Hax1 (1:500).
Acknowledgments
This study was supported by a grant from the Canadian Institute of Health Research to LP (MOP-82718). DVJ is a CIHR Banting and Best Canada Graduate Scholar. LP is a FRSQ Chercheur-Boursier Senior scholar.
Abbreviations
- MOMP
mitochondrial outer membrane permeabilization
- IMS
mitochondrial intermembrane space
- OMM
outer mitochondrial membrane
- IMM
inner mitochondrial membrane
References
- 1.Henry-Mowatt J, Dive C, Martinou JC, James D. Role of mitochondrial membrane permeabilization in apoptosis and cancer. Oncogene. 2004;23:2850–60. doi: 10.1038/sj.onc.1207534. [DOI] [PubMed] [Google Scholar]
- 2.Bernardi P, Petronilli V, Di Lisa F, Forte M. A mitochondrial perspective on cell death. Trends Biochem Sci. 2001;26:112–7. doi: 10.1016/s0968-0004(00)01745-x. [DOI] [PubMed] [Google Scholar]
- 3.Martinou JC, Youle RJ. Which came first, the cytochrome c release or the mitochondrial fission? Cell Death Differ. 2006;13:1291–5. doi: 10.1038/sj.cdd.4401985. [DOI] [PubMed] [Google Scholar]
- 4.Jourdain A, Martinou JC. Mitochondrial outer-membrane permeabilization and remodelling in apoptosis. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Scorrano L. Opening the doors to cytochrome c: Changes in mitochondrial shape and apoptosis. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 7.Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–84. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- 8.Germain M, Shore GC. Cellular distribution of Bcl-2 family proteins. Sci STKE. 2003;2003:pe10. doi: 10.1126/stke.2003.173.pe10. [DOI] [PubMed] [Google Scholar]
- 9.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–84. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Chao JR, Parganas E, Boyd K, Hong CY, Opferman JT, Ihle JN. Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature. 2008;452:98–102. doi: 10.1038/nature06604. [DOI] [PubMed] [Google Scholar]
- 11.Martinou I, Desagher S, Eskes R, Antonsson B, André E, Fakan S, Martinou JC. The release of cytochrome c from mitochondria during apoptosis of NGF-deprived sympathetic neurons is a reversible event. J Cell Biol. 1999;144:883–9. doi: 10.1083/jcb.144.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–25. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 13.James DI, Martinou JC. Mitochondrial dynamics and apoptosis: a painfulseparation. Dev Cell. 2008;15:341–3. doi: 10.1016/j.devcel.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Herzig S, Martinou JC. Mitochondrial dynamics: to be in good shape to survive. Curr Mol Med. 2008;8:131–7. doi: 10.2174/156652408783769625. [DOI] [PubMed] [Google Scholar]
- 15.Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA, Korsmeyer SJ. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 16.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–89. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–75. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 18.Vande Walle L, Lamkanfi M, Vandenabeele P. The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ. 2008;15:453–60. doi: 10.1038/sj.cdd.4402291. [DOI] [PubMed] [Google Scholar]
- 19.Pellegrini L, Scorrano L. A cut short to death: Parl and Opa1 in the regulation of mitochondrial morphology and apoptosis. Cell Death Differ. 2007;14:1275–84. doi: 10.1038/sj.cdd.4402145. [DOI] [PubMed] [Google Scholar]
- 20.Klein C, Grudzien M, Appaswamy G, Germeshausen M, Sandrock I, Schäffer AA, et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nat Genet. 2007;39:86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- 21.Carlsson G, Aprikyan AA, Tehranchi R, Dale DC, Porwit A, Hellström-Lindberg E, et al. Kostmann syndrome: severe congenital neutropenia associated with defective expression of Bcl-2, constitutive mitochondrial release of cytochrome c, and excessive apoptosis of myeloid progenitor cells. Blood. 2004;103:3355–61. doi: 10.1182/blood-2003-04-1011. [DOI] [PubMed] [Google Scholar]
- 22.Sharp TV, Wang HW, Koumi A, Hollyman D, Endo Y, Ye H, et al. K15 protein of Kaposi's sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J Virol. 2002;76:802–16. doi: 10.1128/JVI.76.2.802-816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Y, Zhang L, Hu T, Shen X, Ding J, Chen K, et al. A conserved hydrophobic core at Bcl-x(L) mediates its structural stability and binding affinity with BH3-domain peptide of pro-apoptotic protein. Arch Biochem Biophys. 2009;484:46–54. doi: 10.1016/j.abb.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Petros AM, Olejniczak ET, Fesik SW. Structural biology of the Bcl-2 family of proteins. Biochim Biophys Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 25.White SH, von heijne G. Transmembrane helices before, during, and after insertion. Curr Opin Struct Biol. 2005;15:378–86. doi: 10.1016/j.sbi.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Stojanovski D, Müller JM, Milenkovic D, Guiard B, Pfanner N, Chacinska A. The MIA system for protein import into the mitochondrial intermembrane space. Biochim Biophys Acta. 2008;1783:610–7. doi: 10.1016/j.bbamcr.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeyaraju DV, Xu L, Letellier MC, Bandaru S, Zunino R, Berg EA, et al. Phosphorylation and cleavage of presenilin-associated rhomboid-like protein (PARL) promotes changes in mitochondrial morphology. Proc Natl Acad Sci U S A. 2006;103:18562–7. doi: 10.1073/pnas.0604983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemberg MK, Freeman M. Functional and evolutionary implications of enhancedgenomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–46. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc Natl Acad Sci U S A. 2005;102:1883–8. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urban S. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 2006;20:3054–68. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- 33.Sík A, Passer BJ, Koonin EV, Pellegrini L. Self-regulated cleavage of the mitochondrial intramembrane-cleaving protease PARL yields Pbeta, a nuclear-targeted peptide. J Biol Chem. 2004;279:15323–9. doi: 10.1074/jbc.M313756200. [DOI] [PubMed] [Google Scholar]
- 34.Freeman M. Rhomboid proteases and their biological functions. Annu Rev Genet. 2008;42:191–210. doi: 10.1146/annurev.genet.42.110807.091628. [DOI] [PubMed] [Google Scholar]
- 35.Moisoi N, Klupsch K, Fedele V, East P, Sharma S, Renton A, et al. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 2009;16:449–64. doi: 10.1038/cdd.2008.166. [DOI] [PubMed] [Google Scholar]
- 36.Vafiadaki E, Sanoudou D, Arvanitis DA, Catino DH, Kranias EG, Kontrogianni-Konstantopoulos A. Phospholamban interacts with HAX-1, a mitochondrial protein with anti-apoptotic function. J Mol Biol. 2007;367:65–79. doi: 10.1016/j.jmb.2006.10.057. [DOI] [PubMed] [Google Scholar]
- 37.Rizzuto R, Duchen MR, Pozzan T. Flirting in little space: the ER/mitochondria Ca2+ liaison. Sci STKE. 2004;2004:re1. doi: 10.1126/stke.2152004re1. [DOI] [PubMed] [Google Scholar]
- 38.Giacomello M, Drago I, Pizzo P, Pozzan T. Mitochondrial Ca2+ as a key regulator of cell life and death. Cell Death Differ. 2007;14:1267–74. doi: 10.1038/sj.cdd.4402147. [DOI] [PubMed] [Google Scholar]
- 39.Pizzo P, Pozzan T. Mitochondria-endoplasmic reticulum choreography: structure and signaling dynamics. Trends Cell Biol. 2007;17:511–7. doi: 10.1016/j.tcb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Szabadkai G, Simoni AM, Bianchi K, De Stefani D, Leo S, Wieckowski MR, Rizzuto R. Mitochondrial dynamics and Ca2+ signaling. Biochim Biophys Acta. 2006;1763:442–9. doi: 10.1016/j.bbamcr.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Pinton P, Rizzuto R. Bcl-2 and Ca2+ homeostasis in the endoplasmic reticulum. Cell Death Differ. 2006;13:1409–18. doi: 10.1038/sj.cdd.4401960. [DOI] [PubMed] [Google Scholar]
- 42.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–10. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 43.Rimessi A, Giorgi C, Pinton P, Rizzuto R. The versatility of mitochondrial calcium signals: from stimulation of cell metabolism to induction of cell death. Biochim Biophys Acta. 2008;1777:808–16. doi: 10.1016/j.bbabio.2008.05.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeyaraju DV, Cisbani G, Pellegrini L. Calcium regulation of mitochondria motility and morphology. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbabio.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic andbiochemical update. Nat Rev Cancer. 2005;5:857–66. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 46.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–82. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 47.Frezza C, Gottlieb E. Mitochondria in cancer: not just innocent bystanders. Semin Cancer Biol. 2009;19:4–11. doi: 10.1016/j.semcancer.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Schapira AH. Mitochondrial dysfunction in Parkinson's disease. Cell Death Differ. 2007;14:1261–6. doi: 10.1038/sj.cdd.4402160. [DOI] [PubMed] [Google Scholar]
- 49.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–95. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 51.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 53.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–5. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 54.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rost B, Yachdav G, Liu J. The Predict Protein server. Nucleic Acids Res. 2004;32:W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–7. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–31. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–86. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 59.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- 60.Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]