Abstract
AIM: To investigate the genes regulated in mesenchymal stem cells (MSCs) and diffuse-type gastric cancer (GC), gene expression was analyzed.
METHODS: Gene expression of MSCs and diffuse-type GC cells were analyzed by microarray. Genes related to stem cells, cancer and the epithelial-mesenchymal transition (EMT) were extracted from human gene lists using Gene Ontology and reference information. Gene panels were generated, and messenger RNA gene expression in MSCs and diffuse-type GC cells was analyzed. Cluster analysis was performed using the NCSS software.
RESULTS: The gene expression of regulator of G-protein signaling 1 (RGS1) was up-regulated in diffuse-type GC cells compared with MSCs. A panel of stem-cell related genes and genes involved in cancer or the EMT were examined. Stem-cell related genes, such as growth arrest-specific 6, musashi RNA-binding protein 2 and hairy and enhancer of split 1 (Drosophila), NOTCH family genes and Notch ligands, such as delta-like 1 (Drosophila) and Jagged 2, were regulated.
CONCLUSION: Expression of RGS1 is up-regulated, and genes related to stem cells and NOTCH signaling are altered in diffuse-type GC compared with MSCs.
Keywords: Mesenchymal stem cells, Gastric cancer, Stem cells, Gene, Epithelial-mesenchymal transition
Core tip: Recent studies have revealed that epithelial-mesenchymal transition (EMT) regulators play important roles in cellular phenotypes. This study has shown that EMT-related genes are regulated in diffuse-type gastric cancer and mesenchymal stem cells (MSCs). Regulator of G-protein signaling 1 (RGS1) was significantly up-regulated in diffuse-type GC compared with MSCs. These results are interesting and provide insights about the mechanisms of the stem cell phenotype transition and gastric cancer progression. These insights will examine the novel role of EMT-related genes and RGS1.
INTRODUCTION
It is a mystery how the epithelial-mesenchymal transition (EMT) and cancer stem cells (CSCs) are correlated[1,2]. Although it is known that the EMT is related to cancer migration and shares similar molecular characteristics with CSCs, whether the EMT occurs in CSCs or if there is any threshold for this phenotype between stem and cancer cells is largely unknown. The EMT is the cellular process involving the cell phenotype transitioning from an epithelial phenotype to a mesenchymal phenotype. Cancer cells experience the EMT when adhesion is less important, and the cells can freely migrate throughout the body. This mechanism is thought to be important in cancer cell invasion and migration and CSC development. The stem cell phenotype transforms from a differentiated stage to an undifferentiated stage and vice versa during the development of tissues and organs and during different disease states. Investigating stem cell-regulated genes is a highly important issue to elucidate the mechanisms of CSCs.
It has been shown that mesenchymal stem cells (MSCs) can promote the expression of an EMT phenotype in cancer cells via CD44, lysyl oxidase and TWIST[3]. The relationship between MSCs and cancer cells, including the mechanism of stemness exhibited by cancer cells, remains unclear. Moreover, the self-renewal of colorectal cancer initiating cells is considered to be a new therapeutic approach[4]. It is important to uncover the gene signature of stem and cancer cells to further understand the stemness of these cells. Studies to reveal the stemness phenotype have already been performed. A marker of the putative stem cell region of human intestinal crypts has been identified using a Fourier transform infrared microspectroscopy approach[5-7]. Genes, such as necdin homolog (mouse) (NDN), EPH receptor A5 (EPHA5), nephroblastoma overexpressed gene (NOV) and runt-related transcription factor 2 (RUNX2), have been correlated with MSC passage numbers[8]. Molecular signatures have also been investigated, and a prediction model of bone marrow stromal cell senescence has been discovered[9]. Even gene expression at the single cell level can be determined using a recent technique[10]. These new approaches, including a physical theory, may be important for the advancement of stem cell therapeutics.
Gastric cancer is classified into diffuse and intestinal types[11,12]. Diffuse-type gastric carcinomas supposedly originate from nonmetaplastic gastric epithelium, and the intestinal type originates from the regenerating epithelium in chronic atrophic gastritis[12]. Diffuse-type gastric cancer (GC) are known to correlate with the EMT[13]. We have previously shown that diffuse-type GC can be distinguished with combination of cadherin 1, type 1, E-cadherin (epithelial) (CDH1) and cadherin 2, type 1, N-cadherin (neuronal) (CDH2) gene expression in MSC and diffuse-type GC associated with the EMT[14]. Considering that hepatoma cells are converted into human induced pluripotent stem cell-like cells using microRNAs, the difference between stem cells and cancer cells should be deeply investigated[15]. Breast CSCs show both mesenchymal-like and epithelial-like phenotypes and transition between EMT- and mesenchymal-epithelial transition-like states[16]. Diffuse-type GC has been reported to be involved in the remodeling of the pulmonary artery, which suggests cancer niche formation by diffuse-type GC[17]. Considering the finding that MSCs provide microenvironment for CSCs, the features of MSC and cancer cells should be investigated[18].
In this article, MSC gene expression has been investigated and compared with diffuse-type GC cells that exhibit a mesenchymal phenotype more than intestinal-type GC cells do, and panels of regulated genes in MSCs and diffuse-type GC have been generated to clarify the important molecular patterns that distinguish stem and cancer cells.
MATERIALS AND METHODS
Cell culture
MSCs from human bone marrow were cultured according to the manufacturer’s protocol (Lonza Walkersville, Inc., Walkersville, MD, United States). Detailed methods have been previously described[8]. Briefly, MSCs were cultured in vitro in mesenchymal stem cell growth medium (Lonza Catalog #PT-3001; mesenchymal stem cell basal medium plus SingleQuots™ of growth Supplements) at 37 °C in a CO2 (5%) incubator. Each number in the MSC sample name indicates the passage number.
Total RNA extraction
Diffuse-type GC samples were surgically resected and obtained from the National Cancer Center Research Institute. Detailed information on total RNA extraction has been previously described[14,19]. Briefly, total RNA was obtained by suspending the cells in ISOGEN lysis buffer from surgically resected samples, which were provided by the Central Hospital or East Hospital at the National Cancer Center[14,19]. Total RNA of MSCs were purified using RNeasy kit (QIAGEN, Düsseldorf, Germany) as previously described[8].
Microarray analysis
MSC gene expression data are available to the public via the NCBI Gene Expression Omnibus (GEO)[8]. Diffuse-type GC gene expression data are also available via the NCBI GEO[14]. The accession numbers of data used for gene expression analysis are GSE7888 and GSE42252 for MSCs and diffuse-type GC, respectively. Briefly, total RNA from MSCs and diffuse-type GC was analyzed using a GeneChip® Human Genome U133 Plus 2.0 Array (Affymetrix, Inc., Santa Clara, CA, United States). Probe set and Gene Ontology information is based on annotation version na34. Cluster analysis was performed using NCSS 2007 (NCSS, LLC., Kaysville, UT, United States).
Statistical analysis
All data are presented as the mean ± SE. Statistical significance was calculated with GraphPad Prism® 6 or Microsoft® Excel® using the unpaired Student’s t test or one-way ANOVA with Bonferroni’s multiple comparisons test.
RESULTS
Regulated genes in diffuse-type GC and MSCs
Genes regulated in diffuse-type GC and MSCs were investigated and analyzed using cluster analysis (NCSS) (Figure 1). Probe sets that have a more than a 50-fold significant difference between MSCs and diffuse-type GC (P < 10-9 in the equal variance hypothesis and P < 0.01 in the non-equal variance hypothesis) are shown in Figure 1. Each panel shows the gene expression in 30 probe sets. Panels A, B and C show the up-regulated probe sets in diffuse-type GC compared with MSCs, whereas panel D shows 29 up-regulated probe sets and one down-regulated gene, pentraxin 3, long (PTX3). In this analysis, regulator of G-protein signaling 1 (RGS1) was the most up-regulated gene in diffuse-type GC compared with MSCs (Figure 2A). RGS4 gene expression was down-regulated in diffuse-type GC compared with MSCs (Figure 2B). The regulated RGS family genes are shown in Table 1. Additional up-regulated genes in diffuse-type GC compared to MSCs included spleen tyrosine kinase (SYK) and CDH1, which have been suggested to be mutated in cancer, based on an assessment of the public database Cancer Gene Census (http://cancer.sanger.ac.uk/cancergenome/projects/census/).
Figure 1.
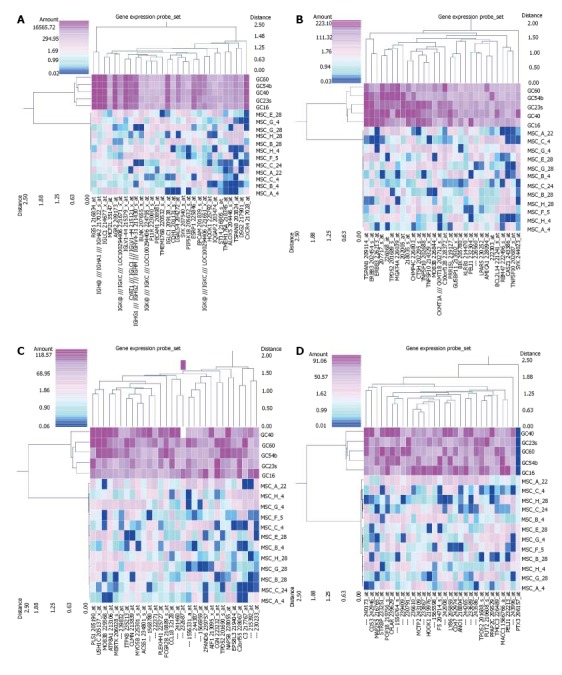
Genes regulated in mesenchymal stem cells and diffuse-type gastric cancer. A total of 120 probe sets that had more than a 50-fold change in mesenchymal stem cells (MSCs) and diffuse-type gastric cancer (GC) are shown. Each panel (A to D) shows 30 probe sets up-regulated or down-regulated in diffuse-type GC compared with MSCs. (Fold change > 50, P < 0.01 and P < 10-9 using Student’s t test in the non-equal variance hypothesis and equal variance hypothesis, respectively).
Figure 2.
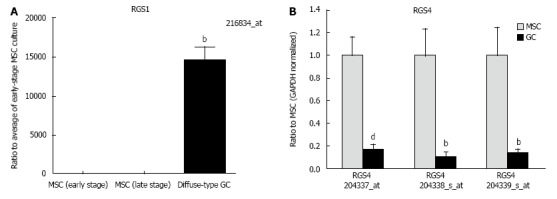
Regulator of G-protein signaling 1 and regulator of G-protein signaling 4 gene expression was up-regulated and down-regulated, respectively, in diffuse-type gastric cancer. A: regulator of G-protein signaling 1 (RGS1) gene expression was up-regulated in diffuse-type gastric cancer (GC) compared with early-stage mesenchymal stem cell (MSC) cultures [n = 5 in early-stage MSC cultures (B#4, C#4, F#5, G#4, and H#4), n = 5 in late-stage MSC cultures (B#28, C#24, E#28, G#28, and H#28), and n = 5 in diffuse-type GC (GC16, GC40, GC54b, GC60, and GC23s)]. Results represent the mean ± SE. bP < 0.01 using a one-way analysis of variance with Bonferroni’s multiple comparisons test; B: RGS4 gene expression was down-regulated in diffuse-type GC compared with MSCs. Results represent the mean ± SE (n = 12 in MSC cultures and n = 5 in diffuse-type GC). bP < 0.01 and dP < 0.001 using Student’s t test with the non-equal variance hypothesis.
Table 1.
Gene expression alterations of regulator of G-protein signaling family genes in mesenchymal stem cells and diffuse-type gastric cancer
| Gene symbol | Detailed description | P value | Ratio of diffuse-type GC to MSCs | Probe set ID |
| RGS1 | Immune response / signal transduction / adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway / negative regulation of signal transduction / termination of G-protein coupled receptor signaling pathway / positive regulation of GTPase activity | 0.013 | 1004 | 202988_s_at |
| 0.001 | 13808 | 216834_at | ||
| RGS2 | Regulation of translation / cell cycle / spermatogenesis / regulation of G-protein coupled receptor protein signaling pathway / negative regulation of signal transduction / negative regulation of phospholipase activity / negative regulation of cardiac muscle hypertrophy / termination of G-protein coupled receptor signaling pathway / negative regulation of mitogen-activated protein (MAP) kinase activity / positive regulation of GTPase activity / negative regulation of G-protein coupled receptor protein signaling pathway / brown fat cell differentiation / relaxation of cardiac muscle / relaxation of vascular smooth muscle / positive regulation of cardiac muscle contraction / regulation of adrenergic receptor signaling pathway | 0.007 | 14.51 | 202388_at |
| RGS3 | Inactivation of mitogen-activated protein kinase (MAPK) activity / signal transduction / regulation of G-protein coupled receptor protein signaling pathway / negative regulation of signal transduction / termination of G-protein coupled receptor signaling pathway / positive regulation of GTPase activity | 1.00E-05 | 0.39 | 203823_at |
| RGS4 | Inactivation of MAPK activity / regulation of G-protein coupled receptor protein signaling pathway / negative regulation of signal transduction / termination of G-protein coupled receptor signaling pathway / positive regulation of GTPase activity | 3.00E-04 | 0.17 | 204337_at |
| 0.003 | 0.11 | 204338_s_at | ||
| 0.004 | 0.14 | 204339_s_at | ||
| RGS5 | Signal transduction / regulation of G-protein coupled receptor protein signaling pathway / negative regulation of signal transduction / termination of G-protein coupled receptor signaling pathway / positive regulation of GTPase activity | 0.01 | 47.79 | 209070_s_at |
| 0.008 | 81.82 | 209071_s_at | ||
| 0.009 | 95.52 | 218353_at | ||
| RGS10 | Negative regulation of signal transduction / termination of G-protein coupled receptor signaling pathway / positive regulation of GTPase activity | 0.003 | 3.92 | 204319_s_at |
| RGS19 | Autophagy / G-protein coupled receptor signaling pathway / small GTPase mediated signal transduction / negative regulation of signal transduction / termination of G-protein coupled receptor signaling pathway / positive regulation of GTPase activity | 9.00E-04 | 2.45 | 204336_s_at |
RGS family genes that have significant alterations in MSCs and diffuse-type GC are shown. RGS1, RGS2, RGS5, RGS10 and RGS19 were up-regulated in diffuse-type GC compared with MSCs. RGS3 and RGS4 were down-regulated in diffuse-type GC compared with MSCs. RGS: Regulator of G-protein signaling; MSCs: Mesenchymal stem cells; GC: Gastric cancer.
Stem cell gene panel
The expression of genes related to stem cells was investigated (Table 2). Considering that gene combination is important to understand stem cell phenotype, several genes involving to stem cells have been analyzed[20]. NANOG and SUZ12, which have been found by gene expression analysis to be involved in MSC invasion was up-regulated in diffuse-type GC compared to MSCs[21]. Gene expression of tumor necrosis factor receptor superfamily, member 19 (TNFRSF19), tended to decrease in diffuse-type GC compared with MSCs, but it was not significantly altered in MSCs and diffuse-type GC (Figure 3). The gene expression of TNFRSF8, also known as CD30, was slightly up-regulated in diffuse-type GC compared with MSCs. The gene expression of growth arrest-specific 6 (GAS6), musashi RNA-binding protein 2 (MSI2) and hairy and enhancer of split 1, (Drosophila) (HES1), which have stem cell terms in the Gene Ontology biological process, were regulated in diffuse-type GC and MSCs (Figure 4).
Table 2.
Gene expression alterations of stem cell-related genes in mesenchymal stem cells and diffuse-type gastric cancer
| Gene symbol | Gene name | P value | Ratio of diffuse-type GC to MSCs | Probe set ID |
| NANOG | Nanog homeobox | 0.0037 | 14.9 | 220184_at |
| SUZ12 | SUZ12 polycomb repressive complex 2 subunit | 0.0022 | 35.4 | 1566191_at |
| TNFRSF8 | Tumor necrosis factor receptor superfamily, member 8 | 0.0126 | 1.98 | 206729_at |
| CD9 | CD9 molecule | 0.0012 | 3.44 | 201005_at |
| ICAM3 | Intercellular adhesion molecule-3 | 0.0143 | 2.69 | 204949_at |
| CD200 | CD200 molecule | 0.0013 | 4.79 | 209583_s_at |
| 0.0002 | 4.35 | 209582_s_at | ||
| THY1 | Thy-1 cell surface antigen | 0.0081 | 0.61 | 208850_s_at |
| 0.0301 | 0.79 | 208851_s_at | ||
| 0.0024 | 0.67 | 213869_x_at |
Genes related to stem cells in the literature are shown with their gene expression alterations in MSCs and diffuse-type GC. NANOG and SUZ12, which have been found by gene expression analysis to be involved in MSC invasion was up-regulated in diffuse-type GC compared to MSCs. RGS: Regulator of G-protein signaling; MSCs: Mesenchymal stem cells; GC: Gastric cancer.
Figure 3.
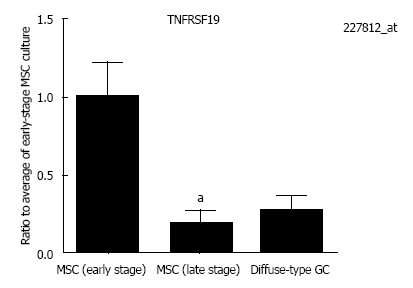
Tumor necrosis factor receptor superfamily, member 19 gene expression. Tumor necrosis factor receptor superfamily, member 19 (TNFRSF19) gene expression tended to decrease in diffuse-type gastric cancer (GC) but was not significantly altered compared with early-stage mesenchymal stem cell (MSC) cultures. TNFRSF19 gene expression in late-stage MSC cultures was significantly down-regulated compared with early-stage MSC cultures. Results represent the mean ± SE [n = 5 in early-stage MSC cultures (B#4, C#4, F#5, G#4, and H#4), n = 5 in late-stage MSC cultures (B#28, C#24, E#28, G#28, and H#28), and n = 5 in diffuse-type GC (GC16, GC40, GC54b, GC60, and GC23s)]. aP < 0.05 using a one-way ANOVA with Bonferroni’s multiple comparisons test.
Figure 4.
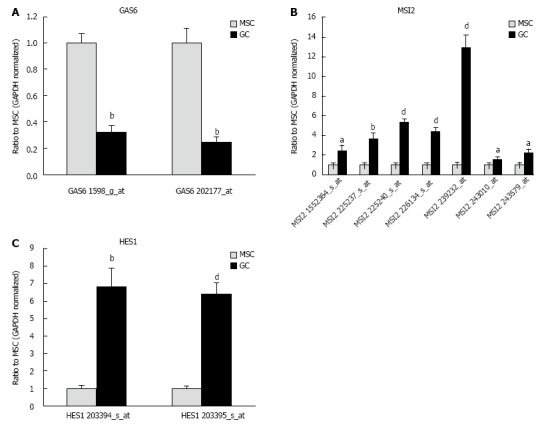
Gene expression of stem cell-related genes. A: The gene expression of growth arrest-specific 6 (GAS6) was down-regulated in diffuse-type gastric cancer (GC) compared with mesenchymal stem cell (MSCs). Results represent the mean ± SE (n = 12 in MSC and n = 5 in diffuse-type GC). bP < 0.001 using Student’s t test; B: The gene expression of musashi RNA-binding protein 2 (MSI2) was up-regulated in diffuse-type GC compared with MSCs. Results represent the mean ± SE (n = 12 in MSC and n = 5 in diffuse-type GC). aP < 0.05, bP < 0.01, and dP < 0.001 using Student’s t test; C: The gene expression of hairy and enhancer of split 1 (Drosophila) (HES1) was up-regulated in diffuse-type GC compared with MSCs. Results represent the mean ± SE (n = 12 in MSC and n = 5 in diffuse-type GC). bP < 0.01 and dP < 0.001 using Student’s t test. All three genes have stem cell terms in the Gene Ontology biological process.
Investigation of EMT markers
To examine the cellular phenotype of cancer and stem cells, the expression levels of EMT-related genes were investigated (Table 3). The gene expression of the TWIST, ZEB and SNAI families were examined because these genes have been reported to be associated with the EMT and cancer regulation[22]. The gene expression of twist family bHLH transcription factor 1 (TWIST1) and paired related homeobox 1 (PRRX1) were down-regulated in diffuse-type GC compared with MSCs. The gene expression of zinc finger E-box binding homeobox 2 (ZEB2) was up-regulated in diffuse-type GC compared with MSCs. Previously, high mobility group AT-hook 2 (HMGA2) has been shown to be involved in the EMT, WNT/β-catenin signaling and malignant cancer[23,24]. HMGA2 gene expression was down-regulated in diffuse-type GC compared with MSCs. The gene expression of one of the probe sets for catenin (cadherin-associated protein), beta 1, 88 kDa (CTNNB1) was up-regulated in diffuse-type GC compared with MSCs. The gene expression of the other two probe sets for CTNNB1 was stable. Sp1 transcription factor (SP1) and lymphoid enhancer-binding factor 1 (LEF1) were up-regulated in diffuse-type GC compared with MSCs. SP1 is known to be an important factor for cell cycle regulation and is up-regulated in various cancers[25]. The AKT gene family is known to be important in cancer and has been suggested to regulate the EMT and stem cells in cancer cells[26]. Three AKT isoforms were examined, and the gene expression of v-akt murine thymoma viral oncogene homolog 2 (AKT2) was up-regulated. However, some of the probe sets recognizing AKT3 showed a down-regulation of AKT3 gene expression. The gene expression of stratifin (SFN); F11 receptor (F11R); gap junction protein, beta 2, 26 kDa (GJB2); occluding (OCLN); coxsackie virus and adenovirus receptor (CXADR); and keratin 8 (KRT8) were up-regulated in diffuse-type GC compared with MSCs.
Table 3.
Gene expression alterations of epithelial-mesenchymal transition-related genes in mesenchymal stem cells and diffuse-type gastric cancer
| Gene symbol | Gene name | P value | Ratio of diffuse-type GC to MSCs | Probe set ID |
| TWIST1 | Twist family bHLH transcription factor 1 | 0.0004 | 0.2 | 213943_at |
| PRRX1 | Paired related homeobox 1 | 1.00E-05 | 0.3 | 226695_at |
| ZEB2 | Zinc finger E-box binding homeobox 2 | 0.027 | 2.1 | 203603_s_at |
| 0.03 | 1.7 | 228333_at | ||
| 6.00E-05 | 3.9 | 235593_at | ||
| TJP1 | Tight junction protein 1 | 0.184 | 1.2 | 202011_at |
| 0.795 | 1 | 214168_s_at | ||
| CTNNB1 | Catenin (cadherin-associated protein), beta 1, 88 kDa | 0.001 | 9.6 | 223679_at |
| VIM | Vimentin | 0.031 | 4.7 | 1555938_x_at |
| 0.016 | 0.7 | 201426_s_at | ||
| FN1 | Fibronectin 1 | 0.204 | 0.6 | 1558199_at |
| 0.015 | 0.7 | 210495_x_at | ||
| 0.004 | 0.7 | 211719_x_at | ||
| 0.001 | 0.6 | 212464_s_at | ||
| 4.00E-08 | 0.04 | 214701_s_at | ||
| 1.00E-10 | 0.02 | 214702_at | ||
| 0.008 | 0.7 | 216442_x_at | ||
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 | 0.187 | 1.2 | 207163_s_at |
| AKT2 | v-akt murine thymoma viral oncogene homolog 2 | 8.00E-09 | 3.1 | 225471_s_at |
| 0.002 | 4.4 | 226156_at | ||
| 0.0006 | 5.6 | 236664_at | ||
| 0.0006 | 7.6 | 1560689_s_at | ||
| AKT3 | v-akt murine thymoma viral oncogene homolog 3 | 0.0006 | 6.2 | 242876_at |
| 0.008 | 0.6 | 212607_at | ||
| 0.549 | 1.2 | 212609_s_at | ||
| 0.385 | 0.8 | 219393_s_at | ||
| 0.772 | 1.1 | 222880_at | ||
| 0.0003 | 0.5 | 242879_x_at | ||
| SFN | Stratifin | 0.003 | 66 | 33322_i_at |
| 0.008 | 111 | 33323_r_at | ||
| EPCAM | Epithelial cell adhesion molecule | 0.0009 | 725 | 201839_s_at |
| F11R | F11 receptor | 0.0004 | 35 | 221664_s_at |
| 0.002 | 21 | 222354_at | ||
| 0.0009 | 201 | 223000_s_at | ||
| 0.012 | 25 | 224097_s_at | ||
| GJB2 | Gap junction protein, beta 2, 26 kDa | 0.023 | 69 | 223278_at |
| LEF1 | Lymphoid enhancer-binding factor 1 | 0.002 | 26 | 221558_s_at |
| JUN | Jun proto-oncogene | 0.042 | 4.8 | 201464_x_at |
| 0.022 | 5.1 | 201466_s_at | ||
| 0.079 | 3.8 | 201465_s_at | ||
| 0.019 | 4.1 | 213281_at | ||
| TGFB1 | Transforming growth factor, beta 1 | 0.002 | 2 | 203085_s_at |
| TGFB2 | Transforming growth factor, beta 2 | 0.07 | 0.4 | 209909_s_at |
| 0.055 | 0.6 | 220407_s_at | ||
| 0.588 | 0.8 | 228121_at | ||
| TGFB3 | Transforming growth factor, beta 3 | 0.015 | 6.8 | 209747_at |
| SP1 | Sp1 transcription factor | 0.0002 | 3.4 | 224754_at |
| 2.00E-05 | 2.7 | 224760_at | ||
| HMGA2 | High mobility group AT-hook 2 | 0.003 | 0.3 | 208025_s_at |
| ITGA5 | Integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | 0.007 | 0.5 | 201389_at |
| OCLN | Occludin | 0.0008 | 8 | 231022_at |
| 0.012 | 9.4 | 235937_at | ||
| 0.027 | 11 | 209925_at | ||
| 0.0045 | 11 | 227492_at | ||
| CXADR | Coxsackie virus and adenovirus receptor | 0.013 | 265 | 203917_at |
| 0.014 | 297 | 226374_at | ||
| 0.021 | 75 | 239155_at | ||
| KRT8 | Keratin 8 | 0.012 | 128 | 209008_x_at |
RGS: Regulator of G-protein signaling; MSCs: Mesenchymal stem cells; GC: Gastric cancer.
Messenger RNA expression of NOTCH signaling-related genes
NOTCH signaling-related genes are involved in cancer, and NOTCH signaling is one of the target signaling pathways of CSCs[27-30]. Next, NOTCH messenger RNA (mRNA) expression was examined (Table 4). NOTCH1, 3 and 4 gene expression was up-regulated, whereas NOTCH2 gene expression was down-regulated in diffuse-type GC compared with MSCs (Figure 5A). The gene expression of delta-like 1 (Drosophila) (DLL1) and Jagged 2 (JAG2) were up-regulated in diffuse-type GC compared with MSCs (Figure 5B and 5C). The scheme for NOTCH signaling and cross-talk between cells is presented in Figure 6A. The Gene Ontology biological process of the NOTCH family genes are shown in Tables 5.
Table 4.
Gene expression alterations of NOTCH signaling-related genes in mesenchymal stem cells and diffuse-type gastric cancer
| Gene symbol | Gene name | P value | Ratio of diffuse-type GC to MSCs | Probe set ID |
| NOTCH1 | Notch 1 | 0.004 | 6.9 | 218902_at |
| NOTCH2 | Notch 2 | 0.001 | 0.7 | 202445_s_at |
| 1.00E-05 | 0.5 | 202443_x_at | ||
| 1.00E-05 | 0.5 | 212377_s_at | ||
| DLL1 | Delta-like 1 (Drosophila) | 0.007 | 31 | 224215_s_at |
| JAG2 | Jagged 2 | 0.003 | 11 | 32137_at |
| JAG1 | Jagged 1 | 0.003 | 1.5 | 209099_x_at |
| 0.022 | 1.4 | 216268_s_at | ||
| 0.49 | 1.1 | 209098_s_at | ||
| WNT9A | Wingless-type MMTV integration site family, member 9A | 0.027 | 5.4 | 230643_at |
| WNT2B | Wingless-type MMTV integration site family, member 2B | 0.026 | 4.4 | 206458_s_at |
| WNT5A | Wingless-type MMTV integration site family, member 5A | 8.00E-05 | 0.3 | 213425_at |
| 3.00E-05 | 0.2 | 205990_s_at | ||
| WNT5B | Wingless-type MMTV integration site family, member 5B | 0.0002 | 0.2 | 221029_s_at |
RGS: Regulator of G-protein signaling; MSCs: Mesenchymal stem cells; GC: Gastric cancer.
Figure 5.
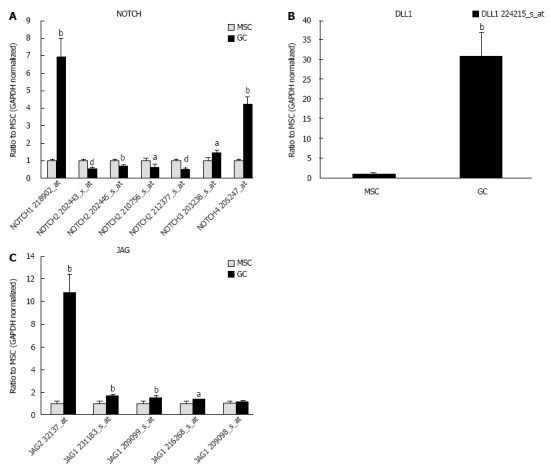
Gene expression of NOTCH, delta-like 1 (Drosophila) (DLL1) and JAG. A: The gene expression of NOTCH1, 3 and 4 was up-regulated in diffuse-type gastric cancer (GC) compared with mesenchymal stem cell (MSCs), whereas NOTCH2 was down-regulated. Results represent the mean ± SE (n = 12 in MSC cultures and n = 5 in diffuse-type GC). aP < 0.05, bP < 0.01, and dP < 0.001 using Student’s t test; B: DLL1, a NOTCH ligand, was up-regulated in diffuse-type GC compared with MSCs. Results represent the mean ± SE (n = 12 in MSC cultures and n = 5 in diffuse-type GC). bP < 0.01 using Student’s t test; C: Jagged 2 (JAG2), a NOTCH ligand, was up-regulated in diffuse-type GC compared with MSCs. Jagged 1 (JAG1) was up-regulated in diffuse-type GC compared with MSCs. Results represent the mean ± SE (n = 12 in MSC cultures and n = 5 in diffuse-type GC). aP < 0.05 and bP < 0.01 using Student’s t test.
Figure 6.
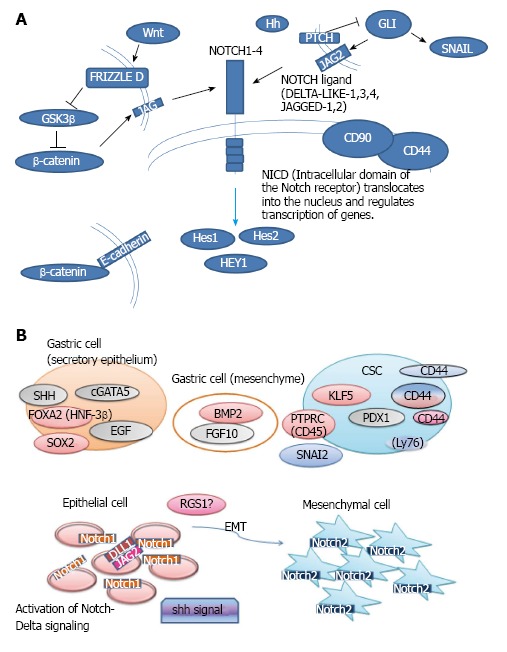
Notch signaling and the epithelial-mesenchymal transition hypothesis. A: Upon cellular stimulation by the Notch ligand, the intracellular domain of the Notch receptor (NICD) translocates into the nucleus and regulates gene transcription; B: In the EMT hypothesis, epithelial-phenotype cells expressing NOTCH1 transition to mesenchymal-phenotype cells expressing NOTCH2. RGS1: Regulator of G-protein signaling 1; EMT: Epithelial-mesenchymal transition; JAG2: Jagged 2; CSCs: Cancer stem cells.
Table 5.
Gene ontology of NOTCH family genes
| Gene symbol | Gene ontology biological process |
| NOTCH1 | 0000122 // negative regulation of transcription from RNA polymerase II promoter // inferred from sequence or structural similarity /// 0001525 // angiogenesis // inferred from electronic annotation /// 0001701 // in utero embryonic development // inferred from electronic annotation /// 0001708 // cell fate specification // inferred from electronic annotation /// 0001837 // epithelial to mesenchymal transition // inferred from sequence or structural similarity /// 0001889 // liver development // inferred from electronic annotation /// 0001947 // heart looping // inferred from sequence or structural similarity /// 0002040 // sprouting angiogenesis // inferred from electronic annotation /// 0002052 // positive regulation of neuroblast proliferation // inferred from electronic annotation /// 0002437 // inflammatory response to antigenic stimulus // inferred from electronic annotation /// 0003157 // endocardium development // inferred from sequence or structural similarity /// 0003160 // endocardium morphogenesis // inferred from sequence or structural similarity /// 0003162 // atrioventricular node development // inferred from electronic annotation /// 0003169 // coronary vein morphogenesis // inferred from sequence or structural similarity /// 0003180 // aortic valve morphogenesis // inferred from mutant phenotype /// 0003181 // atrioventricular valve morphogenesis // inferred from sequence or structural similarity /// 0003184 // pulmonary valve morphogenesis // inferred from mutant phenotype /// 0003192 // mitral valve formation // inferred from mutant phenotype /// 0003198 // epithelial to mesenchymal transition involved in endocardial cushion formation // inferred from sequence or structural similarity /// 0003203 // endocardial cushion morphogenesis // inferred from sequence or structural similarity /// 0003207 // cardiac chamber formation // inferred from sequence or structural similarity /// 0003208 // cardiac ventricle morphogenesis // inferred from sequence or structural similarity /// 0003209 // cardiac atrium morphogenesis // inferred from sequence or structural similarity /// 0003213 // cardiac right atrium morphogenesis // inferred from sequence or structural similarity /// 0003214 // cardiac left ventricle morphogenesis // inferred from sequence or structural similarity /// 0003219 // cardiac right ventricle formation // inferred from electronic annotation /// 0003222 // ventricular trabecula myocardium morphogenesis // inferred from sequence or structural similarity /// 0003241 // growth involved in heart morphogenesis // inferred from sequence or structural similarity /// 0003256 // regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation // inferred from sequence or structural similarity /// 0003264 // regulation of cardioblast proliferation // inferred from electronic annotation /// 0003270 // Notch signaling pathway involved in regulation of secondary heart field cardioblast proliferation // inferred from electronic annotation /// 0003273 // cell migration involved in endocardial cushion formation // inferred from sequence or structural similarity /// 0003344 // pericardium morphogenesis // inferred from sequence or structural similarity /// 0006351 // transcription, DNA-dependent // inferred from electronic annotation /// 0006355 // regulation of transcription, DNA-dependent // traceable author statement /// 0006357 // regulation of transcription from RNA polymerase II promoter // inferred from electronic annotation /// 0006367 // transcription initiation from RNA polymerase II promoter // traceable author statement /// 0006955 // immune response // non-traceable author statement /// 0006959 // humoral immune response // inferred from electronic annotation /// 0007219 // Notch signaling pathway // inferred from mutant phenotype /// 0007219 // Notch signaling pathway // traceable author statement /// 0007220 // Notch receptor processing // traceable author statement /// 0007221 // positive regulation of transcription of Notch receptor target // inferred from sequence or structural similarity /// 0007275 // multicellular organismal development // inferred from electronic annotation /// 0007283 // spermatogenesis // inferred from electronic annotation /// 0007368 // determination of left/right symmetry // inferred from sequence or structural similarity /// 0007386 // compartment pattern specification // inferred from electronic annotation /// 0007409 // axonogenesis // inferred from electronic annotation /// 0007440 // foregut morphogenesis // inferred from electronic annotation /// 0007492 // endoderm development // inferred from electronic annotation /// 0007507 // heart development // inferred from mutant phenotype /// 0008284 // positive regulation of cell proliferation // inferred from direct assay /// 0008284 // positive regulation of cell proliferation // inferred from mutant phenotype /// 0008285 // negative regulation of cell proliferation // inferred from direct assay /// 0008544 // epidermis development // inferred from electronic annotation /// 0009790 // embryo development // inferred from electronic annotation /// 0009912 // auditory receptor cell fate commitment // inferred from electronic annotation /// 0010001 // glial cell differentiation // inferred from electronic annotation /// 0010467 // gene expression // traceable author statement /// 0010468 // regulation of gene expression // inferred from electronic annotation /// 0010718 // positive regulation of epithelial to mesenchymal transition // inferred from mutant phenotype /// 0010812 // negative regulation of cell-substrate adhesion // inferred from direct assay /// 0010832 // negative regulation of myotube differentiation // inferred from sequence or structural similarity /// 0014031 // mesenchymal cell development // inferred from sequence or structural similarity /// 0014807 // regulation of somitogenesis // inferred from electronic annotation /// 0021515 // cell differentiation in spinal cord // inferred from electronic annotation /// 0021915 // neural tube development // inferred from electronic annotation /// 0030154 // cell differentiation // inferred from electronic annotation /// 0030182 // neuron differentiation // inferred from electronic annotation /// 0030216 // keratinocyte differentiation // inferred from electronic annotation /// 0030279 // negative regulation of ossification // inferred from sequence or structural similarity /// 0030324 // lung development // inferred from electronic annotation /// 0030326 // embryonic limb morphogenesis // inferred from electronic annotation /// 0030334 // regulation of cell migration // inferred from electronic annotation /// 0030335 // positive regulation of cell migration // inferred from sequence or structural similarity /// 0030513 // positive regulation of BMP signaling pathway // inferred from sequence or structural similarity /// 0030514 // negative regulation of BMP signaling pathway // inferred from sequence or structural similarity /// 0030900 // forebrain development // inferred from electronic annotation /// 0031069 // hair follicle morphogenesis // inferred from electronic annotation /// 0031100 // organ regeneration // inferred from electronic annotation /// 0031960 // response to corticosteroid stimulus // inferred from electronic annotation /// 0032495 // response to muramyl dipeptide // inferred from electronic annotation /// 0032496 // response to lipopolysaccharide // inferred from electronic annotation /// 0035116 // embryonic hindlimb morphogenesis // inferred from electronic annotation /// 0035148 // tube formation // inferred from mutant phenotype /// 0035914 // skeletal muscle cell differentiation // inferred from electronic annotation /// 0035924 // cellular response to vascular endothelial growth factor stimulus // inferred from direct assay /// 0042127 // regulation of cell proliferation // inferred from electronic annotation /// 0042246 // tissue regeneration // inferred from electronic annotation /// 0042640 // anagen // inferred from electronic annotation /// 0043065 // positive regulation of apoptotic process // inferred from electronic annotation /// 0043066 // negative regulation of apoptotic process // inferred from electronic annotation /// 0043086 // negative regulation of catalytic activity // inferred from sequence or structural similarity /// 0045165 // cell fate commitment // inferred from electronic annotation /// 0045596 // negative regulation of cell differentiation // inferred from electronic annotation /// 0045603 // positive regulation of endothelial cell differentiation // inferred from electronic annotation /// 0045618 // positive regulation of keratinocyte differentiation // inferred from electronic annotation /// 0045662 // negative regulation of myoblast differentiation // inferred from mutant phenotype /// 0045665 // negative regulation of neuron differentiation // inferred from electronic annotation /// 0045668 // negative regulation of osteoblast differentiation // inferred from sequence or structural similarity /// 0045687 // positive regulation of glial cell differentiation // inferred from electronic annotation /// 0045892 // negative regulation of transcription, DNA-dependent // inferred from sequence or structural similarity /// 0045893 // positive regulation of transcription, DNA-dependent // inferred from sequence or structural similarity /// 0045944 // positive regulation of transcription from RNA polymerase II promoter // inferred from direct assay /// 0045944 // positive regulation of transcription from RNA polymerase II promoter // inferred from sequence or structural similarity /// 0045955 // negative regulation of calcium ion-dependent exocytosis // inferred from electronic annotation /// 0046427 // positive regulation of JAK-STAT cascade // inferred from sequence or structural similarity /// 0046533 // negative regulation of photoreceptor cell differentiation // inferred from electronic annotation /// 0048103 // somatic stem cell division // inferred from electronic annotation /// 0048663 // neuron fate commitment // inferred from electronic annotation /// 0048708 // astrocyte differentiation // inferred from electronic annotation /// 0048709 // oligodendrocyte differentiation // inferred from electronic annotation /// 0048711 // positive regulation of astrocyte differentiation // inferred from sequence or structural similarity /// 0048715 // negative regulation of oligodendrocyte differentiation // inferred from sequence or structural similarity /// 0048754 // branching morphogenesis of an epithelial tube // inferred from electronic annotation /// 0050678 // regulation of epithelial cell proliferation // inferred from electronic annotation /// 0050679 // positive regulation of epithelial cell proliferation // inferred from electronic annotation /// 0050767 // regulation of neurogenesis // inferred from electronic annotation /// 0050768 // negative regulation of neurogenesis // inferred from sequence or structural similarity /// 0050793 // regulation of developmental process // inferred from electronic annotation /// 0055008 // cardiac muscle tissue morphogenesis // inferred from sequence or structural similarity /// 0060038 // cardiac muscle cell proliferation // inferred from electronic annotation /// 0060045 // positive regulation of cardiac muscle cell proliferation // inferred from sequence or structural similarity /// 0060253 // negative regulation of glial cell proliferation // inferred from sequence or structural similarity /// 0060317 // cardiac epithelial to mesenchymal transition // inferred from sequence or structural similarity /// 0060411 // cardiac septum morphogenesis // inferred from sequence or structural similarity /// 0060412 // ventricular septum morphogenesis // inferred from mutant phenotype /// 0060528 // secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development // inferred from electronic annotation /// 0060548 // negative regulation of cell death // inferred from electronic annotation /// 0060740 // prostate gland epithelium morphogenesis // inferred from electronic annotation /// 0060768 // regulation of epithelial cell proliferation involved in prostate gland development // inferred from electronic annotation /// 0060842 // arterial endothelial cell differentiation // inferred from sequence or structural similarity /// 0060843 // venous endothelial cell differentiation // inferred from sequence or structural similarity /// 0060948 // cardiac vascular smooth muscle cell development // inferred from sequence or structural similarity /// 0060956 // endocardial cell differentiation // inferred from sequence or structural similarity /// 0060979 // vasculogenesis involved in coronary vascular morphogenesis // inferred from sequence or structural similarity /// 0060982 // coronary artery morphogenesis // inferred from sequence or structural similarity /// 0061314 // Notch signaling involved in heart development // inferred from mutant phenotype /// 0061384 // heart trabecula morphogenesis // inferred from sequence or structural similarity /// 0061419 // positive regulation of transcription from RNA polymerase II promoter in response to hypoxia // inferred from sequence or structural similarity /// 0070986 // left/right axis specification // inferred from electronic annotation /// 0071372 // cellular response to follicle-stimulating hormone stimulus // inferred from direct assay /// 0072017 // distal tubule development // inferred from electronic annotation /// 0072044 // collecting duct development // inferred from electronic annotation /// 0072144 // glomerular mesangial cell development // inferred from electronic annotation /// 0072602 // interleukin-4 secretion // inferred from electronic annotation /// 0090051 // negative regulation of cell migration involved in sprouting angiogenesis // inferred from direct assay /// 0090090 // negative regulation of canonical Wnt receptor signaling pathway // inferred from electronic annotation /// 0097150 // neuronal stem cell maintenance // inferred from expression pattern /// 1901201 // regulation of extracellular matrix assembly // inferred from sequence or structural similarity /// 2000737 // negative regulation of stem cell differentiation // inferred from mutant phenotype /// 2000811 // negative regulation of anoikis // inferred from mutant phenotype /// 2000974 // negative regulation of pro-B cell differentiation // inferred from sequence or structural similarity /// 2001027 // negative regulation of endothelial cell chemotaxis // inferred from direct assay |
| NOTCH2 | 0001701 // in utero embryonic development // inferred from electronic annotation /// 0001709 // cell fate determination // traceable author statement /// 0001890 // placenta development // inferred from electronic annotation /// 0002011 // morphogenesis of an epithelial sheet // inferred from electronic annotation /// 0002437 // inflammatory response to antigenic stimulus // inferred from electronic annotation /// 0003184 // pulmonary valve morphogenesis // inferred from mutant phenotype /// 0006351 // transcription, DNA-dependent // inferred from electronic annotation /// 0006355 // regulation of transcription, DNA-dependent // traceable author statement /// 0006367 // transcription initiation from RNA polymerase II promoter // traceable author statement /// 0006915 // apoptotic process // traceable author statement /// 0006917 // induction of apoptosis // traceable author statement /// 0006959 // humoral immune response // inferred from electronic annotation /// 0007050 // cell cycle arrest // inferred from direct assay /// 0007219 // Notch signaling pathway // traceable author statement /// 0007220 // Notch receptor processing // traceable author statement /// 0007275 // multicellular organismal development // non-traceable author statement /// 0007368 // determination of left/right symmetry // inferred from electronic annotation /// 0007399 // nervous system development // non-traceable author statement /// 0008285 // negative regulation of cell proliferation // inferred from direct assay /// 0009887 // organ morphogenesis // inferred from expression pattern /// 0010467 // gene expression // traceable author statement /// 0016049 // cell growth // inferred from direct assay /// 0019827 // stem cell maintenance // traceable author statement /// 0030097 // hemopoiesis // traceable author statement /// 0030154 // cell differentiation // inferred from electronic annotation /// 0030326 // embryonic limb morphogenesis // inferred from electronic annotation /// 0042246 // tissue regeneration // inferred from electronic annotation /// 0043065 // positive regulation of apoptotic process // inferred from electronic annotation /// 0043066 // negative regulation of apoptotic process // traceable author statement /// 0046579 // positive regulation of Ras protein signal transduction // inferred from direct assay /// 0046849 // bone remodeling // inferred from mutant phenotype /// 0050793 // regulation of developmental process // inferred from electronic annotation /// 0060413 // atrial septum morphogenesis // inferred from mutant phenotype /// 0060674 // placenta blood vessel development // inferred from electronic annotation /// 0061314 // Notch signaling involved in heart development // inferred by curator /// 0072602 // interleukin-4 secretion // inferred from electronic annotation |
| NOTCH3 | 0006351 // transcription, DNA-dependent // inferred from electronic annotation /// 0006355 // regulation of transcription, DNA-dependent // inferred from electronic annotation /// 0006367 // transcription initiation from RNA polymerase II promoter // traceable author statement /// 0007219 // Notch signaling pathway // traceable author statement /// 0007220 // Notch receptor processing // traceable author statement /// 0007275 // multicellular organismal development // inferred from electronic annotation /// 0010467 // gene expression // traceable author statement /// 0030154 // cell differentiation // inferred from electronic annotation /// 0030900 // forebrain development // inferred from electronic annotation /// 0045596 // negative regulation of cell differentiation // inferred from electronic annotation /// 0045665 // negative regulation of neuron differentiation // inferred from electronic annotation /// 0048661 // positive regulation of smooth muscle cell proliferation // inferred from electronic annotation /// 0048663 // neuron fate commitment // inferred from electronic annotation /// 0050793 // regulation of developmental process // inferred from electronic annotation /// 0072104 // glomerular capillary formation // inferred from electronic annotation |
| NOTCH4 | 0001569 // patterning of blood vessels // inferred from sequence or structural similarity /// 0001709 // cell fate determination // traceable author statement /// 0001763 // morphogenesis of a branching structure // inferred from sequence or structural similarity /// 0001886 // endothelial cell morphogenesis // inferred from electronic annotation /// 0006351 // transcription, DNA-dependent // inferred from electronic annotation /// 0006355 // regulation of transcription, DNA-dependent // inferred from electronic annotation /// 0006367 // transcription initiation from RNA polymerase II promoter // traceable author statement /// 0007219 // Notch signaling pathway // traceable author statement /// 0007220 // Notch receptor processing // traceable author statement /// 0007221 // positive regulation of transcription of Notch receptor target // traceable author statement /// 0007275 // multicellular organismal development // inferred from electronic annotation /// 0009790 // embryo development // inferred from sequence or structural similarity /// 0010467 // gene expression // traceable author statement /// 0030097 // hemopoiesis // traceable author statement /// 0030154 // cell differentiation // non-traceable author statement /// 0030879 // mammary gland development // inferred from direct assay /// 0045446 // endothelial cell differentiation // inferred from electronic annotation /// 0045596 // negative regulation of cell differentiation // non-traceable author statement /// 0045602 // negative regulation of endothelial cell differentiation // inferred from sequence or structural similarity /// 0045893 // positive regulation of transcription, DNA-dependent // traceable author statement /// 0050793 // regulation of developmental process // inferred from electronic annotation |
RGS1 expression in normal tissues
To confirm that the up-regulation of RGS1 was not stomach specific, RGS1 gene expression in normal tissues was investigated using various data in the database, such as the GEO database. From data analysis of various types of normal human tissues (GSE2361 and GSE2193)[31,32], RGS1 has been shown to be expressed in normal stomach, bladder, colon, corpus, liver, lung, placenta, prostate, skin, spinal cord, spleen, thymus, thyroid, and trachea, whereas RGS1 gene expression is relatively high in the thymus and spinal cord but not in the stomach.
DISCUSSION
Gastric cancer progression is associated with Helicobacter pylori and gastric stem cells are suggested to involve in the progression from chronic atrophic gastritis to gastric cancer[33]. It may be assumed that RGS1 plays a role in regulating G protein signaling and mitogen-activated protein (MAP) kinase signaling during gastric cancer progression, which may provide some hypotheses in which RGS1 promotes stem cell transition into CSCs, CSCs induce RGS1 expression in gastric cancer, or RGS1 orchestrates with other molecules in EMT. In gastric cancer cells, CDH1 isoform is found to increase gastric cancer cell invasion and angiogenesis, and some inflammatory mechanism is suggested to be involved in CDH1 regulation[34]. Analysis of Rgs1(-/-) has shown that B cell responses to chemokines, plasma cell localization, and immune tissue architecture are disturbed when Rgs1 is deleted[35]. In our studies, RGS1 was up-regulated in diffuse-type GC compared to MSC, which may suggest that RGS1 plays a role to response cancer cells and indicate the possibility to involve in cancer immunity. It may be possible that RGS1 protects the cells from cancer progression by enhancing anti-cancer immunity, or promote cell phenotype transition to cancer. The possibility that diffuse-type GC sample contains a variety of cells in cancer microenvironment should be considered. It may also be possible that RGS1 is up-regulated in some cancer microenvironment cells. Because the dedifferentiation of epithelial cells into stem cells may play a role in regenerative medicine or diseases, including cancer, investigating the genes regulated in stem and cancer cells is important[36]. Changes in the gene expression profiles of early- and late-stage MSC cultures have provided evidence regarding the importance of studying gene combinations to understand cellular phenotypes[8]. Cell surface antigens, such as SSEA4 or TRA-1-81, should also be included for future quality control in stem cells[37-39]. Furthermore, cell type-specific transcriptional profiles have been suggested for neural lineage cells, hepatic lineage cells, embryonic stem cells and induced pluripotent stem cells (iPSCs)[40]. Interestingly, insulin-like growth factor-1 receptor signaling and EMT crosstalk is sufficient to down-regulate erlotinib function in cells, while exon 19 of the erlotinib-sensitive epidermal growth factor receptor is mutated in non-small cell lung carcinomas[41]. Many cell-based iPSC models that are derived from patient cells have been investigated[42], and successful genetic or epigenetic reprogramming of cellular identity is crucial for the development of these stem-cell based disease models[42]. The investigation of the molecular combinations associated with different cell types is important for determining cellular phenotypes.
RGS1 has been shown to be related to cancer in some studies[43-45]. In late-stage non-small cell lung cancer, single nucleotide polymorphisms in RGS1 have been shown to be associated with overall survival in patients who received chemotherapy[43]. Silencing of RGS1 and RGS13 mRNAs resulted in enhanced responsiveness to chemoattractants in a human B lymphoma line[44]. RGS1 has been suggested to be involved in regulatory T cell trafficking[45]. Furthermore, RGS1 is up-regulated in T cells from the human gut compared with T cells from peripheral blood, and RGS1 regulates gut T cell trafficking[46]. Cancer cells interacting with the immune system may be future targets for anti-cancer therapy[47]. Considering that RGS1 is up-regulated in diffuse-type GC compared with MSCs, RGS1 may be a potential target for cancer treatment. RGS1 is involved in an adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway, immune response, positive regulation of GTPase activity, and termination of a G-protein coupled receptor signaling pathway in the Gene Ontology biological process [Traceable author statement (TSA); Inferred from electronic annotation (IEA)] (NCBI). From the investigation of EMT-related genes and RGS1, this study may suggest the possibility of future gene applications, including RGS1, to evaluate cancer cells to their sensitivity to treatment. microRNAs have been reported to be involved in RGS5 regulation in neuroblastoma[48]. Further assessments are needed to examine the possibility that RGS1 could be a marker for responsiveness to cancer treatment.
NOTCH gene expression was altered in diffuse-type GC and MSCs. Gene expression of NOTCH1, 3 and 4 was up-regulated in diffuse-type GC compared with MSCs, whereas NOTCH2 was down-regulated in diffuse-type GC. Upon Notch ligand cellular stimulation, the intracellular domain of the Notch receptor (NICD) translocates into the nucleus and regulates gene transcription, which leads to cell communication in signaling[49,50]. The regulation of NOTCH family gene expression may play an important role in the transcriptional regulation of cell phenotype transitions. NOTCH1 expression is increased in gastric cancer[51]. It has been shown that NOTCH1 is associated with gastric cancer with non-cardia location and diffuse type[51]. The up-regulation of NOTCH1 in diffuse-type GC compared to MSCs in our studies is consistent with the results in previous reports. NOTCH2 has been reported to be a candidate marker for prognosis of neoadjuvant-treated gastric cancer[52]. Combination of expression of GSK3B, CTNNB1, NOTCH2 was suggested to be correlated with patient survival. The previous study indicated that NOTCH2 may be involved in chemotherapy resistance in GC[52]. Considering that CSCs may be involved in drug resistance, the role of NOTCH2 should be investigated in CSCs or EMT. NOTCH2 expression was increased in post-therapeutic tumors[52]. Since NOTCH2 was down-regulated in diffuse-type GC compared to MSCs, NOTCH2 may mark some cancer phenotype in different stages. NOTCH3 was found to be increased in diffuse-type GC[51,53]. The previous reports are consistent with the results in this study indicating that NOTCH3 is up-regulated in diffuse-type GC compared to MSCs. Interestingly, NOTCH4 is suggested to be related to gastric carcinogenesis targeted by microRNA miR-181c[54]. Our data indicating NOTCH4 up-regulation in diffuse-type GC compared to MSCs is consistent with the previous findings suggesting involvement of NOTCH4 in gastric carcinogenesis. NOTCH1 and NOTCH2 exhibit opposite functions and play important roles in cancer as oncogenes or cancer suppressors depending on the cancer species. The relationship with NOTCH signaling and gastric cancer signaling should be investigated in the future (Figure 6A). In the EMT hypothesis, it is possible that epithelial-phenotype cells expressing NOTCH1 transition to mesenchymal-phenotype cells that express NOTCH2 (Figure 6B). Integrin beta 1 expression is important for the cancer cell population, and the insights into the mechanisms of cell phenotype conversion by gene regulation may provide fundamental understanding of cancer development[55]. Several pathways, including the claudin family genes and MAP kinase signaling pathways, have been investigated by gene set enrichment analysis, which may be the next step for the precise investigation of cancer drug response signaling[56].
ACKNOWLEDGMENTS
The authors acknowledge the support from the people who are involved in this study.
COMMENTS
Background
Mesenchymal stem cells (MSCs) exhibit mesenchymal features and are useful in regenerative medicine. Gastric cancer (GC) can be classified into two types, including diffuse-type GC that has mesenchymal features and intestinal-type GC that has epithelial features. The epithelial-mesenchymal transition (EMT) is an important mechanism in cancer development. The EMT is known to be involved in cancer cell migration and malignancy. Cancer stem cells (CSCs) are known to be resistant to anti-cancer drugs and involved in cancer recurrence.
Research frontiers
Gene regulation in MSCs and diffuse-type GC may provide new insights into CSCs. In this study, regulator of G-protein signaling (RGS) has been identified to be up-regulated in diffuse-type GC compared with MSCs. It is a novel hypothesis where RGS signaling may be involved in the EMT pathway.
Innovations and breakthroughs
Recent progress in genome-wide analysis in cancer has revealed that the gene signature is important for cancer diagnosis and exploring the targets of cancer therapy. CSCs are one of the targets for cancer treatment because these cells tend to be resistant to anti-cancer drugs and are involved in cancer recurrence. This study is the first to demonstrate that RGS1 may distinguish diffuse-type GC and MSCs.
Applications
By understanding how RGS1 is involved in the EMT and CSC signaling, this study represents potential future strategies for GC including diffuse-type GC treatment and prediction.
Terminology
RGS1 is protein involved in G-protein signaling and the regulation of GDP dissociation from the G alpha subunit and GTP binding to form trimetric G proteins. The EMT is the mechanism by which epithelial cells change into mesenchymal cells with a migratory capacity. Several adhesion molecules are involved in the EMT, and RGS1 may contribute to the changes in signaling.
Peer review
The authors described a global gene expression analysis in comparison between MSCs and diffuse-type GC cells. The authors found that RGS1 gene was upregulated in diffuse-type GC cells compared with MSCs, and that genes related to stem cells and NOTCH signaling were altered in diffuse-type GC cells compared with MSCs. The results may provide some insights in correlation between the EMT and CSCs. This paper summarizes important and actual data, it fulfils all criteria and demands put on articles published in World Journal of Stem Cells.
Footnotes
P- Reviewer: Kamihira M, Mokry J, Papaccio G S- Editor: Gong XM L- Editor: A E- Editor: Lu YJ
Supported by Cancer Research from the Ministry of Health, Labour and Welfare
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: July 19, 2014
First decision: September 4, 2014
Article in press: December 16, 2014
References
- 1.Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 2.Kong D, Li Y, Wang Z, Sarkar FH. Cancer Stem Cells and Epithelial-to-Mesenchymal Transition (EMT)-Phenotypic Cells: Are They Cousins or Twins? Cancers (Basel) 2011;3:716–729. doi: 10.3390/cancers30100716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas C, Karnoub AE. Lysyl oxidase at the crossroads of mesenchymal stem cells and epithelial-mesenchymal transition. Oncotarget. 2013;4:376–377. doi: 10.18632/oncotarget.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreso A, van Galen P, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, Cao L, Baiazitov R, Du W, Sydorenko N, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med. 2014;20:29–36. doi: 10.1038/nm.3418. [DOI] [PubMed] [Google Scholar]
- 5.Walsh MJ, Fellous TG, Hammiche A, Lin WR, Fullwood NJ, Grude O, Bahrami F, Nicholson JM, Cotte M, Susini J, et al. Fourier transform infrared microspectroscopy identifies symmetric PO(2)(-) modifications as a marker of the putative stem cell region of human intestinal crypts. Stem Cells. 2008;26:108–118. doi: 10.1634/stemcells.2007-0196. [DOI] [PubMed] [Google Scholar]
- 6.Walsh MJ, Hammiche A, Fellous TG, Nicholson JM, Cotte M, Susini J, Fullwood NJ, Martin-Hirsch PL, Alison MR, Martin FL. Tracking the cell hierarchy in the human intestine using biochemical signatures derived by mid-infrared microspectroscopy. Stem Cell Res. 2009;3:15–27. doi: 10.1016/j.scr.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Martin FL, Kelly JG, Llabjani V, Martin-Hirsch PL, Patel II, Trevisan J, Fullwood NJ, Walsh MJ. Distinguishing cell types or populations based on the computational analysis of their infrared spectra. Nat Protoc. 2010;5:1748–1760. doi: 10.1038/nprot.2010.133. [DOI] [PubMed] [Google Scholar]
- 8.Tanabe S, Sato Y, Suzuki T, Suzuki K, Nagao T, Yamaguchi T. Gene expression profiling of human mesenchymal stem cells for identification of novel markers in early- and late-stage cell culture. J Biochem. 2008;144:399–408. doi: 10.1093/jb/mvn082. [DOI] [PubMed] [Google Scholar]
- 9.Ren J, Stroncek DF, Zhao Y, Jin P, Castiello L, Civini S, Wang H, Feng J, Tran K, Kuznetsov SA, et al. Intra-subject variability in human bone marrow stromal cell (BMSC) replicative senescence: molecular changes associated with BMSC senescence. Stem Cell Res. 2013;11:1060–1073. doi: 10.1016/j.scr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 11.Laurén P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 12.Laurén P. Histogenesis of intestinal and diffuse types of gastric carcinoma. Scand J Gastroenterol Suppl. 1991;180:160–164. [PubMed] [Google Scholar]
- 13.Ohta H, Aoyagi K, Fukaya M, Danjoh I, Ohta A, Isohata N, Saeki N, Taniguchi H, Sakamoto H, Shimoda T, et al. Cross talk between hedgehog and epithelial-mesenchymal transition pathways in gastric pit cells and in diffuse-type gastric cancers. Br J Cancer. 2009;100:389–398. doi: 10.1038/sj.bjc.6604846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanabe S, Aoyagi K, Yokozaki H, Sasaki H. Gene expression signatures for identifying diffuse-type gastric cancer associated with epithelial-mesenchymal transition. Int J Oncol. 2014;44:1955–1970. doi: 10.3892/ijo.2014.2387. [DOI] [PubMed] [Google Scholar]
- 15.Tsuno S, Wang X, Shomori K, Hasegawa J, Miura N. Hsa-miR-520d induces hepatoma cells to form normal liver tissues via a stemness-mediated process. Sci Rep. 2014;4:3852. doi: 10.1038/srep03852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishiwatari T, Okubo Y, Tochigi N, Wakayama M, Nemoto T, Kobayashi J, Shinozaki M, Aki K, Sasai D, Yamamoto Y, et al. Remodeling of the pulmonary artery induced by metastatic gastric carcinoma: a histopathological analysis of 51 autopsy cases. BMC Cancer. 2014;14:14. doi: 10.1186/1471-2407-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura K, Semba S, Aoyagi K, Sasaki H, Yokozaki H. Mesenchymal stem cells provide an advantageous tumor microenvironment for the restoration of cancer stem cells. Pathobiology. 2012;79:290–306. doi: 10.1159/000337296. [DOI] [PubMed] [Google Scholar]
- 19.Aoyagi K, Minashi K, Igaki H, Tachimori Y, Nishimura T, Hokamura N, Ashida A, Daiko H, Ochiai A, Muto M, et al. Artificially induced epithelial-mesenchymal transition in surgical subjects: its implications in clinical and basic cancer research. PLoS One. 2011;6:e18196. doi: 10.1371/journal.pone.0018196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanabe S. Perspectives of gene combinations in phenotype presentation. World J Stem Cells. 2013;5:61–67. doi: 10.4252/wjsc.v5.i3.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathews LA, Hurt EM, Zhang X, Farrar WL. Genomic Analysis of Invasive Human Bone Marrow Derived Mesenchymal Stem Cells. J Bone Marrow Res. 2013;1:122. doi: 10.4172/2329-8820.1000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 23.Wu J, Liu Z, Shao C, Gong Y, Hernando E, Lee P, Narita M, Muller W, Liu J, Wei JJ. HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res. 2011;71:349–359. doi: 10.1158/0008-5472.CAN-10-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wend P, Runke S, Wend K, Anchondo B, Yesayan M, Jardon M, Hardie N, Loddenkemper C, Ulasov I, Lesniak MS, et al. WNT10B/β-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol Med. 2013;5:264–279. doi: 10.1002/emmm.201201320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chae JI, Jeon YJ, Shim JH. Downregulation of Sp1 is involved in honokiol-induced cell cycle arrest and apoptosis in human malignant pleural mesothelioma cells. Oncol Rep. 2013;29:2318–2324. doi: 10.3892/or.2013.2353. [DOI] [PubMed] [Google Scholar]
- 26.Iliopoulos D, Polytarchou C, Hatziapostolou M, Kottakis F, Maroulakou IG, Struhl K, Tsichlis PN. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal. 2009;2:ra62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takebe N, Ivy SP. Controversies in cancer stem cells: targeting embryonic signaling pathways. Clin Cancer Res. 2010;16:3106–3112. doi: 10.1158/1078-0432.CCR-09-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, Miele L. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010;16:3141–3152. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ilagan MX, Kopan R. SnapShot: notch signaling pathway. Cell. 2007;128:1246. doi: 10.1016/j.cell.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Ge X, Yamamoto S, Tsutsumi S, Midorikawa Y, Ihara S, Wang SM, Aburatani H. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86:127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Shyamsundar R, Kim YH, Higgins JP, Montgomery K, Jorden M, Sethuraman A, van de Rijn M, Botstein D, Brown PO, Pollack JR. A DNA microarray survey of gene expression in normal human tissues. Genome Biol. 2005;6:R22. doi: 10.1186/gb-2005-6-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giannakis M, Chen SL, Karam SM, Engstrand L, Gordon JI. Helicobacter pylori evolution during progression from chronic atrophic gastritis to gastric cancer and its impact on gastric stem cells. Proc Natl Acad Sci USA. 2008;105:4358–4363. doi: 10.1073/pnas.0800668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinheiro H, Carvalho J, Oliveira P, Ferreira D, Pinto MT, Osório H, Licastro D, Bordeira-Carriço R, Jordan P, Lazarevic D, et al. Transcription initiation arising from E-cadherin/CDH1 intron2: a novel protein isoform that increases gastric cancer cell invasion and angiogenesis. Hum Mol Genet. 2012;21:4253–4269. doi: 10.1093/hmg/dds248. [DOI] [PubMed] [Google Scholar]
- 35.Moratz C, Hayman JR, Gu H, Kehrl JH. Abnormal B-cell responses to chemokines, disturbed plasma cell localization, and distorted immune tissue architecture in Rgs1-/- mice. Mol Cell Biol. 2004;24:5767–5775. doi: 10.1128/MCB.24.13.5767-5775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews PW, Banting G, Damjanov I, Arnaud D, Avner P. Three monoclonal antibodies defining distinct differentiation antigens associated with different high molecular weight polypeptides on the surface of human embryonal carcinoma cells. Hybridoma. 1984;3:347–361. doi: 10.1089/hyb.1984.3.347. [DOI] [PubMed] [Google Scholar]
- 38.Wright A, Andrews N, Bardsley K, Nielsen JE, Avery K, Pewsey E, Jones M, Harley D, Nielsen AR, Moore H, et al. Mapping the stem cell state: eight novel human embryonic stem and embryonal carcinoma cell antibodies. Int J Androl. 2011;34:e175–e187; discussion e175-e187. doi: 10.1111/j.1365-2605.2011.01185.x. [DOI] [PubMed] [Google Scholar]
- 39.Henderson JK, Draper JS, Baillie HS, Fishel S, Thomson JA, Moore H, Andrews PW. Preimplantation human embryos and embryonic stem cells show comparable expression of stage-specific embryonic antigens. Stem Cells. 2002;20:329–337. doi: 10.1634/stemcells.20-4-329. [DOI] [PubMed] [Google Scholar]
- 40.Hikichi T, Matoba R, Ikeda T, Watanabe A, Yamamoto T, Yoshitake S, Tamura-Nakano M, Kimura T, Kamon M, Shimura M, et al. Transcription factors interfering with dedifferentiation induce cell type-specific transcriptional profiles. Proc Natl Acad Sci USA. 2013;110:6412–6417. doi: 10.1073/pnas.1220200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vazquez-Martin A, Cufí S, Oliveras-Ferraros C, Torres-Garcia VZ, Corominas-Faja B, Cuyàs E, Bonavia R, Visa J, Martin-Castillo B, Barrajón-Catalán E, et al. IGF-1R/epithelial-to-mesenchymal transition (EMT) crosstalk suppresses the erlotinib-sensitizing effect of EGFR exon 19 deletion mutations. Sci Rep. 2013;3:2560. doi: 10.1038/srep02560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherry AB, Daley GQ. Reprogramming cellular identity for regenerative medicine. Cell. 2012;148:1110–1122. doi: 10.1016/j.cell.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai J, Gu J, Lu C, Lin J, Stewart D, Chang D, Roth JA, Wu X. Genetic variations in the regulator of G-protein signaling genes are associated with survival in late-stage non-small cell lung cancer. PLoS One. 2011;6:e21120. doi: 10.1371/journal.pone.0021120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han JI, Huang NN, Kim DU, Kehrl JH. RGS1 and RGS13 mRNA silencing in a human B lymphoma line enhances responsiveness to chemoattractants and impairs desensitization. J Leukoc Biol. 2006;79:1357–1368. doi: 10.1189/jlb.1105693. [DOI] [PubMed] [Google Scholar]
- 45.Derycke MS, Charbonneau B, Preston CC, Kalli KR, Knutson KL, Rider DN, Goode EL. Toward understanding the genetics of regulatory T cells in ovarian cancer. Oncoimmunology. 2013;2:e24535. doi: 10.4161/onci.24535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibbons DL, Abeler-Dörner L, Raine T, Hwang IY, Jandke A, Wencker M, Deban L, Rudd CE, Irving PM, Kehrl JH, et al. Cutting Edge: Regulator of G protein signaling-1 selectively regulates gut T cell trafficking and colitic potential. J Immunol. 2011;187:2067–2071. doi: 10.4049/jimmunol.1100833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bianchi ME. Killing cancer cells, twice with one shot. Cell Death Differ. 2014;21:1–2. doi: 10.1038/cdd.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumps C, Fieuw A, Mestdagh P, Menten B, Lefever S, Pattyn F, De Brouwer S, Sante T, Schulte JH, Schramm A, et al. Focal DNA copy number changes in neuroblastoma target MYCN regulated genes. PLoS One. 2013;8:e52321. doi: 10.1371/journal.pone.0052321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L, Tan J, Zhang Y, Han N, Di X, Xiao T, Cheng S, Gao Y, Liu Y. DLK1 promotes lung cancer cell invasion through upregulation of MMP9 expression depending on Notch signaling. PLoS One. 2014;9:e91509. doi: 10.1371/journal.pone.0091509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh M, Katoh M. Transcriptional regulation of WNT2B based on the balance of Hedgehog, Notch, BMP and WNT signals. Int J Oncol. 2009;34:1411–1415. [PubMed] [Google Scholar]
- 51.Du X, Cheng Z, Wang YH, Guo ZH, Zhang SQ, Hu JK, Zhou ZG. Role of Notch signaling pathway in gastric cancer: a meta-analysis of the literature. World J Gastroenterol. 2014;20:9191–9199. doi: 10.3748/wjg.v20.i27.9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bauer L, Langer R, Becker K, Hapfelmeier A, Ott K, Novotny A, Höfler H, Keller G. Expression profiling of stem cell-related genes in neoadjuvant-treated gastric cancer: a NOTCH2, GSK3B and β-catenin gene signature predicts survival. PLoS One. 2012;7:e44566. doi: 10.1371/journal.pone.0044566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang H, An HJ, Song JY, Kim TH, Heo JH, Ahn DH, Kim G. Notch3 and Jagged2 contribute to gastric cancer development and to glandular differentiation associated with MUC2 and MUC5AC expression. Histopathology. 2012;61:576–586. doi: 10.1111/j.1365-2559.2012.04274.x. [DOI] [PubMed] [Google Scholar]
- 54.Hashimoto Y, Akiyama Y, Otsubo T, Shimada S, Yuasa Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis. 2010;31:777–784. doi: 10.1093/carcin/bgq013. [DOI] [PubMed] [Google Scholar]
- 55.Kato T, Enomoto A, Watanabe T, Haga H, Ishida S, Kondo Y, Furukawa K, Urano T, Mii S, Weng L, et al. TRIM27/MRTF-B-dependent integrin β1 expression defines leading cells in cancer cell collectives. Cell Rep. 2014;7:1156–1167. doi: 10.1016/j.celrep.2014.03.068. [DOI] [PubMed] [Google Scholar]
- 56.Bateman AR, El-Hachem N, Beck AH, Aerts HJ, Haibe-Kains B. Importance of collection in gene set enrichment analysis of drug response in cancer cell lines. Sci Rep. 2014;4:4092. doi: 10.1038/srep04092. [DOI] [PMC free article] [PubMed] [Google Scholar]


