Abstract
Chaperone-mediated autophagy (CMA) is a selective type of autophagy responsible for the lysosomal degradation of soluble cytosolic proteins. In contrast to other forms of autophagy where cargo is sequestered and delivered to lysosomes through membrane fusion/excision, CMA substrates reach the lysosomal lumen after direct translocation across the lysosomal membrane. CMA is part of the cellular quality control systems and as such, essential for the cellular response to stress. CMA activity decreases with age, likely contributing to the accumulation of altered proteins characteristic in tissues from old organisms. Furthermore, impairment of CMA underlies the pathogenesis of certain human pathologies such as neurodegenerative disorders. These findings have drawn renewed attention to CMA and a growing interest in the measurement of changes in CMA activity under different physiological and pathological conditions. In this chapter we review the different experimental approaches utilized to assess CMA activity both in cells in culture and in different organs from animals.
Introduction
Chaperone-mediated autophagy (CMA) is a type of autophagy responsible for the degradation of a subset of cytosolic proteins bearing in their amino acid sequence a consensus motif, biochemically related to KFERQ, that targets them for lysosomal degradation (Dice, 1990). This motif is recognized by a cytosolic chaperone, the heat shock cognate protein of 70 kDa (hsc70), in complex with its cochaperones (Chiang et al., 1989). The substrate/chaperone complex is delivered to the surface of the lysosomes where it binds to a CMA receptor, the lysosome-associated membrane protein type–2A (LAMP-2A) (Cuervo and Dice, 1996). After unfolding, the substrate protein is translocated across the lysosomal membrane in an ATP-dependent manner, assisted by a resident lysosomal chaperone (lys-hsc70) (Agarraberes et al., 1997). Once in the lysosomal lumen, CMA substrates are rapidly degraded (in 5–10 min) by the broad array of lysosomal proteases. These two features, the selectivity towards substrate proteins and their direct translocation across the lysosomal membrane, make CMA distinct from the other types of autophagy in mammalian cells, namely macroautophagy and microautophagy, where cargo is typically delivered in bulk to lysosomes through processes involving vesicular fusion (macroautophagy) and/or membrane excision (microautophagy) (Cuervo, 2004a; Levine and Klionsky, 2004; Shintani and Klionsky, 2004).
Approximately 30% of soluble cytosolic proteins contain the CMA-targeting motif (Dice, 1990). They constitute a very heterogeneous pool of intracellular proteins including among others some glycolytic enzymes (glyceraldehyde-3-phosphate dehydrogenase, aldolase, phosphoglyceromutase), particular transcription factors and inhibitors of transcription factors (c-fos, the inhibitor of NFκB (IκB)), calcium-binding proteins (Annexins I, II, IV and VI), vesicular trafficking proteins (α-synuclein), cytosolic forms of secretory proteins (α-2-microglobulin) and even some of the catalytic and regulatory subunits of the proteasome, the major cytosolic protease (reviewed in Dice, 2007; Majeski and Dice, 2004; Massey et al., 2004; Massey et al., 2006b). Based on the broad nature of CMA substrate proteins and their participation in many different intracellular processes it is easy to infer that changes in the activity of this pathway may have major consequences on cell functioning.
Although some level of basal CMA activity is detectable in almost all cells, CMA is maximally up-regulated under stress conditions, such as prolonged nutrient deprivation (serum removal in cultured cells or starvation in rodents) (Cuervo et al., 1995a; Wing et al., 1991), mild oxidative stress (Kiffin et al., 2004) and exposure to toxins (Cuervo et al., 1999). The selectivity that characterizes CMA may be beneficial during prolonged starvation as it will allow the degradation of non-essential proteins to provide amino acids for the synthesis of proteins required to guarantee cell survival under those stressful circumstances. Activation of CMA in conditions associated with protein damage, such as oxidative stress, may also facilitate the selective removal of altered proteins without disturbing neighboring functional ones.
The signaling mechanisms leading to CMA activation/inactivation are presently unknown, but most of the regulation of this pathway takes place at the lysosomal compartment (Bandyopadhyay et al., 2008; Cuervo and Dice, 2000a; Cuervo and Dice, 2000b; Kaushik et al., 2006). Binding of substrate proteins to the CMA receptor, LAMP-2A is rate limiting for this pathway. In fact, levels of LAMP-2A at the lysosomal membrane are tightly regulated and they directly correlate with CMA activity (Cuervo and Dice, 2000a; Cuervo and Dice, 2000b). In addition, the presence of hsc70 in the lysosomal lumen is also necessary to attain substrate translocation into lysosomes via CMA (Agarraberes et al., 1997). In fact, although all lysosomes contain LAMP-2A in their membrane, only a subset of lysosomes contain hsc70 in their lumen, and they are the only ones competent for CMA (Cuervo et al., 1997). Under conditions such as prolonged starvation, both levels of LAMP-2A at the lysosomal membrane and of hsc70 in the lumen increase gradually, resulting in a progressive increase in CMA activity (Agarraberes et al., 1997; Cuervo et al., 1995a). If starvation persists beyond 3 days in rodents, part of the pool of lysosomes normally unable to perform CMA acquire the lumenal chaperone (hsc70) and become competent for this pathway. During maximal CMA activation, the pool of lysosomes active for this pathway relocates from the cell periphery toward the perinuclear region, although the reasons for this redistribution remain unclear (Cuervo and Dice, 2000b).
A decrease in CMA activity has been reported both in senescent cells in culture and in different organs from old rodents (Cuervo and Dice, 2000c; Dice, 1982). This age-related decline in CMA activity is mainly due to a gradual decrease in the levels of LAMP-2A at the lysosomal membrane because of its increased instability with age (Cuervo and Dice, 2000c; Kiffin et al., 2007). Reduced CMA activity thus contributes to the accumulation of abnormal proteins in the cytosol and it is probably in part responsible for the higher susceptibility to stressors – a characteristic of old organisms (Massey et al., 2006a). Malfunctioning of CMA has also been described in different pathologies such as some lysosomal storage disorders (Cuervo et al., 2003), certain toxic-induced nephropathies (Cuervo et al., 1999), the hypertrophic kidney secondary to diabetes (Sooparb et al., 2004) and in familial forms of Parkinson’s disease (Cuervo et al., 2004). The important roles of CMA as part of the cellular response to stress and the association of its malfunctioning with human pathologies have increased the interest in assessing CMA activity in different physiological and pathological conditions.
Here we describe the experimental models commonly used to study CMA, the characteristics of CMA substrates and the different methods developed by our and other groups to monitor CMA activity: i) measurement of rates of long-lived protein degradation; ii) monitoring changes in the levels of key CMA components in isolated lysosomes; iii) analysis of the subcellular location of lysosomes active for CMA; and iv) measurement of the translocation of known CMA substrates into isolated lysosomes via an in vitro assay.
Experimental models for the study of CMA
CMA has been identified so far in mammalian cells only. Yeast have a somehow related process known as the vacuolar import and degradation pathway in which substrate proteins are first translocated in a chaperone-dependent manner into small vesicles that then fuse with the yeast vacuole where their cargo is degraded (Brown et al., 2003). Although this process resembles a combination of CMA and macroautophagy, the proteins involved in the translocation of substrates to the vesicles are different from those that participate in CMA. In fact, LAMP-2A, the spliced variant of the lamp-2 gene that is essential for CMA, is not conserved in yeast. In species phylogenetically lower than mammals, such as worms, flies or fish, a transcript with 40–50% homology to LAMP-2A has been identified, but this homology is lost in the transmembrane and cytosolic regions of the protein, those that differentiate the three spliced variants of the lamp-2 gene (Konecki et al., 1995). The LAMP-2 isoform conserved in these species seems to correspond to LAMP-2B, for which a role in macroautophagy but not in CMA is proposed (Eskelinen et al., 2005; Eskelinen et al., 2003). Thus the LAMP-2A variant, required for CMA, appears much later in evolution, being described for the first time in avians and above (Konecki et al., 1995).
Although at different levels, CMA activity has been detected in many different types of transformed cells - NIH3T3 (mouse fibroblasts), 293HEK (human kidney epithelial cells), CHO (Chinese hamster ovary cells), Rat-1 (rat kidney epithelial cells), RALA (rat hepatoblastoma), Huh7 (human hepatoblastoma), astrocytome, several human lung cancer cell lines (H820, A549, H460) and in several primary cells in culture (human skin fibroblasts, dopaminergic neurons, cortical neurons, astrocytes, dendritic cells, macrophages and CD4+ naïve T cells). Among the different tissues in rodents in which CMA activity has been detected (liver, kidney, heart, spleen, lung), liver is by far the tissue in which this pathway has been better characterized (Dice, 2007; Massey et al., 2006b). Although based on the absence of changes in the levels of KFERQ-containing proteins in response to starvation it was initially proposed that CMA is not active in brain (Wing et al., 1991), recent studies with isolated astrocytes and dopaminergic and cortical neurons support the presence of CMA activity in these cells, although it is unresponsive to changes in the nutritional status (Cuervo, 2004a; Martinez-Vicente et al., 2008).
Currently there are no knock-out mouse models with impaired CMA. A complete LAMP-2 knock-out mouse was developed several years ago (Tanaka et al., 2000). These animals present alterations in macroautophagy that manifest as an accumulation of autophagic vacuoles in different tissues, and inefficient lysosomal biogenesis, that probably contributes to the observed decrease in protein degradation, and abnormal cholesterol metabolism and impaired vesicular trafficking (Eskelinen et al., 2002; Huynh et al., 2007). Surprisingly, mouse embryonic fibroblasts from these animals do not show changes in protein degradation, suggesting possible activation of compensatory mechanisms in undifferentiated cells but not in non-dividing differentiated cells (Eskelinen et al., 2004).
Our laboratory has developed a bi-transgenic mouse model with regulated expression of LAMP-2A in liver (Zhang and Cuervo, 2008). As in cultured cells, overexpression of LAMP-2A in liver results in an increase in CMA activity. Using this model, we have recently analyzed the consequences of maintaining normal levels of LAMP-2A until advanced ages in liver, by activating the expression of the exogenous form of LAMP-2A once the levels of the endogenous protein start to decrease. We found that livers of old transgenic mice contain lower levels of altered proteins (oxidized, aggregate, etc.), respond more efficiently to different stressors, and show a significant improvement in liver function, supporting the critical role of CMA in maintenance of cellular homeostasis (Zhang and Cuervo, 2008). To determine possible tissue-specific differences in the requirements for functional CMA and whether or not restoration of CMA in a broad number of tissues will have a positive effect on life-span, we are currently developing novel transgenic models with regulatable expression of LAMP-2A in different tissues.
Properties of CMA substrates
An often asked question when analyzing CMA is whether or not a protein is a substrate for this pathway. Proteins can follow different proteolytic pathways depending on changes in the protein itself (posttranslational modifications) or in the cellular conditions that result in activation/inhibition of particular proteolytic pathways (Cuervo, 2004b). Consequently, whether or not a protein is a substrate for CMA needs to be experimentally analyzed.
Table I summarizes the accepted criteria that a protein has to fulfill to be considered a CMA substrate. Briefly, the candidate protein has to bear in its amino acid sequence a CMA-targeting motif. The presence of this motif is necessary and sufficient to target proteins to CMA (Dice, 1990). Thus, proteins that do not carry a CMA-targeting motif can be directed to lysosomes via CMA when the sequence is incorporated as a fusion tag in the proteins. However, the fact a protein contains the motif in its sequence indicates that it can be degraded via CMA, but it does not necessarily imply that the protein is undergoing degradation through CMA, as often the targeting motifs are only exposed on the surface of the protein after conformational modifications. As with other proteins degraded via lysosomes, CMA substrates have usually long half-lives (ranging from >10 h up to several days), and their half-life changes with changes in CMA activity (increases when CMA is blocked or decreases if CMA is maximally activated). All CMA substrates interact with the two major components of this pathway, hsc70, the chaperone in the cytosol, and the LAMP-2A receptor at the lysosomal membrane. Finally, the ultimate evidence of a protein being a bona fide CMA substrate is if it can be translocated into isolated lysosomes in an ATP- and hsc70-dependent manner (see below). Of the substrate proteins identified until date, most of them are soluble cytosolic proteins. In fact, although it is plausible to think that this could be also a biogenic pathway for the delivery of enzymes into lysosomes, none of the known lysosomal hydrolases contain the CMA-targeting motif.
Table I.
Requirements for a protein to be considered as a CMA substrate
| REQUIREMENT | ASSAY | REFERENCES |
|---|---|---|
| Presence of KFERQ-like motif in its sequence | Sequence analysis | (Dice, 1990) |
| Long half-life | Metabolic labeling/immunoprecipitation | (Cuervo et al., 1998b; Cuervo et al., 2004) |
| Increases with CMA blockage Decreases with CMA activation | ||
| Binding to cytosolic hsc70 | Co-immunoprecipitation from cytosol | (Cuervo et al., 1999; Cuervo et al., 1998b) |
| Binding to LAMP-2A at the lysosomal membrane | Co-immunoprecipitation from isolated lysosomes | (Cuervo et al., 2004) |
| Translocation into isolated lysosomes | In vitro translocation/degradation assays | (Cuervo et al., 1999; Cuervo et al., 1998a; Cuervo et al., 1995b; Cuervo et al., 1994; Terlecky et al., 1992; Terlecky and Dice, 1993) |
| ATP/hsc70-dependent Competed by other CMA substrates |
Methods to measure CMA
The applicability of the methods described in the following sections depends on the experimental model. The four procedures described here can be used to track CMA in cultured cells, although it is true that the number of cells required for the isolation of lysosomes for the in vitro assays can be a limitation for some types of primary cells in culture. For animal tissues, such as the liver, where it is relatively easy to prepare a homogenous culture of hepatocytes, all procedures can be applied. However, when cell culture is not possible, measurement of protein degradation is not a straightforward procedure and measurement of CMA relies on the other three procedures.
Measurement of protein degradation rates
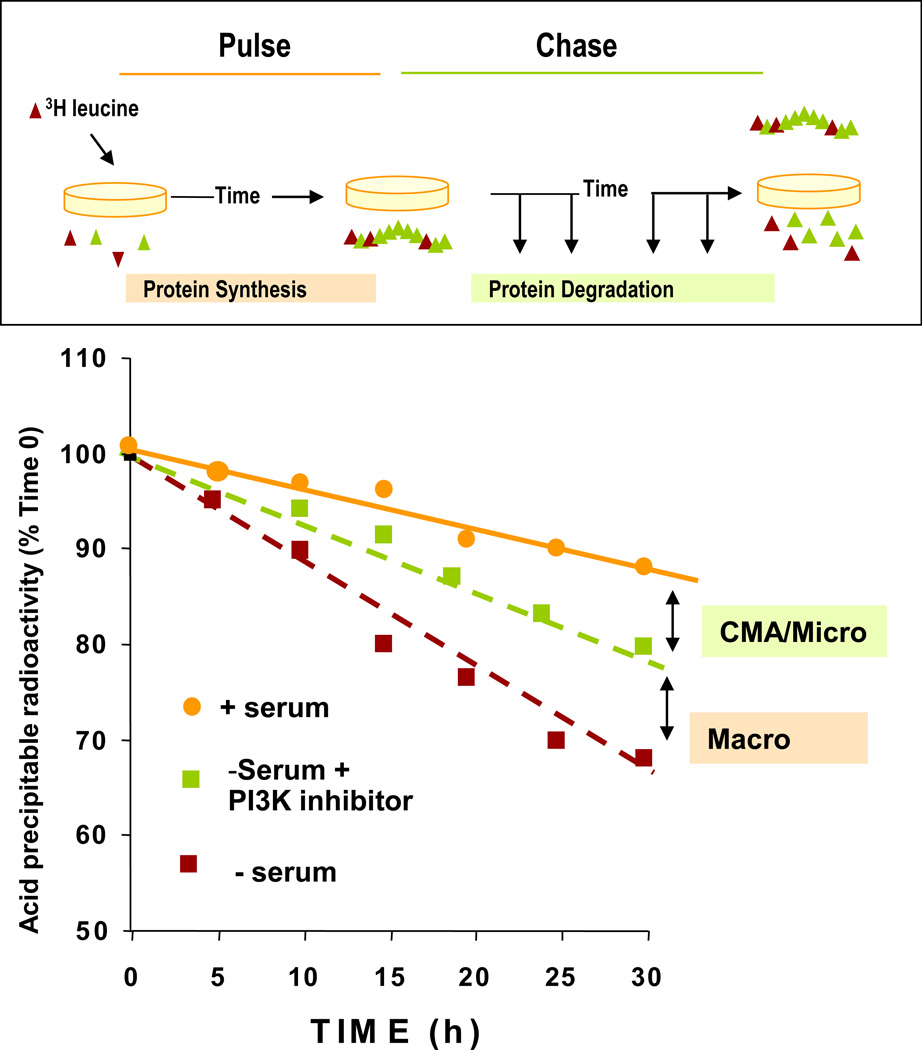
With some exceptions, proteins degraded in lysosomes have long half-lives (Cuervo, 2004b). Consequently, measurement of the rates of degradation of long-lived protein in cultured cells through metabolic labeling in pulse/chase experiments can be used as a good assessment of lysosomal function (Fig. 1) (Auteri et al., 1983). This procedure relies on the incorporation of a radiolabeled amino acid in the proteins synthesized during the labeling period (pulse) and tracking of the release of radiolabeled amino acid into the medium as the labeled proteins undergo degradation (chase). Separation of amino acid and small peptides from intact proteins in the medium is attained by precipitation of the proteins in acid.
Figure 1. Measurement of long-lived protein degradation.
Top: Confluent cells in culture are incubated with a radiolabeled amino acid for 48 h and after extensive washing the amount of acid-soluble radioactivity (amino acids and small peptides) released into the medium at different times is determined. Bottom: Typical example of rates of degradation of long-lived proteins in cultured cells due to CMA, micro- or macroautophagy. CMA activity is calculated as the increase in protein degradation during serum deprivation sensitive to lysosomal protease inhibition and insensitive to the effect of PI3-K type III inhibitors.
Pulse and chase experiments
Plate the cells to approximately 40% of confluence in 12 well plates in the culture medium used for normal maintenance of that particular cell type.
Pulse: When cells reach 60–70% confluence, label the cells with 2 µCi/ml [3H]leucine or [3H]valine.
Incubate the cells at 37°C for 48 h to maximize labeling of long-lived proteins.
-
Chase: At the end of the labeling, aspirate the medium, wash cells profusely (5 times) with Hanks’ solution, and plate the cells in 0.5–0.7 ml of chase medium (chase medium contains 50 times the molar concentration of the unlabeled form of the radiolabeled amino acid to prevent reutilization of the labeled amino acid into proteins synthesized during the chase period). In half of the cells, the serum-free chase medium should be supplemented with serum, to be able to analyze changes in protein degradation in response to serum removal, one of the best characterized stimuli of CMA.
Note: Some caution in the use of leucine for the labeling has been recommended because of the inhibitory effect of this amino acid on macroautophagy in different cell types. However, high specific activity of the radiolabeled amino acid allows for the use of very low concentrations of leucine in the chase medium (30 times below the described inhibitory concentrations), making it feasible for most cell types. [3H]valine has been proposed as a good alternative for labeling. However, the fact that the regulatory effect of amino acids on autophagy is not universal brings about the same concerns regarding the use of valine as a replacement for leucine. Consequently, for cells in which the inhibitory role of amino acids has been studied, it is recommended to use the amino acid with less inhibitory ability, whereas for cells where this effect is unknown it is recommended to at least verify if the result obtained with one radiolabeled amino acid is also reproducible using the other, or to directly assess the effect of both amino acids on macroautophagy in those cells using the procedures described in other chapters (see the chapter by Bauvy et al., in this volume).
- Collection of samples: To measure the amount of free radiolabeled amino acid released into the medium upon intracellular degradation of the proteins synthesized during the pulse:
- Incubate the cells in a CO2 incubator at 37°C.
- At the desired times (e.g., 0, 4, 12, 20, 24 h) collect aliquots (50–70 µl) of the medium from each well. It is important that only medium without floating cells is collected. If cell detachment is a problem, the 12 well plate should be centrifuged (500 g for 5 min) before taking the medium, or the medium aliquots should be collected in separate microcentrifuge tubes, spun down using the same conditions, and the supernatant fractions transferred to a clean tube for precipitation. Otherwise, samples can be directly placed in a 0.45-µm pore filter-bottom 96 well plate (Millipore Multiscreen Assay System, Millipore, Bedford, MA, MSVMHTS00) containing half of the final volume of 20% trichloroacetic acid (TCA) (for a final concentration of 10%).
- Precipitation is facilitated by addition of 0.5 mg/ml final concentration of bovine serum albumin (BSA, standard grade powder (protease free)) dissolved in water) followed by incubation at 4°C for at least 1 h.
- After taking the last time point, wash the cells twice with Hanks’ solution and add 1–2 ml of solubilization buffer (0.1 N NaOH, 0.1% sodium deoxycholate,Sigma-Aldrich, D6750) per well. These samples will be used to calculate the total amount of radioactivity incorporated by the cells during the labeling time, as indicated in 6d.
- Incubate the plate at 37°C for 2–6 h, until cells are solubilized.
- Sample processing: To count the amount of radiolabeled amino acid released into the medium (following protein degradation) and present inside the cells (incorporated into proteins):
- Collect the acid-soluble fraction of the aliquots taken from the medium (containing amino acids and short peptides) by vacuum filtration using a Millipore manifolder into a 96 well plate.
- Transfer the collected flow-through (approx. 200 µl) into individual scintillation vials, add scintillation liquid and read in a Beta scintillation counter.
- If desired, the filters can be left to air-dry and then punched into scintillation vials to account for the total amount of undegraded protein released into the medium.
- Take 50 µl of the solubilized cells and count in scintillation vials (this will be the total radioactivity still present inside cells).
- Count all the samples (flow through, filters and cells) as disintegrations per minute in a liquid scintillation analyzer by correcting for quenching using an external standard.
Inhibition of different autophagic pathways
In order to discriminate the pool of long-lived proteins degraded in lysosomes from those degraded in other proteolytic systems (i.e., ubiquitin/proteasome, calpains, etc.) and the contribution of each type of autophagy to the degradation of the long-lived proteins, blockers of lysosomal proteolysis are commonly used in these pulse and chase experiments. Weak bases, such as ammonium chloride or chloroquine, accumulate inside lysosomes neutralizing their intrinsic acid pH, required for maximal activity of the lysosomal proteases (Klionsky et al., 2007). Of the two weak bases, the former is commonly preferred since chloroquine has been shown to also affect protein synthesis. One of the limitations of the use of ammonium chloride is that its neutralizing effect rarely lasts more than 12 h, requiring periodic refreshing of the treatment if the chase is extended beyond this time. To overcome this problem, the group of Erwin Knecht has recently shown that a combination of 20 mM ammonium chloride with 0.1 mM leupeptin results in blocking lysosomal-dependent degradation most effectively without affecting other proteolytic systems (Fuertes et al., 2003). By comparing the degradation in cells supplemented or not with this cocktail it is possible to discriminate lysosomal-dependent degradation from that through other intracellular pathways.
The use in these studies of phosphatidylinositol-3-kinase (PI3K) inhibitors such as 3-methyladenine, makes it possible to separate the percentage of lysosomal degradation (sensitive to ammonium chloride/leupeptin) that occurs via macroautophagy (sensitive to 3-methyladenine), since PI3K are required in this type of autophagy. The remaining lysosomal degradation (insensitive to 3-methyladenine) can be attributed in most cell types to the other autophagic pathways (namely microautophagy and CMA) (see below). As a side note, 3-methyladenine is used in most cell types at 10 mM final concentration and prepared as a 2× stock in the growing medium, as solubility of this compound is dependent on pH.
Calculations
Proteolysis is calculated as the amount of acid-precipitable radioactivity (protein) transformed to acid-soluble radioactivity (peptides and amino acids) at each time during the incubation and it is expressed in percentage. To this purpose, the radioactivity in the aliquot collected from the medium needs to be corrected by the final volume of medium remaining in the well (as the amount of chasing medium decreases with each time point). The total amount of radiolabeled protein in each well is calculated by adding the amount of radioactivity present in the solubilized cells, plus the amount of radioactivity taken in each aliquot (acid-soluble in the flow through and -precipitable in the filters), plus the amount of radioactivity present in the medium in the last time point.
P = acid precipitable radioactivity
St = acid soluble radioactivity in aliquot of the medium
S0 = acid soluble radioactivity in aliquot of the medium at time 0
V0 = total volume of medium at time 0
v = volume of the aliquot taken from the medium
T = radioactivity in the aliquot taken from the solubilized cells
VT = total volume that cells were solubilized in
n = number of aliquots taken before a given time point
a = volume aliquot taken from solubilized cells
Although a certain level of basal CMA is present in all cells, CMA is maximally up-regulated in confluent cultured fibroblasts at approximately 10 h after serum removal. Hence, the inducible form of this pathway can be measured as the percentage of long-lived proteins degraded after removal of serum, inhibited by ammonium chloride (inhibitor of all types of autophagy), but insensitive to phosphatidylinositol-3-kinase inhibitors (Finn et al., 2005; Finn and Dice, 2005; Massey et al., 2006c) (Fig. 1). The two major limitations of this approach is that in certain cells there is considerable basal CMA activity, and consequently, considering only the percentage of degradation responsive to serum removal as CMA underestimates the contribution of this autophagic pathway to protein degradation. On the other hand, the lack of methods to quantify microautophagy in mammals or of selective inhibitors for this pathway makes it difficult to separate CMA-dependent degradation from that occurring via microautophagy, as this pathway contributes to lysosomal degradation both in the presence and absence of serum. Consequently, measurements of long-lived protein degradation should be complemented with other methods to analyze CMA.
Measurement of levels of key CMA components
Levels of hsc70, the cytosolic chaperone responsible for targeting CMA substrates to lysosomes, remain constant in most conditions, as this is the constitutive member of the hsp70 family of molecular chaperones (Cuervo et al., 1995a). However, lysosomal levels of LAMP-2A and lysosomal-hsc70 increase with the increase in CMA activity (Agarraberes et al., 1997; Cuervo and Dice, 2000b). It is thus possible to monitor the increase in CMA activity via immunoblot for LAMP-2A and lys-hsc70 in lysosomes isolated from the tissues/cells of interest. Note that total cellular levels of LAMP-2A and hsc70 often remain constant as most of the changes occur in the particular group of lysosomes involved in CMA (approximately 30–60% of total lysosomes depending on cell type and cell conditions). Although there are circumstances with extreme changes in CMA activity in which an increase or decrease of total cellular levels of LAMP-2A can be observed, it is advisable, when possible, to analyze the levels of this protein in the lysosomal fraction. As described in more detail below, one of the limitations for the isolation of lysosomes competent for CMA is the large number of cultured cells or starting tissue required. An alternative that has been shown to be valid in some cases is the use of the light mitochondrial/lysosomal fraction obtained by differential centrifugation instead (see below) as this can be prepared from as few as 2×106 cells or 0.1 g of tissue. In this case, because both lysosomes and mitochondria are highly enriched in this fraction it is important to normalize the results to the levels of some abundant mitochondrial protein (e.g., cytochrome c, GRP78) to compensate for changes in the mitochondrial content.
Isolation of lysosomes
We describe in this section a procedure to purify lysosomes active for CMA from different tissue samples. A detailed protocol for the isolation of lysosomes from cultured cells can be found in the literature (Storrie and Madden, 1990). A critical point in this procedure is the way in which cells are disrupted, because the intrinsic fragility of this organelle requires the use of nitrogen cavitation, rather than other common physical or mechanical procedures for cell disruption, in order to get intact lysosomes. The lysosomal fraction obtained through this method is highly enriched in lysosomes active for CMA (those containing hsc70 in their lumen) reaching levels of approximately 80% of total lysosomes in that fraction (Agarraberes et al., 1997). Although other procedures also render a highly purified lysosomal fraction (Marzella et al., 1982), the percentage of CMA-active lysosomes in that fraction is considerably lower (20–40% of total lysosomes, depending on the cell type).
The following protocol for the isolation of lysosomes with high and low CMA activity from tissue samples (Cuervo et al., 1997) was developed through modification of a previously published method of lysosomal purification from rat liver (Wattiaux et al., 1978). In addition to liver, we have successfully applied this protocol for the purification of lysosomes from kidney, spleen, lung and different brain regions (grey-matter).
Extensively wash the tissue of interest with cold 0.25 M sucrose (4–5 times with at least 3 times the volume of the tissue). After weighing and mincing it, homogenize it in 0.25 M sucrose (3 volumes/g) in a motorized Teflon-glass homogenizer with 8–10 strokes at maximum speed.
Filter the homogenates through double gauze (common gauze cheesecloth) (to remove some of the interfering lipids), add four volumes of cold 0.25 M sucrose and centrifuge at 6,800 g for 5 min at 4°C.
Collect the supernatant fraction in another tube (ensure that you are not collecting the heavy mitochondria-enriched fraction, the white floating material close to the pellet, which also contains any unbroken cells, nuclei and red blood cells). Resuspend the pellet fraction with a ‘cold finger’ (dry glass test tube filled with ice) in the starting volume of 0.25 M sucrose and centrifuge under the same conditions.
Pool the two supernatant fractions and centrifuge at 17,000 g for 10 min at 4°C.
Resuspend the pellet fraction with the ‘cold finger’, add 3.5 volumes of 0.25 M sucrose/g tissue and centrifuge under the same conditions (wash step).
-
Resuspend the pellet (light mitochondria and lysosome-enriched fraction, (ML fraction)) with the ‘cold finger’, add 1 ml 0.25 M sucrose/3 g tissue and 2 volumes of 85.6% Metrizamide (AK Scientific, Inc., #69696) and mix gently. Load every 10 ml of this final 57% Metrizamide ML fraction at the bottom of an ultracentrifuge tube and layer on top a discontinuous Metrizamide gradient: 6 ml 32.8% Metrizamide, 10 ml 26.3% Metrizamide and 11 ml 19.8% Metrizamide (all diluted in water, pH 7.3). Fill the tube with 0.25 M sucrose and centrifuge in a SW 28 rotor at 141,000 g for 1 h at 4°C.
Note: The pellet of the second 17,000 g, 10 min spin can be used as an alternative to purified lysosomes in conditions in which the amount of starting tissue is limiting.
After the centrifugation, white to brownish material (depending on the original tissue) is visible at each of the interphases (IP) enriched in the following fractions (from bottom to top): IP1—mitochondria; IP2—mixture of mitochondria and lysosomes; IP3—CMA-active and -inactive lysosomes; IP4—CMA-active lysosomes. Collect IP3 and IP4 separately with a Pasteur pipette (in approximately 2–4 ml), dilute with at least five volumes of 0.25 M sucrose and centrifuge at 37,000 g for 15 min at 4°C.
Carefully resuspend the pellet of IP3 with a blunt Pasteur pipette in 1 ml 0.25 M sucrose and centrifuge at 10,000 g for 5 min at 4°C. This pellet is enriched in secondary lysosomes with low CMA activity (lacking lys-hsc70).
-
Use the supernatant fraction of this step to resuspend the pellet of the IP4 fraction to get the fraction enriched in secondary lysosomes with high CMA activity (enriched in lys-hsc70).
Note: To guarantee the reproducibility of any future studies performed with the isolated fractions it is important to systematically evaluate for purity, recovery and enrichment of lysosomal enzymes and for integrity of the lysosomal membrane. Purity can be determined by measuring specific activity of enzyme markers of the main contaminant intracellular components: succinic-dehydrogenase (mitochondria), catalase (peroxisomes) and lactate-dehydrogenase (cytosol) as described before (Storrie and Madden, 1990). Measurement of total and specific activity of β-hexosaminidase or β-N-acetyl-glucosaminidase, two well characterized lysosomal enzymes, are used routinely to determine the recovery (percentage of total cellular lysosomes recovered in the isolated fraction) and the enrichment (fold increase in specific activity of lysosomal markers in the isolated fraction) of lysosomes in the isolated fraction (Storrie and Madden, 1990). Lastly, β-hexosaminidase latency (percentage of β-hexosaminidase activity detected in the incubation medium when lysosomes are incubated in an isotonic buffer for increasing periods of time) is used to assess the integrity of the lysosomal membrane (Storrie and Madden, 1990). Preparations of lysosomes with more than 10% broken lysosomes should be discarded.
For studies requiring separate analysis of lysosomal membranes and matrices (lysosomal content), these two fractions can be isolated after disrupting lysosomes with a hypotonic shock. Briefly, collect the isolated lysosomes by centrifugation (25,000 g for 10 min), resuspend the pellet fraction in a hypotonic buffer (0.025 M sucrose) and after 30 min incubation on ice, spin the samples at 150,000 g for 30 min to recover the membrane fraction in the pellet and the lysosomal content in the supernatant fraction (Ohsumi et al., 1983).
Immunoblot for CMA components
Since most of the changes in the levels of CMA components that occur with changes in the activity of this pathway are not transcriptionally regulated, direct measurement of the protein of interest by immunoblot is the most used method for analysis. The choice of antibody for detection of CMA components is important as although both hsc70 and hsp70 share high homology and most of the available antibodies recognize both proteins, only hsc70 but not hsp70 participates in CMA (Chiang et al., 1989). Thus, antibodies that recognize both chaperones should be avoided. The group of Fred Dice extensively characterized the mouse monoclonal IgM antibody clone 13D3 (now available from different commercial sources, e.g., Abcam Inc. ab2788) as highly selective for hsc70 (Agarraberes et al., 1997). Regarding LAMP-2A, only levels of the A splice variant of the lamp2 gene correlate with CMA activity (Cuervo and Dice, 1996; Cuervo and Dice, 2000a; Cuervo and Dice, 2000b). Most of the commercially available antibodies against LAMP-2 have been developed against the lumenal part of the protein, also shared by the other two isoforms (Carlsson et al., 1988). It is important thus, to use antibodies against the cytosolic tail that will discriminate between each of these isoforms. We developed an antibody against the cytosolic tail of the rat LAMP-2A that also recognizes the mouse isoform, but it does not cross-react with the human protein (now commercially available through Zymed Laboratories, Invitrogen, 51–2200). To the best of our understanding, antibodies selective against human LAMP-2A are not currently available.
An increase in the levels of lysosomal LAMP-2A is usually observed in conditions when CMA is activated. Likewise, decreased levels of LAMP-2A in lysosomes are a good indication of diminished CMA activity. However, although reduced levels of lys-hsc70 will result in decreased lysosomal ability for CMA, because binding to LAMP-2A is the limiting step of this pathway, increased levels of lys-hsc70, do not necessarily transduce into increased CMA. Thus, in conditions such as aging in which a decrease of lysosomal LAMP-2A has been well reported, levels of lys-hsc70 are higher than in control cells, probably reflecting some type of compensatory mechanism (Cuervo and Dice, 2000c).
Analysis of the subcellular location of CMA-active lysosomes
As pointed out in the introduction, only a subset of lysosomes is competent for CMA (Cuervo et al., 1997). These are distinguishable because they have LAMP-2A in their membrane (as do most lysosomes) and also contain detectable levels of hsc70 in their lumen. This pool of lysosomes can be tracked with the antibodies specific for these two proteins both by immunofluorescence with secondary antibodies conjugated to fluorophores or by the use of two differently-sized immunogold-conjugated secondary antibodies in electron microscopy sections of cells or tissues (Cuervo et al., 1997). Using these procedures it was found that in conditions such as prolonged starvation, the number of lysosomes containing hsc70 in their lumen increases gradually with the increase in CMA activity (Cuervo et al., 1997). Similar procedures were used to identify that the above described increase in lys-hsc70 in aging originated indeed from an increase in the number of CMA-competent lysosomes rather than a net increase in the amount of hsc70 per lysosome (Cuervo and Dice, 2000c).
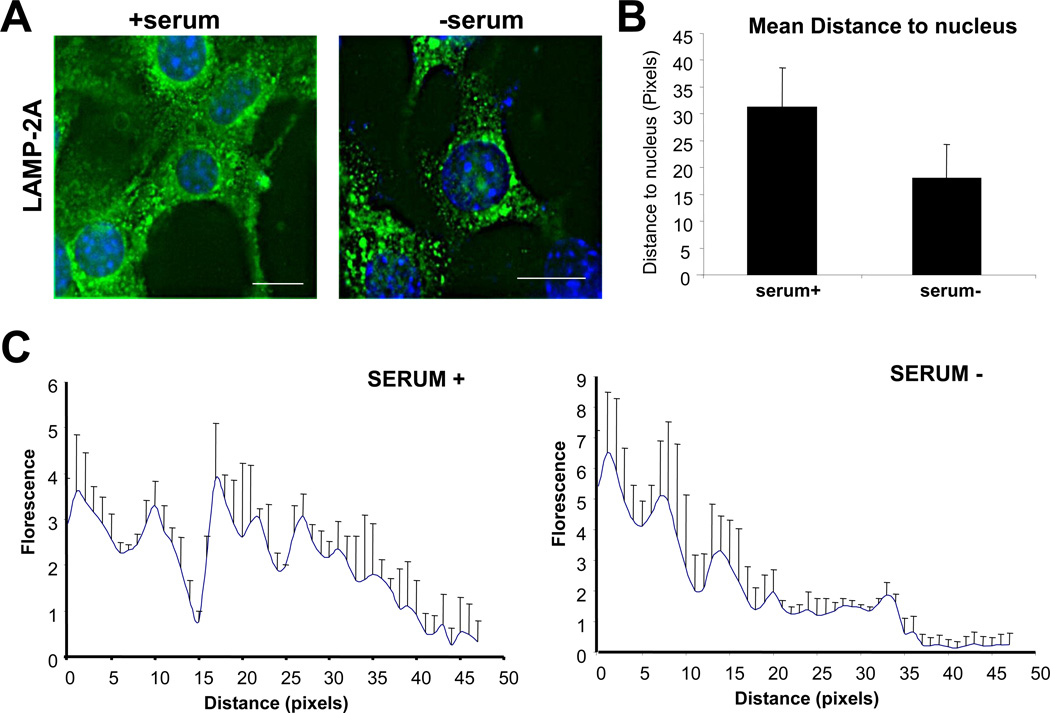
Interestingly, for reasons still unknown, activation of CMA is associated with the mobilization of hsc70/LAMP-2A–enriched lysosomes to the perinuclear region of the cells (Cuervo and Dice, 2000b) (Fig. 2). Visualizing the increase in the number of hsc70/LAMP-2A lysosomes and their subcellular location are also indirect ways to monitor CMA in cells in culture and in tissue sections.
Figure 2. Intracellular redistribution of CMA-active lysosomes.
A. Indirect immunofluorescence for LAMP-2A in cultured mouse fibroblasts maintained in the presence/absence of serum. Bar; 10 µm. B. Mean distance of the fluorescent puncta (lysosomes) to the nucleus C. Graph representing the intracellular distribution of fluorescent puncta with respect to the nucleus in the two indicated conditions. Values are mean + standard error of 4 different cells in each condition.
Immunofluorescence for CMA-active lysosomes
Colocalization of LAMP-2A and hsc70 by immunofluorescence is used often to identify the CMA-active lysosomes in cultured cells. This procedure can be performed following a standard immunofluorescence protocol, with the exception that fixation of the samples should be done using methanol. Methanol fixation will eliminate the soluble form of hsc70 (very abundant in the cytosol) allowing for the detection of the low percentage vesicle-associated hsc70.
Grow cells on cover slips at the bottom of 6-well plates in the complete (serum supplemented) medium that the particular cell type may usually require, until they reach semiconfluence (40–60% confluence). At this point, replace the medium with serum-free medium in half of the cover slips and culture in these conditions for 12–20 h to maximally activate CMA in the serum-deprived cells.
Wash twice with serum-free medium (to remove all remaining IgGs) and fix in −20°C cooled methanol (20% in PBS) for 1 min at room temperature.
Add 2 ml of blocking solution (0.2% (w/v) powdered nonfat milk, 2% newborn calf serum, 0.1 M glycine, 1% BSA, and 0.01% Triton X-100 in PBS) per well and incubate at room temperature for 30 min.
Aspirate the blocking solution and wash with PBS three times.
Dilute the primary antibody (rabbit IgG anti-LAMP-2A 1:100 in filtered 0.1% BSA in PBS). Layer a Petri plate with Parafilm, place the cover slips on it (cells facing up) and put 25 µl of diluted primary antibody on top; incubate in a humidified chamber at room temperature for 1 h.
Extensively wash the cover slips with PBS (by successive immersion of cover slips held by blunt-end forceps in beakers containing PBS).
Incubate the cover slips with 25 µl of the fluorophore-conjugated secondary antibody (diluted in filtered 0.1% BSA in PBS) for 30 min at room temperature, as described in step 5.
After extensive washes (as described in step 6), incubate the cover slips in 25 µl of the other primary antibody (mouse IgM anti-hsc70) diluted 1:150 in filtered 0.1% BSA in PBS, under the same conditions as in step 5.
After extensive washes (as described in step 6), incubate the cover slips in 25 µl of the fluorophore-conjugated secondary antibody (diluted in filtered 0.1% BSA in PBS) for 30 min at room temperature as described in step 5, wash (as described in step 5) and mount by placing them (cells facing down) on top of 15 µl of DAPI (4’,6-diamidino-2-phenylindole)-containing anti-fade mounting medium (Invitrogen, P-7481) spotted on a glass slide, and seal with nail polish to prevent drying.
-
Visualize the slides using a fluorescence microscope (Axiovert 200, Carl Zeiss Ltd.), and deconvolute the captured images using the manufacturer’s software.
Note: Standard immunofluorescence control slides (incubated only with secondary antibodies, or incubated only with one primary antibody and the secondary for the other primary antibody) should be included.
The following parameters can be analyzed as direct indication of CMA activity in the cultured cells:
Colocalization of the two antibodies can be quantified using the Just Another Colocalization Plugin of the ImageJ program (NIH) after setting the appropriate threshold.
The mean distance of the vesicles positive for each antibody to the nucleus is calculated with the “straight lane tool” and the “analyze particles” function of the ImageJ program, by drawing straight lines from the most distant vesicle positive for each antibody to the nucleus and computing the particle distribution (distance and density). An average of six different radial lines per cell and 20 cells per field is usually calculated to determine changes in the intracellular distribution of CMA-active lysosomes (Fig. 2). Although changes in lysosomal localization may be due to many different reasons (e.g., alterations in vesicular trafficking, problems with microtubule polymerization), decreased distance to the nuclear region of the CMA-active lysosomes associated with positive values in any of the other procedures described in this work are good support for CMA activation.
Immunogold and electron microscopy for CMA-active lysosomes
Immunogold staining for LAMP-2A and hsc70 in tissue sections, cultured cells or in isolated lysosomes can also be used to assess changes in the amount of CMA-active lysosomes, and consequently in CMA activity (Cuervo et al., 1997; Cuervo et al., 1995b).
Fix tissue, cultured cells or isolated lysosomes in 4% paraformaldehyde, 0.05% glutaraldehyde and 0.1 M cacodylate in 0.25 M sucrose and process for electron microscopy analysis following standard procedures, with the exception that the sections should be subjected to only one staining step to minimize masking of the gold particles (Cuervo et al., 1997; Cuervo et al., 1995b) (see also the chapter by Ylä-Anttila et al., in this volume).
Perform immunogold staining on ultra thin sections mounted onto copper grids. Incubate the grids with the anti-LAMP-2A and anti-hsc70 antibodies (diluted 1:100) for 8–10 h at room temperature in a humidified chamber, followed by incubation with different-sized gold-conjugated secondary antibodies (Electron microscopy sciences, EM grade) (1:100) for 2 h at room temperature.
Rinse the grids extensively in water.
Negatively stain with 1% uranyl acetate.
-
Capture images using a JEOL 100CX II transmission electron microscope at 80kV.
Note: Required control samples are the same as indicated for immunofluorescence.
Morphometric analysis of digital images of the sections can be done using ImageJ after drawing the lysosomal profiles, and after applying the “clearing outside” function determining the “number of particles” per lysosome for each size gold particle and the number of lysosomes containing both sizes of gold particles.
In vitro assay to measure translocation of CMA substrates
The most unequivocal method to measure CMA activity is by monitoring the direct translocation of known CMA substrates in lysosomes isolated from the tissues or cultured cells of interest. This method requires the isolation of intact highly purified CMA-active lysosomes from these samples (as described in the Isolation of lysosomes section). The light mitochondria/lysosomal fraction proposed as an alternative to purified lysosomes for the immunoblot assays is not usually a good replacement for purified lysosomes in these assays due to the high consumption of ATP by the contaminant mitochondria in the fraction and the difficulty to normalize for mitochondria content in these assays. The ideal fraction for these assays is that of lysosomes enriched in hsc70 in their lumen (CMA+ lysosomes), although a pool of secondary lysosomes with differing hsc70 content can also be used for transport assays (Cuervo et al., 1997).
The purity of the lysosomal fraction and the integrity of the lysosomal membrane are both critical for proper assessment of CMA substrate translocation (Storrie and Madden, 1990). Thus, if lysosomal enzymes leak from the lumen during the experiment, they could degrade the substrate proteins outside lysosomes and provide the erroneous idea that this degradation is taking place after lysosomal translocation. Accurate measurements can only be attained with a tight control of lysosomal membrane integrity and of contaminant components in the fraction as described before (Isolation of lysosomes section). Despite the exorbitant cost of Metrizamide, the density medium used for the isolation of CMA-competent lysosomes, we still strongly recommend its use as this is the density medium that guarantees higher purity and lower lysosomal breakage during isolation (researchers are strongly discouraged from the use of sucrose as a density medium as it is actively transported into lysosomes and results in a high percentage of lysosomal breakage by hyperosmotic shock). Likewise, a rigorous control of the osmolarity of the incubation medium and all the additions to the medium is required to preserve lysosomal integrity during the incubation.
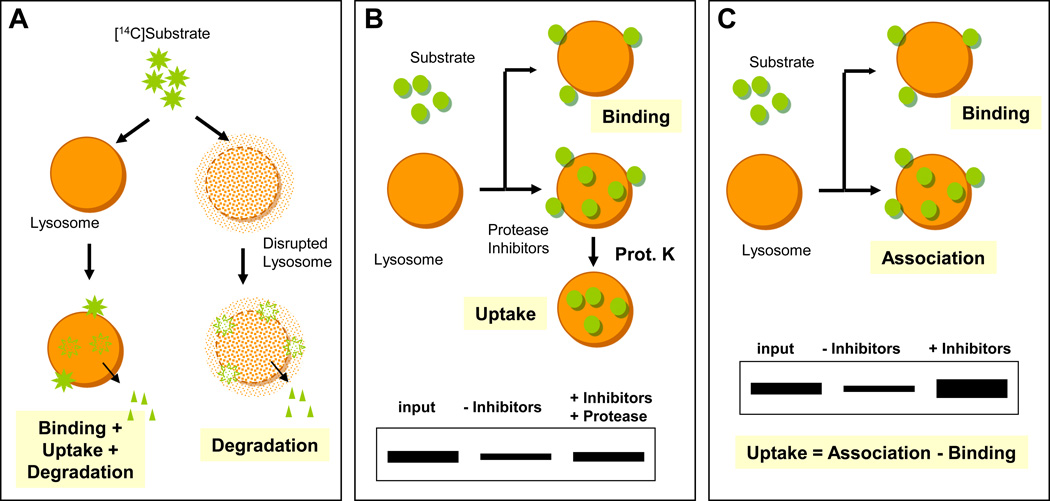
There are two procedures to track CMA substrate translocation into lysosomes. We routinely apply both methods as they allow analyzing different steps in CMA. Measurement of the degradation of radiolabeled CMA substrates (e.g., [14C]GAPDH) by isolated lysosomes recapitulates the three main lysosomal steps of CMA: binding, translocation and proteolysis (Terlecky and Dice, 1993) (Fig. 3A). Parallel experiments with lysosomes disrupted by hypotonic shock (to allow free access of the enzymes to the CMA substrate) permits determining possible changes in proteolysis independent of binding/uptake. This method is quantitatively accurate and the use of a 96-well plate-based filtration device allows rapid processing of a large number of samples (Terlecky and Dice, 1993).
Figure 3. In vitro assays for the direct quantification of CMA.
A. Incubation of intact or broken lysosomes with radiolabeled CMA substrates allows quantification of the amount of protein processed into soluble amino acids (degradation). In intact lysosomes, substrates need to bind to the lysosomal membrane and translocate before they can be degraded. B-C. Incubation of CMA substrates with intact lysosomes treated or not with protease inhibitors allows determination of the amount of substrate bound to the lysosomal membrane via immunoblot against the substrate after collecting the lysosomes by centrifugation. The amount of substrate translocated into the lysosomal lumen can be calculated in lysosomes treated with protease inhibitors after degradation of the substrate bound to the cytosolic side of the lysosomal membrane with an exogenous protease (B) or by subtracting the amount of “bound” substrate from the total amount of substrate associated with the protease-inhibited lysosomes (C).
Radiolabeling of CMA substrates
Purified CMA substrates are radiolabeled with [14C]formaldehyde by reductive methylation. We routinely label glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Sigma-Aldrich G5262) and ribonuclease A (RNase A: Rockland Immunochemicals, Inc., MB-113-0005) because both are commercially available as purified proteins.
Dissolve the protein in reaction buffer (10 mM MES, pH 7) to a final concentration of 3 mg/ml.
Add [14C]formaldehyde (250 µCi) (PerkinElmer NEC039H001MC) and sodium cyanoborohydrate (Sigma-Aldrich, 156159) (final concentration of 1.8 mg/ml in reaction buffer) and incubate this reaction mixture in a final volume of 500 µl at 25°C for 1 h.
Separate the radiolabeled protein from the unincorporated radioisotope by gel filtration through an appropriate Sephadex matrix (according to the size of the protein). We routinely use mini-spin columns packed with the matrix (we prepare 1 ml columns with insulin syringes filled with the matrix, but there are also some commercially available alternatives (e.g., Pierce 89849) previously blocked with 20 mg/ml BSA (5 vol, for 30 min at 25°C) (to prevent nonspecific binding) and equilibrated with the reaction buffer (10 vol). Spinning time is adjusted depending on the protein of interest and the characteristics of the mini-spin column (for most mini-spin columns and proteins in the 100-30kDa range, the spin time varies from 1 to 5 minutes).
Collect the eluted radiolabeled protein in separate aliquots.
Measure the amount of radiolabeled protein and free radioisotope in each aliquot by determining the radioactivity associated with the acid-precipitable fraction (radiolabeled protein) and acid-soluble fraction (free radioisotope) using TCA precipitation (as described in the Pulse and chase experiments section).
Protein degradation with isolated lysosomes
Incubate freshly isolated intact lysosomes (25 µg protein in 10 µl final volume after dilution in proteolysis buffer (10 mM 3-(N-morpholino) propanesulfonic acid (MOPS), pH 7.3, 0.3 M sucrose, 5.4 µM cysteine, 1 mM DTT) with 10 µl of radiolabeled protein (260 nM, 2,000 dpm/µl in the same buffer), 1 µl of the 6× energy-regenerating system (10 mM MgCl2, 10 mM ATP, 2 mM phosphocreatine (Sigma-Aldrich, P-6502), 50g/ml creatinephosphokinase (Sigma-Aldrich, C-3755) and 10 µg/ml of GST-hsc70 or hsc70 purified from bovine brain or liver (as described previously (Welch and Feramisco, 1985)) in a final volume of 60 µl (adjusted with proteolysis buffer) for 30 min at 37°C in a 0.22-µm 96-well filter plate (previously wet with sterile water for 10 min at room temperature and rinsed out).
Include a blank well containing all the reagents except for the lysosomes, to account for the amount of protein spontaneously cleaved, and the possible contaminant amount of free radiolabeled amino acids present in the purified labeled protein fraction.
Stop the reaction by adding TCA (10% final concentration) and BSA to a final concentration of 0.5 mg/ml to favor protein precipitation.
After incubation at 4°C for at least 30 min, collect the acid-soluble flow-through using the Millipore multiscreen vacuum system (as described in the Pulse and chase experiments section) and transfer to 5 ml vials, add scintillation liquid and count in a liquid scintillation counter.
Proteolysis is calculated as the amount of acid-precipitable radioactivity (protein) transformed into acid-soluble radioactivity (amino acids and small peptides) at the end of the incubation: [((dpm flow-through sample – dpm flow-through blank)/dpm pellet at time 0)*100].
Protease protection assay
The previous assay measures proteolysis of substrates translocated into lysosomes by CMA and recapitulates thus binding, uptake and translocation. However, individual CMA steps cannot be separately analyzed by this procedure. To dissect the two initial steps of CMA, a second in vitro assay with isolated lysosomes was developed in which association of substrate proteins to lysosomes is quantified by immunoblot (Aniento et al., 1993). The original version of this assay was based on the protease-protection assays widely used for the study of translocation of proteins into different organelles (Fig. 3B). Briefly, incubation of substrates with intact lysosomes results in their translocation and rapid degradation in the lumen. Consequently, when pulled down, the only lysosome-associated substrate protein is that bound to the cytosolic side of the lysosomal membrane. However, if the substrate proteins are incubated with lysosomes previously treated with protease inhibitors, the translocated protein remains inside the lumen. To quantify the protein present in the lumen of the lysosomes, an exogenous protease is added. The protease degrades the substrate bound to the cytosolic side of the lysosomal membrane but cannot access the one present in the lumen. A recent variation of this experiment (to avoid problems associated with resistance to cleavage by the exogenous protease) has been proposed in which the amount of internalized protein is calculated by subtracting the amount of “bound” substrate from the amount of substrate that is associated with lysosomes treated with protease inhibitors (bound + internalized) (Fig. 3C) (Salvador et al., 2000). Finally, binding and uptake can also be distinguished by modifying the incubation temperature. At temperatures below 10°C binding occurs but substrates do not translocate, whereas at temperatures above this both binding and uptake via CMA are coupled (Aniento et al., 1993). For all these assays, it is essential to include strict controls with lysosomes incubated alone (to account for any endogenous lysosomal protein recognized by the antibody against the substrate), lysosomes in which the membrane is disrupted with detergent (to demonstrate that the exogenous protease can degrade the substrate if access is allowed), and incubations in the presence of other CMA substrate and non-substrate proteins (to account for any translocation not mediated by CMA). To control for the amount of degradation of substrate due to lysed or leaky lysosomes, the same amount of lysosomes added per reaction should be centrifuged (25,000 g for 5 min at 4°C) and the supernatant fraction (that will contain the enzymes leaking from the lysosomes) should be incubated with the substrate protein under the same conditions.
Incubate freshly isolated intact lysosomes with a (x100) protease inhibitor cocktail (10 mM leupeptin, 10 mM AEBSF, 1 mM pepstatin and 100 mM EDTA) for 10 min on ice.
In 0.5-ml microcentrifuge tubes, add freshly isolated intact lysosomes (100 µg protein) pretreated or not with protease inhibitor, along with 10–50 µg of CMA substrate (GAPDH, RNase A or any other protein of interest), 5 µl of (6×) energy regenerating system and 10 µg/ml GST-hsc70 or purified brain or liver bovine hsc70 in 30 µl final volume of incubation buffer (10 mM MOPS, pH 7.3, 0.3 M sucrose).
Incubate the samples for 20 min at 37°C.
At the end of the incubation, cool down half of the tubes pretreated with protease inhibitors on ice (1 min) and add proteinase K (5 µl of a 1 mg/ml stock in 1mM CaCl2, −50 mM Tris-HCl pH 8). The proteinase K solution should be made fresh or kept frozen to prevent self-degradation.
Incubate the samples on ice for 10 min, add AEBSF (5 µl) and centrifuge all samples at 25,000 g for 5 min at 4°C.
Aspirate the supernatant fractions and wash the pellet fractions twice with 100 µl of incubation buffer to eliminate any protein bound non-specifically to the lysosome surface.
Resuspend the final pellets in Laemmli buffer (Laemmli, 1970) with protease inhibitors, boil for 5 min at 95°C and perform SDS-PAGE and immunoblot with antibodies specific for the substrate of choice. Include in the gel a lane containing 1/10 of the amount of substrate added to the incubation to use it as reference in the calculations of the amount of substrate bound or translocated.
Calculations: Using densitometry of the immunoblotted membranes calculate substrate binding and uptake as follows:
binding = the percentage of total added substrate associated with lysosomes untreated with protease inhibitors
association = the percentage of substrate recovered in lysosomes treated with protease inhibitors
uptake = either the difference between association and binding or the percentage of substrate associated with lysosomes treated with protease inhibitors after proteinase K treatment.
Although lysosomal internalization of proteins by other autophagic pathways (macroautophagy or microautophagy) cannot be reproduced in vitro, at least in the conditions used in this assay, as new proteins are considered as possible CMA substrate candidates we cannot discard the possibility of the existence of other yet to be identified mechanisms for direct translocation of soluble proteins in lysosomes. Consequently, to confirm that the binding/uptake/degradation of putative CMA substrates assayed by the two methods described above is certainly occurring via CMA, it is advisable that one or both of the following assays are performed:
Competition assays with well-characterized CMA substrates: If the putative substrate is translocated into lysosomes by CMA, addition of equimolar concentrations of GAPDH or RNase A (two well-characterized CMA substrates) to any of the incubations indicated above should decrease binding, uptake and degradation of the tested substrate (as they compete for the same lysosomal machinery for degradation) (Cuervo et al., 1994; Terlecky and Dice, 1993).
Blockage of the CMA receptor: Binding of CMA substrates to the cytosolic tail of LAMP-2A is required for their lysosomal translocation. Consequently, pre-incubation of the lysosomes added in the above assays with the specific antibody against the cytosolic tail of LAMP-2A or supplementation of the incubation medium with a peptide of the same amino acid composition as the cytosolic tail of LAMP-2A should reduce CMA of the putative substrate (Cuervo and Dice, 1996).
Concluding Remarks
In conclusion, using the battery of assays described in this work it is possible to evaluate changes in CMA activity in cultured cells and different tissues from rodents under different physiological and pathological conditions. The most accurate assays to measure possible changes in CMA activity in different samples are those reproducing in vitro the translocation of known CMA substrates in lysosomes, as the activity of other proteolytic pathways will not be measured in those assays. However, the large amount of cultured cells required for the isolation of lysosomes active for CMA and the training (to some extent) required to become proficient in these procedures (the characteristic instability of the lysosomal membrane upon isolation makes it necessary to perform all of these procedures rapidly in minimal amount of time) relegates often the in vitro uptake assays as a final confirmatory assay once evidence suggestive of changes in CMA have been gathered using several of the other assays.
Future efforts are oriented to the development of image-based reporters incorporated in cells or in animals (i.e., transgenic mouse models with the CMA reporter expressed in all tissues) that allow tracking changes in CMA through changes in the association of the reporter with lysosomes and/or its degradation in this cellular compartment.
Acknowledgements
Work in our laboratory is supported by grants from the National Institute of Health AG021904, AG031782 and DK041918 and by a Glenn Award.
References
- Agarraberes F, et al. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniento F, et al. Uptake and degradation of glyceraldehyde-3- phosphate dehydrogenase by rat liver lysosomes. J Biol Chem. 1993;268:10463–10470. [PubMed] [Google Scholar]
- Auteri J, et al. Regulation of intracellular protein degradation in IMR- 90 human diploid fibroblasts. J Cell Physiol. 1983;115:159–166. doi: 10.1002/jcp.1041150210. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay U, et al. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008 doi: 10.1128/MCB.02070-07. [e-pub ahead of publication] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, et al. The Vid vesicle to vacuole trafficking event requires components of the SNARE membrane fusion machinery. J. Biol. Chem. 2003;278:25688–25699. doi: 10.1074/jbc.M210549200. [DOI] [PubMed] [Google Scholar]
- Carlsson SR, et al. Isolation and characterization of human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2. J Biol Chem. 1988;263:18911–18919. [PubMed] [Google Scholar]
- Chiang H, et al. A role for a 70 kDa heat shock protein in lysosomal degradation of intracellular protein. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- Cuervo A. Autophagy: in sickness and in health. Trends Cell Biol. 2004a;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Cuervo A, Dice J. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- Cuervo A, Dice J. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000a;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- Cuervo A, Dice J. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000b;113:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- Cuervo A, et al. A lysosomal population responsible for the hsc73-mediated degradation of cytosolic proteins in lysosomes. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- Cuervo A, et al. Direct lysosomal uptake of α2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 1999;55:529–245. doi: 10.1046/j.1523-1755.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- Cuervo A, et al. IκB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell. 1998a;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A, et al. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995a;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- Cuervo A, et al. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995b;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- Cuervo A, et al. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J Biol Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- Cuervo AM. Autophagy: many pathways to the same end. Mol Cell Biochem. 2004b;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 2000c;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, et al. IκB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell. 1998b;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, et al. Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 2003;22:12–19. doi: 10.1093/emboj/cdg002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, et al. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Dice J. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem. 1982;257:14624–14627. [PubMed] [Google Scholar]
- Dice J. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Dice J. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- Eskelinen E, et al. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002;13:3355–3368. doi: 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E, et al. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell. 2004;15:3132–3145. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL, et al. Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic. 2005;6:1058–1061. doi: 10.1111/j.1600-0854.2005.00337.x. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL, et al. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- Finn P, et al. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005;1:141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem. 2005;280:25864–25870. doi: 10.1074/jbc.M502456200. [DOI] [PubMed] [Google Scholar]
- Fuertes G, et al. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem. J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K, et al. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007:26. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, et al. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R, et al. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R, et al. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- Klionsky D, et al. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Konecki DS, et al. An alternatively spliced form of the human lysosome-associated membrane protein-2 gene is expressed in a tissue-specific manner. Biochem Biophys Res Comm. 1995;215:757–767. doi: 10.1006/bbrc.1995.2528. [DOI] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Majeski A, Dice J. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, et al. Dopamine-modified α-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzella L, et al. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A, et al. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2420–2434. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Massey A, et al. Autophagic defects in aging: looking for an “emergency exit”? Cell Cycle. 2006a;5:1292–1296. doi: 10.4161/cc.5.12.2865. [DOI] [PubMed] [Google Scholar]
- Massey A, et al. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006b;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- Massey AC, et al. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006c;103:5905–5910. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y, et al. A rapid and simplified method for the preparation of lysosomal membranes from rat liver. J Biochem. 1983;93:547–556. [PubMed] [Google Scholar]
- Salvador N, et al. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. Journal of Biological Chemistry. 2000;275:27447–27456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky D. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sooparb S, et al. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- Storrie B, Madden E. Isolation of subcellular organelles. Meth Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, et al. Accumulation of autophagic vacuoles and cardiomyopathy in Lamp-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- Terlecky S, et al. Protein and peptide binding and stimulation of in vitro lysosomal proteolysis by the 73-kDa heat shock cognate protein. J Biol Chem. 1992;267:9202–9209. [PubMed] [Google Scholar]
- Terlecky S, Dice J. Polypeptide import and degradation by isolated lysosomes. J Biol Chem. 1993;268:23490–23495. [PubMed] [Google Scholar]
- Wattiaux R, et al. Isolation of rat liver lysosomes by isopycnic centrifugation in a metrizamide gradient. J Cell Biol. 1978;78:349–368. doi: 10.1083/jcb.78.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch W, Feramisco J. Rapid purification of mammalian 70,000-dalton stress proteins: affinity of the proteins for nucleotides. Mol Cell Biol. 1985;5:1229–1237. doi: 10.1128/mcb.5.6.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing S, et al. Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem J. 1991;275:165–169. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008 doi: 10.1038/nm.1851. [e-pub, ahead of publication] [DOI] [PMC free article] [PubMed] [Google Scholar]