Abstract
Protein prenylation is a ubiquitous covalent post-translational modification found in all eukaryotic cells, comprising attachment of either a farnesyl or a geranylgeranyl isoprenoid. It is essential for the proper cellular activity of numerous proteins, including Ras family GTPases and heterotrimeric G-proteins. Inhibition of prenylation has been extensively investigated to suppress the activity of oncogenic Ras proteins to achieve antitumor activity. Here, we review the biochemistry of the prenyltransferase enzymes and numerous isoprenoid analogs synthesized to investigate various aspects of prenylation and prenyltransferases. We also give an account of the current status of prenyltransferase inhibitors as potential therapeutics against several diseases including cancers, progeria, aging, parasitic diseases, and bacterial and viral infections. Finally, we discuss recent progress in utilizing protein prenylation for site-specific protein labeling for various biotechnology applications.
Protein Prenylation
Protein prenylation was first discovered in fungi in 1978,1 and almost 10 years later, the first prenylated protein in mammalian cells, farnesylated lamin B, was detected.2,3 Since then, this modification has been studied extensively due to its importance for the proper cellular activity of numerous proteins. Protein prenylation is an irreversible covalent post-translational modification found in all eukaryotic cells, comprising farnesylation and geranylgeranylation. Three prenyltransferase enzymes catalyze this modification. Farnesyltransferase (FTase) and geranylgeranyltransferase type 1 (GGTase-I) catalyze attachment of a single farnesyl (15 carbon) or geranylgeranyl (20 carbon) isoprenoid group, respectively, to a cysteine residue located in a C-terminal consensus sequence commonly known as “CaaX box” (Figure 1), where “C” is cysteine, “a” generally represents an aliphatic amino acid, and the “X” residue is largely responsible for determining which isoprenoid is attached to the protein target.4 Geranylgeranyltransferase type 2 (GGTase-II or Rab geranylgeranyltransferase) catalyzes the addition of two geranylgeranyl groups to two cysteine residues in sequences such as CXC or CCXX close to the C-terminus of Rab proteins (Figure 1).4
Figure 1.
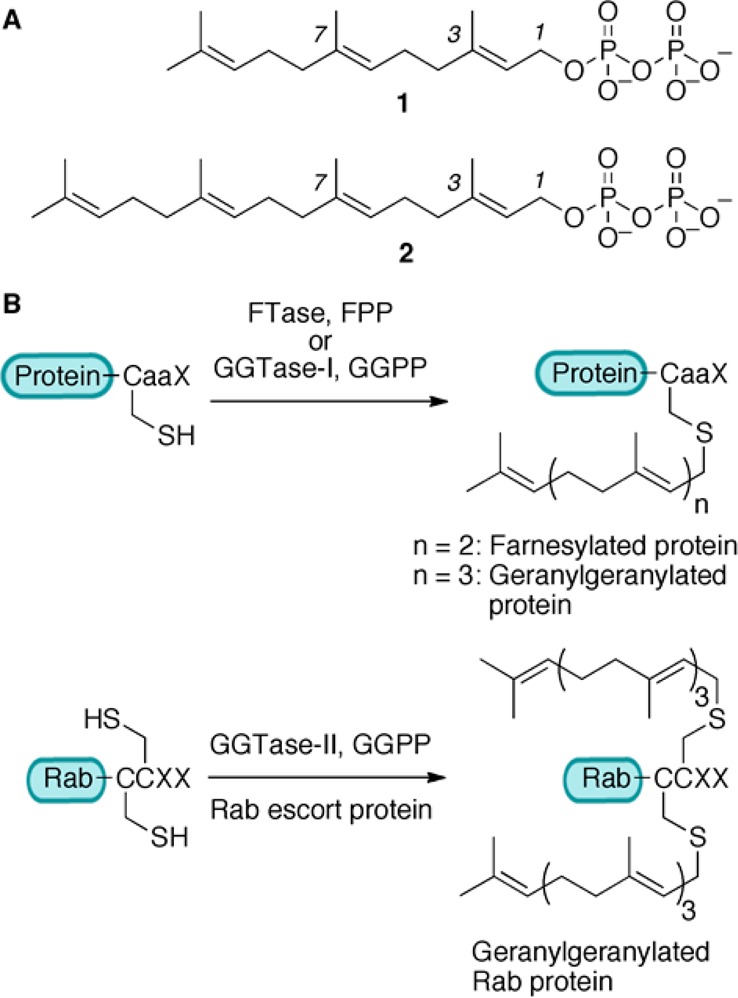
(A) Structures of 1 (farnesyl diphosphate, FPP) and 2 (geranylgeranyl diphosphate, GGPP). (B) Reactions catalyzed by prenyltransferase enzymes.
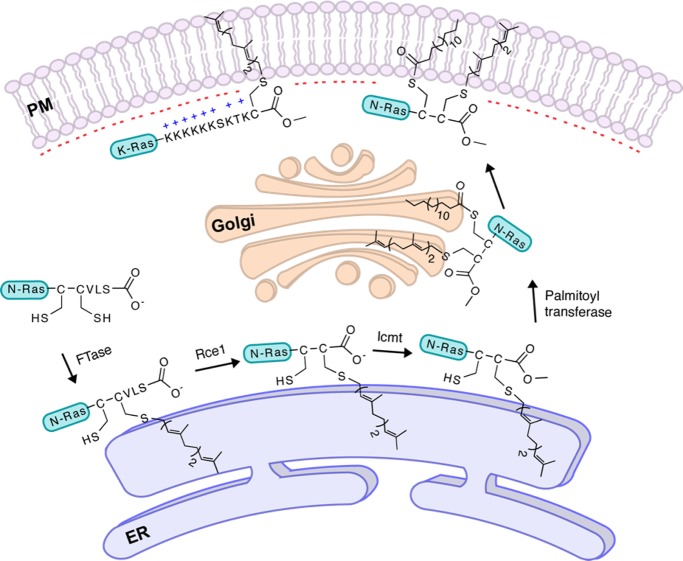
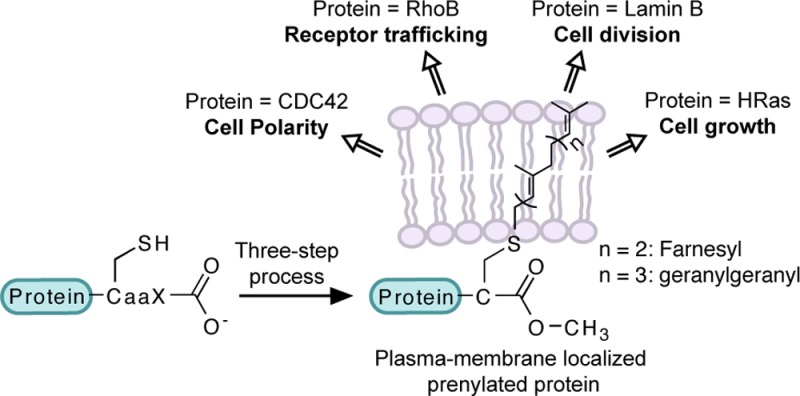
Proteins prenylated with FTase and GGTase-I typically undergo two additional processing steps.5 First, the C-terminal aaX tripeptide is cleaved from the newly prenylated CaaX protein by an endoprotease, either Ras-converting enzyme 1 (Rce1p) or Ste24p (Figure 2). This is followed by methylation of the prenylcysteine residue at the new C-terminus by isoprenylcysteine carboxylmethyltransferase (Icmt, Figure 2). This three-step process increases protein hydrophobicity and often leads to plasma membrane association.5 However, it is been noted that prenylation alone is not sufficient to cause stable membrane association.6 Either the presence of a polybasic domain upstream of the CaaX box (as found in K-Ras4B, for example) or additional lipid modification such as palmitoylation at one or two cysteine residues (such as in H-Ras) supports more stable membrane localization of prenylated proteins (Figure 2).
Figure 2.
Three-step prenylation processing of proteins. Proteins undergo farnesylation and proteolytic cleavage of aaX residues, followed by carboxymethylation, and then get localized at the plasma membrane. Some proteins, shown here N-Ras, undergo palmitoylation and then localize to plasma membrane, while other proteins, shown here K-Ras, have a polybasic sequence upstream of the “CaaX box” facilitating membrane localization.
In normal healthy cells, the function of the Ras superfamily GTPases in diverse cellular processes, such as growth, cell movement, and protein trafficking, critically depends on their presence in the correct cellular membrane.7 Prenylation serves as the first critical step for membrane targeting and binding, as well as mediating protein–protein interactions of a large number of these proteins; heterotrimeric G-proteins also require prenylation for activity.8 Significant interest in studying protein prenylation originally stemmed from the finding that this modification was necessary to maintain malignant activity of oncogenic Ras proteins.9 Inhibition of prenylation has provided an attractive strategy to inhibit oncogenic activity of Ras and achieve antitumor effects. In recent years, however, robust clinical activity against Ras-dependent tumors using prenyltransferase inhibitors has not been generally achieved contrary to the successful preclinical studies.10 Currently, it is unclear why some tumors are sensitive to these inhibitors and others are not. One important conclusion from those studies is that it is essential to completely define the prenylated proteome, and in particular, to identify which proteins are impacted by therapeutic levels of prenyltransferase inhibitors. This review first summarizes studies probing the enzymology of prenyltransferases. Next, it focuses on experiments that probe the specificity of prenyltransferases and work directed at the global identification of the prenylated proteome. A subsequent section gives a glimpse of prenyltransferase inhibitors as anticancer agents and their emerging applications in therapies against progeria and parasitic diseases. Finally, recent advances in utilizing protein prenylation for biotechnological applications, including site-specific protein labeling, are discussed.
Mechanism of Protein Prenylation
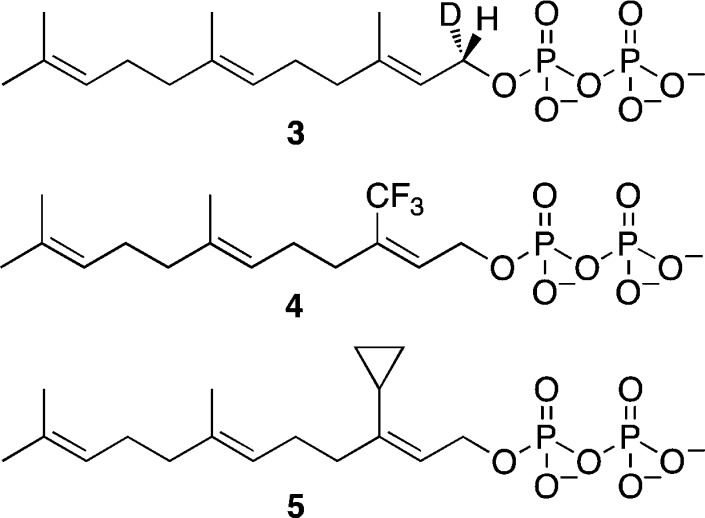
Protein prenylation is catalyzed by three distinct prenyltransferase enzymes that all exist as heterodimers and have very similar topologies (Figure 3A,B). While FTase and GGTase-I share a common α-subunit, the α-subunit of rat GGTase-II has only 22% sequence similarity to the rat FTase α-subunit.11 In rat-derived enzymes, the β-subunit of FTase is only 25% and 32% identical to that of GGTase-I and GGTase-II, respectively.12 The reaction catalyzed by GGTase-II requires an additional escort protein for activity. Most mechanistic analyses have focused on farnesylation with a more limited number of studies probing geranylgeranylation. Early kinetic analysis demonstrated that farnesylation proceeds via an ordered mechanism in which FPP binds first.13,14 After binary complex formation occurs, the CaaX-box-containing substrate binds, and C–S bond formation occurs. At that point, a new FPP molecule binds, while the farnesylated protein remains bound, followed by product dissociation either prior to or concerted with binding of a new CaaX-box substrate protein. All of these intermediates have been observed crystallographically, providing a clear view of the events occurring during catalysis; interestingly, minimal differences in the protein conformation are observed in these different structures.15 Single turnover kinetic experiments and calculations suggest that a conformational change in the enzyme occurs prior to C–S bond formation although no evidence for this has been noted in any of the crystal structures solved to date.16,17 Stereochemical analysis of the enzymatic process using deuterated isotopomers of FPP (3, Figure 4) revealed that the reaction proceeds with clean inversion of configuration of stereochemistry at C-1 of the isoprenoid18,19 suggesting that attack of the sulfur nucleophile is concerted with departure of the diphosphate leaving group. Work with isoprenoid analogs incorporating electron withdrawing fluorine substituents (4) provides evidence that the transition state involves some carbocationic character20 although analogs including 5 designed to trap such intermediates failed to do so.21 Kinetic isotope effect measurements with both 2H- and 13C-isotopomers suggest a transition state that involves participation of the incoming sulfur nucleophile with significant development of positive charge at C-1 of the isoprenoid (Figure 5A–C);22−24 QM/MM computational experiments are in accord with this since no evidence for a discrete carbocationic species was observed.25 For efficient catalysis, kinetic experiments indicate that the enzyme activates the sulfur nucleophile as a Zn-thiolate.26,27 Such a species is consistent with what has been observed crystallographically as well as via EXAFS spectroscopy.28
Figure 3.
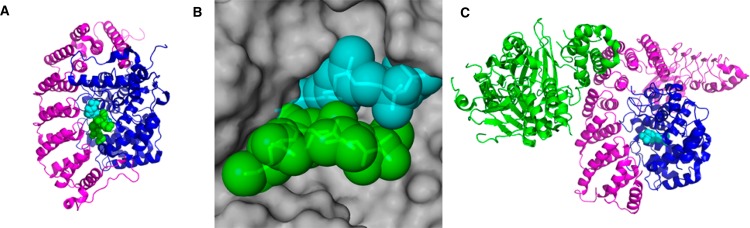
Crystal structures of prenyltransferase enzymes. (A) Crystal structure of FTase in complex with a nonhydrolyzable FPP analog and a peptide substrate based on KRas-4B (PDB 1D8D): magenta, α-subunit; blue, β-subunit; cyan, isoprenoid analog; green, CaaX peptide. (B) Binding pocket of FPP showing interaction of protein and isoprenoid substrates over a large surface area: gray, space-fill structure of β-subunit of FTase; cyan, isoprenoid analog; green, CaaX peptide. (C) Crystal structure of GGTase-II in complex with Rab escort protein and FPP (PDB 1LTX): magenta, GGTase-II α-subunit; blue, GGTase-II β-subunit; green, Rab escort protein; cyan, FPP.
Figure 4.
Structures of isoprenoid analogs used to probe mechanism of prenyltransferase enzymes.
Figure 5.
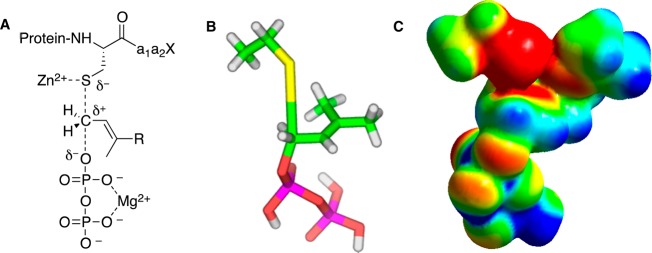
Key features of catalysis by protein farnesyltransferase. (A) Schematic representation of transition state showing thiol activation by Zn2+, diphosphate stabilization by Mg2+, and partial bonding to leaving group and incoming nucleophile (adapted from ref (23)). (B) Structural model for transition state based on kinetic isotope effect measurements and DFT calculations. The model reaction used for computation (shown in these images) employed ethanethiol and dimethylallyl diphosphate. (C) Electrostatic potential map of transition state based on the same model shown in panel B (images B and C images adapted from ref (24)). Color scheme for B: carbon (green), hydrogen (white), oxygen (red), phosphorus (magenta), and sulfur (yellow). Color scheme for C: red represents more negative potential, blue represents less negative potential, and green is intermediate.
The enzymology of GGTase-I is similar to that FTase. The enzyme proceeds via an ordered sequential kinetic mechanism,29 and the reaction proceeds with inversion of stereochemistry at the C-1 position of the isoprenoid.30 Interestingly, GGTase-I does not require Mg2+ for catalysis due to the presence of a lysine residue in the active site, which assists with the departure of the diphosphate leaving group in lieu of the divalent metal.31 The overall structure of GGTase-I12 is very similar to that of FTase although the former has a deeper isoprenoid binding pocket to accommodate the larger substrate.32 The process catalyzed by GGTase-II is more complex since it involves participation of an additional escort protein, Rab escort protein (REP),33 that both recruits the substrate proteins and traffics them following prenylation.33,34 Typically two cysteine residues (CC or CXC) are prenylated in a processive fashion. Several crystal structures of these enzymes have been reported.35,36 Unlike FTase and GGTase-I, GGTase-II has a more extended binding site for the protein target and interacts with the latter over a larger surface rendering short peptides inefficient as substrates. Finally, it should be noted that while the majority of the above enzymological studies have been carried out in vitro, the situation may be more complex in vivo. For example, recent work suggests that some Ras-like proteins may associate with putative chaperone proteins prior to prenylation; hence the true substrates of prenyltransferases in vivo may be protein complexes instead of simply the polypeptide substrate alone.37 This remains an active area of inquiry.
Peptide Substrate Specificity
Early work with prenyltransferases suggested that the X residue in CaaX box determines whether the protein is farnesylated or geranylgeranylated and that CaaX sequences with the X residue being alanine, serine, methionine, or glutamine are preferred by FTase, whereas leucine, isoleucine, and phenylalanine are preferred by GGTase-I. However, these enzymes do not manifest mutually exclusive substrate specificity.38 Some of the examples of overlapping substrate specificity include K-Ras and Rho B proteins. The CVIM sequence from K-Ras protein is normally farnesylated in mammalian cells; however, it can be processed by GGTase-I when FTase is inhibited.10 Rho B, which has a CKVL motif at its C-terminus, is present in both farnesylated and geranylgeranylated forms in cells.10
The human protein database (SwissProt) indicates the presence of 756 unique proteins containing the Cxxx motif at their C-termini.39 Based on the available descriptions of sequence motifs recognized by prenyltransferase enzymes, Eisenhaber and co-workers developed an amino-acid sequence based predictor, the Prenylation Prediction Suite, and showed its utility by predicting which prenylated proteins would be preferentially affected in response to an enzyme-specific prenyltransferase inhibitor.40,41
While that predictive algorithm has proven to be useful for inferring the prenylation status of nonannotated proteins, it was designed using a limited number of known prenylated proteins; importantly, it does not correctly predict the prenylation efficiency of numerous CaaX-box sequences.42 Hence, expanding the training set should increase the accuracy of such bioinformatic methods. The process of testing a large number of sequences for activity against FTase has been possible since the enzyme accepts short peptides, including CaaX tetrapeptides, as substrates with affinity and reactivity comparable to full-length proteins.43,44 Fluorescence-based assays using dansylated peptides have been highly useful in this regard.45,46 Accordingly, several studies with small-scale and large-scale libraries of CaaX peptides have been conducted.47,48 In particular, Gibbs and co-workers utilized a library of 41 peptides with CaaL sequences and found that FTase efficiently processed a number of peptides having leucine at X position.49 Fierke and colleagues screened a library of 80 peptides with CVaX sequences and interestingly found that substrate recognition is not limited to the nature of the X residue.50 They reported that FTase is sensitive to both the size and hydrophobicity of the residue at the a2 position, and the nature of residue at the X position affects the selectivity at the a2 position. Later, the same group carried out an analysis of a library of small peptides representing the CaaX motif of 213 human proteins, and they observed that FTase could catalyze farnesylation of several CaaX sequences that were not computationally predicted to be FTase substrates, with multiple turnover reactivity.42 They also identified a large number of sequences that were processed with single turnover reactivity and noted that at least two of these sequences corresponded to a known in vivo FTase substrate. This indicated that some of the single turnover in vitro peptide substrates could potentially be multiple turnover substrates in a cellular context.51 Results from those studies have allowed for the development of improved bioinformatic programs, such as a FlexPepBind-based prediction protocol.39
Recently, Distefano and co-workers designed a method to synthesize solid-phase peptide libraries with free C-termini for studying the substrate specificity of prenyltransferase enzymes.52 Using this method, they created libraries incorporating 760 peptides (based on CVaX and CCaX sequences), and screening of these libraries revealed numerous sequences that could be processed by mammalian FTase, including sequences occurring in genomes of bacteria and viruses.53 This opens an exciting opportunity for therapeutic intervention against such proteins.
Isoprenoid Analogs
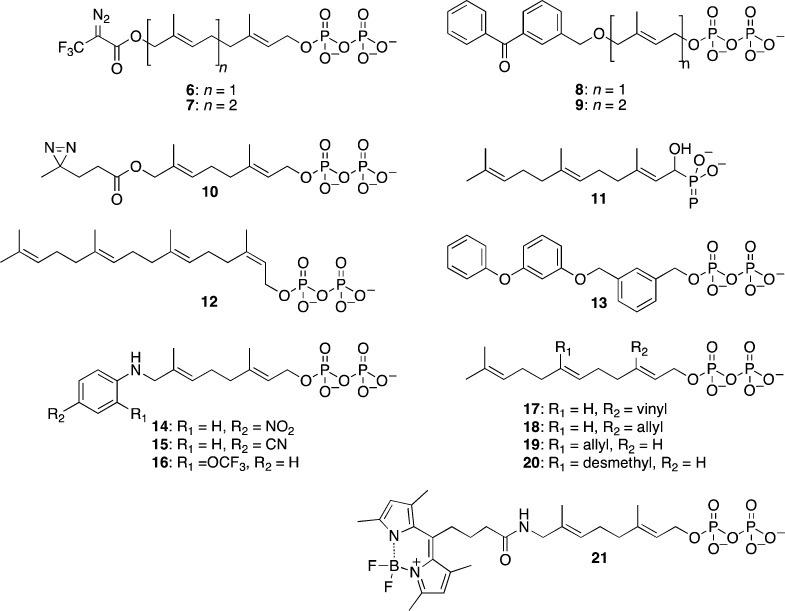
A large number of isoprenoid analogs containing various functionalities have been synthesized to study a variety of aspects of the prenylation reaction and prenyltransferases. Before the crystal structure of FTase was solved, photoaffinity probes such as compounds 6–9 (Figure 6) were used extensively to probe the structural features of yeast and mammalian variants of FTase and GGTase-I.29,54−61 These analogs revealed the role of the β-subunit of FTase and GGTase-I in the recognition and binding of isoprenoid substrates, as well as indicated differences between active site architecture of mammalian and yeast FTases. Later, similar experiments were also carried out with GGTase-II, which led to identification of proteins with which Rab5 interacts via the isoprenoid group.62 Interestingly, it was noted that while 8 was accepted as a substrate by FTase, 9, containing one more isoprenoid unit, was a potent inhibitor of yeast FTase.63,64 Recently, a new photoactive isoprenoid probe containing a diazirine group (10) was reported whose size more closely approximates that of FPP. Peptides incorporating that photoactive isoprenoid were used in cross-linking studies of Icmt.65 Phosphonate 11 and related analogs66 have been particularly useful for crystallographic studies that have revealed that the isoprenoid binds in an extended conformation and that several active site residues undergo rearrangement upon isoprenoid binding compared with the unliganded enzyme.67
Figure 6.
Structures of isoprenoid analogs used to study structure, mechanism, and isoprenoid substrate specificity of FTase and GGTase-I.
Recent studies with isoprenoid analogs have focused on investigating isoprenoid substrate specificity of prenyltransferase enzymes. Gibbs and co-workers synthesized a number of geometric isomers of all-trans FPP and GGPP (such as 12). They found that while most of the analogs were substrates to mammalian FTase, mammalian GGTase-I did not accept them as substrates. In fact, 12 was an inhibitor to GGTase-I, indicating more stringent specificity for the isoprenoid substrates for that enzyme.68 Spielmann and co-workers have reported substantial plasticity in the binding site of FTase. For example, analog 13, where all isoprene units were replaced with aryl groups, was an efficient FTase substrate.69 They reported that the anilinogeranyl-based isoprenoid analogs 14 and 15, which have 2–5 orders of magnitude less hydrophobicity compared with FPP, were substrates for FTase. Proteins modified with these analogs were processed by downstream enzymes Rce1 and Icmt; however, the resulting modified proteins were not biologically active, indicating the importance of increased hydrophobicity upon prenylation.70 Additionally, they noted that some of the anilinogeranyl-based analogs, such as 16, were substrates for FTase when a peptide based on K-Ras C-terminal sequence, dansyl-GCVIM, was used, but remarkably they were potent inhibitors of the enzyme when dansyl-GCVLS (IC50 of 16 was 3.0 nM), a sequence based on C-terminal of H-Ras, was used.71
A large number of analogs having substitutions at the 3- or 7-positions of FPP have been synthesized by Gibbs and co-workers. They concluded that subtle changes in the functionalities incorporated at these positions can lead to large and unexpected differences in incorporation efficiency. For example, they found that the 3-vinyl analog, 17, was a slow FTase substrate in cells, whereas the 3-allyl analog, 18, was an FTase inhibitor.72 During the screening of analogs against a library of eight CaaX sequences, 7-allyl analog 19 could farnesylate only the CVIM sequence, while 20 was an extremely efficient substrate to almost all the sequences in their library.73 These subtle differences likely reflect the fact that the protein and isoprenoid substrates interact with each other over a large surface area when bound to FTase (Figure 3B); thus small perturbations in one of the substrate structures require compensatory changes in the other substrate to obtain optimal complementarity.
Waldmann and co-workers described the synthesis of fluorescent isoprenoid analogs based on NBD and BODIPY groups and demonstrated the use of compound 21 in flow cytometry and imaging for analysis of uptake of these analogs in mammalian cells and zebrafish embryos.74 Coumarin-, anthranilate-, and dansyl-functionalized analogs have also been prepared for both in vitro and cell-based assays.75,76 Finally, several types of FPP analogs have also been explored as FTase inhibitors; however, they were not as successful as other classes of FTIs possibly due to nonselective inhibition of other FPP utilizing enzymes and difficulty in designing cell penetrable analogs containing pyrophosphate group of FPP.77,78
Analysis of the Prenylated Proteome
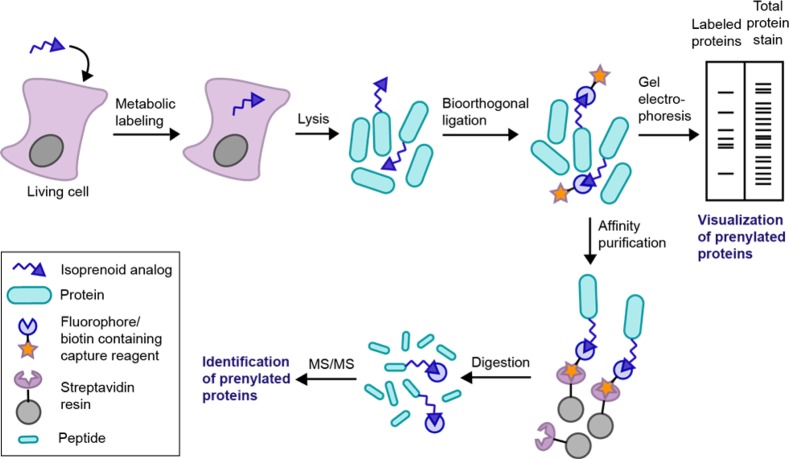
As noted above, there is considerable interest in identifying proteins that are prenylated in a cellular context in order to determine which prenyltransferase protein substrates have their prenylation status affected by FTIs. Chemical proteomic methods have been highly useful in this regard (Figure 7). In this approach, metabolic labeling of living cells is first carried out using isoprenoid analogs to tag prenylated proteins with a reporter group, such as an azide or alkyne. These tagged proteins are then functionalized using bioorthogonal reactions to install either a fluorophore for gel-based proteomic studies or a biotin moiety for enrichment of tagged proteins.
Figure 7.
Chemical proteomic strategy for analysis of the prenylated proteome.
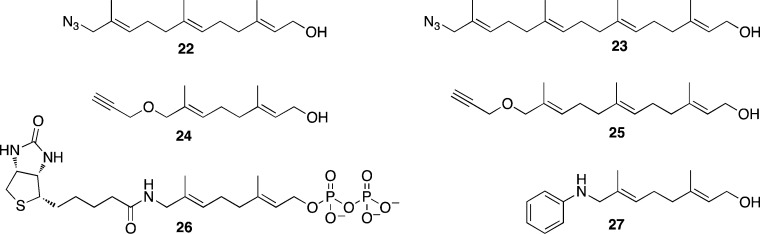
In 2004, Zhao and co-workers reported the first prenylomic study, wherein they employed azide-modified FPP analog 22 (Figure 8) for the metabolic labeling of farnesylated proteins.79 Chemoselective conjugation of labeled proteins to a biotinylated phosphine capture reagent using a Staudinger ligation reaction allowed for affinity purification and mass spectrometric identification of 18 farnesylated proteins. Tamanoi and co-workers incorporated an azide-modified analog of GGPP (23) into geranylgeranylated proteins and subsequently installed a fluorophore via Cu(I)-catalyzed click reaction on the azide-labeled proteins.80 They used pH fractionation coupled with narrow pH range 2D SDS-PAGE to achieve separation of low abundance geranylgeranylated proteins from the rest of the proteins in cells. LC-MS/MS analysis of some of the fluorescent protein spots led to identification of four substrates of GGTase-I and six substrates of GGTase-II. Berry et al. reported a method for detection and affinity purification of geranylgeranylated proteins utilizing an azide-modified GGPP analog and an alkyne capture reagent functionalized with a fluorophore as well as biotin.81
Figure 8.
Structures of isoprenoid analog used to analyze prenylated proteome.
Since alkyne-modified chemical reporters tend to give more sensitive and selective detection compared with their azido counterparts, the Distefano and Hang groups used alkyne-functionalized isoprenoid analogs for analysis of the prenylome.82,83 DeGraw et al. reported two alkyne-modified analogs for metabolic labeling: 24, which is a substrate for FTase,84 and 25, which is a substrate for both FTase and GGTase-I.85 The authors derivatized the proteins labeled by 24 with a fluorophore via the Cu(I)-catalyzed click reaction and separated proteins on a 2D gel. Six prenylated proteins were identified upon MS analysis of some of the fluorescent protein spots. In another study, metabolic labeling with 25 was carried out in the presence and absence of a farnesyltransferase inhibitor. The two corresponding protein samples were then reacted with two spectrally orthogonal azide-functionalized dyes and mixed together.86 This was followed by differential gel electrophoresis (DIGE) to facilitate visualization of several protein spots with altered levels of labeling in the presence of an FTI. LC-MS/MS analysis of some of the protein spots identified 8 proteins with decreased amounts of labeling and 11 proteins with increased amounts of labeling in response to the FTI treatment.
In recent years, several reports indicated that intracellular human pathogens, which lack prenylation machinery, translocate several effector proteins containing CaaX-box motifs into host cell cytosol.87−90 These proteins are prenylated using host cell prenyltransferase enzymes and later form membrane-bound organelles supporting replication of the pathogen.88,89 Hang and colleagues exploited analog 25 for investigating prenylation of a bacterial effector protein as well as immune effector proteins in host cells.83,91 They first detected intracellular prenylation of Salmonella T3SS effector protein SifA upon immunopurification and gel electrophoretic analysis of SifA metabolically labeled with 25 and conjugated to a fluorophore.83 Later they carried out profiling of prenylated proteins in macrophages91 using a Cu(I)-catalyzed click reaction of alkyne-tagged proteins with an azido-biotin reagent containing a chemically cleavable linker for enrichment and selective elution. Tandem mass spectrometric analysis of eluted proteins identified 17 prenylated proteins with high confidence and 5 proteins with medium confidence, along with many other candidate isoprenoid-modified proteins. During this analysis, they discovered isoform-specific farnesylation of zinc-finger antiviral protein (ZAP) and found that farnesylation of this protein was essential for increasing the antiviral activity of this immune effector protein.
Alternative approaches of prenylome analysis bypass the need for a bioorthogonal reaction. Alexandrov and colleagues employed a biotinylated isoprenoid, 26, for in vitro prenylation of proteins using either wt GGTase-II or engineered FTase and GGTase-I, to allow for subsequent enrichment using streptavidin beads.92 Using this approach, they identified 42 Rab GTPases as GGTase-II substrates and also quantitatively analyzed ex vivo effects of prenylation inhibitors in cell culture on the prenylation of these Rab GTPases. In a study carried out by Reuter and co-workers, anilinogeraniol, 27, was used to tag the farnesylated proteome.93 Proteins were separated on 2D gels and tagged proteins were detected by Western blot using antibodies against the anilinogeranyl moiety. Effects of FTI treatments on labeling with 27 were also visualized using this approach.
Porcu et al. utilized a genomic method to globally analyze effects of FTIs on cellular activity.94 They carried out differential labeling of cDNA with two fluorescent dyes by reverse RNA transcription of DNA isolated from FTI-treated or untreated yeast cells. Labeled cDNAs were mixed together and hybridized with an array of 6240 unique yeast ORFs. Expression levels of all the genes in the presence and absence of FTI treatment were quantified by comparing the fluorescence intensity of the two color panels. Utilizing this method, the authors identified downstream effector proteins that get either up- or down-regulated as a result of FTI treatment.
Inhibition and Therapeutic Applications
The initial efforts to develop farnesyltransferase inhibitors (FTIs) targeted the inhibition of oncogenic Ras proteins. This was predicated on the occurrence of Ras mutations in more than 30% of human cancers, and the discovery that farnesylation of Ras proteins is essential for their proper cellular localization and signaling activity.38,95 Initially, FTIs were designed to be competitive inhibitors, either peptidomimetic compounds, isoprenoid analogs or bisubstrate analogs. Later, potent inhibitors were identified from library screening efforts.96
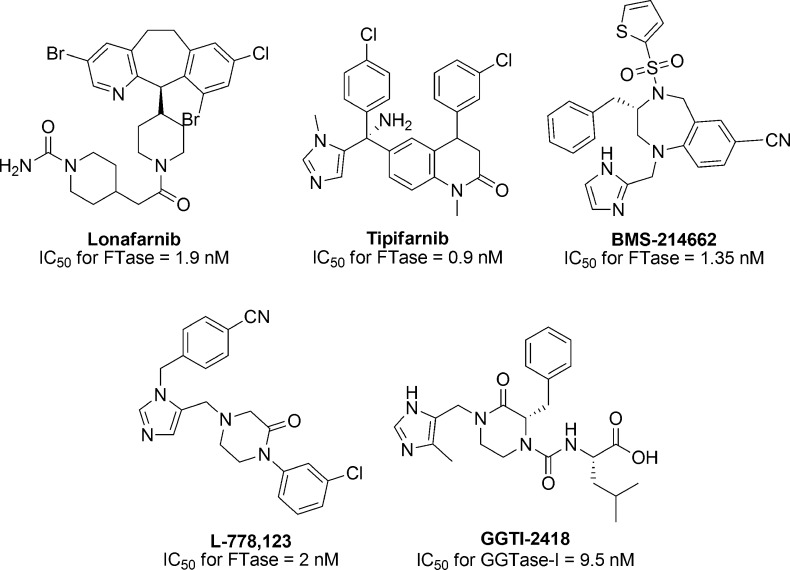
Preclinical studies of FTIs in cancer cell lines as well as animal models were highly successful leading to the advancement of four FTIs (Figure 9) into clinical trials starting in 2000.10 While phase 1 and 2 studies were encouraging, this early stage success did not yield robust anticancer activity.97 The four FTIs were evaluated in at least 75 clinical trials either as monotherapy or in combination with other anticancer drugs. The results were discouraging with >28% trials reporting no objective response and >36% studied showing very little (<15%) response.10 One of the key reasons behind the failure of FTIs in clinical trials is the incorrect selection of patients enrolled in these studies.10,97 K-Ras protein, which is the most frequent isoform of oncogenic Ras proteins, was reported to get geranylgeranylated and remain fully active when FTase activity is inhibited.98,99 And while it was known that K-Ras could escape FTI-mediated inhibition of FTase, phase III clinical trials were performed with patients having advanced or metastatic tumors harboring mutant K-Ras.10 Recently, methods are being developed to predict patient populations that are likely to be susceptible to FTIs. Karp and co-workers developed a two-gene expression assay to predict clinical outcome of tipifarnib (Figure 9) treatment administered to acute myeloid leukemia (AML) patients.100 More clinical studies with patients who are predicted to be responsive to the FTI based on their gene expression profile could potentially identify FTI-based personalized anticancer therapies. Recently, experiments with a caged FTI demonstrated that such compounds may be useful for selective release of an FTI within a defined tissue location.101
Figure 9.
Structures of FTIs and GGTI investigated in clinical trials against cancer or HGPS.
Since K-Ras can be geranylgeranylated and other geranylgeranylated proteins may also be involved in cancer, inhibitors of GGTase-I (GGTIs) have been evaluated as an alternative strategy to achieve anticancer therapies. Through structure–activity relationships, several GGTIs selectively inhibiting GGTase-I over FTase were developed. Only one of the GGTIs (Figure 9) entered a phase I clinical trial in 2009 and that was discontinued due to lack of efficacy in patients.38
The discovery that Hutchinson–Gilford progeria syndrome (HGPS) is caused, at least in part, by the accumulation of a farnesylated protein derived from a mutant form of farnesylated prelamin A generated interest in using FTIs to treat HGPS.38 Following in vitro and mouse model studies, two clinical trials using lonafarnib (Figure 9) as treatment for HGPS were undertaken.102,103 The first phase II clinical trial started in 2007 with 25 patients, where lonafarnib treatment for at least 2 years was well received. It provided preliminary evidence that lonafarnib could potentially improve one or more disease measures related to HGPS.104 The other phase II clinical trial was initiated by Children’s Hospital Boston in 2009, to test a combination lonafarnib and two other compounds for patients with progeria, and it is currently estimated to be completed in 2017.105 The results of this study should help to design combination therapies to treat progeria with prenylation inhibitors.
The pathogenic parasites causing diseases such as malaria, African sleeping sickness, and Chagas disease have their own farnesyltransferase enzyme.106 Inhibition of FTase severely affected the growth of these parasites, indicating FTIs as a useful treatment strategy.106−108 Development of potent FTIs (IC50 ≥ 1 nM) having up to 136-fold selectivity for the Plasmodium falciparum (malaria) FTase over the mammalian counterpart shows promise for potential uses of FTIs in treating parasitic diseases.109 FTIs and GGTIs are also being investigated for several other diseases including multiple sclerosis, osteoporosis, aging disorders, and viral diseases such as hepatitis.110−113
Biotechnological Applications
In recent years, the properties of FTase to specifically modify a single cysteine residue located at the C-terminal CaaX motif and to incorporate isoprenoid analogs containing bioorthogonal functionalities have been exploited for site-specific modifications of proteins. This has been possible since the presence of a CaaX-box at its C-terminus is sufficient to render almost any protein an efficient FTase substrate. Functionalization of the resulting proteins via bioorthogonal reactions provides a convenient methodology for preparing a wide of array of protein conjugates in a site-specific manner.
Both the Poulter and Distefano groups have used azide- and alkyne-functionalized FPP analogs in FTase-catalyzed reactions followed by click reactions or Staudinger ligations for immobilization of proteins (GFP or GST) onto solid surfaces such as glass slides or agarose resin.114−117 Maynard and co-workers utilized a similar strategy for immobilization of mCherry protein tagged with 25 onto a patterned azide-functionalized surface created by microcontact printing.118 A photochemical thiol–ene reaction between farnesylated recombinant proteins and surface-exposed thiols from functionalized surfaces was applied by Waldmann and co-workers for oriented and selective immobilization of functional proteins (mCherry and Ypt1).119 Recently, Poulter and co-workers achieved highly ordered, regioselective immobilization of the glutathione S-transferase enzyme and antibody-binding protein G to self-assembled monolayers on gold surfaces.120 They further created sandwich antibody arrays of immobilized recombinant antibody-binding protein L for capturing antibodies for direct- and sandwich-type immunofluorescent detection of ligands in a microarray format.121 In general, the prenylation-based immobilization strategy has several potential biomedical and biotechnology applications where oriented protein immobilization is required, such as protein arrays, and diagnostic applications based on immunoassays, surface plasmon resonance (SPR), or electrochemical methods.
A simple and efficient method for the derivatization and purification of recombinant proteins (such as YPT7, Rab7, GST) was developed by Alexandrov and co-workers using a fluorescent analog of FPP and phase partitioning.122 Recently, Distefano and co-workers described the use of an aldehyde-functionalized FPP analog in conjunction with a hydrazide resin-based catch-and-release strategy to purify and functionalize proteins with groups like a fluorophore or PEG moiety.123
One important application of the prenylation-based labeling strategy is the formation of site-specific protein modifications such as protein–DNA conjugates, PEGylated proteins, and dually labeled proteins.124 Some of the protein–DNA conjugates that have been synthesized using this method include a nanoscale sized defined tetrahedron architecture composed of four oligonucleotides and four GFP molecules, therapeutically relevant proteins GIP and HIV NC attached to oligonucleotides, and DNA–protein cross-links as DNA lesions to study DNA repair and replication.124−126 Prenylation of a recombinant protein cilliary neurotrophic factor (CNTF) with an isoprenoid analog modified with an aldehyde group followed by oxime ligation-based catch-and-release yielded PEGylated CNTF, where attachment of the PEG group could potentially increase serum half-life of this biomedically important protein.127 In another report, Rashidian et al. describe a multifunctional macromolecular protein self-assembly consisting of an antibody nanoring structure bearing a single chain anti-CD3 antibody as the targeting element, as well as a model cargo protein and a fluorophore. In that case, a trifunctional FPP analog incorporating both aldehyde and alkyne functionality was used to create the key multifunctional fragment consisting of a cargo protein, fluorophore and protein dimerizer. This high-avidity “effector–antibody–fluorophore” conjugate was endocytosed into T-leukemia cells highlighting its potential use in developing protein–drug conjugates for therapeutic protein delivery and tracking.128
Concluding Remarks
Protein prenylation has emerged as an important post-translational modification responsible for the correct cellular localization, activity, and protein–protein interactions of a number of signaling proteins. Over the past 25 years, a large number of isoprenoid analogs have been synthesized and employed to probe the structural and mechanistic features of the prenyltransferase enzymes. With the extensive studies using these analogs and peptide substrates and X-ray crystallographic analysis of the enzymes in complex with the substrates and product, the enzymology of prenyltransferases is now well understood. One of the early hypotheses in the field of prenylation was that prenyltransferase inhibitors could be used to suppress malignant activity of oncogenic Ras proteins. While those inhibitors gave early success in the laboratory, clinical trials proved less promising. Those results make it clear that much remains to be learned concerning the roles of prenylated proteins in living cells, and this remains an intense area of current investigation. Several peptide library screening efforts, proteomic studies, and yeast-based genomic experiments have provided preliminary results toward this end; however, completely defining the prenylated proteome is still an ongoing task; addressing this issue will be central for assessing which patients are the best candidates for treatment with prenylation inhibitors. More work also needs to be carried out to explore the potential of FTIs and GGTIs for the treatment of other afflictions including progeria, multiple sclerosis, parasitic diseases, and bacterial and viral infections. Protein prenylation has also shown promise for site-specific modifications of proteins. While many interesting applications have been demonstrated in recent years, future work must focus on creating protein conjugates including antibody–drug conjugates, PEGylated proteins, and other constructs that are directly applicable to therapeutic studies so that they can be evaluated in clinical contexts. Overall, these challenges suggest that investigation of protein prenylation and will remain an active and vibrant field of inquiry for some time to come.
Acknowledgments
This work was supported by the National Institutes of Health (Grants GM084152 and CA185783), the National Science Foundation (Grant CHE-1308655), and the University of Minnesota.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Kamiya Y.; Sakurai A.; Tamura S.; Takahashi N.; Abe K.; Tsuchiya E.; Fukui S.; Kitada C.; Fujino M. (1978) Structure of rhodotorucine-A, a novel lipopeptide, inducing mating tube formation in rhodoporidium-toruloids. Biochem. Biophys. Res. Commun. 83, 1077–1083. [DOI] [PubMed] [Google Scholar]
- Wolda S. L.; Glomset J. A. (1988) Evidence for modification of Lamin B by a product of mevalonic acid. J. Biol. Chem. 263, 5997–6000. [PubMed] [Google Scholar]
- Farnsworth C. C.; Wolda S. L.; Gelb M. H.; Glomset J. A. (1989) Human Lamin-B contains a farnesylated cysteine residue. J. Biol. Chem. 264, 20422–20429. [PMC free article] [PubMed] [Google Scholar]
- Zhang F. L.; Casey P. J. (1996) Protein prenylation: Molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269. [DOI] [PubMed] [Google Scholar]
- Ghomashchi F.; Zhang X. H.; Liu L.; Gelb M. H. (1995) Binding of prenylated and polybasic peptides to membranes - affinities and intervesicle exchange. Biochemistry 34, 11910–11918. [DOI] [PubMed] [Google Scholar]
- Resh M. D. (2006) Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2, 584–590. [DOI] [PubMed] [Google Scholar]
- Seabra M. C. (1998) Membrane association and targeting of prenylated Ras-like GTPases. Cell. Signalling 10, 167–172. [DOI] [PubMed] [Google Scholar]
- Placzek A. T.; Krzysiak A. J.; Gibbs R. A. (2011) Chemical probes of protein prenylation. Enzymes 30, 91–127. [Google Scholar]
- Jackson J. H.; Cochrane C. G.; Bourne J. R.; Solski P. A.; Buss J. E.; Der C. J. (1990) Farnesol modification of Kristen-ras exon 4B is essential for transformation. Proc. Natl. Acad. Sci. U. S. A. 87, 3042–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt N.; Hamilton A. D.; Sebti S. M. (2011) Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer 11, 775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Seabra M. C.; Deisenhofer J. (2000) Crystal structure of Rab geranylgeranyltransferase at 2.0 angstrom resolution. Structure 8, 241–251. [DOI] [PubMed] [Google Scholar]
- Taylor J. S.; Reid T. S.; Terry K. L.; Casey P. J.; Beese L. S. (2003) Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J. 22, 5963–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompliano D. L.; Schaber M. D.; Mosser S. D.; Omer C. A.; Shafer J. A.; Gibbs J. B. (1993) Isoprenoid diphosphate utilization by recombinant human farnesyl-protein transferase - interactive binding between substrates and a preferred kinetic pathway. Biochemistry 32, 8341–8347. [DOI] [PubMed] [Google Scholar]
- Dolence J. M.; Cassidy P. B.; Mathis J. R.; Poulter C. D. (1995) Yeast protein farnesyltransferase: Steady-state kinetic studies of substrate binding. Biochemistry 34, 16687–16694. [DOI] [PubMed] [Google Scholar]
- Long S. B.; Casey P. J.; Beese L. S. (2002) Reaction path of protein farnesyltransferase at atomic resolution. Nature 419, 645–650. [DOI] [PubMed] [Google Scholar]
- Pickett J. S.; Bowers K. E.; Hartman H. L.; Fu H. W.; Embry A. C.; Casey P. J.; Fierke C. A. (2003) Kinetic studies of protein farnesyltransferase mutants establish active substrate conformation. Biochemistry 42, 9741–9748. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Wang B.; Ucisik M. N.; Cui G.; Fierke C. A.; Merz K. M. Jr. (2012) Insights into the mechanistic dichotomy of the protein farnesyltransferase peptide substrates CVIM and CVLS. J. Am. Chem. Soc. 134, 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y. Q.; Omer C. A.; Gibbs R. A. (1996) On the stereochemical course of human protein-farnesyl transferase. J. Am. Chem. Soc. 118, 117–123. [Google Scholar]
- Edelstein R. L.; Weller V. A.; Distefano M. D.; Tung J. S. (1998) Stereochemical Analysis of the Reaction Catalyzed by Yeast Protein Farnesyltransferase. J. Org. Chem. 63, 5298–5299. [Google Scholar]
- Dolence J. M.; Poulter C. D. (1995) A Mechanism for Posttranslational Modifications of Proteins by Yeast Protein Farnesyltransferase. Proc. Natl. Acad. Sci. U.S.A. 92, 5008–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y. Q.; Gibbs R. A.; Eubanks L. M.; Poulter C. D. (1996) Cuprate-mediated synthesis and biological evaluation of cyclopropyl- and tert-butylfarnesyl diphosphate analogs. J. Org. Chem. 61, 8010–8015. [DOI] [PubMed] [Google Scholar]
- Weller V. A.; Distefano M. D. (1998) Measurement of the alpha-secondary kinetic isotope effect for a prenyltransferase by MALDI mass spectrometry. J. Am. Chem. Soc. 120, 7975–7976. [Google Scholar]
- Pais J. E.; Bowers K. E.; Fierke C. A. (2006) Measurement of the alpha-secondary kinetic isotope effect for the reaction catalyzed by mammalian protein farnesyltransferase. J. Am. Chem. Soc. 128, 15086–15087. [DOI] [PubMed] [Google Scholar]
- Lenevich S.; Xu J.; Hosokawa A.; Cramer C. J.; Distefano M. D. (2007) Transition state analysis of model and enzymatic prenylation reactions. J. Am. Chem. Soc. 129, 5796–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M.-H.; Vivo M. D.; Peraro M. D.; Klein M. L. (2009) Unraveling the Catalytic Pathway of Metalloenzyme Farnesyltransferase through QM/MM Computation. J. Chem. Theory Comput. 5, 1657–1666. [DOI] [PubMed] [Google Scholar]
- Huang C. C.; Casey P. J.; Fierke C. A. (1997) Evidence for a catalytic role of zinc in protein farnesyltransferase - Spectroscopy of Co2+-farnesyltransferase indicates metal coordination of the substrate thiolate. J. Biol. Chem. 272, 20–23. [DOI] [PubMed] [Google Scholar]
- Rozema D. B.; Poulter C. D. (1999) Yeast protein farnesyltransferase. pK(a)s of peptide substrates bound as zinc thiolates. Biochemistry 38, 13138–13146. [DOI] [PubMed] [Google Scholar]
- Tobin D. A.; Pickett J. S.; Hartman H. L.; Fierke C. A.; Penner-Hahn J. E. (2003) Structural Characterization of the Zinc Site in Protein Farnesyltransferase. J. Am. Chem. Soc. 125, 9962–9969. [DOI] [PubMed] [Google Scholar]
- Yokoyama K.; McGeady P.; Gelb M. H. (1995) Mammalian protein geranylgeranyltransferase-I: Substrate specificity, kinetic mechanism, metal requirements, and affinity labeling. Biochemistry 34, 1344–1354. [DOI] [PubMed] [Google Scholar]
- Clausen V. A.; Edelstein R. L.; Distefano M. D. (2001) Stereochemical analysis of the reaction catalyzed by human protein geranylgeranyl transferase. Biochemistry 40, 3920–3930. [DOI] [PubMed] [Google Scholar]
- Hartman H. L.; Bowers K. E.; Fierke C. A. (2004) Lysine b311 of Protein Geranylgeranyltransferase Type I Partially Replaces Magnesium. J. Biol. Chem. 279, 30546–30553. [DOI] [PubMed] [Google Scholar]
- Terry K. L.; Casey P. J.; Beese L. S. (2006) Conversion of protein farnesyltransferase to a geranylgeranyltransferase. Biochemistry 45, 9746–9755. [DOI] [PubMed] [Google Scholar]
- Desnoyers L.; Anant J. S.; Seabra M. C. (1996) Geranylgeranylation of Rab proteins. Biochem. Soc. Trans. 24, 699–703. [DOI] [PubMed] [Google Scholar]
- Witter D. J.; Poulter C. D. (1996) Yeast geranylgeranyltransferase type-II: Steady state kinetic studies of the recombinant enzyme. Biochemistry 35, 10454–10463. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Seabra M. C.; Deisenhofer J. (2000) Crystal Structure of Rab geranylgeranyltransferase at 2.0 A resolution. Structure 8, 241–251. [DOI] [PubMed] [Google Scholar]
- Guo Z.; Wu Y.-W.; Das D.; Delon C.; Cramer J.; Yu S.; Thuns S.; Lupilova N.; Waldmann H.; Brunsveld L.; Goody R. S.; Alexandrov K.; Blankenfeldt W. (2008) Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation. EMBO J. 27, 2444–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuld N. J.; Vervacke J. S.; Lorimer E. L.; Simon N. C.; Hauser A. D.; Barbieri J. T.; Distefano M. D.; Williams C. L. (2014) The chaperone protein SmgGDS interacts with small GTPases entering the prenylation pathway by recognizing the last amino acid in the CAAX motif. J. Biol. Chem. 289, 6862–6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochocki J. D.; Distefano M. D. (2013) Prenyltransferase inhibitors: treating human ailments from cancer to parasitic infections. MedChemComm 4, 476–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N.; Lamphear C. L.; Hougland J. L.; Fierke C. A.; Schueler-Furman O. (2011) Identification of a novel class of farnesylation targets by structure-based modeling of binding specificity. Plos Comput. Biol. 7, e1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S.; Koranda M.; Benetka W.; Schneider G.; Sirota F. L.; Eisenhaber F. (2007) Towards complete sets of farnesylated and geranylgeranylated proteins. Plos Comput. Biol. 3, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S.; Eisenhaber F. (2005) Refinement and prediction of protein prenylation motifs. Genome Biol. 6, R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougland J. L.; Hicks K. A.; Hartman H. L.; Kelly R. A.; Watt T. J.; Fierke C. A. (2010) Identification of novel peptide substrates for protein farnesyltransferase reveals two substrate classes with distinct sequence selectivities. J. Mol. Biol. 395, 176–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss Y.; Stradley S. J.; Gierasch L. M.; Brown M. S.; Goldstein J. L. (1991) Sequence requirement for peptide recognition by rat-brain P21Ras protein farnesyltransferase. Proc. Natl. Acad. Sci. U. S. A. 88, 732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower K. E.; Huang C. C.; Casey P. J.; Fierke C. A. (1998) H-Ras peptide and protein substrates bind protein farnesyltransferase as an ionized thiolate. Biochemistry 37, 15555–15562. [DOI] [PubMed] [Google Scholar]
- Pompliano D. L.; Gomez R. P.; Anthony N. J. (1992) Intramolecular fluorescence enhancement- A continuous assay of Ras farnesyl-protein transferase. J. Am. Chem. Soc. 114, 7945–7946. [Google Scholar]
- Stirtan W. G.; Poulter C. D. (1995) Yeast protein geranylgeranyltransferase type-I - Overproduction, purification, and characterization. Arch. Biochem. Biophys. 321, 182–190. [DOI] [PubMed] [Google Scholar]
- Boutin J. A.; Marande W.; Petit L.; Loynel A.; Desmet C.; Canet E.; Fauchére J. L. (1999) Investigation of S-farnesyl transferase substrate specificity with combinatorial tetrapeptide libraries. Cell. Signalling. 11, 59–69. [DOI] [PubMed] [Google Scholar]
- Krzysiak A. J.; Scott S. A.; Hicks K. A.; Fierke C. A.; Gibbs R. A. (2007) Evaluation of protein farnesyltransferase substrate specificity using synthetic peptide libraries. Bioorg. Med. Chem. Lett. 17, 5548–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak A. J.; Aditya A. V.; Hougland J. L.; Fierke C. A.; Gibbs R. A. (2010) Synthesis and screening of a CaaL peptide library versus FTase reveals a surprising number of substrates. Bioorg. Med. Chem. Lett. 20, 767–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougland J. L.; Lamphear C. L.; Scott S. A.; Gibbs R. A.; Fierke C. A. (2009) Context-dependent substrate recognition by protein farnesyltransferase. Biochemistry 48, 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear C. L.; Zverina E. A.; Hougland J. L.; Fierke C. A. (2011) Global identification of protein prenyltransferase substrates: defining the prenylated proteome. Enzymes 29, 207–234. [Google Scholar]
- Wang Y.-C.; Distefano M. D. (2012) Solid-phase synthesis of C-terminal peptide libraries for studying the specificity of enzymatic protein prenylation. Chem. Commun. 48, 8228–8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-C.; Dozier J. K.; Beese L. S.; Distefano M. D. (2014) Rapid analysis of protein farnesyltransferase substrate specificity using peptide libraries and isoprenoid diphosphate analogues. ACS Chem. Biol. 9, 1726–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer C. A.; Kral A. M.; Diehl R. E.; Prendergast G. C.; Powers S.; Allen C. M.; Gibbs J. B.; Kohl N. E. (1993) Characterization of recombinant human farnesyl-protein transferase: Cloning, expression, farnesyl diphosphate binding, and functional homology with yeast prenyl-protein transferases. Biochemistry 32, 5167–5176. [DOI] [PubMed] [Google Scholar]
- Bukhtiyarov Y. E.; Omer C. A.; Allen C. M. (1995) Photoreactive analogs of prenyl diphosphates as inhibitors and probes of human protein farnesyltransferase and geranylgeranyltransferase type-I. J. Biol. Chem. 270, 19035–19040. [DOI] [PubMed] [Google Scholar]
- Edelstein R. L.; Distefano M. D. (1997) Photoaffinity labeling of yeast farnesyl protein transferase and enzymatic synthesis of a Ras protein incorporating a photoactive isoprenoid. Biochem. Biophys. Res. Commun. 235, 377–382. [DOI] [PubMed] [Google Scholar]
- Gaon I.; Turek T. C.; Distefano M. D. (1996) Farnesyl and geranylgeranyl pyrophosphate analogs incorporating benzoylbenzyl ethers: synthesis and inhibition of yeast protein farnesyltransferase. Tetrahedron Lett. 37, 8833–8836. [Google Scholar]
- Gaon I.; Turek T. C.; Weller V. A.; Edelstein R. L.; Singh S. K.; Distefano M. D. (1996) Photoactive Analogs of Farnesyl Pyrophosphate Containing Benzoylbenzoate Esters: Synthesis and Application to Photoaffinity Labeling of Yeast Farnesyltransferase. J. Org. Chem. 61, 7738–7745. [DOI] [PubMed] [Google Scholar]
- Turek T. C.; Gaon I.; Distefano M. D. (1996) Analogs of farnesyl pyrophosphate incorporating internal benzoylbenzoate esters: Synthesis, inhibition kinetics and photoinactivation of yeast protein farnesyltransferase. Tetrahedron Lett. 37, 4845–4848. [DOI] [PubMed] [Google Scholar]
- Turek T. C.; Gaon I.; Distefano M. D.; Strickland C. L. (2001) Synthesis of Farnesyl Diphosphate Analogues Containing Ether-Linked Photoactive Benzophenones and Their Application in Studies of Protein Prenyltransferases. J. Org. Chem. 66, 3253–3264. [DOI] [PubMed] [Google Scholar]
- Turek T. C.; Gaon I.; Gamache D.; Distefano M. D. (1997) Synthesis and evaluation of benzophenone-based photoaffinity labeling analogs of prenyl pyrophosphates containing stable amide linkages. Bioorg. Med. Chem. Lett. 7, 2125–2130. [Google Scholar]
- Quellhorst G. J.; Allen C. M.; Wessling-Resnick M. (2001) Modification of Rab5 with a photoactivatable analog of geranylgeranyl diphosphate. J. Biol. Chem. 276, 40727–40733. [DOI] [PubMed] [Google Scholar]
- Turek-Etienne T. C.; Strickland C. L.; Distefano M. D. (2003) Biochemical and structural studies with prenyl diphosphate analogues provide insights into isoprenoid recognition by protein farnesyl transferase. Biochemistry 42, 3716–3724. [DOI] [PubMed] [Google Scholar]
- Hovlid M. L.; Edelstein R. L.; Henry O.; Ochocki J.; DeGraw A.; Lenevich S.; Talbot T.; Young V. G.; Hruza A. W.; Lopez-Gallego F.; Labello N. P.; Strickland C. L.; Schmidt-Dannert C.; Distefano M. D. (2010) Synthesis, properties, and applications of diazotrifluropropanoyl-containing photoactive analogs of farnesyl diphosphate containing modified linkages for enhanced stability. Chem. Biol. Drug Des. 75, 51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervacke J. S.; Funk A. L.; Wang Y.-C.; Strom M.; Hrycyna C. A.; Distefano M. D. (2014) Diazirine-containing photoactivatable isoprenoid: synthesis and application in studies with isoprenylcysteine carboxyl methyltransferase. J. Org. Chem. 79, 1971–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGraw A. J.; Zhao Z.; Hsieh J.; Jefferies M.; Distefano M. D.; Strickland C. L.; Shintani D.; Nural H.; McMahan C.; Xie W. (2007) A photoactive isoprenoid diphosphate analogue containing a stable phosphonate linkage: synthesis and structural biochemical studies with prenyltransferases. J. Org. Chem. 72, 4587–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland C. L.; Windsor W. T.; Syto R.; Wang L.; Bond R.; Wu Z.; Schwartz J.; Le H. V.; Beese L. S.; Weber P. C. (1998) Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue. Biochemistry 37, 16601–16611. [DOI] [PubMed] [Google Scholar]
- Zahn T. J.; Whitney J.; Weinbaum C.; Gibbs R. A. (2001) Synthesis and evaluation of GGPP geometric isomers: Divergent substrate specificities of FTase and GGTase I. Bioorg. Med. Chem. Lett. 11, 1605–1608. [DOI] [PubMed] [Google Scholar]
- Subramanian T.; Pais J. E.; Liu S.; Troutman J. M.; Suzuki Y.; Subramanian K. L.; Fierke C. A.; Andres D. A.; Spielmann H. P. (2012) Farnesyl Diphosphate Analogues with Aryl Moieties Are Efficient Alternate Substrates for Protein Farnesyltransferase. Biochemistry 51, 8307–8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. J.; Troutman J. M.; Chehade K. A. H.; Cha H. C.; Kao J. P. Y.; Huang X.; Zhan C.-G.; Peterson Y. K.; Subramanian T.; Kamalakkannan S.; Andres D. A.; Spielmann H. P. (2006) Hydrophilic anilinogeranyl diphosphate prenyl analogues are Ras function inhibitors. Biochemistry 45, 15862–15872. [DOI] [PubMed] [Google Scholar]
- Troutman J. M.; Subramanian T.; Andres D. A.; Spielmann H. P. (2007) Selective modification of CaaX peptides with ortho-substituted anilinogeranyl lipids by protein farnesyl transferase: Competitive substrates and potent inhibitors from a library of farnesyl diphosphate analogues. Biochemistry 46, 11310–11321. [DOI] [PubMed] [Google Scholar]
- Gibbs B. S.; Zahn T. J.; Mu Y. Q.; Sebolt-Leopold J. S.; Gibbs R. A. (1999) Novel farnesol and geranylgeraniol analogues: A potential new class of anticancer agents directed against protein prenylation. J. Med. Chem. 42, 3800–3808. [DOI] [PubMed] [Google Scholar]
- Krzysiak A. J.; Rawat D. S.; Scott S. A.; Pais J. E.; Handley M.; Harrison M. L.; Fierke C. A.; Gibbs R. A. (2007) Combinatorial Modulation of Protein Prenylation. ACS Chem. Biol. 2, 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D.; Tnimov Z.; Nguyen U. n. T. T.; Thimmaiah G.; Lo H.; Abankwa D.; Wu Y.; Goody R. S.; Waldmann H.; Alexandrov K. (2012) Flexible and general synthesis of functionalized phosphoisoprenoids for the study of prenylation in vivo and in vitro. ChemBioChem 13, 674–683. [DOI] [PubMed] [Google Scholar]
- Dozier J. K.; Khatwani S. L.; Wollack J. W.; Wang Y.-C.; Schmidt-Dannert C.; Distefano M. D. (2014) Engineering protein farnesyltransferase for enzymatic protein labeling applications. Bioconjugate Chem. 25, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. K.; Kleckley T. S.; Wiemer A. J.; Holstein S. A.; Hohl R. J.; Wiemer D. F. (2004) Synthesis and activity of fluorescent isoprenoid pyrophosphate analogues. J. Org. Chem. 69, 8186–8193. [DOI] [PubMed] [Google Scholar]
- Placzek A. T.; Krzysiak A. J.; Gibbs R. A. (2011) Chemical probes of protein prenylation, in Protein Prenylation, Part B (Hrycyna C. A., Bergo M. O., and Tamanoi F., Eds.), Enzymes, Vol. 30, pp 91–127, Elsevier, Amsterdam.
- Patel D. V.; Schmidt R. J.; Biller S. A.; Gordon E. M.; Robinson S. S.; Manne V. (1995) Farnesyl Diphosphate-Based Inhibitors of Ras Farnesyl Protein Transferase. J. Med. Chem. 38, 2906–2921. [DOI] [PubMed] [Google Scholar]
- Kho Y.; Kim S. C.; Jiang C.; Barma D.; Kwon S. W.; Cheng J.; Jaunbergs J.; Weinbaum C.; Tamanoi F.; Falck J.; Zhao Y. (2004) A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. U. S. A. 101, 12479–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L. N.; Hart C.; Guo L.; Nyberg T.; Davies B. S. J.; Fong L. G.; Young S. G.; Agnew B. J.; Tamanoi F. (2009) A novel approach to tag and identify geranylgeranylated proteins. Electrophoresis 30, 3598–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A. F. H.; Heal W. P.; Tarafder A. K.; Tolmachova T.; Baron R. A.; Seabra M. C.; Tate E. W. (2010) Rapid multilabel detection of geranylgeranylated proteins by using bioorthogonal ligation chemistry. ChemBioChem 11, 771–773. [DOI] [PubMed] [Google Scholar]
- DeGraw A. J.; Palsuledesai C.; Ochocki J. D.; Dozier J. K.; Lenevich S.; Rashidian M.; Distefano M. D. (2010) Evaluation of Alkyne-Modified Isoprenoids as Chemical Reporters of Protein Prenylation. Chem. Biol. Drug Des. 76, 460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G.; Tsou L. K.; Maguire W.; Yount J. S.; Hang H. C. (2011) Alkynyl-farnesol reporters for detection of protein S-prenylation in cells. Mol. BioSyst. 7, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth B. P.; Zhang Z.; Hosokawa A.; Distefano M. D. (2007) Selective labeling of proteins by using protein farnesyltransferase. ChemBioChem 8, 98–105. [DOI] [PubMed] [Google Scholar]
- Hosokawa A.; Wollack J. W.; Zhang Z. Y.; Chen L.; Barany G.; Distefano M. D. (2007) Evaluation of an alkyne-containing analogue of farnesyl diphosphate as a dual substrate for protein-prenyltransferases. Int. J. Pept. Res. Ther. 13, 345–354. [Google Scholar]
- Palsuledesai C. C.; Ochocki J. D.; Markowski T. W.; Distefano M. D. (2014) A combination of metabolic labeling and 2D-DIGE analysis in response to a farnesyltransferase inhibitor facilitates the discovery of new prenylated proteins. Mol. BioSyst. 10, 1094–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinicke A. T.; Hutchinson J. L.; Magee A. I.; Mastroeni P.; Trowsdale J.; Kelly A. P. (2005) A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J. Biol. Chem. 280, 14620–14627. [DOI] [PubMed] [Google Scholar]
- Price C. T. D.; Al-Quadan T.; Santic M.; Jones S. C.; Abu Kwaik Y. (2010) Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J. Exp. Med. 207, 1712–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S. S.; Charron G.; Hang H. C.; Roy C. R. (2010) Lipidation by the host prenyltransferase machinery facilitates membrane localization of legionella pneumophila effector proteins. J. Biol. Chem. 285, 34686–34698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakasova E. S.; Akhmatova N. K.; Amaya M.; Eisenhaber B.; Eisenhaber F.; van Hoek M. L.; Baranova A. V. (2013) Prenylation: From bacteria to eukaryotes. Mol. Biol. 47, 622–633. [PubMed] [Google Scholar]
- Charron G.; Li M. M. H.; MacDonald M. R.; Hang H. C. (2013) Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Proc. Natl. Acad. Sci. U. S. A. 110, 11085–11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen U. T. T.; Guo Z.; Delon C.; Wu Y.; Deraeve C.; Fraenzel B.; Bon R. S.; Blankenfeldt W.; Goody R. S.; Waldmann H.; Wolters D.; Alexandrov K. (2009) Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat. Chem. Biol. 5, 227–235. [DOI] [PubMed] [Google Scholar]
- Onono F. O.; Morgan M. A.; Spielmann H. P.; Andres D. A.; Subramanian T.; Ganser A.; Reuter C. W. M. (2010) A tagging-via-substrate approach to detect the farnesylated proteome using two-dimensional electrophoresis coupled with Western blotting. Mol. Cell. Proteomics 9, 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcu G.; Wilson C.; Di Giandomenico D.; Ragnini-Wilson A. (2010) A yeast-based genomic strategy highlights the cell protein networks altered by FTase inhibitor peptidomimetics. Mol. Cancer 9, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer W. R.; Kim R.; Sterne R.; Thorner J.; Kim S. H.; Rine J. (1989) Genetic and pharmacological suppression of oncogenic mutations in Ras genes of yeast and humans. Science 245, 379–385. [DOI] [PubMed] [Google Scholar]
- Reiss Y.; Goldstein J. L.; Seabra M. C.; Casey P. J.; Brown M. S. (1990) Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides. Cell 62, 81–88. [DOI] [PubMed] [Google Scholar]
- Bishop W. R.; Doll R.; Kirschmeier P. (2011) Farnesyl transferase inhibitors: From targeted cancer therapeutic to a potential treatment for progeria. Enzymes 29, 275–303. [Google Scholar]
- Bos J. L. (1989) Ras oncogenes in human cancer - a review. Cancer Res. 49, 4682–4689. [PubMed] [Google Scholar]
- Lerner E. C.; Zhang T. T.; Knowles D. B.; Qian Y. M.; Hamilton A. D.; Sebti S. M. (1997) Inhibition of the prenylation of K-Ras, but not H- or N-Ras, is highly resistant to CAAX peptidomimetics and requires both a farnesyltransferase and a geranylgeranyltransferase I inhibitor in human tumor cell lines. Oncogene 15, 1283–1288. [DOI] [PubMed] [Google Scholar]
- Raponi M.; Lancet J. E.; Fan H.; Dossey L.; Lee G.; Gojo I.; Feldman E. J.; Gotlib J.; Morris L. E.; Greenberg P. L.; Wright J. J.; Harousseau J.-L.; Loewenberg B.; Stone R. M.; De Porre P.; Wang Y.; Karp J. E. (2008) A 2-gene classifier for predicting response to the famesyltransferase inhibitor tipifarnib in acute myeloid leukemia. Blood 111, 2589–2596. [DOI] [PubMed] [Google Scholar]
- Abate-Pella D.; Zeliadt N. A.; Ochocki J. D.; Warmka J. K.; Dore T. M.; Blank D. A.; Wattenberg E. V.; Distefano M. D. (2012) Photochemical Modulation of Ras-Mediated Signal Transduction using Caged Farnesyltransferase Inhibitors: Activation via One- and Two-Photon Excitation. ChemBioChem 13, 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. H.; Bergo M. O.; Toth J. I.; Qiao X.; Hu Y.; Sandoval S.; Meta M.; Bendale P.; Gelb M. H.; Young S. G.; Fong L. G. (2005) Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc. Natl. Acad. Sci. U. S. A. 102, 10291–10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong L. G.; Frost D.; Meta M.; Qiao X.; Yang S. H.; Coffinier C.; Young S. G. (2006) A protein farnesyltransferase inhibitor ameliorates disease in a mouse model of progeria. Science 311, 1621–1623. [DOI] [PubMed] [Google Scholar]
- Gordon L. B.; Kleinman M. E.; Miller D. T.; Neuberg D. S.; Giobbie-Hurder A.; Gerhard-Herman M.; Smoot L. B.; Gordon C. M.; Cleveland R.; Snyder B. D.; Fligor B.; Bishop W. R.; Statkevich P.; Regen A.; Sonis A.; Riley S.; Ploski C.; Correia A.; Quinn N.; Ullrich N. J.; Nazarian A.; Liang M. G.; Huh S. Y.; Schwartzman A.; Kieran M. W. (2012) Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 109, 16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston C. s. H. (2009) Study of zoledronic acid, pravastatin and lonafarnib for patients with progeria; US National Institute of Health, http://clinicaltrials.gov/ct2/show/record/NCT00916747.
- Kraus J. M.; Tatipaka H. B.; McGuffin S. A.; Chennamaneni N. K.; Karimi M.; Arif J.; Verlinde C. L. M. J.; Buckner F. S.; Gelb M. l. H. (2010) Second generation analogues of the cancer drug clinical candidate tipifarnib for anti-Chagas disease drug discovery. J. Med. Chem. 53, 3887–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrico D.; Ohkanda J.; Kendrick H.; Yokoyama K.; Blaskovich M. A.; Bucher C. J.; Buckner F. S.; Van Voorhis W. C.; Chakrabarti D.; Croft S. L.; Gelb M. H.; Sebti S. M.; Hamilton A. D. (2004) In vitro and in vivo antimalarial activity of peptidomimetic protein farnesyltransferase inhibitors with improved membrane permeability. Bioorg. Med. Chem. 12, 6517–6526. [DOI] [PubMed] [Google Scholar]
- Yokoyama K.; Trobridge P.; Buckner F. S.; Scholten J.; Stuart K. D.; Van Voorhis W. C.; Gelb M. H. (1998) The effects of protein farnesyltransferase inhibitors on trypanosomatids: inhibition of protein farnesylation and cell growth. Mol. Biochem. Parasitol. 94, 87–97. [DOI] [PubMed] [Google Scholar]
- Fletcher S.; Cummings C. G.; Rivas K.; Katt W. P.; Horney C.; Buckner F. S.; Chakrabarti D.; Sebti S. M.; Gelb M. l. H.; Van Voorhis W. C.; Hamilton A. D. (2008) Potent, plasmodium-selective farnesyltransferase inhibitors that arrest the growth of malaria parasites: Structure-activity relationships of ethylenediamine-analogue scaffolds and homology model validation. J. Med. Chem. 51, 5176–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters C. E.; Pryce G.; Hankey D. J. R.; Sebti S. M.; Hamilton A. D.; Baker D.; Greenwood J.; Adamson P. (2002) Inhibition of Rho GTPases with protein prenyltransferase inhibitors prevents leukocyte recruitment to the central nervous system and attenuates clinical signs of disease in an animal model of multiple sclerosis. J. Immunol. 168, 4087–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coxon F. P.; Helfrich M. H.; Van’t Hof R.; Sebti S.; Ralston S. H.; Hamilton A.; Rogers M. J. (2000) Protein geranylgeranylation is required for osteoclast formation, function, and survival: Inhibition by bisphosphonates and GGTI-298. J. Bone Mineral Res. 15, 1467–1476. [DOI] [PubMed] [Google Scholar]
- Bordier B. B.; Marion P. L.; Ohashi K.; Kay M. A.; Greenberg H. B.; Casey J. L.; Glenn J. S. (2002) A prenylation inhibitor prevents production of infectious hepatitis delta virus particles. J. Virol. 76, 10465–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Meray R. K.; Grammatopoulos T. N.; Fredenburg R. A.; Cookson M. R.; Liu Y.; Logan T.; Lansbury P. T. Jr. (2009) Membrane-associated farnesylated UCH-L1 promotes alpha-synuclein neurotoxicity and is a therapeutic target for Parkinson’s disease. Proc. Natl. Acad. Sci. U. S. A. 106, 4635–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth B. P.; Xu J.; Taton T. A.; Guo A.; Distefano M. D. (2006) Site-specific, covalent attachment of proteins to a solid surface. Bioconjugate Chem. 17, 967–974. [DOI] [PubMed] [Google Scholar]
- Gauchet C.; Labadie G. R.; Poulter C. D. (2006) Regio- and chemoselective covalent immobilization of proteins through unnatural amino acids. J. Am. Chem. Soc. 128, 9274–9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth B. P.; Zhang Z. Y.; Hosokawa A.; Distefano M. D. (2007) Selective labeling of proteins by using protein farnesyltransferase. ChemBioChem 8, 98–105. [DOI] [PubMed] [Google Scholar]
- Xu J.; DeGraw A. J.; Duckworth B. P.; Lenevich S.; Tann C.-M.; Jenson E. C.; Gruber S. J.; Barany G.; Distefano M. D. (2006) Synthesis and reactivity of 6,7-dihydrogeranylazides: Reagents for primary azide incorporation into peptides and subsequent Staudinger ligation. Chem. Biol. Drug Des. 68, 85–96. [DOI] [PubMed] [Google Scholar]
- Tolstyka Z. P.; Richardson W.; Bat E.; Stevens C. J.; Parra D. P.; Dozier J. K.; Distefano M. D.; Dunn B.; Maynard H. D. (2013) Chemoselective Immobilization of Proteins by Microcontact Printing and Bio-orthogonal Click Reactions. ChemBioChem 14, 2464–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich D.; Lin P.-C.; Jonkheijm P.; Nguyen U. T. T.; Schroeder H.; Niemeyer C. M.; Alexandrov K.; Goody R.; Waldmann H. (2010) Oriented Immobilization of Farnesylated Proteins by the Thiol-Ene Reaction. Angew. Chem., Int. Ed. 49, 1252–1257. [DOI] [PubMed] [Google Scholar]
- Choi S.-r.; Seo J.-s.; Bohaty R. F. H.; Poulter C. D. (2014) Regio- and chemoselective immobilization of proteins on gold surfaces. Bioconjugate Chem. 25, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.-s.; Poulter C. D. (2014) Sandwich antibody arrays using recombinant antibody-binding Protein L. Langmuir 30, 6629–6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dursina B.-E.; Reents R.; Niculae A.; Veligodsky A.; Breitling R.; Pyatkov K.; Waldmann H.; Goody R. S.; Alexandrov K. (2005) A genetically encodable microtag for chemo-enzymatic derivatization and purification of recombinant proteins. Protein Expression Purif. 39, 71–81. [DOI] [PubMed] [Google Scholar]
- Mahmoodi M. M.; Rashidian M.; Dozier J. K.; Distefano M. D. (2013) Chemoenzymatic site-specific reversible immobilization and labeling of proteins from crude cellular extract without prior purification using oxime and hydrazine ligation. Curr. Protoc. Chem. Biol. 5, 89–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth B. P.; Chen Y.; Wollack J. W.; Sham Y.; Mueller J. D.; Taton T. A.; Distefano M. D. (2007) A universal method for the preparation of covalent protein-DNA conjugates for use in creating protein nanostructures. Angew. Chem., Int. Ed. 46, 8819–8822. [DOI] [PubMed] [Google Scholar]
- Khatwani S. L.; Kang J. S.; Mullen D. G.; Hast M. A.; Beese L. S.; Distefano M. D.; Taton T. A. (2012) Covalent protein-oligonucleotide conjugates by copper-free click reaction. Bioorg. Med. Chem. 20, 4532–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo J. E.; Wickramaratne S.; Khatwani S.; Wang Y.-C.; Vervacke J.; Distefano M. D.; Tretyakova N. Y. (2014) Synthesis of Site-Specific DNA–Protein Conjugates and Their Effects on DNA Replication. ACS Chem. Biol. 9, 1860–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidian M.; Mahmoodi M. M.; Shah R.; Dozier J. K.; Wagner C. R.; Distefano M. D. (2013) A Highly Efficient Catalyst for Oxime Ligation and Hydrazone–Oxime Exchange Suitable for Bioconjugation. Bioconjugate Chem. 24, 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidian M.; Kumarapperuma S. C.; Gabrielse K.; Fegan A.; Wagner C. R.; Distefano M. D. (2013) Simultaneous Dual Protein Labeling Using a Triorthogonal Reagent. J. Am. Chem. Soc. 135, 16388–16396. [DOI] [PMC free article] [PubMed] [Google Scholar]