ABSTRACT
Although CD8+ T cells are important for the control of HIV-1 in vivo, the precise correlates of immune efficacy remain unclear. In this study, we conducted a comprehensive analysis of viral sequence variation and T-cell receptor (TCR) repertoire composition across multiple epitope specificities in a group of antiretroviral treatment-naive individuals chronically infected with HIV-1. A negative correlation was detected between changes in antigen-specific TCR repertoire diversity and CD8+ T-cell response magnitude, reflecting clonotypic expansions and contractions related to alterations in cognate viral epitope sequences. These patterns were independent of the individual, as evidenced by discordant clonotype-specific transitions directed against different epitopes in single subjects. Moreover, long-term asymptomatic HIV-1 infection was characterized by evolution of the TCR repertoire in parallel with viral replication. Collectively, these data suggest a continuous bidirectional process of adaptation between HIV-1 and virus-specific CD8+ T-cell clonotypes orchestrated at the TCR-antigen interface.
IMPORTANCE We describe a relation between viral epitope mutation, antigen-specific T-cell expansion, and the repertoire of responding clonotypes in chronic HIV-1 infection. This work provides insights into the process of coadaptation between the human immune system and a rapidly evolving lentivirus.
INTRODUCTION
CD8+ T cells are key determinants of immune efficacy in human immunodeficiency virus type 1 (HIV-1) infection (reviewed in reference 1). Although simple quantitative correlates of protection are generally lacking, previous studies have identified parameters that typically associate with effective HIV-specific CD8+ T-cell responses, including targeting specificity and breadth, antigen sensitivity, recall proliferation, and polyfunctionality (reviewed in references 2 to 4). Nonetheless, these and other potentially important properties cannot fully explain the different disease outcomes associated with infection.
The inherent quality of a CD8+ T-cell response depends on the arsenal of T-cell receptor (TCR) clonotypes deployed to engage the targeted peptide-human leukocyte antigen class I (pHLA-I) complex. Current paradigms hold that diverse and/or cross-reactive TCR repertoires are beneficial in the face of rapidly evolving RNA viruses because they enable early recognition of emerging epitope variants (5–7). Indeed, restricted TCR diversity is associated with immune escape in hepatitis C virus (HCV) infection (8). Moreover, diverse but highly biased repertoires can facilitate escape due to a lack of variant recognition (9). On the other hand, CD8+ T-cell repertoires that incorporate highly cross-reactive clonotypes are associated with delayed disease progression in simian immunodeficiency virus (SIV) and HIV-1 infection (10–12). Nonetheless, a broad TCR repertoire per se is not necessarily protective, implicating additional clonotypic determinants of CD8+ T-cell efficacy (13). In this light, it has been shown previously that superior viral control can associate with enhanced antigen-specific clonal turnover, reflecting continual replenishment of the response with effective T-cell clonotypes (14–16). However, repertoire evolution is a variable phenomenon, even within CD8+ T-cell responses directed against the same viral epitope (16). In addition, clonotype persistence has similarly been linked with long-term asymptomatic HIV-1 infection (17). These contrasting observations underscore the fact that the HIV-specific CD8+ T-cell response is highly heterogeneous.
Most antigen-specific repertoire studies to date in the HIV-1 field have focused on a single epitope, with limited information on the circulating viral quasispecies. In contrast, we conducted a comprehensive analysis of cognate TCR sequences and viral epitope variation across four targeted specificities in a group of antiretroviral treatment-naive individuals with chronic HIV-1 infection. All subjects carried the highly prevalent HLA-I alleles A*02 and B*08, enabling simultaneous analysis of more than one epitope-specific CD8+ T-cell response over time. A delicate balance was observed between HIV-1 variation and the virus-specific TCR repertoire, whereby only a few clonotypes reacted to changes in the viral milieu. These so-called “clonotypic shifts” markedly affected CD8+ T-cell response magnitude in an antigen-driven manner. Moreover, long-term asymptomatic HIV-1 infection was achieved when the TCR repertoire adapted in response to viral replication.
MATERIALS AND METHODS
Study population.
Eight initial participants with known seroconversion dates were selected from the Amsterdam Cohort Studies on HIV-1 infection and AIDS based on the presence of both HLA-A*02 and HLA-B*08; three individuals also carried the protective HLA-B*27 allele. All subjects were antiretroviral therapy naive prior to and during the time of sample collection. Peripheral blood mononuclear cell (PBMC) and serum samples were drawn from two time points per person: (i) early (time point 1, t1), with a median of 218 days postseroconversion (range, 169 to 568 days), and (ii) late (time point 2, t2), with a median of 1,133 days postseroconversion (range, 986 to 1,226 days) (Table 1). Further PBMC samples were collected from three participants, subjects 5, 6, and 9, an additional seroprevalent individual exclusively selected for these extra analyses (see Table SI in the supplemental material). All individuals were in the asymptomatic, chronic phase of infection.
TABLE 1.
Subject characteristics
| Subject no. | ACS no.a | HLA-I profile | t1 (days post-SC)b | t2 (days post-SC) | CD4+ T-cell count (cells/μl) |
CD8+ T-cell count (cells/μl) |
Viral load (copies/ml)c |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| t1 | t2 | t1 | t2 | t1 | t2 | |||||
| 1 | 19957 | A*01, A*02, B*0801, B*15 | 202 | 1,226 | 470 | 330 | 650 | 790 | 12,000 | ∼58,000* (38,000 to 98,000) |
| 2 | 19885 | A*01, A*02, B*0801, B*27 | 234 | 1,188 | 550 | 410 | 750 | 730 | ∼49,000* (86,000 to 5,800) | 1,200 |
| 3 | 19861 | A*01, A*02, B*0801, B*51 or B*52 | 184 | 1,092 | 620 | 380 | 500 | 960 | 26,000 | 12,000 |
| 4 | 19453 | A*01, A*02, B*0801, B*38 | 169 | 986 | 1,020 | 920 | 650 | 640 | 15,000 | <1,000* |
| 5 | 19342 | A*01, A*02, B*0801, B*40 | 536 | 1,166 | 490 | 800 | 670 | 860 | ∼17,000* (22,000 to 7,600) | ∼7,500* (9,600 to <1,000) |
| 6 | 18840 | A*02, A*02, B*0801, B*27 | 568 | 1,169 | 480 | 690 | 600 | 910 | ∼1,400* (<1,000 to 1,640) | 5,400 |
| 7 | 18839 | A*0207, B*0801, B*27 | 266 | 1,100 | 360 | 420 | 440 | 850 | ∼33,000* (48,000 to 19,000) | 15,000 |
| 8 | 18785 | A*01, A*02, B*0801, B*07 | 189 | 1,080 | 400 | 380 | 510 | 690 | 40,000 | 210,000 |
| Median (range) for group | 218 (169 to 568) | 1,133 (986 to 1,226) | 485 (360 to 1,020) | 415 (330 to 920) | 625 (440 to 750) | 820 (640 to 960) | ||||
ACS, Amsterdam Cohort Study.
SC, seroconversion.
*, trend viral load (range), estimated from the values of the closest adjacent time points.
Flow-assisted sorting of antigen-specific CD8+ T cells.
As no prescreening information was available on the presence/absence of measurable epitope-specific CD8+ T-cell responses, we selected well-defined, dominant epitopes for the three HLA-I alleles of interest: B*08-FLKEKGGL (B8-FL8), B*08-EIYKRWII (B8-EI8), A*02-SLYNTVATL (A2-SL9), and B*27-KRWIILGLNK (B27-KK10). Antigen-specific CD8+ T cells were labeled with pretitrated concentrations of the respective fluorochrome-conjugated pHLA-I tetrameric complexes: (i) B*0801-FL8 and B*0801-EI8 (monomers produced in-house as described previously with minor modifications [18]), conjugated with QD705 and QD605 (Life Technologies), respectively; (ii) A*0201-SL9-APC (where APC is allophycocyanin) (Sanquin); and, as applicable, (iii) B*2705-KK10-PE (where PE is phycoerythrin) (Sanquin). Nonviable cells were eliminated from the analysis using Live/Dead Aqua (Life Technologies). Cells were then washed and surface stained with the following monoclonal antibodies (MAbs): anti-CD3-APC-H7, anti-CD4-PE-Cy5.5, anti-CD8-PE-Cy7, anti-CD14-Alexa Fluor 700, and anti-CD19-AmCyan (where AmCyan is the cyan fluorescent protein from Anemonia majano) (Caltag/Invitrogen). After exclusion of nonviable/CD14+/CD19+ cells, up to four CD3+ CD8+ tetramer-positive populations were sorted in parallel at >98% purity directly into RNAlater solution (Life Technologies) using a customized FACSAria II flow cytometer (BD Biosciences) and stored at −80°C for subsequent TCRβ clonotype analysis.
TCRβ clonotype analysis.
Clonotype analysis was performed as described previously with minor modifications (19). Briefly, mRNA from sorted CD8+ T-cell populations was extracted using a μMACS mRNA isolation kit (Miltenyi Biotec). An anchored template-switch reverse transcription-PCR (RT-PCR) was then used to amplify all expressed TCRβ chains linearly. Amplified products were ligated into the pGEM-T Easy vector (Promega) and transformed into chemically competent Escherichia coli bacteria. Subcloned products were amplified using M13 primers and sequenced via capillary electrophoresis with a BigDye Terminator cycle kit, version 3.1, cycle kit (Life Technologies). Analysis of each TCRβ sequence and assignment of gene usage were performed using Web-based software from ImMunoGeneTics (20). At least 50 TCRβ sequences were successfully analyzed for each sample, a cutoff widely considered appropriate for antigen-specific memory T-cell responses (21).
Sequence analysis of HIV-1 epitopes.
For Gag, viral RNA was isolated from serum using a Viral RNA Minikit (Qiagen) or silica particles as described previously (22). A combined cDNA synthesis and first-round PCR was then performed in 30-μl reaction mixtures using a Titan One-Tube RT-PCR kit (Roche). The following parameters were used: (i) 50°C for 30 min to synthesize cDNA; (ii) 95°C for 2 min to melt; (iii) 40 cycles of 95°C for 15 s, 57°C for 30 s, and 68°C for 2.5 min (increased by 5 s per cycle for the last 30 cycles) to amplify; and (iv) 72°C for 10 min to complete extension. The second, nested PCR was performed using 5 μl of the first-round product in 30-μl reaction mixtures with an Expand High Fidelity PCR System (Roche). The following parameters were used: (i) 95°C for 2 min to melt; (ii) 30 cycles of 95°C for 15 s, 58°C for 30 s, and 68°C for 2.5 min to amplify; and (iii) 72°C for 10 min to complete extension. Primers KVL064 (forward, 5′-GTTGTGTGACTCTGGTAACTAGAGATCCCTCAGA-3′) and NCrev-2 (reverse, 5′-CCTTCCTTTCCACATTTCCAACAG-3′) were used for the combined cDNA synthesis/first-round PCR, and primers KVL066 (forward, 5′-TCTCTAGCAGTGGCGCCCGAACAG-3′) and NCrev-3 (reverse, 5′-CTTTTTCCTAGGGGCCCTGCAATTT-3′) were used for the second, nested PCR.
For Nef, viral RNA was isolated from serum using a Viral RNA Minikit (Qiagen). cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen) using a Nef-specific primer (Nef rv1, 5′-GCTTATATGCAGGATCTGAGG-3′) and purified on silica-based columns (Macherey-Nagel). Template-specific amplification was performed as described previously (23).
Amplified Gag and Nef products were gel purified (Macherey-Nagel), A-tailed, and ligated using a pGEM-T Easy vector system (Promega). Ligated products were then transformed into chemically competent E. coli bacteria and sequenced as described above (4 to 48 clones per sample).
TCRβ diversity analysis.
A T-cell clonotype was defined as a TCRβ chain encoded by a unique nucleotide sequence. Sample clonality was estimated by counting the relative number of distinct clonotypes and by using Simpson's diversity index (DS) (21). This index is defined according to the following equation:
where ni is the clonal size of the ith clonotype (i.e., the number of copies of a specific clonotype), c is the number of different clonotypes, and n is the total number of analyzed TCRβ sequences. This index uses the relative frequency of each clone to calculate a diversity index ranging between 0 and 1, indicating minimal and maximal diversity, respectively. To account for differences in sample size (i.e., the number of successfully analyzed TCRβ sequences), all samples were normalized by random sampling (without replacement) to an equal number of sequences (n = 50) prior to the calculation of TCRβ diversity (i.e., the relative number of unique clonotypes and Simpson's diversity index). This process was repeated 1,000 times, after which median values of TCRβ diversity were determined and used for subsequent analyses.
Statistical analysis.
Sample normalization and statistical analyses were performed using SPSS, version 20.0.0 (SPSS, Inc.). A P value of ≤0.05 was considered statistically significant. Graphics were generated using GraphPad Prism, version 5.04 (GraphPad Software, Inc.). Note that in some analyses (see Fig. 2A and 3), data from multiple T-cell populations per individual and different epitope specificities were pooled; in these instances, the data points cannot be considered fully independent of each other.
FIG 2.
CD8+ T-cell repertoire diversity is not related to viral epitope variation. Antigen-specific TCRβ repertoire diversity (Simpson's diversity index) and viral epitope sequences were determined in parallel for a subset of samples (n = 22). The percentage of wild-type (WT) epitope sequences (A) and the number of variant epitopes present at the time of analysis (B) were used as measures of epitope composition in the autologous viral quasispecies. All plots include data derived from the early and late time points. Correlation testing for data in panel A was performed using the Spearman rank test. Note that Simpson's diversity index was determined after data normalization for appropriate diversity comparisons (see Materials and Methods for details).
FIG 3.
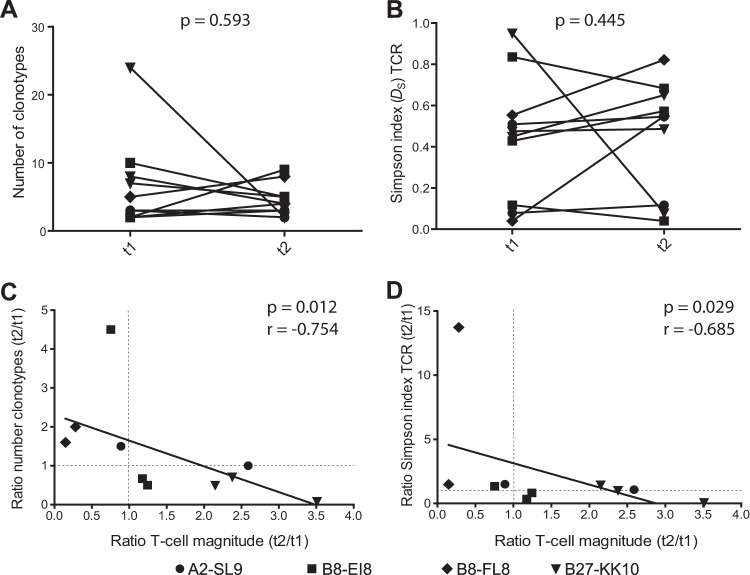
Longitudinal variations in CD8+ T-cell response magnitude correlate negatively with TCRβ diversity. (A and B) TCRβ diversity was quantified at the early (t1) and late (t2) time points for a subset of antigen-specific CD8+ T-cell responses (n = 10) using the relative number of clonotypes (A) and Simpson's diversity index (B). Statistical analyses were performed using the Wilcoxon signed-rank test. (C and D) Changes in CD8+ T-cell response magnitude (ratio of t2 to t1) were related to differences in TCRβ diversity (ratio of t2 to t1) determined by the relative number of clonotypes (C) and Simpson's diversity index (D). Each dot represents a CD8+ T-cell response analyzed at two time points. Correlation testing was performed using the Spearman rank test.
RESULTS
Isolation and analysis of antigen-specific CD8+ T cells.
Eight treatment-naive individuals with chronic HIV-1 infection were selected for coexpression of the HLA-A*02 and HLA-B*08 alleles. Each subject was studied at two time points, approximately 1 (0.5 to 1.5) and 3 (2.7 to 3.5) years postseroconversion (Table 1). Initially, we used pHLA-I tetramers to characterize CD8+ T-cell responses directed against the frequently targeted epitopes A*02-SLYNTVATL (A2-SL9; p17-Gag), B*08-EIYKRWII (B8-EI8; p24-Gag), B*08-FLKEKGGL (B8-FL8; Nef), and B*27-KRWIILGLNK (B27-KK10; p24-Gag). Response magnitude varied as a function of specificity, with B27-KK10 and A2-SL9 eliciting the biggest and smallest CD8+ T-cell responses, respectively (Fig. 1A).
FIG 1.
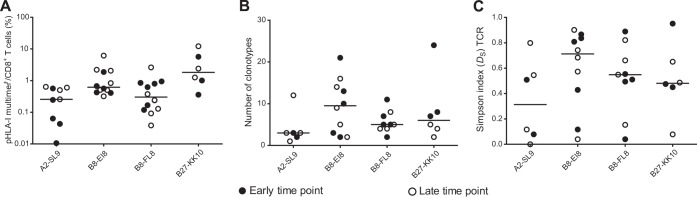
Analysis of CD8+ T-cell populations directed against A2-SL9, B8-EI8, B8-FL8, and B27-KK10. (A) Antigen-specific CD8+ T cells were labeled with pHLA-I tetramers and quantified by flow cytometry. Response magnitude is shown as the frequency of tetramer-positive (multimer+) events in the total CD8+ T-cell population. (B and C) TCRβ diversity was quantified using the relative number of clonotypes (B) and Simpson's diversity index (C). Data are shown for each individual at both the early and late time points.
To compare the clonotypic composition of distinct antigen-specific CD8+ T-cell populations, we used a template-switch anchored RT-PCR to amplify all expressed TCRβ chains from pHLA-I tetramer-positive cells sorted by flow cytometry to high levels of purity. Two measures of diversity were calculated: (i) the relative number of clonotypes (i.e., each unique TCRβ nucleotide sequence after normalization) and (ii) Simpson's diversity index (DS), which accounts for clonotype frequency (21). CD8+ T-cell populations directed against B8-EI8 and A2-SL9 selected repertoires with the highest and lowest degrees of diversity, respectively (Fig. 1B and C; see also Fig. S1 and S2 in the supplemental material). Of note, highly focused and polyclonal responses were observed within each specificity; these patterns were not associated with either the time of sampling (Fig. 1, filled versus open symbols) or the magnitude of the CD8+ T-cell population (data not shown).
Viral epitope variation does not correlate directly with CD8+ T-cell repertoire diversity.
Previous studies from our group and others have suggested that the T-cell repertoire is shaped primarily by the presented epitope (24, 25). To assess the relationship between TCRβ diversity and viral epitope mutation, we conducted an extensive analysis of autologous HIV-1 sequences in targeted regions of the viral genome. Modest variations were detected across viral populations. A single epitope sequence was usually dominant, deviating from the wild type (WT) most prominently in the B8-FL8 and B27-KK10 regions (Tables 2 and 3). Of note, the majority of these variants were predicted to bind their respective HLA-I molecules (73% according to the NetMHC 3.4 server; 82% according to the NetMHCpan server) (see Table SII in the supplemental material) (26, 27). No significant correlations were detected between the frequency of the WT epitope and the diversity of the corresponding TCRβ repertoire, either in terms of Simpson's diversity index (Fig. 2A) or the relative number of clonotypes (see Fig. S3 in the supplemental material). Similarly, there were no associations between TCRβ diversity and either the presence of epitope variants or the number of different epitope sequences (Fig. 2B; see also Fig. S3). A direct analysis of viral epitope variation quantified using Simpson's diversity index also failed to reveal significant correlations with TCRβ diversity (data not shown). These findings indicate that the composition of the viral epitope population does not necessarily associate with TCRβ repertoire diversity at any given time point, although a relationship between these parameters cannot be excluded from the current data.
TABLE 2.
HIV-1 sequence analysis of epitopes restricted by HLA-A*02 and HLA-B*08
| Subject no. | ACS no.a | HLA type(s) | Time point | A2-SL9 |
B8-EI8 |
B8-FL8 |
|||
|---|---|---|---|---|---|---|---|---|---|
| SLYNTVATL | Frequency (%)b | EIYKRWII | Frequency (%) | FLKEKGGL | Frequency (%) | ||||
| 1 | 19957 | A*01, A*02, B*0801, B*15 | t1 | --------- | 100 | -------- | 100 | ----T--- | 79.2 |
| -------- | 19.2 | ||||||||
| ----R--- | 1.6 | ||||||||
| t2 | -----I--- | 100 | -------- | 100 | ----M--- | 93.4 | |||
| -------- | 6.6 | ||||||||
| 3 | 19861 | A*01, A*02, B*0801, B*51 or B*52 | t1 | --------- | 100 | -------- | 100 | -------- | 100 |
| t2 | NDc | ND | ND | ||||||
| 4 | 19453 | A*01, A*02, B*0801, B*38 | t1 | --F------ | 51.7 | ND | ND | ||
| --------- | 48.3 | ||||||||
| t2 | --------- | 100 | -------- | 100 | ----M--- | 86.7 | |||
| -------- | 13.3 | ||||||||
| 5 | 19342 | A*01, A*02, B*0801, B*40 | t1 | ND | ND | ND | |||
| t2 | --F------ | 100 | D------- | 100 | ----E--- | 100 | |||
| 8 | 18785 | A*01, A*02, B*0801, B*07 | t1 | --------- | 100 | -------- | 100 | --N----- | 86.1 |
| ----E--- | 14.0 | ||||||||
| t2 | --------- | 100 | ND | --N----- | 93.9 | ||||
| L-N----- | 3.0 | ||||||||
| -------- | 3.0 | ||||||||
ACS, Amsterdam Cohort Study.
Percentage of sequences in which the epitope occurs.
ND, not detected.
TABLE 3.
HIV-1 sequence analysis of epitopes restricted by HLA-A*02, HLA-B*08, and HLA-B*27
| Subject no. | ACS no.a | HLA type | Time point | A2-SL9 |
B8-EI8 |
B8-FL8 |
B27-KK10 |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SLYNTVATL | Frequency (%)b | EIYKRWII | Frequency (%) | FLKEKGGL | Frequency (%) | KRWIILGLNK | Frequency (%) | ||||
| 2 | 19885 | A*01, A*02, B*0801, B*27 | t1 | --F-A--V- | 100 | -------- | 96.8 | --R----- | 100 | ---------- | 93.6 |
| ---E---- | 3.2 | E--------- | 3.2 | ||||||||
| -----P---- | 3.2 | ||||||||||
| t2 | --F----V- | 93.8 | -------- | 85.7 | ND | ---------- | 85.7 | ||||
| --F--A-V- | 6.3 | -------V | 14.3 | ----VM---- | 14.3 | ||||||
| 6 | 18840 | A*02, A*02, B*0801, B*27 | t1 | --------- | 94.7 | -------- | 100 | -------- | 48.9 | -----M---- | 68 |
| P-------- | 5.3 | --R----- | 51.1 | ---------- | 28 | ||||||
| -----I---- | 4 | ||||||||||
| t2 | NDc | ND | ND | ND | |||||||
| 7 | 18839 | A*0207, A*0207, B*0801, B*27 | t1 | --------- | 100 | -------- | 100 | -------- | 64 | -----M---- | 100 |
| ----R--- | 36 | ||||||||||
| t2 | --------- | 90.3 | -------- | 94.7 | ----R--- | 97.6 | ---------- | 52.6 | |||
| -----I--- | 9.7 | ---R---- | 5.3 | ----E--- | 2.4 | -----M---- | 42.1 | ||||
| R--------- | 5.3 | ||||||||||
ACS, Amsterdam Cohort Study.
Percentage of sequences in which the epitope occurs.
ND, not detected.
Parallel evolution of the TCR repertoire, viral quasispecies, and CD8+ T-cell responses.
It is well established that CD8+ T-cell response magnitude, TCR diversity, and viral epitope sequences can evolve significantly during the course of HIV-1 infection (16). Accordingly, we examined a subset of HIV-specific CD8+ T-cell responses (n = 10) over time. Repertoire diversity varied between the early and late time points without a common tendency to increase or decrease (Fig. 3A and B). Next, we studied how changes in TCRβ diversity (measured as the ratio of the number of normalized clonotypes or the Simpson index at time point 2 divided by the corresponding values at time point 1) related with longitudinal changes in CD8+ T-cell response magnitude (measured as the ratio of the response magnitude at time point 2 divided by the corresponding value at time point 1) (Fig. 3C and D). A significant negative correlation was observed between these two parameters, indicating that the repertoire became less diverse with CD8+ T-cell expansion and more diverse with CD8+ T-cell contraction. Similar correlations were observed between absolute changes in response magnitude and absolute changes in TCRβ diversity (data not shown). These observations suggest that shifts in response magnitude over time were associated with inflation and deflation of particular clonotypes.
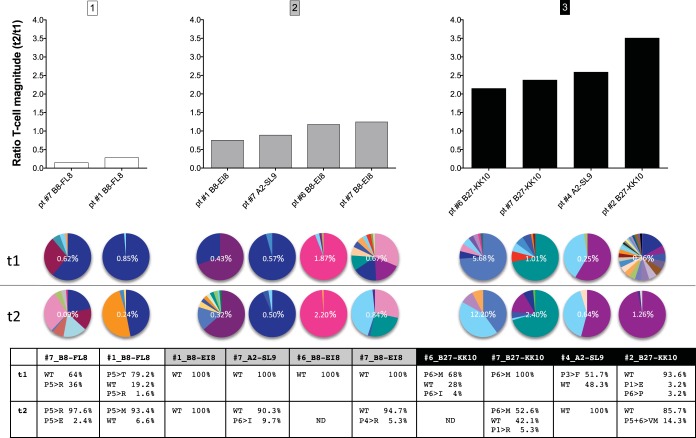
On this basis, we examined the relationship between clonotypic stability and viral epitope diversity. All longitudinal CD8+ T-cell responses were first stratified according to changes in magnitude over time (ratio of t2 to t1): (i) large decreases in magnitude (ratio of ≤0.5); (ii) conservation of magnitude (ratio of >0.5 and <2.0); and (iii) large increases in magnitude (ratio of ≥2.0) (Fig. 4, upper panels). The corresponding TCRβ repertoires (Fig. 4, middle panels) and circulating viral epitopes (Fig. 4, lower panel) were then compared across categories. Interestingly, the observed shifts in CD8+ T-cell response magnitude were often linked with changes in the TCRβ repertoire and viral epitope over time. For example, the two decreasing responses (subject 1 and subject 7, B8-FL8) were accompanied by deflation of one (subject 1) or two (subject 7) dominant clonotypes (Fig. 4A). Similarly, two increasing responses (subject 2 and subject 6, B27-KK10) were paralleled by inflation of previously subdominant clonotypes (Fig. 4C). In both scenarios, viral epitope sequences changed over time. For CD8+ T-cell responses that remained relatively stable, however, fewer mutations were detected in the targeted viral epitopes and TCRβ repertoire composition remained largely unchanged (Fig. 4B).
FIG 4.
The relationship between CD8+ T-cell response magnitude, TCRβ diversity, and viral epitope variation. Antigen-specific CD8+ T-cell responses were stratified according to changes in magnitude over time (ratio of t2 to t1). The bar graphs at the top represent CD8+ T-cell responses that subsided over time (A), remained stable over time (B), or increased over time (C). The clonotypic composition of each CD8+ T-cell population is illustrated in the pie charts (middle), with each color illustrating a unique clonotype, and the respective viral epitope sequences are shown in the chart at the bottom (where P5>R, for example, indicates the substitution of an R residue at position 5). Response magnitudes are indicated in the pie charts as the frequency of tetramer-positive events in the total CD8+ T-cell population. Pie chart colors match clonotypes for each epitope pair but do not correspond between pairs. pt, patient.
To validate these observations, we calculated changes in the absolute numbers of tetramer-binding CD8+ T cells (see Table SIII in the supplemental material). Based on these values, the majority of subjects adhered to the categories determined above with respect to changes in response magnitude. The only exception was the B8-EI8-specific response in subject 7, where the ratio in absolute numbers fell below 2.0. Notably, more pronounced shifts were apparent in the corresponding TCRβ repertoire than in other HIV-specific CD8+ T-cell responses in this category.
Together, these results suggest that preferential inflation or deflation of specific clonotypes within the available repertoire may relate to viral epitope mutations and drive changes in the magnitude of the antigen-specific CD8+ T-cell response.
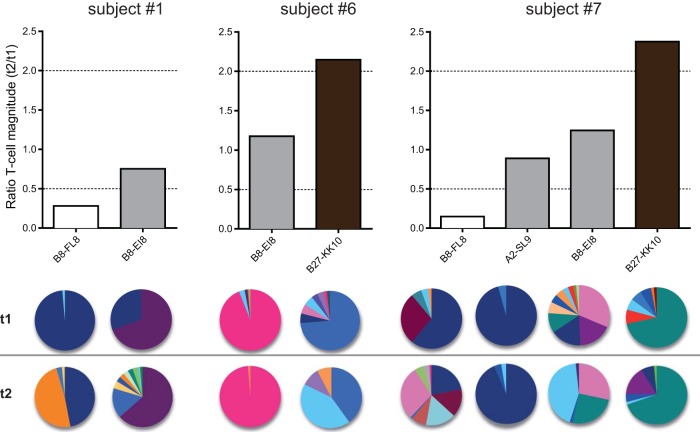
Discordant evolution of CD8+ T-cell responses within individuals.
Next, we conducted a longitudinal analysis of HIV-specific CD8+ T-cell responses in three individuals (Fig. 5). Differences in response magnitude over time (upper panels) were again linked with TCRβ repertoire stability (lower panels), now stratified per person. Discordant patterns of evolution across epitope specificities were also apparent in each donor. For example, the B8-EI8-specific response in subject 1 showed minimal changes in magnitude and clonotypic representation over time, whereas the corresponding B8-FLK-specific response varied substantially across both parameters. Similarly, some responses were stable in subject 6 (B8-EI8) and subject 7 (A2-SL9 and B8-EI8), while others changed dramatically in terms of magnitude and TCRβ repertoire composition (subject 6, B27-KRW; subject 7, B8-FL8 and B27-KK10). Together, these data indicate that HIV-specific CD8+ T-cell responses evolve independently of the host, most likely driven by TCR-antigen interactions.
FIG 5.
CD8+ T-cell response evolution is not dependent on the individual. Antigen-specific CD8+ T-cell responses were stratified according to subject origin. Bar colors in the upper panel are adapted from Fig. 4 and represent decreasing (white), stable (gray), and increasing (black) response magnitudes over time. The clonotypic composition of each CD8+ T-cell population is illustrated in the pie charts. Each color in a pie chart represents a unique clonotype. Pie chart colors match clonotypes for each epitope pair but do not correspond between pairs.
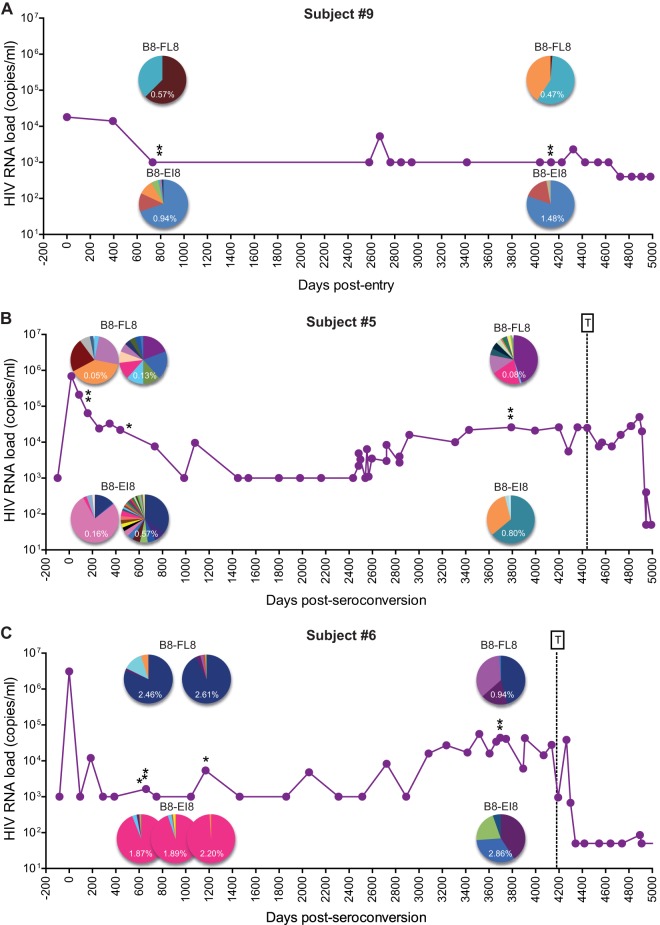
TCR repertoire evolution and viral load dynamics.
A minority of individuals infected with HIV-1 maintain control of viral load at low or undetectable levels. To determine the long-term impact of such low-level viral replication and antigen presentation, we analyzed TCRβ repertoire composition in CD8+ T-cell populations specific for B8-FL8 and B8-EI8 using additional samples from subjects 5, 6, and 9 (Fig. 6), all of whom showed signs of delayed disease progression (asymptomatic with stable viral loads and CD4+ T-cell counts of >300 cells/μl at least 7 years after seroconversion).
FIG 6.
TCR repertoire evolution and viral load dynamics during long-term asymptomatic infection. Viral load trajectories and CD8+ T-cell repertoires specific for B8-FL8 and B8-EI8 are shown for subject 9 (A), subject 5 (B), and subject 6 (C). Each color in a pie chart illustrates a unique clonotype. Pie chart colors match clonotypes for each epitope in each individual but do not correspond between epitopes or individuals. Single asterisks correspond to the time points described in Table 1. Double asterisks correspond to additional time points (see Table SI in the supplemental material). T denotes the commencement of antiretroviral therapy.
Subject 9 maintained undetectable viral loads 14 years after entry into the cohort (Fig. 6A). Subjects 5 and 6 similarly controlled viral loads to low or undetectable levels after acute infection, although progressive increases approximately 7 years after seroconversion warranted subsequent antiretroviral therapy (Fig. 6B and C). Different patterns of TCRβ repertoire evolution were observed in these individuals. Clonotypic representation remained stable in some epitope-specific CD8+ T-cell populations (B8-EI8 in subject 9; B8-FL8 and B8-EI8 in subject 6 during early infection), whereas considerable changes in the constituent TCRβ clonotypes were observed in others (B8-FL8 and B8-EI8 in subject 5; B8-FL8 and B8-EI8 in subject 6 during late infection). Moreover, these clonotypic characteristics often paralleled viral load trajectories. Subject 6 displayed stable viral loads during early infection in conjunction with largely constant TCRβ repertoires specific for B8-FL8 and B8-EI8. As viral loads increased during late infection, however, dramatic changes in clonotypic composition were apparent for both specificities. Similar patterns were observed in subject 5. In this case, epitope-specific TCRβ repertoire instability mirrored viral load fluctuations during both early and late infection. Conversely, the B8-EI8-specific TCRβ repertoire in subject 9 remained stable in the presence of undetectable viral loads, although clear clonotypic shifts were observed in the B8-FL8-specific CD8+ T-cell population. Collectively, these data suggest that the HIV-specific TCR repertoire evolves more rapidly with changes in viral load. Thus, viremic control can be associated with relatively conserved repertoires, whereas higher levels of viral replication tend to drive clonotypic turnover.
DISCUSSION
Although it is widely accepted that HIV-1 evades CD8+ T-cell immunity via epitope mutation, the clonotypic correlates of this phenomenon remain poorly understood. Accordingly, we investigated antigen-specific CD8+ T-cell repertoire dynamics in relation to viral epitope variation in antiretroviral therapy-naive seroconverters with asymptomatic HIV-1 infection. In line with previous work, we found no time-matched correlations between viral epitope composition and clonotypic diversity within the cognate HIV-specific CD8+ T-cell response (28). However, longitudinal analyses revealed a more nuanced picture. A negative correlation across multiple specificities was initially detected between changes in TCR repertoire diversity and CD8+ T-cell response magnitude. More detailed investigations showed that this association reflected clonotype-specific expansions and contractions related to alterations in cognate viral epitope sequences. These patterns were discordant within individuals, suggesting an antigen-driven process. Moreover, clonotype turnover was related to viral load, as noted previously (16). Although tempered by sampling limitations, these data suggest a continuous bidirectional process of adaptation between HIV-1 and virus-specific CD8+ T-cell clonotypes that could ultimately govern immune efficacy and the outcome of infection.
Previous studies have highlighted such dynamic interplay between the TCR repertoire and lentiviral pathogens. This is perhaps best exemplified in the B27-KK10 system, where the early mobilization of public TRBV4-3/TRBJ1-3 clonotypes drives the emergence of TCR escape mutations, which can subsequently be controlled by cross-reactive TRBV6-5/TRBJ1-1 clonotypes in some individuals (11, 29). Adaptive plasticity in the B27-KK10-specific repertoire may even underlie the protective phenotype conferred by this HLA-I allele (30). Similarly cross-reactive clonotypes are also thought to confer preferential outcomes in the context of nonprotective HLA-I alleles. Indeed, a B8-FL8-specific TCR previously associated with long-term nonprogressive disease was detected in this study (12). Nonetheless, it is possible that clonotypic adaptation represents a double-edged sword, in some cases exhausting immune resources without demonstrable benefit. Further detailed studies will be required to clarify these issues in relation to specific epitopes and restriction elements.
In silico analysis predicted sufficient affinities for the majority of epitope variants detected in this study to bind the relevant HLA-I molecules. It therefore seems likely that many of these mutations arose to circumvent TCR recognition. However, the efficacy of this evasion strategy in the presence of a potentially vast cognate TCR repertoire is almost certainly limited to specific scenarios, in contrast to viral mutations that abrogate epitope presentation via effects on antigen processing and/or HLA-I binding. Nonetheless, our data suggest that such “shifting” epitope structures shape the virus-specific TCR repertoire over time. It is notable in this context that clonotypic overlap between CD8+ T-cell populations directed against WT and variant epitopes has been reported previously and may even be commonplace (17, 28).
Despite the primary roles of antigen quantity and quality as determinants of TCR repertoire dynamics, it is important to note that other factors are implicated by the heterogeneous patterns observed in our study. For example, the A2-SL9-specific TCR repertoire in subject 4 remained stable despite substantial sequence variation in the cognate viral epitope and changes in response magnitude. In this case, it seems likely that both dominant clonotypes were equally responsive to the emerging variant, suggesting the operation of additional selection pressures during viral evolution. Conversely, the B8-FL8-specific TCR repertoire in subject 9 shifted substantially over time despite a consistently undetectable viral load. In contrast, the corresponding B8-EI8-specific response remained clonotypically stable over the same prolonged time period. Thus, antigen drive alone does not fully explain the evolutionary patterns observed across distinct epitope-specific TCR repertoires in this study.
In summary, our data show that the antigen-specific CD8+ T-cell repertoire is intimately linked with viral load and epitope variation during chronic HIV-1 infection. This complex dynamic interplay confounds simplistic interpretation and hinders the search for clonotypic determinants of CD8+ T-cell efficacy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the AIDS Fonds Netherlands (number 2011033) and a Fellowship from the Fundação para a Ciência e Tecnologia. D.A.P. is a Wellcome Trust Senior Investigator.
We are grateful to Gerrit Spierenburg and Koos Gaiser for technical assistance.
A.I.C. and D.K. performed experiments, analyzed data, and wrote the manuscript. K.L. performed experiments and analyzed data. J.E.M. performed experiments. B.P.X.G. assisted with data normalization. I.M.M.S. and C.K. critically revised the manuscript. P.V.H. assisted with experiments. M.N. assisted with experiments and revised the manuscript. J.A.M.B. and D.A.P. wrote the manuscript. D.V.B. designed the study and experiments, analyzed data, and wrote the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01765-14.
REFERENCES
- 1.Goulder PJ, Watkins DI. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol 8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seder RA, Darrah PA, Roederer M. 2008. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol 8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 3.Appay V, Douek DC, Price DA. 2008. CD8+ T cell efficacy in vaccination and disease. Nat Med 14:623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 4.Bangham CR. 2009. CTL quality and the control of human retroviral infections. Eur J Immunol 39:1700–1712. doi: 10.1002/eji.200939451. [DOI] [PubMed] [Google Scholar]
- 5.Turner SJ, La Gruta NL, Kedzierska K, Thomas PG, Doherty PC. 2009. Functional implications of T cell receptor diversity. Curr Opin Immunol 21:286–290. doi: 10.1016/j.coi.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikolich-Zugich J, Slifka MK, Messaoudi I. 2004. The many important facets of T-cell repertoire diversity. Nat Rev Immunol 4:123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 7.Davenport MP, Price DA, McMichael AJ. 2007. The T cell repertoire in infection and vaccination: implications for control of persistent viruses. Curr Opin Immunol 19:294–300. doi: 10.1016/j.coi.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Meyer-Olson D, Shoukry NH, Brady KW, Kim H, Olson DP, Hartman K, Shintani AK, Walker CM, Kalams SA. 2004. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med 200:307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, Letvin NL, Franchini G, Wolinsky SM, Koup RA, Douek DC. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity 21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Price DA, Asher TE, Wilson NA, Nason MC, Brenchley JM, Metzler IS, Venturi V, Gostick E, Chattopadhyay PK, Roederer M, Davenport MP, Watkins DI, Douek DC. 2009. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med 206:923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladell K, Hashimoto M, Iglesias MC, Wilmann PG, McLaren JE, Gras S, Chikata T, Kuse N, Fastenackels S, Gostick E, Bridgeman JS, Venturi V, Arkoub ZA, Agut H, van Bockel DJ, Almeida JR, Douek DC, Meyer L, Venet A, Takiguchi M, Rossjohn J, Price DA, Appay V. 2013. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity 38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 12.Dong T, Stewart-Jones G, Chen N, Easterbrook P, Xu X, Papagno L, Appay V, Weekes M, Conlon C, Spina C, Little S, Screaton G, van der Merwe A, Richman DD, McMichael AJ, Jones EY, Rowland-Jones SL. 2004. HIV-specific cytotoxic T cells from long-term survivors select a unique T cell receptor. J Exp Med 200:1547–1557. doi: 10.1084/jem.20032044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendoza D, Royce C, Ruff LE, Ambrozak DR, Quigley MF, Dang T, Venturi V, Price DA, Douek DC, Migueles SA, Connors M. 2012. HLA B*5701-positive long-term nonprogressors/elite controllers are not distinguished from progressors by the clonal composition of HIV-specific CD8+ T cells. J Virol 86:4014–4018. doi: 10.1128/JVI.06982-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med 204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC. 2005. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med 202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer-Olson D, Brady KW, Bartman MT, O'Sullivan KM, Simons BC, Conrad JA, Duncan CB, Lorey S, Siddique A, Draenert R, Addo M, Altfeld M, Rosenberg E, Allen TM, Walker BD, Kalams SA. 2006. Fluctuations of functionally distinct CD8+ T-cell clonotypes demonstrate flexibility of the HIV-specific TCR repertoire. Blood 107:2373–2383. doi: 10.1182/blood-2005-04-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Bockel DJ, Price DA, Munier ML, Venturi V, Asher TE, Ladell K, Greenaway HY, Zaunders J, Douek DC, Cooper DA, Davenport MP, Kelleher AD. 2011. Persistent survival of prevalent clonotypes within an immunodominant HIV Gag-specific CD8+ T cell response. J Immunol 186:359–371. doi: 10.4049/jimmunol.1001807. [DOI] [PubMed] [Google Scholar]
- 18.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 19.Quigley MF, Almeida JR, Price DA, Douek DC. 2011. Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. Curr Protoc Immunol Chapter 10:Unit 10.33. doi: 10.1002/0471142735.im1033s94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giudicelli V, Brochet X, Lefranc MP. 2011. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc 2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 21.Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. 2007. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods 321:182–195. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. 1990. Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28:495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navis M, Schellens IM, van Swieten P, Borghans JA, Miedema F, Kootstra NA, van Baarle D, Schuitemaker H. 2008. A nonprogressive clinical course in HIV-infected individuals expressing human leukocyte antigen B57/5801 is associated with preserved CD8+ T lymphocyte responsiveness to the HW9 epitope in Nef. J Infect Dis 197:871–879. doi: 10.1086/528695. [DOI] [PubMed] [Google Scholar]
- 24.Koning D, Costa AI, Hoof I, Miles JJ, Nanlohy NM, Ladell K, Matthews KK, Venturi V, Schellens IM, Borghans JA, Kesmir C, Price DA, van Baarle D. 2013. CD8+ TCR repertoire formation is guided primarily by the peptide component of the antigenic complex. J Immunol 190:931–939. doi: 10.4049/jimmunol.1202466. [DOI] [PubMed] [Google Scholar]
- 25.Turner SJ, Kedzierska K, Komodromou H, La Gruta NL, Dunstone MA, Webb AI, Webby R, Walden H, Xie W, McCluskey J, Purcell AW, Rossjohn J, Doherty PC. 2005. Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations. Nat Immunol 6:382–389. doi: 10.1038/ni1175. [DOI] [PubMed] [Google Scholar]
- 26.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. 2008. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Roder G, Peters B, Sette A, Lund O, Buus S. 2007. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai KL, Karandikar NJ, Casazza JP, Koup RA. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol 168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- 29.Iglesias MC, Almeida JR, Fastenackels S, van Bockel DJ, Hashimoto M, Venturi V, Gostick E, Urrutia A, Wooldridge L, Clement M, Gras S, Wilmann PG, Autran B, Moris A, Rossjohn J, Davenport MP, Takiguchi M, Brander C, Douek DC, Kelleher AD, Price DA, Appay V. 2011. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood 118:2138–2149. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, Cesa KT, Sela J, Cung TD, Toth I, Pereyra F, Yu XG, Douek DC, Kaufmann DE, Allen TM, Walker BD. 2012. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol 13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.