ABSTRACT
The precise role(s) and topological organization of different factors in the hepatitis C virus (HCV) RNA replication complex are not well understood. In order to elucidate the role of viral and host proteins in HCV replication, we have developed a novel in vitro replication system that utilizes a rolling-circle RNA template. Under close-to-physiological salt conditions, HCV NS5BΔ21, an RNA-dependent RNA polymerase, has poor affinity for the RNA template. Human replication protein A (RPA) and HCV NS5A recruit NS5BΔ21 to the template. Subsequently, NS3 is recruited to the replication complex by NS5BΔ21, resulting in RNA synthesis stimulation by helicase. Both RPA and NS5A(S25-C447), but not NS5A(S25-K215), enabled the NS5BΔ21-NS3 helicase complex to be stably associated with the template and synthesize RNA product in a highly processive manner in vitro. This new in vitro HCV replication system is a useful tool that may facilitate the study of other replication factors and aid in the discovery of novel inhibitors of HCV replication.
IMPORTANCE The molecular mechanism of hepatitis C virus (HCV) replication is not fully understood, but viral and host proteins collaborate in this process. Using a rolling-circle RNA template, we have reconstituted an in vitro HCV replication system that allows us to interrogate the role of viral and host proteins in HCV replication and delineate the molecular interactions. We showed that HCV NS5A(S25-C447) and cellular replication protein A (RPA) functionally cooperate as a processivity factor to stimulate HCV replication by HCV NS5BΔ21 polymerase and NS3 helicase. This system paves the way to test other proteins and may be used as an assay for discovery of HCV inhibitors.
INTRODUCTION
Hepatitis C virus (HCV) is a blood-borne pathogen that infects an estimated 130 million to 200 million people worldwide and constitutes a global health problem (1). A majority of people exposed to HCV (∼85%) go on to develop chronic hepatitis, which can result in liver cirrhosis, liver failure, or hepatocellular carcinoma (2).
HCV is a positive-stranded RNA virus belonging to the Flaviviridae family. It has an ∼9.6-kb genome which encodes a single polyprotein that is co- and posttranslationally processed by cellular and viral proteases to give rise to structural proteins C, E1, and E2 and nonstructural (NS) proteins P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B, which are involved in polyprotein processing, genome replication, or virion packaging (3). HCV replication is carried out by a membrane-associated ribonucleoprotein complex which is comprised of HCV nonstructural proteins and host factors (4, 5). While the functions of some of the HCV proteins, such as NS3 protease, NS3 helicase, and NS5B RNA-dependent RNA polymerase (RdRp), are well understood, the functions of others, such as NS4B and NS5A, are less clear (3). In addition, it is unknown how the various viral and host proteins assemble into a replication complex and participate in the replication process (6).
NS3 is a bifunctional protein with an N-terminal serine protease and a C-terminal NTPase/helicase domain. NS4A is a 54-amino-acid (aa) cofactor that is essential for optimal proteolytic activity and forms a tight complex with NS3 to form NS3-4A. The proteolytic activity of NS3-4A is required to process cleavage sites between NS3 and NS4A, NS4A and NS4B, NS4B and NS5A, and NS5A and NS5B in the HCV polyprotein to release components of the HCV replicase (5, 7, 8). The structure of the NS3 protease domain alone and in complex with the NS4A cofactor has been solved and reveals that the central region of NS4A is buried deeply in the core of NS3, forming a tight noncovalent complex with the NS3 protease catalytic domain, and assists in organizing the active site of the enzyme (9, 10).
The NS3 helicase activity is essential for HCV replication in cultured cells, and it can unwind RNA/RNA, RNA/DNA, and DNA/DNA substrates in a 3′-to-5′ direction (11–16). The unwinding activity of the NS3 helicase domain has been shown to be exquisitely sensitive to salt concentrations. The helicase activity was reduced to 10% in the presence of 50 mM NaCl, and no activity was detected at 150 mM NaCl (17). In other studies, maximal activity was obtained in the absence of potassium ions (11, 15). Similarly, strand displacement by the full-length NS3 was maximal in the absence of sodium ions (18). The NS3 helicase sensitivity to inhibition by salt is unusual; most DNA and RNA helicases are stimulated by salt. For example, the optimal salt concentration for Rep52 DNA helicase is 50 to100 mM NaCl, and that for RNA helicase A is 50 to 100 mM KCl (19, 20).
The structures of the NS3 helicase/NTPase domain alone and in complex with an oligonucleotide have been published previously (21–27). In a crystal structure of a single-chain fusion of NS4A with full-length NS3, the helicase domain is separated from the protease domain by a flexible polypeptide linker (28). A comparison of the unwinding activity of the single-chain NS4A-NS3 fusion with that of the helicase domain or full-length NS3 revealed that the protease domain and the core peptide of NS4A are important for optimal HCV helicase activity (29–32). A β-strand located at the C terminus of NS3 that interacts with the NS3 protease domain serves as a toggle in inducing a large conformational change in NS3 that presumably allows NS3 to switch between its protease and helicase activities (33).
The NS4A protein cofactor of the protease domain has been shown to promote RNA-coupled ATP hydrolysis by the NS3 helicase in NS3-4A (34, 35). There have been various reports that the HCV helicase is modulated by the NS5B polymerase and that the effect differs depending on whether the helicase domain or full-length NS3 is used (32). Other investigators have demonstrated a direct interaction between NS3 and NS5B that is primarily mediated through the protease domain of NS3 (36).
NS5B is an RdRp whose structure adopts a unique shape in the polymerase family due to extensive interactions between the finger and thumb subdomains that encircle the active site of the enzyme (37–39). NS5B appears to lack specificity, has poor affinity for HCV RNA, and can copy heterologous nonviral RNA or DNA templates (40–46). Compared to other well-studied RNA- and DNA-dependent polymerases, NS5B has very poor catalytic activity in vitro on primed single-stranded templates (46, 47). This lack of specificity for HCV RNA and the poor catalytic activity of NS5B in vitro may reflect the possibility that additional viral and host proteins are required for specific protein-protein and protein-RNA interactions that are essential for initiation and elongation of HCV RNA synthesis (48–53).
NS5A is an ∼450-amino-acid, membrane-associated zinc metalloprotein that is essential for viral replication and virion production and may serve as a molecular switch between these two aspects of the HCV life cycle (3, 5, 54). The protein is phosphorylated and organized into three domains (5, 54–59). The structure of full-length NS5A has not yet been solved; however, three different crystal structures of N-terminal domain I of NS5A have been published (60–62). The structure published by Tellinghuisen et al. (60) shows domain I as a homodimer with a positively charged cleft that can serve as a potential RNA binding groove. In contrast, the cleft is absent in the second published structure (61), and the proposed RNA binding surfaces of monomers that formed the cleft in the first structure are exposed to the solvent and face away from each other. In the most recent crystal structure of NS5A domain I, two new dimeric forms of this domain were observed (62). The biological significance of these configurations of NS5A dimeric structure is unknown. While multiple functions have been ascribed to NS5A, its precise roles in HCV replication have not been delineated (5).
The two aims of this study were to (i) create an in vitro HCV replication assay that utilized both HCV NS5B RdRp and NS3 helicase and (ii) use the system to elucidate the roles of other viral and host proteins that might be involved in HCV RNA replication. We describe the development of a novel in vitro HCV replication system that utilizes a rolling-circle RNA template and provide experimental evidence supporting a role for HCV NS5A and human replication protein A (RPA) in HCV replication. RPA is a single-stranded binding (SSB) protein comprised of a three-subunit protein complex (70 kDa, 32 kDa, and 14 kDa) that has multiple essential activities in eukaryotic DNA replication and signaling pathways (63–65). NS5BΔ21 alone has no activity on the rolling-circle template and requires RPA, truncated NS5A(S25-K215) containing domain 1 alone, or NS5A(S25-C447) for RNA synthesis to occur. The addition of NS3 helicase increases the rate of RNA synthesis. Under close-to-physiological salt conditions, RPA and almost full-length NS5A(S25-C447) or truncated NS5A(S25-K215) form protein-protein interactions with NS5BΔ21 and recruit the polymerase to the RNA template, thus stimulating RNA synthesis. However, the presence of both RPA and NS5A(S25-C447), but not truncated NS5A(S25-K215), was required for the replication complex containing NS5BΔ21 and NS3 helicase to stably interact with the rolling-circle RNA template in a highly processive manner. Thus, NS5A(S25-C447) and human RPA show functional cooperativity and behave as a processivity factor for HCV RNA polymerase.
MATERIALS AND METHODS
Materials.
Labeled nucleotides, [35S]methionine (1,000 Ci/mmol), and [32P]UTP (specific activity, 5,000 cpm/pmol) were obtained from PerkinElmer (Waltham, MA); unlabeled nucleotides and T4 gene 32 protein (gp32) were obtained from Amersham Pharmacia Biotech (GE Healthcare Biosciences, Pittsburgh, PA). T7 RNA polymerase and RNasin were obtained from Promega (Madison, WI). M13mp18 single-stranded DNA (ssDNA) and restriction enzymes were obtained from New England BioLabs (Ipswich, MA). Unless mentioned otherwise, all other chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Expression and purification of HCV genotype 1a NS3 helicase domain.
The HCV NS3 RNA helicase domain (S1207 to T1657 of the HCV polyprotein sequence; GenBank accession no. ABV46164.2) was cloned into pET-BS(+). The resulting plasmid, pET-BS(+)/HCV/NS3 helicase, contains a six-histidine tag fused to the N terminus of the NS3 helicase domain to facilitate protein purification. Escherichia coli BL21(DE3) cells, freshly transformed with the pET-BS(+)/HCV/NS3 helicase plasmid, were grown at 30°C in Luria broth (LB) supplemented with 50 μg/ml of carbenicillin. When an optical density at 600 nm (OD600) of 1.0 was reached, the cells were induced for 3 h at 30°C by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.8 mM. After induction, the cells were harvested and stored frozen at −70°C until purification.
Cell paste containing the cloned NS3 helicase domain (NS3h) was lysed in buffer A (50 mM HEPES [pH 8.0], 200 mM NaCl, 10% glycerol, 2.5 mM β-mercaptoethanol [β-ME]) containing 0.1% beta-octoglucoside and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). The lysate was treated with DNase to eliminate DNA bound to the helicase and centrifuged at 54,000 × g to remove debris. The protein was batch adsorbed overnight to Talon resin (Clontech Laboratories, Mountain View, CA). The resin was washed to baseline with buffer A to remove detergent and eluted with buffer A containing 50 mM imidazole adjusted to pH 7.5. The protein was precipitated from the pool by the addition of solid (NH2)SO4 to 45% (2 M) for 30 min on ice. The precipitate was harvested by centrifugation at 54,000 × g. The pellet was resolubilized in buffer B (50 mM HEPES [pH 8.0], 100 mM NaCl, 10% glycerol, 2 mM dithiothreitol [DTT]) and run over a Sephacryl S-100 column (GE Healthcare Biosciences) in buffer B. The eluted peak was treated with thrombin (2 U/mg for 2 h at room temperature [RT]) to remove the histidine tag. The NaCl was diluted to 30 mM and the helicase protein was purified to homogeneity by exchange on a Mono Q ion-exchange column (GE Healthcare Biosciences). The protein was eluted at approximately 150 mM NaCl.
Expression and purification of HCV genotype 1a NS3-4A (protease-helicase domain and NS4A).
The region encoding the 631-amino-acid NS3 full-length protein sequence (A1027 to T1657 of HCV genotype 1a polyprotein sequence; GenBank accession no. ABV46164.2) with the 54-amino-acid NS4A (S1658 to C1711) at its C terminus was cloned into the baculovirus transfer vector pVL1392 (Life Technologies, Grand Island, NY). The resulting NS3-4A (695-aa) plasmid construct containing a six-histidine tag fused to the N terminus of the NS3 protease domain was transfected into Sf9 cells to generate plaque-purified high-titer baculovirus stock, and recombinant NS3-4A was expressed for purification as described previously (66).
Cell paste containing the expressed NS3-4A protein was resuspended in 5 volumes of buffer A (50 mM Na2HPO4 [pH 8.0], 10% glycerol, 300 mM NaCl, 5 mM β-mercaptoethanol, 0.2 mM PMSF, 2.5 μg/ml of leupeptin, 1.0 μg/ml of E64, and 2.0 μg/ml of pepstatin). The suspension was passed once through a microfluidizer (Microfluidics Corporation, Newton, MA), and the lysate was centrifuged at 100,000 × g for 30 min at 4°C. The pellet was resuspended in buffer B (buffer A containing 0.5% n-dodecyl-β-d-maltoside [Affymetrix, Santa Clara, CA]; 2.5 ml/g of paste), Dounce homogenized, and mixed at 4°C for 3 h before centrifugation at 100,000 × g for 30 min at 4°C. The imidazole concentration of the supernatant fraction was adjusted to 10 mM and batch adsorbed by mixing overnight with nickel-nitrilotriacetic acid (NTA) agarose metal affinity resin (Qiagen, Valencia, CA; 1 ml of resin/5 mg of expected NS3-4A protein). The resin was poured into a gravity flow column (Kontes FlexColumn; 2.5 cm by 10 cm) and washed with 10 volumes of buffer C (buffer A containing 0.1% n-dodecyl-β-d-maltoside and 10 mM imidazole). NS3-4A was eluted on ice in 3 to 4 column volumes of buffer D (buffer C containing 300 mM imidazole), and fractions were evaluated for the presence of NS3-4A by SDS-PAGE. Fractions containing NS3-4A were pooled and stored at −70°C until the next purification step.
The pooled NS3-4A protein-containing fraction from nickel affinity chromatography was purified further on a Superdex 200 26/60 column (GE Healthcare Biosciences) that was equilibrated with buffer D (20 mM HEPES [pH 8.0], 10% glycerol, 300 mM NaCl, 10 mM β-mercaptoethanol, and 0.05% n-dodecyl-β-d-maltoside). Column fractions were analyzed by SDS-PAGE, and NS3-4A-containing fractions were pooled and stored at −70°C.
Expression and purification of HCV genotype 1b C-terminally truncated NS5A(S25-K215).
The N-terminal domain I of NS5A (S25-K215; HCV 1b; GenBank accession no. KC124831.1) lacking the N-terminal amphipathic helix (24 amino acids) was cloned into pET30b (EMD Millipore, Billerica, MA) with a C-terminal polyhistidine tag and enterokinase cleavage site and expressed in 6 liters of BL21(DE3) cells. An overnight seed culture grown in LB medium containing 30 μg/ml of kanamycin was used to inoculate 6 1-liter cultures. The cells were grown at 37°C shaking at 250 rpm to an OD600 of 0.55 in brain heart infusion medium (BD Biosciences, Franklin Lakes, NJ) and induced with 1 mM IPTG for 5 h at 25°C. Cells were harvested by centrifugation at 7,000 × g for 10 min, and the pellets were stored at −80°C. All purification steps, except for the polyhistidine tag cleavage, were performed on ice or at 4°C. Cell paste from 6 liters of culture producing recombinant NS5A(S25-K215) was resuspended in lysis buffer (50 mM HEPES containing 10% glycerol, 250 mM NaCl, 5 mM imidazole-HCl, 5 mM β-ME, 20 μl/liter of Benzonase nuclease [EMD Millipore], 2 μg/liter of leupeptin, 1 μg/liter of pepstatin [Roche Diagnostics Corp., Indianapolis, IN], 20 μM diisopropylfluoro-phosphate [DFP; pH 7.9; Sigma-Aldrich, St. Louis, MO]) at a ratio of 10 ml per g of cell paste. The suspension was passed two times through a microfluidizer (Microfluidics Corporation, Newton MA), and the lysate was centrifuged at 54,000 × g for 1 h. The supernatant fraction was batch adsorbed by mixing overnight with 5 ml of His-Select nickel affinity gel (Sigma-Aldrich). The resin containing bound NS5A(S25-K215) was packed in a column, washed with wash buffer (50 mM HEPES containing 10% glycerol, 250 mM NaCl, 5 mM imidazole-HCl, and 5 mM β-ME [pH 7.9]), and eluted with 4 column volumes of elution buffer (wash buffer containing 250 mM imidazole-HCl [pH 8.0]). The eluate was collected into 75 ml of buffer A (20 mM HEPES containing 10% glycerol and 2 mM DTT [pH 8.0]) and loaded onto a 5-ml HiTrap Q HP column (GE Healthcare Biosciences) that had been preequilibrated in buffer B (buffer A containing 50 mM NaCl). The HiTrap Q column was washed with buffer B and eluted with a 40-column-volume linear gradient from 50 mM NaCl to 400 mM NaCl. Fractions containing NS5A(S25-K215) were pooled, concentrated using a centrifugal concentrator (VS2002; Sartorius Stedim Biotech, Bohemia, NY), and incubated overnight at room temperature with recombinant enterokinase (New England BioLabs, Ipswich, MA) to remove the polyhistidine tag. The cleaved protein was further purified on a 5-ml HiTrap Q HP column followed by a Superdex 75 column (GE Healthcare Biosciences) that had been preequilibrated in buffer C (20 mM HEPES containing 250 mM NaCl and 2 mM DTT [pH 8.0]). The peak monodisperse fractions were pooled, concentrated to 1.15 mg/ml, and stored at −80°C.
Expression and purification of HCV genotype 1b NS5A(S25-C447).
NS5A(S25-C447) (HCV 1b; GenBank accession no. KC124831.1) containing a deletion of the N-terminal amphipathic helix was cloned in pET30b (EMD Millipore) containing a C-terminal polyhistidine tag and an enterokinase cleavage site and expressed at the 1-liter scale in 2.5 liters of ultrayield baffled flasks (Thomson Instrument Company, Oceanside, CA), via autoinduction, using Overnight Express instant TB medium (EMD Millipore) containing 30 μg/ml of kanamycin. Cultures were inoculated with 10 ml of an overnight LB medium seed and grown at 37°C for 4 h with shaking at 250 rpm; the temperature was then dropped to 15°C for an additional 48 h. Cells were harvested via centrifugation (5,000 × g) for 15 min, and the cell paste was frozen at −80°C until purification.
All purification steps were performed on ice or at 4°C. Cell paste from 1 liter of culture producing recombinant NS5A(S25-C447) was resuspended in lysis buffer (50 mM potassium phosphate containing 10 mM Tris-HCl, 5% glycerol, 500 mM NaCl, 0.25% Tween 20, 5 mM imidazole-HCl, 5 mM β-ME, 20 μl/liter of Benzonase nuclease, 2 μg/liter of leupeptin, 1 μg/liter of pepstatin, and 20 μM DFP [pH 8.0]) at a ratio of 10 ml per g of cell paste. The resuspension was lysed by sonication, and the lysate was centrifuged at 43,000 × g for 1 h. The supernatant fraction was batch incubated overnight with 4 ml of His-Select nickel affinity gel (Sigma-Aldrich, St. Louis, MO) with mixing. The resin containing the bound NS5A(S25-C447) was packed into a column, washed with 20 column volumes of wash buffer (50 mM potassium phosphate containing 10 mM Tris-HCl, 5% glycerol, 500 mM NaCl, 0.25% Tween 20, 5 mM imidazole-HCl, and 5 mM β-ME), and eluted with 4 column volumes of wash buffer containing 300 mM imidazole-HCl. The eluate was immediately loaded onto a HiPrep 26/10 desalting column (GE Healthcare Biosciences) that had been preequilibrated in buffer B (20 mM HEPES containing 10% glycerol and 200 mM NaCl). Fractions containing NS5A(S25-C447) were pooled and concentrated in a centrifugal concentrator and loaded onto a Superdex 200 10/300 GL column (GE Healthcare Biosciences). The peak monodisperse fractions were collected and stored at −80°C.
Cloning, expression, and purification of HCV genotype 1b NS5BΔ21 polymerase.
NS5B polymerase (aa 2420 to 2989 of the HCV polyprotein) from hepatitis C virus subtype 1b (GenBank accession no. AAK08509) containing a hexahistidine tag at the N terminus and lacking the C-terminal 21-amino-acid amphipathic helix was cloned into a pET21B vector and transformed into E. coli BL21(DE3) cells using a standard protocol. Freshly transformed cells were grown at 37°C in brain heart infusion medium (Difco Laboratories, Detroit, MI) supplemented with 100 μg/ml of ampicillin. Cells were grown at 37°C up to an optical density of 0.7 at 600 nm, and expression was induced at 28°C with 0.2 mM IPTG. Cells were harvested via centrifugation 4 h postinduction and flash frozen at −80°C prior to purification.
Frozen cell pellets (∼30 g) were thawed in 10 volumes of buffer A (40 mM potassium phosphate buffer [pH 7.4], 200 mM NaCl, 10% [vol/vol] glycerol, 3 mM β-ME, and 5 mM imidazole) containing 50 μM DFP, 1 μg/ml of E-64 protease inhibitor (Sigma-Aldrich), 1 μg/ml of leupeptin, and 10 μg/ml of pepstatin (Roche Diagnostics Corp., Indianapolis, IN) and lysed in a microfluidizer (Microfluidics, Newton, MA). The lysate was centrifuged at 54,000 × g for 45 min, and the supernatant was incubated with 0.5 ml of nickel metal affinity resin (Sigma-Aldrich) per g of cell paste overnight at 4°C. The resin was washed with 20 column volumes of buffer B (40 mM potassium phosphate buffer [pH 7.4], 500 mM NaCl, 10% [vol/vol] glycerol, 3 mM β-ME, and 5 mM imidazole), followed by a wash with 5 column volumes of buffer A and elution with buffer A containing 200 mM imidazole. The fractions containing the polymerase were pooled and desalted into buffer C (25 mM HEPES [pH 7.5], 0.25 mM EDTA, 10% glycerol, and 1 mM DTT) containing 200 mM NaCl using a Sephadex G-25 16/60 column and loaded onto a 5-ml HiTrap SP Sepharose column (Thermo Fisher Scientific, Waltham, MA) that had been preequilibrated with buffer C plus 200 mM NaCl. The column was washed with 2 column volumes of buffer C containing 200 mM NaCl, followed by a steep wash gradient from 200 to 500 mM NaCl in buffer C over 2 column volumes. The polymerase was eluted with a 500 to 700 mM NaCl gradient in buffer C over 10 column volumes, concentrated by ultrafiltration using a Vivaspin with a 30-kDa molecular mass cutoff (Thermo Fisher Scientific) and loaded onto a Superdex 200 column (60 by 2.6 cm; Thermo Fisher Scientific) equilibrated in buffer D (10 mM Tris-HCl [pH 7.5], 600 mM NaCl, 10% [vol/vol] glycerol, and 5 mM DTT). Fractions were pooled based on SDS-PAGE and stored at −80°C until further use.
Purification of RPA endogenous complex from HeLa cells.
HeLa cells from a 3-liter suspension cell culture (∼2 × 109 cells) were used for the purification procedure. The cells were collected by centrifugation for 10 min at 3,000 rpm in a Beckman JS-4.2 rotor at 4°C. The cell pellets were resuspended in 40 ml of ice-cold phosphate-buffered saline (PBS), transferred into a conical 50-ml centrifugation tube, and pelleted in a Beckman CS-6R centrifuge for 10 min at 1,000 rpm at 4°C. The cell pellet was resuspended in 1.5 times the packed cell volume of ice-cold hypotonic buffer H (20 mM HEPES [pH 7.5], 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 1 mM PMSF, and Roche complete mini-cocktail protease inhibitors at the recommended concentrations) and centrifuged immediately for 10 min at 2,000 rpm in a Beckman CS-6R centrifuge (Beckman Coulter Inc., Brea, CA) at 4°C. The cells were resuspended in 12 ml (3 times the column volume) of ice-cold hypotonic buffer and kept on ice for 20 min, allowing them to swell. The swollen cells were transferred into a glass Dounce homogenizer and homogenized with a glass B pestle until the release of nuclei was seen under the microscope by trypan blue (Invitrogen, Carlsbad, CA) inclusion in about 80% of cells (30 to 50 up-and-down strokes). While the cell homogenate was stirred on ice, the salt concentration was adjusted to 0.2 M by dropwise additions of 5 M NaCl, and the cell homogenate was kept on ice with occasional swirling for an additional 20 min (nuclear extraction). The cell homogenate was cleared from nuclei by centrifugation in a Beckman Avanti J-25 (Beckman Coulter Inc.) centrifuge with a JA-25.50 rotor at 20,000 rpm (17,217 × g) for 30 min at 4°C. The supernatants were collected and centrifuged again at 100,000 × g in a 45Ti rotor in a Beckman Optima LE-80K ultracentrifuge for 30 min at 4°C. The supernatant containing the cell extract was dialyzed overnight against buffer A (20 mM Tris [pH 7.5], 0.1 mM EDTA, 50 mM NaCl, 0.01% NP-40, 10% glycerol, Roche complete mini-cocktail protease inhibitors at the recommended concentrations) at a volume ratio of 1:200 at 4°C. The dialyzed cell extract was collected and precipitated by dropwise addition of saturated ammonium sulfate salt solution. The concentration of ammonium sulfate in the cell extract was adjusted to 35% of saturation at 4°C (0.56 ml of saturated ammonium sulfate per 1 ml of cell extract), and the precipitates were pelleted by centrifugation at 10,000 × g for 30 min. The precipitates were resuspended in 4 ml of ice-cold buffer B (20 mM Tris [pH 7.5], 0.1 mM EDTA, 500 mM NaCl, 0.01% NP-40, 10% glycerol, and Roche complete mini-cocktail protease inhibitors at the recommended concentrations) and, after filtration through a Whatman filter paper, loaded onto a 0.5-ml ssDNA-cellulose column (Sigma-Aldrich). After being washed with 20 ml of buffer B, the RPA-containing fractions were eluted with 2 ml of buffer C (20 mM Tris [pH 7.5], 0.1 mM EDTA, 2 M NaCl, 0.01% NP-40, 10% glycerol, Roche complete mini-protease inhibitor cocktail). The eluate was dialyzed overnight against 4 liters of buffer A at 4°C. The next day, the sample was loaded onto a Q-Sepharose column (HiTrap Q HP; GE Healthcare Biosciences) equilibrated with buffer A. After washing with 5 column volumes of equilibration buffer, the bound proteins were eluted with a linear 50 to 400 mM NaCl gradient in 25 ml. The fractions of gradient were collected in 0.5-ml volumes. RPA came off the column at 125 to 150 mM NaCl. The presence of RPA in the gradient fractions was determined by SDS-PAGE and Western blotting with specific antibody against the largest p70 subunit of RPA (H-7; Santa Cruz Biotechnology Inc., Dallas, TX). The fractions containing RPA were combined, concentrated in protein concentrators with a molecular mass cutoff of 20 kDa (Pierce; Thermo Fisher Scientific) up to 0.25 μg per ml, dialyzed against buffer D (20 mM Tris [pH 7.5], 0.1 mM EDTA, 150 mM NaCl, 20% glycerol), aliquoted in 100-μl volumes, and stored at −80°C. The purity of isolated RPA, as estimated by silver staining of SDS-PAGE gel, was around 70%.
Structural comparison of HCV NS5A and CMV UL44.
The dimeric structure of HCV NS5A domain I (PDB code 1ZH1) was overlaid with the dimeric structure of the cytomegalovirus (CMV) DNA polymerase subunit UL44 (PDB code 1T6L), which has a C clamp shape and is known to act as a processivity-promoting factor in CMV replication. The PyMOL Molecular Graphics System (Schrödinger, LLC) was used for overlaying the two structures. As there are no similarities in amino acid sequence or secondary-structure elements of the two structures, the 3-dimensional (C clamp) shape of the two structures was used to guide the manual overlay of the two structures.
Preparation of RNA rolling-circle template.
Briefly, two RNA oligonucleotides (74 nucleotides [nt]and 106 nucleotides) were synthesized with T7 RNA polymerase on a parental DNA template primed with a T7 promoter primer. The 74-nt RNA oligonucleotide was hybridized to a bridging DNA oligonucleotide and ligated with DNA ligase. The double-stranded RNA rolling-circle template was constructed by hybridization of the RNA circle and the 106-nucleotide RNA oligonucleotide. Four DNA oligonucleotides were synthesized to generate the rolling-circle template for in vitro transcription reactions: DNA1 oligonucleotide, GAGTGGTATAGTGGAGTGAAGTGAGGTGAAGGGTTGATGGTGAATATTGGGGAGGGTGGAGGTTATGGTGTCTCCCTATAGTGAGTCGTATTA; DNA2 oligonucleotide, CTCCCCAATATTCACCATCAACCCTTCACCTCACTTCACTCCACTATACCACTCGGGAGACACCATAACCTCCACATAAAAAAAAAAAAAAAAAAAAAAAAATCTCCCTATAGTGAGTCGTATTA; T7 promoter oligonucleotide, TAATACGACTCACTATAGGGAGA; and bridge oligonucleotide, CCCTCTGTGGTAGAGTGGTATAGT. Specifically, 20 μg of DNA1 or DNA2 was incubated in 20 μl of annealing buffer (20 mM Tris-HCl [pH 7.5] and 100 mM NaCl) in the presence of 200 μg of the T7 promoter oligonucleotide for 1 min at 90°C and then cooled slowly to 24°C. Twelve micrograms of each annealed oligonucleotide was used to prepare short RNA transcripts using the Ambion MEGAshortscript transcription kit (Life Technologies, Grand Island, NY). After incubation for 3 h at 37°C, the reaction mixtures were incubated with DNase for 15 min at 37°C, cooled to 4°C, extracted with phenol-chloroform, and precipitated with ethanol as per standard protocols. RNA oligonucleotide 1 (the transcript of DNA1; 5.16 nmol) and bridge DNA oligonucleotide (25.8 nmol) were incubated in 60 μl of buffer A for 1 min at 65°C and then cooled slowly to 16°C. DNA ligase (10,000 U; New England BioLabs) was added, and incubation was continued for an additional 12 h at 16°C. After DNase treatment for 30 min at 37°C, circular RNA was purified by 10% urea-PAGE. A total of 270 pmol of circle RNA was mixed with 11,382 pmol of RNA oligonucleotide 2 (the transcript of DNA2) in 50 μl of annealing buffer and incubated for 5 min at 75°C, after which the mixture was cooled slowly to 4°C.
RNA synthesis assays using the rolling-circle template.
Sixty-six nanograms (1 pmol) of rolling-circle RNA was incubated with 2 pmol (126 ng) (or as indicated) of NS5BΔ21, 4 pmol (200 ng) of NS3h (when present), and 16.7 pmol (2 μg) of RPA (when present) in 25 μl of replication buffer (20 mM Tris-HCl [pH 7.5], 8 mM MgCl2, 2 mM ATP, 250 μM [each] CTP, GTP, and UTP, 0.1 mM EDTA, 5 mM DTT, 4% glycerol, 40 μg/ml of bovine serum albumin [BSA], 1 μg/ml of RNasin, [32P]UTP [5,000 cpm/pmol], and 150 mM NaCl) for 2 h at 30°C. At different time points, aliquots of 4 μl were removed and analyzed as described below.
RNA synthesis assays using a poly(A)/oligo(dT) template.
A 2.5-μg quantity of poly(A)/oligo(dT) (Life Technologies, Grand Island, NY) was incubated in 50 μl of replication buffer (described above) at 30°C. At different time points, aliquots of 4 μl were removed, 50 μg of carrier yeast tRNA (Sigma-Aldrich, St. Louis, MO) was added, and the mixtures were precipitated in 1 ml of 30% trichloroacetic acid (TCA) and spotted onto Whatman GF/F filters (Thermo Fisher Scientific); the filters were washed with 10% TCA followed by 70% ethanol, dried, and counted in a liquid scintillation counter.
Interaction of 35S-labeled proteins with RPA bound to ssDNA.
35S-labeled proteins were prepared using a commercially available transcription/translation kit (TnT Quick Coupled Transcription/Translation System; Promega, Madison, WI) using DNA templates encoding proteins under the control of a T7 promoter as per the manufacturer's instructions. Reactions were performed in 150 μl containing 800 fmol of M13mp18 ssDNA, 50 μg of RPA (when present) or 75 μg of gp32 (when present), and 25 fmol of 35S-labeled protein in column buffer (20 mM Tris-HCl [pH 7.5], 8 mM MgCl2, 5 mM DTT, 5% glycerol, 50 μg/ml of BSA) containing 150 mM NaCl. Reaction mixtures were incubated for 10 min at 30°C and then applied to 5-ml agarose BioGel A15 columns (Bio-Rad Laboratories, Hercules, CA) equilibrated with column buffer at 4°C. Fractions of 200 μl were collected, and 150-μl aliquots were TCA precipitated and analyzed for 35S-protein using liquid scintillation counter.
SPR.
Immobilization of NS5BΔ21 (5,900 resonance units [RU]) and NS3h (3,900 RU) was performed on Biacore CM dextran matrix-coated sensor chip CM5 (GE Healthcare Biosciences). After each analysis, the surface was regenerated by a 15-min wash with 100 mM ethanolamine without decreasing the capacity of the immobilized proteins. Surface plasmon resonance (SPR) analysis was performed by passing 35 μl of RPA, NS3h, or NS5BΔ21 (40 nM protein solution) over a chip containing immobilized protein. All proteins were dialyzed against SPR buffer (10 mM HEPES-NaOH [pH 7.4], 150 mM NaCl, 3.4 mM EDTA, and 0.005% Tween 20) to remove buffer-related artifacts. All proteins were tested for interaction with a sensor chip containing no protein.
Gel filtration analysis of 35S-NS3h and NS5BΔ21.
The mixtures contained 0.3 mg of NS5BΔ21 and 600 fmol of 35S-NS3h or 35S-NS3h alone in a volume of 200 μl of column buffer (20 mM Tris-HCl [pH 7.5], 8 mM MgCl2, 5 mM DTT, 5% glycerol, 50 μg/ml of BSA) containing 200 mM NaCl. Proteins were incubated at 30°C for 10 min before injection of the mixture into an HR 10/30 Superose 12 column equilibrated with column buffer containing 200 mM NaCl. Fractions of 200 μl were collected, and 5-μl aliquots were analyzed for 35S-protein using liquid scintillation counting.
Processivity competition experiment.
A standard rolling-circle reaction with 66 ng (1 pmol) of the rolling-circle RNA template, 2 pmol (126 ng) of NS5BΔ21, 4 pmol (200 ng) of NS3h, 16.7 pmol (2 μg) of RPA (when present), and 3 pmol of NS5A (when present) was used. Protein mixtures were premixed in the replication buffer (20 mM Tris-HCl [pH 7.5], 8 mM MgCl2, 2 mM ATP, 250 μM GTP, 0.1 mM EDTA, 5 mM DTT, 4% glycerol, 40 μg/ml of BSA, 1 μg/ml of RNasin [Promega, Madison, WI], and 150 mM NaCl) and incubated on ice for 30 min before addition of the rolling-circle RNA template, [32P]UTP (250 μM), and CTP (250 μM). Reaction mixtures were warmed up to 30°C for 5 min, and 5 μg of poly(A)/oligo(U) was added. The mixtures were incubated for an additional 2 min, and synthesis was initiated by addition of GTP and [32P]UTP (5,000 cpm/pmol). At the desired time points, the reactions were stopped with 0.5 M EDTA and 10 μg of yeast tRNA. After phenol-chloroform extractions and ethanol precipitation, the reaction mixtures were diluted in 1× NorthernMax formaldehyde loading dye (Life Technologies, Grand Island, NY), electrophoresed on a formaldehyde-0.5% agarose gel, and autoradiographed.
RESULTS
Purification of HCV proteins and human RPA.
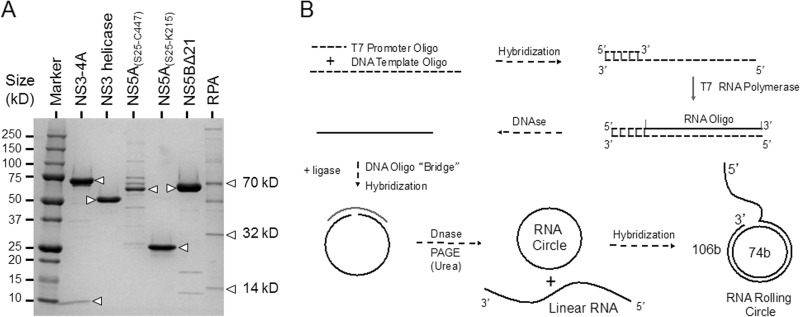
In order to set up an in vitro HCV replication system, NS3-4A, the NS3 helicase domain, NS5A(S25-K215), NS5A(S25-C447), and NS5BΔ21 polymerase of HCV were cloned and expressed as described in Materials and Methods. Native human RPA was purified from HeLa cell extracts as described in Materials and Methods. The protein preparations were visualized by SDS-PAGE (Fig. 1A), and their identities were confirmed by Western blot analyses (data not shown).
FIG 1.
Setting up an in vitro HCV rolling-circle replication system. (A) Purification of HCV proteins and RPA. HCV proteins NS3-4A, NS3 helicase, NS5A(S25-C447), NS5A(S25-K215), NS5BΔ21 polymerase and the native human RPA were purified as described in Materials and Methods and analyzed using SDS-polyacrylamide gel electrophoresis and Coomassie blue staining. The arrowheads indicate the bands corresponding to individual proteins or protein subunits in the case of NS3-4A and RPA. (B) Design of the rolling-circle RNA template. Two RNA oligonucleotides (74 nt and 106 nt) were synthesized with T7 RNA polymerase on the primed DNA templates with T7 promoter sequence primers as described in Materials and Methods. The 74-nt RNA oligonucleotide was hybridized with the DNA oligonucleotide “bridge” and ligated with DNA ligase to create a RNA circle. The rolling-circle RNA template was constructed by hybridization of the RNA circle and the 106-nt RNA oligonucleotide to complete the formation of the rolling-circle RNA template.
Design of a rolling-circle RNA template.
Rolling-circle systems are well established for the T4 and E. coli in vitro DNA replication systems (67, 68). In these systems, a helicase unwinds the DNA duplex at the fork of the template and the replicative DNA polymerase is in constant contact with the helicase as it incorporates nucleotides at the 3′ end of the annealed strand. The product of the synthesis is a continuously growing, high-molecular-weight, 5′ DNA tail. Previous studies have shown that NS5B can utilize both RNA and DNA templates (40–46). The NS3 helicase can unwind RNA/RNA, RNA/DNA, and DNA/DNA substrates in a 3′-to-5′ direction (11, 13–15). To incorporate both the RNA polymerase activity of NS5B and the RNA helicase activity of NS3 into one assay, we prepared a rolling-circle RNA template as described in Materials and Methods. In our system and in contrast to the E. coli and the T4 DNA replication systems described above, the product of synthesis on a rolling-circle RNA template by RNA-dependent RNA polymerase will result in a continuously growing, high-molecular-weight 5′ RNA tail (Fig. 1B).
RNA synthesis by NS5BΔ21 on a rolling-circle RNA template requires RPA and is stimulated by NS3 helicase.
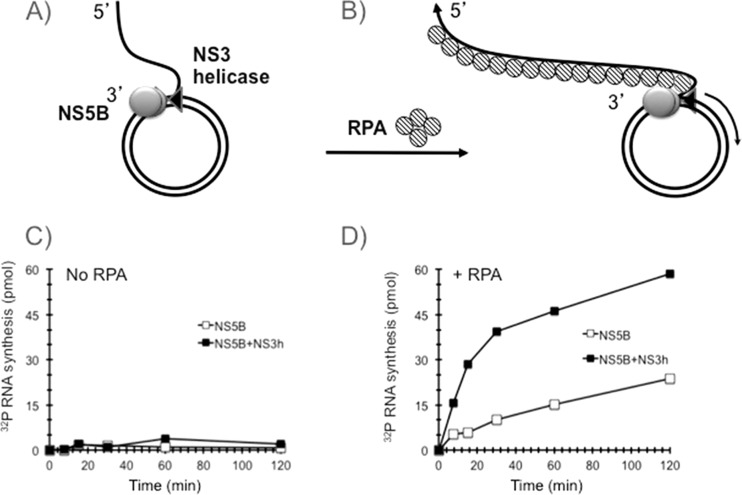
In order to test whether NS5BΔ21 can synthesize RNA on a rolling-circle RNA template under close-to-physiological salt conditions, we added 2 pmol of NS5BΔ21 and 4 pmol of NS3 helicase domain individually and in combination to the rolling-circle template and initiated synthesis by adding NTP in the presence of 100 mM NaCl. These reactions were carried out in both the absence and presence of 2 μg of human RPA (schematically shown in Fig. 2A and B, respectively). In the absence of RPA, no significant RNA synthesis was observed with NS5BΔ21 alone or in combination with the NS3 helicase domain (Fig. 2C). In contrast, the addition of RPA stimulated RNA synthesis by NS5BΔ21 (Fig. 2D). The addition of NS3 helicase domain significantly increased RNA synthesis, from 15 to 50 pmol of RNA product per hour per pmol of the rolling-circle RNA template (Fig. 2D). The continuous synthesis of RNA product by HCV NS5BΔ21 and the NS5BΔ21-NS3 helicase domain in the presence of RPA did not reach a plateau after 2 h, suggesting that RNA synthesis was robust and stable and that the accumulation of product did not inhibit the reaction. The exquisite sensitivity of the unwinding activity of the HCV NS3 helicase domain to salt inhibition has been a conundrum because the helicase has to be active at physiological salt conditions during infection. Using a rolling-circle RNA template, we observed significant stimulation of RNA synthesis by the NS3 helicase domain in the presence of NS5BΔ21 and RPA in the presence of 100 mM NaCl.
FIG 2.
RPA is required for RNA rolling-circle synthesis. (A and B) Schematic representations of the templates that were used in the reactions. (C and D) NS5BΔ21 (open squares) or NS5BΔ21-NS3h (filled squares) were assembled onto a rolling-circle template without RPA (C) or in the presence of RPA (D). Synthesis was initiated by adding NTP, aliquots were withdrawn at the indicated time points, and [32P]UMP incorporation was determined as described in Materials and Methods.
NS5BΔ21 has poor activity on a multiply primed linear template, and activity is not stimulated by the addition of RPA, T4 gp32, or NS3 helicase.
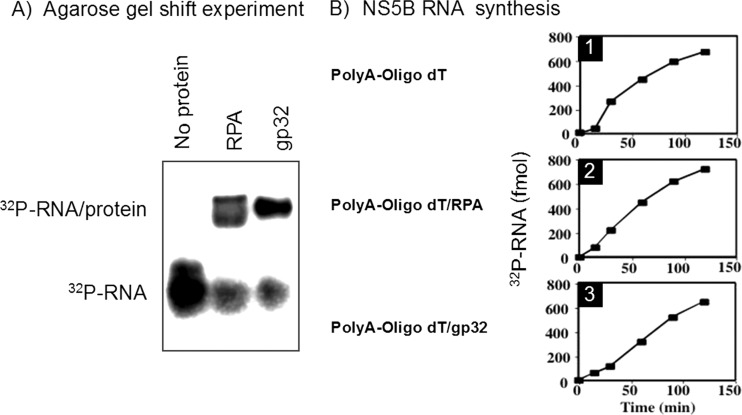
One way in which RPA might stimulate NS5B RNA synthesis is by melting secondary structures through its interaction with single-stranded RNA. If this hypothesis is true, any SSB-like protein that interacts with RNA and prevents secondary-structure formation should be able to substitute for RPA in the rolling-circle reaction. Another SSB protein that fits these requirements is the bacteriophage T4 protein gp32. T4 gp32 is a component of the T4 DNA replicase and binds RNA with an affinity similar to that of RPA (69, 70). First we demonstrated that gp32 and RPA were able to bind a 32P-labeled RNA oligonucleotide comparably in a gel mobility shift assay (Fig. 3A). Next we tested the ability of both RNA-binding SSB proteins to support HCV RNA synthesis on a multiply primed poly(A)/oligo(dT) linear template. It is important to note that a single rolling-circle template has only one 3′ end, whereas the poly(A)/oligo(dT) template contains multiple oligo(dT) primers hybridized to a single poly(A) molecule. NS5BΔ21 had very poor activity (<1 pmol of RNA product in 2 h) on the linear template (Fig. 3B, top) that was not increased by the addition of RPA (Fig. 3B, middle) or T4 gp32 (Fig. 3B, bottom).
FIG 3.
RPA and gp32 interact with RNA. (A) 32P-RNA gel shift assay. 32P-RNA was preincubated with RPA or gp32 and loaded onto a 10% agarose gel. The two proteins interact with the RNA with similar affinities, producing a supershifted band. (B1) NS5BΔ21 synthesis on a poly(A)/oligo(dT) template: The reaction was started by the addition of NTP, and the incorporation [32P]UTP into the synthesized product was measured in samples obtained at the indicated time points. (B2) NS5BΔ21 synthesis in the presence of RPA on the poly(A)/oligo(dT) template. Conditions were as described above. (B3) NS5BΔ21 synthesis in the presence of gp32 on the poly(A)/oligo(dT) template. Conditions were as described above.
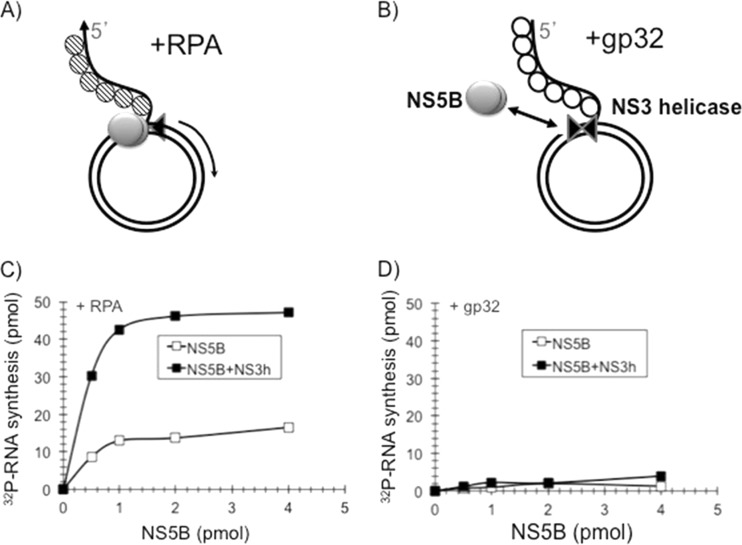
NS5BΔ21 RNA synthesis on a rolling-circle template is stimulated by RPA but not by T4 gp32.
Next we compared the efficiencies of RNA synthesis using a rolling-circle RNA template coated either with RPA (schematically shown in Fig. 4A) or with gp32 (schematically shown in Fig. 4B) in the presence of increasing concentrations of NS5BΔ21 and a constant amount of the NS3 helicase domain. As shown in Fig. 4C, RPA but not T4 gp32 (Fig. 4D) was able to support RNA synthesis by NS5BΔ21 on the rolling-circle template, while in the absence of NS5B, no detectable RNA synthesis was observed. The inability of another RNA binding SSB (T4 gp32) to substitute for RPA suggests that the stimulation of NS5BΔ21 activity by RPA is not likely to be mediated through the nonspecific melting of the RNA secondary structure. Based on these data, we hypothesized that RPA facilitates the binding of NS5B to the rolling-circle template through specific protein-protein contacts, increasing the initiation rate of RNA synthesis by NS5B.
FIG 4.
RPA is specifically required for rolling-circle RNA synthesis. The rolling-circle RNA template (1 picomole) was coated with RPA as shown schematically in panel A or gp32 as shown schematically in panel B. (C and D) The templates were tested for the ability to support RNA synthesis in the presence of NS5BΔ21 (open squares) or NS5BΔ21-NS3h (filled squares) for 60 min with increasing amounts of NS5B as described in Materials and Methods. Robust synthesis of RNA was observed only in the presence of RPA (C) and not in the presence of gp32 (D).
RPA forms a stable complex with NS5BΔ21 and ties it to the template.
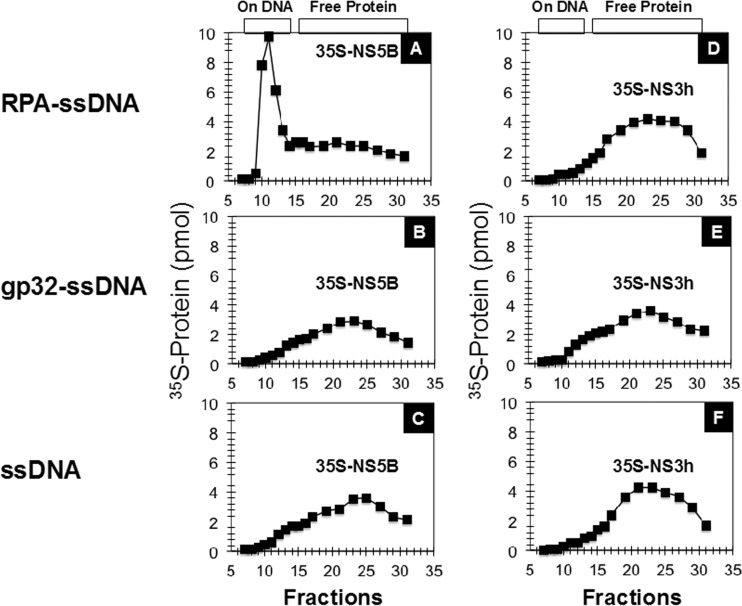
In order to test whether RPA ties NS5BΔ21 to the template, the NS3 helicase domain and NS5BΔ21 were individually labeled with [35S]methionine using an in vitro translation system and purified by gel filtration chromatography. 35S-labeled NS5BΔ21 and the NS3 helicase domain were incubated with ssDNA alone or coated with RPA or T4 gp32 and assayed for stable association by gel filtration chromatography (Fig. 5). Complexes of proteins bound to ssDNA were excluded from the column in the void volume (fractions 10 to 15), whereas unbound proteins were eluted with the included column volume (fractions 16 to 32). Thus, the gel filtration column separates proteins bound to DNA from proteins that are not bound to DNA. As shown in Fig. 5, 35S-NS5BΔ21 stably bound to RPA-coated ssDNA (Fig. 5A) but not to gp32-coated DNA (Fig. 5B) or to ssDNA alone (Fig. 5C) under physiological salt conditions (150 mM NaCl). As shown in Fig. 5, under the same conditions, the 35S-labeled NS3 helicase domain did not stably interact with either ssDNA coated with RPA (Fig. 5D), gp32 (Fig. 5E), or ssDNA alone (Fig. 5F). These studies support the hypothesis that RPA ties NS5BΔ21 to the template and suggest that there may be specific protein-protein interactions between NS5BΔ21 and RPA.
FIG 5.
RPA ties NS5BΔ21 to the template. 35S-labeled NS5BΔ21 and NS3h, individually synthesized in transcription/translation reactions in vitro in the presence of [35S]methionine, were incubated with circular ssM13mp18 DNA coated with RPA (A and D) or gp32 (B and E) or alone (C and F) and run on a BioGel A100 5-ml gel filtration column to resolve protein bound to DNA (fractions 10 to 15) from free protein (fractions 16 to 30). 35S-NS5BΔ21 stably interacted with ssDNA coated with RPA but not with ssDNA alone or ssDNA coated with T4 gp32. 35S-NS3h, on the other hand, did not show a stable interaction with ssDNA alone or when coated with RPA or T4 gp32.
NS5BΔ21 interacts directly with RPA.
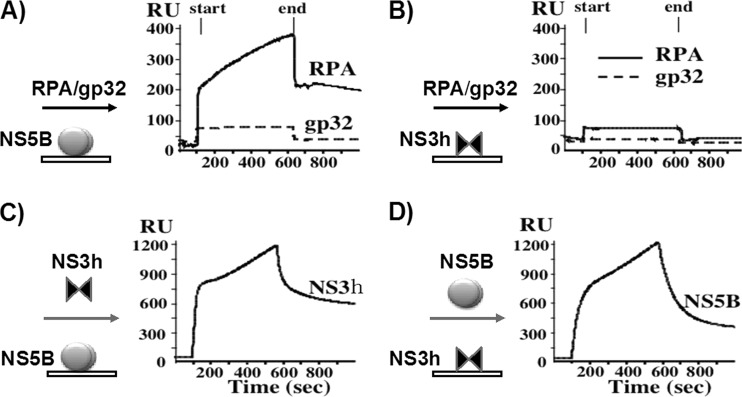
In order to further investigate whether NS5BΔ21 forms protein-protein interactions with RPA in the absence of ssDNA, we performed surface plasmon resonance (SPR) analysis. In the SPR technique, one macromolecule is immobilized on a sensor chip and a solution containing another molecule is passed over the chip. If a stable interaction is formed between the two macromolecules, the resultant increase in mass on the sensor chip is detected as an increase in surface plasmon resonance that can be measured in response or resonance units (RU). NS5BΔ21 (Fig. 6A) or the NS3 helicase domain (Fig. 6B) was immobilized on a chip and a solution of RPA or gp32 was passed over the chip. These results showed that RPA, but not gp32, can stably interact directly with NS5BΔ21. In contrast, neither RPA nor gp32 could stably interact with the NS3 helicase domain.
FIG 6.
NS5BΔ21 directly interacts with RPA and NS3. Panels A through D show surface plasmon resonance analysis of interactions between NS5BΔ21, NS3h, RPA, and T4 gp32. NS5BΔ21 (A) or NS3h (B) was immobilized to the sensor chip, and solutions of either RPA or T4 gp32 were passed over the chip. NS5BΔ21 directly interacted with RPA but not with T4 gp32, whereas NS3h showed no interaction with either RPA or T4 gp32. Panels C and D show NS3h passed over immobilized NS5BΔ21 (C) and NS5BΔ21 passed over immobilized NS3h (D). In both configurations, NS5BΔ21 showed direct interaction with NS3h.
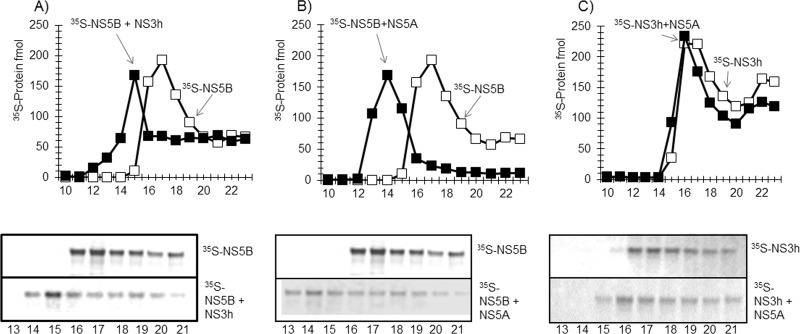
The NS3 helicase domain interacts directly with NS5BΔ21 and forms a stable complex.
Our studies have shown that although the NS3 helicase domain stimulates the polymerase activity of NS5BΔ21 in the presence of RPA on a rolling-circle template, it does not interact with ssDNA alone or with ssDNA coated with RPA in the presence of 150 mM NaCl. In order to investigate how NS3 helicase is recruited to the replication complex, we used SPR to examine interactions between NS5BΔ21 and the NS3 helicase domain. We found that the NS3 helicase domain and NS5BΔ21 strongly interacted with each other, independent of which partner was immobilized on the chip, with a Kd (dissociation constant) below 4.8 × 10−9 M (Fig. 6C and D).
The NS3 helicase domain and NS5BΔ21 form a stable complex that can bind ssDNA.
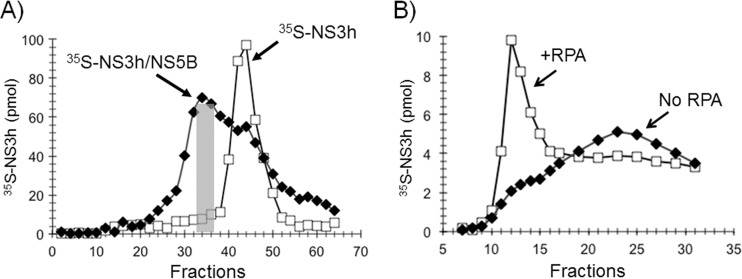
In order to investigate whether the NS3 helicase domain and NS5BΔ21 can form a stable complex that is capable of binding ssDNA stably, we incubated 35S-labeled NS3 helicase domain with NS5BΔ21 and isolated the complex on a gel filtration column. The 35S-NS3 helicase domain NS3h-NS5BΔ21 complex was eluted from the column in the void volume earlier than the 35S-NS3 helicase domain alone, which was eluted in the included volume, indicating that NS3 helicase domain and NS5BΔ21 form a stable protein complex (Fig. 7A). Fractions 32 to 36 (Fig. 7A, gray bar) containing the 35S-NS3h–NS5BΔ21 complex were pooled and analyzed for the ability of the complex to interact with ssDNA alone or ssDNA coated with RPA (Fig. 7B). As expected, the 35S-NS3h–NS5BΔ21 complex was able to stably interact with ssDNA only when it was coated with RPA and not with ssDNA alone or ssDNA coated with T4 gp32 (data not shown).
FIG 7.
Preformed NS3h-NS5BΔ21 complex is a stable functional complex. (A) 35S-NS3h–NS5BΔ21 complex formation and isolation. Superose 12 gel filtration chromatography profiles of 35S-NS3h alone (open squares) and with preincubation with cold NS5BΔ21 (filled diamonds). The gray box shows fractions 32 to 38 of 35S-NS3h–NS5BΔ21 complex, which were pooled and examined for interaction. (B) 35S-NS3h–NS5BΔ21 complex interaction with ssDNA (filled diamonds) and with ssDNA coated with RPA (open squares) on a BioGel A100 gel filtration column.
NS5A stimulates NS5BΔ21 RNA synthesis on a rolling-circle template.
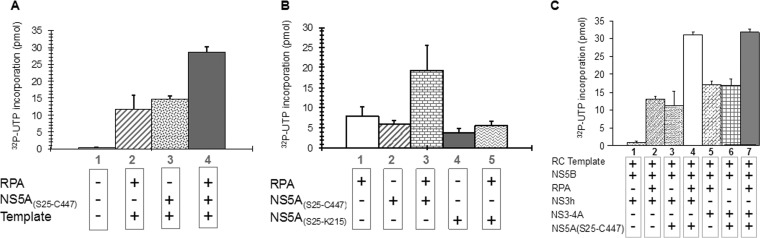
In order to investigate the role of HCV NS5A, a truncated NS5A(S25-K215) containing domain I and almost full-length NS5A(S25-C447), both lacking the N-terminal 24-amino-acid amphipathic domain, were preincubated with NS5BΔ21 and incubated with the rolling-circle RNA template and NS3h in the presence or absence of RPA (Fig. 8). NS5BΔ21 alone exhibited no activity, but both NS5A(S25-C447) (Fig. 8A) and domain I-containing truncated NS5A(S25 to K215) (Fig. 8B) were able to stimulate NS5BΔ21 RNA synthesis similar to that observed with RPA (Fig. 8A). It was previously reported that preincubation of NS5B and NS5A, prior to template addition and initiation of reaction, was needed to observe stimulation of RNA synthesis by NS5B (71). We observed a similar preincubation requirement using the rolling-circle RNA template (data not shown). Addition of NS5A(S25-C447), but not the truncated NS5A(S25-K215) containing domain I, to RPA resulted in an additive increase in RNA synthesis on the rolling-circle RNA template (Fig. 8A and B). Two minor contaminants, E. coli GroEL and DnaK, that copurified with NS5A(S25-C447) did not stimulate RNA synthesis (data not shown). It is interesting that there was no difference in stimulation by the helicase domain alone compared with NS3-4A containing both the protease and helicase domains and NS4A (Fig. 8C).
FIG 8.
NS5A stimulates NS5BΔ21 RNA synthesis on a rolling-circle template. (A) NS5A(S25-C447) and RPA stimulate RNA synthesis by NS5BΔ21 and show additive stimulatory effects in combination. (B) Domain I of NS5A(S25-K215) does not stimulate RNA synthesis by NS5BΔ21 in the presence of RPA. (C) The native NS3-4A complex comprising the 631-amino-acid NS3 protein containing the protease and the helicase domain complexed with the full-length 54-amino-acid NS4A protein stimulated RNA synthesis by NS5BΔ21 polymerase on a rolling-circle RNA template that was comparable to that of the NS3 helicase (NS3h) domain both in the presence and absence of NS5A(S25-C447) and human RPA.
RPA and NS5A(S25-C447) confer high processivity on the NS5BΔ21 replication complex on a rolling-circle template.
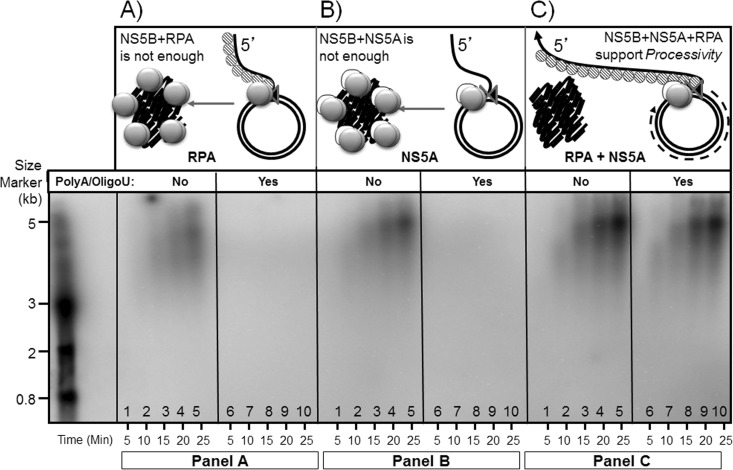
In order to test whether the combination of NS5A(S25-C447) and RPA ties NS5BΔ21 more tightly to the rolling-circle template than NS5A(S25-C447) or RPA alone, a classical template competition experiment was performed. Nonprocessive replication complexes continuously fall off the template and have to reinitiate. Highly processive replication complexes tend to stay on the template, resulting in high-molecular-weight product synthesis. HCV replication assays were carried out in the presence or absence of a large molar excess of a poly(A)/oligo(U) template which acts as a sponge and sequesters the replication complex when it falls off the rolling-circle template, thus preventing the reinitiation of RNA synthesis. Radiolabeled RNA products resulting from the addition of either RPA or NS5A(S25-C447) or RPA-NS5A(S25-C447) to reaction mixtures containing NS5BΔ21 polymerase and the NS3 helicase domain were analyzed by polyacrylamide gel electrophoresis and autoradiography. RNA synthesis from the rolling-circle template in the presence of RPA (Fig. 9A) or NS5A(S25-C447) (Fig. 9B) was completely abolished in the presence of the competing poly(A)/oligo(U) template. These data suggest that the addition of either RPA or NS5A(S25-C447) separately to the NS5BΔ21-NS3 helicase domain complex on a rolling-circle template results in an increased rate of synthesis of RNA but not in an increased length of RNA synthesis or processivity (the replicase falls off the template and gets trapped by the excess poly(A)/oligo(U) templates). In contrast, when NS5A(S25-C447) and RPA were both present in the reaction, robust synthesis of a high-molecular-weight RNA product occurred from the rolling-circle template even in the presence of the poly(A)/oligo(U) template (Fig. 9C). These data suggest that NS5A(S25-C447) and RPA functionally cooperate in imparting high processivity to the HCV replicase containing NS5BΔ21 and the NS3 helicase domain on a rolling-circle RNA template.
FIG 9.
RPA and NS5A cooperate to form RNA polymerase processivity factor. The radiolabeled RNA products resulting from in vitro reactions carried out as described in Materials and Methods were analyzed by polyacrylamide gel electrophoresis and autoradiographed. (A) RNA synthesis by NS5BΔ21-NS3h from the rolling-circle template in the presence of RPA without and with poly(A)/oligo(U). (B) RNA synthesis by NS5BΔ21-NS3h from the rolling-circle template in the presence of NS5A(S25-C447) without and with poly(A)/oligo(U). (C) RNA synthesis by NS5BΔ21-NS3h from the rolling-circle template in the presence of RPA and NS5A(S25-C447), without and with poly(A)/oligo(U).
NS5A(S25-C447) forms a stable complex with NS5BΔ21 but not with the NS3 helicase domain.
As described above, an interaction between NS3 helicase and NS5BΔ21 was previously demonstrated using SPR (Fig. 6C and D). In order to examine the interactions between NS5A(S25-C447) with NS5BΔ21 and the NS3 helicase domain, gel filtration chromatography experiments were performed. Consistent with previous results (Fig. 6 and 7), incubation of 35S-NS5BΔ21 with NS3 helicase resulted in the formation of a higher-molecular-weight complex that was eluted from the gel filtration column faster than 35S-NS5BΔ21 alone (Fig. 10A). Similarly, incubation of 35S-NS5BΔ21 with NS5A also resulted in the formation of a higher-molecular-weight complex, indicating that NS5B and NS5A interact with each other (Fig. 10B). Interestingly, the elution profile of 35S-NS3 helicase domain plus NS5A(S25-C447) was similar to that of 35S-NS3 helicase domain alone, indicating that these two proteins do not directly interact with each other (Fig. 10C).
FIG 10.
Protein-protein interaction between NS5BΔ21, NS5A(S25-C447), and NS3h. NS5BΔ21 and NS3h form a complex (A), NS5BΔ21 and NS5A(S25-C447) form a stable protein complex (B), and NS3h and NS5A(S25-C447) do not interact directly (C). 35S-labeled proteins were incubated with the high excess of “cold” protein to form a stable protein complex. Proteins were separated on a Superose 12 gel filtration column, and fractions were collected and analyzed using a scintillation counter and by polyacrylamide gel electrophoresis followed by autoradiography.
DISCUSSION
Development of an HCV in vitro replication system.
The goal of this study was to create an in vitro HCV replication assay to study viral and cellular proteins that constitute the HCV replication complex and influence the activity and processivity of HCV polymerase on an RNA template. All replication systems utilize a complex of different proteins that are required to deliver certain core functions required for RNA or DNA synthesis. In the case of viral replication, the functions can come from the virus or the host. Based on a comparison of different viral in vitro replication systems, we hypothesized that a minimum of six replication functions are required: (i) an origin recognition protein, (ii) a polymerase, (iii) a priming mechanism, (iv) a single-stranded binding protein, (v) an unwinding protein, and (vi) one or more processivity factors. Based on our assumption that the polymerase and helicase must form part of a replication complex, we set the minimum criteria for our in vitro replication system to be one where polymerase activity is stimulated by the presence of NS3 helicase. We could readily identify the RNA-dependent RNA polymerase for NS5B and the helicase for the C-terminal region of NS3. Using a rolling-circle RNA template and these two purified proteins as our starting points, we looked for other proteins of the HCV replication complex, such as processivity factors.
HCV replication has been shown to be dependent on human proteins or factors (72). We hypothesized that at least one of those host factors could be the human single-stranded binding (SSB) protein also known as replication protein A (RPA) (63). In the in vitro replication system, we showed that RPA strongly stimulated RNA synthesis by NS5BΔ21 on a rolling-circle template in the presence of NS3 helicase. We believe that this is the first example of a viral RNA polymerase that uses human RPA as a processivity factor. Finally, we investigated the mechanism of action of NS5A in the in vitro replication system, which revealed that NS5A(S25-C447) and RPA interact with NS5B polymerase, forming a highly processive complex that includes NS3 helicase on a rolling-circle RNA template, resulting in the synthesis of high-molecular-weight RNA product. These results help to extend our understanding into at least one of the roles of NS5A, a complex multifunctional protein, in the HCV replication complex.
Multiple roles for RPA and other SSBs in human and viral DNA replication in vitro.
The requirement of RPA for human and viral DNA replication has been well established (63). Although not much is known about the function of RPA in RNA metabolism, it has been shown that direct interactions between RPA and the RdRp QDE-1 are essential for RNA synthesis (73). T4 gp32, the SSB protein of bacteriophage T4, plays important roles in phage DNA replication, repair, and recombination, facilitates its activity at the replication fork through binding to ssDNA, mediates specific protein-protein interaction with the major components of the DNA replication complex, including DNA polymerase (T4 gp43) activity, and recruits primase-helicase (T4 gp59) assembly to the primosome complex T4 gp61-41 (74–77).
Role of SSB protein in leading- and lagging-strand DNA synthesis in vitro.
In previous studies, rolling-circle templates have been used in DNA replication experiments to elucidate the requirements for coordinated leading- and lagging-strand replication. In the E. coli and T7 bacteriophage rolling-circle DNA replication systems, the presence of E. coli SSB protein or the T7 SSB gp2.5, respectively, is not required for leading-strand synthesis but is essential for lagging-strand synthesis, in which SSB protein plays a role in protein-protein switching (67, 78, 79). The simian virus 40 (SV40) proliferating cell nuclear antigen (PCNA)-dependent dipolymerase replication system and the eukaryotic human DNA replication system follow the same rules whereby RPA is specifically required for protein-protein switching on the lagging strand through direct interactions with components of the replication complex but is not essential for synthesis of the leading strand because it can be replaced with E. coli SSB protein in the primer extension reaction (80, 81).
Role of RPA in the HCV rolling-circle replication system.
On a multiprimed template [poly(A)/oligo(dT)], NS5BΔ21 alone was able to synthesize a small amount of product (0.7 pmol) in 2 h and RPA did not show any stimulatory effects on RNA synthesis (Fig. 3B). In contrast, on a rolling-circle RNA template, RPA was absolutely required for the activity of NS5BΔ21 (Fig. 2). The inability of gp32, which has SSB protein-like properties, to substitute for RPA (Fig. 4), despite its high affinity to bind RNA (Fig. 3), suggests that RPA plays an additional role other than melting RNA secondary structures. RPA is known to be a nuclear factor for host and viral DNA replication, although RPA is available in the cytoplasm, where HCV replicates (63). Competition studies using ssDNA saturated with RPA in the presence of competitor RNA have shown that the Kd of RPA for RNA in the presence of ssDNA is 10−6 to 10−7 M. Although the affinity of RPA for ssDNA is 3 orders of magnitude stronger than for RNA, there is no ssDNA in the cytoplasm to compete with HCV RNA for RPA (63–65).
Role of HCV helicase in the HCV rolling-circle replication system.
The addition of NS3 helicase to HCV polymerase and RPA on a rolling-circle template increased RNA synthesis from ∼7.0 to ∼50 pmol in 2 h. Other investigators have demonstrated various effects of the protease domain on the helicase domain alone, in the full-length NS3 (631 amino acids containing the HCV protease and helicase domains), or in the NS3-4A complex (29). In contrast, we observed that NS3-4A had activity in rolling-circle replication similar to that of the NS3 helicase domain alone (Fig. 8), suggesting that the protease domain does not participate in the interaction between helicase and NS5B.
HCV helicase activity has been shown to be extremely sensitive to NaCl concentrations approaching physiological levels (82). This led to a hypothesis that NS3 helicase is associated with other HCV nonstructural proteins or host factors as part of a replication complex in vivo, and these additional factors are needed to ensure the functionality of HCV helicase and efficient replication of HCV in cells (16). Consistent with this hypothesis, the stimulatory activity of the helicase domain or NS3-4A in the replication complex on a rolling-circle template is significantly less sensitive to salt and HCV helicase is able to form a stable complex with HCV polymerase under physiological salt conditions. Taken together, these data suggest that the presence of NS5B and RPA stabilizes helicase activity under physiological salt conditions either through an induced conformation of the helicase or by excluding salt from the active site of the helicase.
Role of NS5A in the HCV rolling-circle replication system.
Most viral RdRps are not processive enzymes and can switch templates or jump off the template during RNA synthesis (83). Previous studies have shown that HCV NS5B alone is not a processive enzyme in vitro (40–46). NS5A has been shown to interact with RNA (84) and NS5B (85) and to play a role in HCV replication (86) In this study, NS5A(S25-C447) and RPA formed a functional cooperative interaction which enabled RNA synthesis by NS5BΔ21 to be highly processive on a rolling-circle RNA template.
NS5A has been predicted to comprise three domains, and while the crystal structure of domain 1 has been solved, the structures of domains 2 and 3 are unknown; nuclear magnetic resonance (NMR) studies suggest that they are intrinsically disordered, at least in experimentally manipulated proteins (87–90). Interestingly, in our studies, functional cooperativity between NS5A and RPA was observed with the almost full-length NS5A(S25-C447) but not with a truncated NS5A(S25-K215) comprising solely domain 1, suggesting a role for the other domains of native NS5A in HCV replication (Fig. 8). It has been shown previously that domain 3 plays a role in virus assembly and can be deleted without affecting RNA replication (54, 58, 91). Our results that show that NS5A(S25-C447) and NS5BΔ21 form a stable complex are consistent with previous findings regarding RNA and NS5B interaction domains in NS5A (54, 55, 85, 92, 93).
Protein-protein interactions within the replicase.
The interactions between replicative DNA polymerases and helicases are known from studies in the T7 and E. coli DNA replication systems (67, 78). We have shown that NS5BΔ21 binds poorly to nucleic acid and to a rolling-circle template by itself, and previous studies have also shown that NS3 helicase binds poorly to an ssDNA template at physiological salt levels (∼150 mM) (82). Our studies suggest that human RPA is required to bring NS5BΔ21 to the template through specific NS5BΔ21-RPA interactions (Fig. 5) and that NS3 helicase, in turn, is brought to the template through specific NS3 helicase-NS5BΔ21 interactions (Fig. 6 and 7). The interaction between the NS3 helicase domain and NS5B was predicted from in vivo studies, and it was shown that the NS3 helicase domain modulates template recognition by NS5B and increases the amount of product synthesized (94, 95). Similar results were observed with NS3-4A, consistent with their distinct roles in HCV life cycle. Thus, the interactions of the NS3 helicase domain with NS5B and between NS5B and RPA are crucial for formation of the replication complex and might represent new antiviral drug targets.
NS5A and RPA functionally cooperate to tie the replication complex to the template.
Our biochemical data demonstrate that functional cooperativity exists between NS5A(S25-C447) and RPA in tying the replication complex to the template and promoting RNA synthesis by NS5BΔ21 on a rolling-circle RNA template (Fig. 9). While RPA and NS5A(S25-C447) proteins can independently stimulate RNA synthesis by NS5BΔ21 polymerase in the presence of NS3 helicase, both of these replication complexes were easily competed away from the rolling-circle RNA template by the addition of a large molar excess of poly(A)/oligo(U). In contrast, in the presence of both NS5A(S25-C447) and RPA, the replication complex was highly processive and could not be competed off the rolling-circle template by a large molar excess of poly(A)/oligo(U).
Structural similarity between HCV NS5A domain I and known processivity factors.
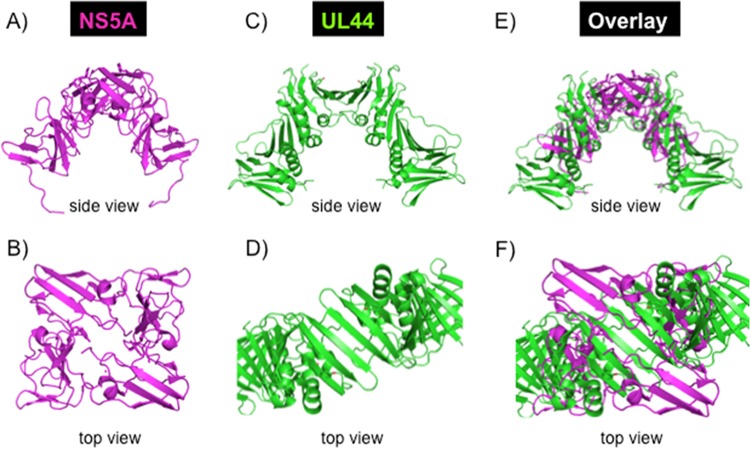
We assessed the similarities in the 3-dimensional structures between the N-terminal NS5A, determined by Tellinghuisen and colleagues, and known DNA processivity factors such as herpes simplex virus (HSV) UL42, cytomegalovirus (CMV) UL44, and human PCNA (60, 96–98). Significant differences exist between the DNA processivity factors; UL42 binds to DNA as a monomer, UL44 forms a dimer, and PCNA forms a trimeric ring structure (99). NS5A has no similarity with UL42, UL44, or PCNA at the monomeric level. However, the shape of the dimeric structure that UL44 uses to bind to DNA is similar to that of the NS5A dimer described by Tellinghuisen et al. (60). The structure of the NS5A domain I dimer was overlaid with the CMV UL44 (Fig. 11). A general similarity in the overall conformations of the two structures was observed, despite the fact that both the structural elements and the assembly of the dimer head-to-head interfaces are quite different for each protein. The lining of basic residues in the RNA binding motif on the inner surface of the NS5A groove is similar to the basic amino acids in the DNA binding motif of UL44, suggesting that the structure of the nucleic acid binding domain (but not the amino acid sequence) is conserved. It is also interesting that the DNA binding cavity is larger for UL44 than the proposed RNA binding cavity of NS5A, consistent with the larger packing volume of DNA than of RNA.
FIG 11.
Structural comparison of NS5A and UL44. Shown are side (A) and top (B) views of HCV NS5A (PDB code 1ZH1) and side (C) and top (D) views of CMV UL44 (PDB code 1T6L) processivity factor, as well as side (E) and top (F) views of the structural overlays of NS5A and UL44. The similarity of dimeric forms of HCV NS5A and CMV UL44 is interesting. The lining of basic residues in the cavity is observed for the two structures. However, the cavity is larger for UL44 than NS5A. The broader similarities may be used to infer a processivity role for NS5A. However, the C clamps are very different from each other. There is no similarity between the structural elements of the two proteins. Head-to-head interfaces for the two proteins are assembled quite differently.
Differences observed in studying the HCV replication complex versus isolated components.
We constrained the replication system we developed by the requirement for an HCV replication complex whose RNA synthesis was NS5B dependent, stimulated by NS3 helicase, and functional in close-to-physiological salt concentrations. NS5B, NS3 helicase, and NS5A are proteins that have been highly characterized separately and apart from the replication complex. We found many differences between the properties of the isolated proteins and the proteins in the replication complex on a rolling-circle template, such as (i) the ability of NS3 helicase to stimulate HCV replication in the presence of 100 mM NaCl, (ii) the lack of an effect of the protease domain on NS3 helicase domain stimulation of HCV RNA synthesis, (iii) the inability of NS3 helicase or NS5BΔ21 to bind to the template in the presence of 100 mM NaCl, (iv) the requirement for human RPA for robust activity of NS5BΔ21 on a rolling-circle template, and (v) the requirement for an almost full-length NS5A(S25-C447), and not just NS5A(S25-K215) comprising domain I, for functional cooperativity with RPA in making the NS5BΔ21 RNA replication complex highly processive. These results suggest that extreme caution must be used when studying components of protein complexes in isolation or even parts of proteins in isolation.
A model for HCV RNA replication.
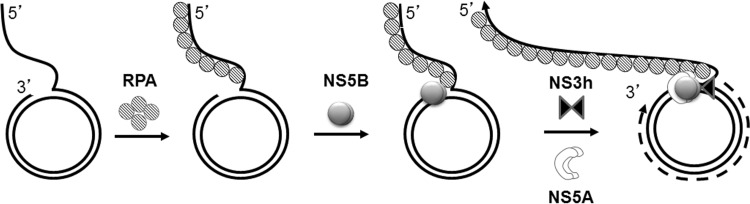
Based on these studies, we propose a working model for RNA replication by HCV NS5B (Fig. 12). RPA, owing to its single-stranded nucleic acid binding property, directly interacts with the rolling-circle RNA template and HCV polymerase (NS5B), recruiting NS5B to the template to initiate formation of the replicase complex. NS5B forms protein-protein interactions with NS5A and NS3, resulting in an increase in activity and processivity of NS5B polymerase. The results from our SPR and gel filtration experiments suggest a model in which NS5B is the nucleation center, linking together the critical components of the replicase complex through specific protein-protein interactions with RPA, NS5A, and NS3 helicase. HCV is thought to initiate RNA synthesis through unprimed, de novo synthesis. Further studies are needed to confirm this model in a cellular context where the HCV replication complex is membrane anchored.
FIG 12.
An in vitro model for HCV RNA replication. RPA, owing to its single-strand binding property, directly interacts with the rolling-circle RNA template and serves to recruit HCV polymerase (NS5B) to the template. NS3 helicase increases polymerase activity of NS5B, although it does not directly interact with RPA or NS5A and both NS5A and NS3 are recruited to the RNA template via a series of protein-protein interactions with NS5B to form a stable replication complex that directs a highly processive RNA synthesis reaction in vitro.
In conclusion, we have developed an in vitro replication system for HCV that allows for highly processive RNA synthesis reaction on a rolling-circle RNA template by NS5BΔ21 polymerase in the presence of NS3 helicase, RPA, and NS5A(S25-C447). This system may be used to interrogate the role of other viral or host factors involved in HCV replication and study potential antiviral compounds that block formation or function of the HCV replication complex.
ACKNOWLEDGMENTS
We acknowledge helpful discussions with Charles Rice (Rockefeller University) and Jerard Hurwitz (Memorial Sloan Kettering).
O.Y., J.T.C., J.B., K.S., J.R.F., and J.A.L. contributed to cloning, expression, and purification of proteins; A.Y. performed experiments; A.Y., N.M., R.R., and A.D.K. contributed to experimental design and project planning; B.G.R. performed computational analyses; and N.M., A.Y., and A.D.K. drafted the manuscript with help from all coauthors.
J.T.C., J.B., K.S., J.R.F., and J.A.L. are current employees of Vertex Pharmaceuticals and own company stock. N.M., R.R., A.Y., O.Y., B.G.R., and A.D.K. are former employees of Vertex Pharmaceuticals and may own company stock.
REFERENCES
- 1.Gravitz L. 2011. Introduction: a smouldering public-health crisis. Nature 474:S2–S4. doi: 10.1038/474S2a. [DOI] [PubMed] [Google Scholar]
- 2.Lavanchy D, McMahon B. 2000. Worldwide prevalence and prevention of hepatitis C, p 185–202. In Liang TJ, Hoofnagle JH (ed), Hepatitis C. Academic Press, San Diego, CA. [Google Scholar]
- 3.Moradpour D, Penin F. 2013. Hepatitis C virus proteins: from structure to function. Curr Top Microbiol Immunol 369:113–142. doi: 10.1007/978-3-642-27340-7_5. [DOI] [PubMed] [Google Scholar]
- 4.Rice CM. 2011. New insights into HCV replication: potential antiviral targets. Top Antiviral Med 19:117–120. [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Lohmann V, Penin F. 2013. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol 11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 6.Gu M, Rice CM. 2013. Structures of hepatitis C virus nonstructural proteins required for replicase assembly and function. Curr Opin Virol 3:129–136. doi: 10.1016/j.coviro.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raney KD, Sharma SD, Moustafa IM, Cameron CE. 2010. Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J Biol Chem 285:22725–22731. doi: 10.1074/jbc.R110.125294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwong AD, Kim JL, Rao G, Lipovsek D, Raybuck SA. 1998. Hepatitis C virus NS3/4A protease. Antiviral Res 40:1–18. doi: 10.1016/S0166-3542(98)00043-6. [DOI] [PubMed] [Google Scholar]
- 9.Love RA, Parge HE, Wickersham JA, Hostomsky Z, Habuka N, Moomaw EW, Adachi T, Hostomska Z. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331–342. doi: 10.1016/S0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 10.Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O'Malley ET, Harbeson SL, Rice CM, Murcko MA, Caron PR, Thomson JA. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343–355. doi: 10.1016/S0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 11.Gwack Y, Kim DW, Han JH, Choe J. 1996. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem Biophys Res Commun 225:654–659. doi: 10.1006/bbrc.1996.1225. [DOI] [PubMed] [Google Scholar]
- 12.Lam AM, Frick DN. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J Virol 80:404–411. doi: 10.1128/JVI.80.1.404-411.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gwack Y, Kim DW, Han JH, Choe J. 1997. DNA helicase activity of the hepatitis C virus nonstructural protein 3. Eur J Biochem 250:47–54. doi: 10.1111/j.1432-1033.1997.00047.x. [DOI] [PubMed] [Google Scholar]
- 14.Hong Z, Ferrari E, Wright-Minogue J, Chase R, Risano C, Seelig G, Lee CG, Kwong AD. 1996. Enzymatic characterization of hepatitis C virus NS3/4A complexes expressed in mammalian cells by using the herpes simplex virus amplicon system. J Virol 70:4261–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tai CL, Chi WK, Chen DS, Hwang LH. 1996. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J Virol 70:8477–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwong AD, Kim JL, Lin C. 2000. Structure and function of hepatitis C virus NS3 helicase. Curr Top Microbiol Immunol 242:171–196. doi: 10.1007/978-3-642-59605-6_9. [DOI] [PubMed] [Google Scholar]
- 17.Wardell AD, Errington W, Ciaramella G, Merson J, McGarvey MJ. 1999. Characterization and mutational analysis of the helicase and NTPase activities of hepatitis C virus full-length NS3 protein. J Gen Virol 80(Part 3):701–709. [DOI] [PubMed] [Google Scholar]
- 18.Gallinari P, Brennan D, Nardi C, Brunetti M, Tomei L, Steinkuhler C, De Francesco R. 1998. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J Virol 72:6758–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith RH, Kotin RM. 1998. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol 72:4874–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee CG, Hurwitz J. 1992. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J Biol Chem 267:4398–4407. [PubMed] [Google Scholar]
- 21.Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber PC. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol 4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 22.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89–100. doi: 10.1016/S0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 23.Gu M, Rice CM. 2010. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci U S A 107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho HS, Ha NC, Kang LW, Chung KM, Back SH, Jang SK, Oh BH. 1998. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J Biol Chem 273:15045–15052. [DOI] [PubMed] [Google Scholar]
- 25.Mackintosh SG, Lu JZ, Jordan JB, Harrison MK, Sikora B, Sharma SD, Cameron CE, Raney KD, Sakon J. 2006. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J Biol Chem 281:3528–3535. doi: 10.1074/jbc.M512100200. [DOI] [PubMed] [Google Scholar]
- 26.Appleby TC, Anderson R, Fedorova O, Pyle AM, Wang R, Liu X, Brendza KM, Somoza JR. 2011. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J Mol Biol 405:1139–1153. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure 7:1353–1363. doi: 10.1016/S0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 28.Taremi SS, Beyer B, Maher M, Yao N, Prosise W, Weber PC, Malcolm BA. 1998. Construction, expression, and characterization of a novel fully activated recombinant single-chain hepatitis C virus protease. Protein Sci 7:2143–2149. doi: 10.1002/pro.5560071011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howe AY, Chase R, Taremi SS, Risano C, Beyer B, Malcolm B, Lau JY. 1999. A novel recombinant single-chain hepatitis C virus NS3-NS4A protein with improved helicase activity. Protein Sci 8:1332–1341. doi: 10.1110/ps.8.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frick DN, Rypma RS, Lam AM, Gu B. 2004. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J Biol Chem 279:1269–1280. doi: 10.1074/jbc.M310630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beran RK, Pyle AM. 2008. Hepatitis C viral NS3-4A protease activity is enhanced by the NS3 helicase. J Biol Chem 283:29929–29937. doi: 10.1074/jbc.M804065200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Cai Z, Kim YC, Kumar R, Yuan F, Shi PY, Kao C, Luo G. 2005. Stimulation of hepatitis C virus (HCV) nonstructural protein 3 (NS3) helicase activity by the NS3 protease domain and by HCV RNA-dependent RNA polymerase. J Virol 79:8687–8697. doi: 10.1128/JVI.79.14.8687-8697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding SC, Kohlway AS, Pyle AM. 2011. Unmasking the active helicase conformation of nonstructural protein 3 from hepatitis C virus. J Virol 85:4343–4353. doi: 10.1128/JVI.02130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beran RK, Lindenbach BD, Pyle AM. 2009. The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase. J Virol 83:3268–3275. doi: 10.1128/JVI.01849-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiryaev SA, Chernov AV, Shiryaeva TN, Aleshin AE, Strongin AY. 2011. The acidic sequence of the NS4A cofactor regulates ATP hydrolysis by the HCV NS3 helicase. Arch Virol 156:313–318. doi: 10.1007/s00705-010-0838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jennings TA, Chen Y, Sikora D, Harrison MK, Sikora B, Huang L, Jankowsky E, Fairman ME, Cameron CE, Raney KD. 2008. RNA unwinding activity of the hepatitis C virus NS3 helicase is modulated by the NS5B polymerase. Biochemistry 47:1126–1135. doi: 10.1021/bi701048a. [DOI] [PubMed] [Google Scholar]
- 37.Kamer G, Argos P. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res 12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol 6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 39.Choi KH, Rossmann MG. 2009. RNA-dependent RNA polymerases from Flaviviridae. Curr Opin Struct Biol 19:746–751. doi: 10.1016/j.sbi.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 40.Behrens SE, Tomei L, De Francesco R. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J 15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 41.Lohmann V, Korner F, Herian U, Bartenschlager R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol 71:8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrari E, Wright-Minogue J, Fang JW, Baroudy BM, Lau JY, Hong Z. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol 73:1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamashita T, Kaneko S, Shirota Y, Qin W, Nomura T, Kobayashi K, Murakami S. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem 273:15479–15486. doi: 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]
- 44.Oh JW, Ito T, Lai MM. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J Virol 73:7694–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Francesco R, Behrens SE, Tomei L, Altamura S, Jiricny J. 1996. RNA-dependent RNA polymerase of hepatitis C virus. Methods Enzymol 275:58–67. doi: 10.1016/S0076-6879(96)75006-1. [DOI] [PubMed] [Google Scholar]
- 46.Lohmann V, Roos A, Korner F, Koch JO, Bartenschlager R. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108–118. doi: 10.1006/viro.1998.9311. [DOI] [PubMed] [Google Scholar]
- 47.Tomei L, Vitale RL, Incitti I, Serafini S, Altamura S, Vitelli A, De Francesco R. 2000. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J Gen Virol 81:759–767. [DOI] [PubMed] [Google Scholar]
- 48.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 49.Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 50.Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol 75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo JT, Bichko VV, Seeger C. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J Virol 75:8516–8523. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kishine H, Sugiyama K, Hijikata M, Kato N, Takahashi H, Noshi T, Nio Y, Hosaka M, Miyanari Y, Shimotohno K. 2002. Subgenomic replicon derived from a cell line infected with the hepatitis C virus. Biochem Biophys Res Commun 293:993–999. doi: 10.1016/S0006-291X(02)00342-X. [DOI] [PubMed] [Google Scholar]
- 53.Gu B, Gates AT, Isken O, Behrens SE, Sarisky RT. 2003. Replication studies using genotype 1a subgenomic hepatitis C virus replicons. J Virol 77:5352–5359. doi: 10.1128/JVI.77.9.5352-5359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog 4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foster TL, Belyaeva T, Stonehouse NJ, Pearson AR, Harris M. 2010. All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J Virol 84:9267–9277. doi: 10.1128/JVI.00616-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitz U, Tan SL. 2008. NS5A—from obscurity to new target for HCV therapy. Recent Pat Antiinfect Drug Discov 3:77–92. doi: 10.2174/157489108784746597. [DOI] [PubMed] [Google Scholar]
- 57.Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog 4:e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J Virol 82:1073–1083. doi: 10.1128/JVI.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J Biol Chem 279:48576–48587. doi: 10.1074/jbc.M407787200. [DOI] [PubMed] [Google Scholar]
- 60.Tellinghuisen TL, Marcotrigiano J, Rice CM. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love RA, Brodsky O, Hickey MJ, Wells PA, Cronin CN. 2009. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J Virol 83:4395–4403. doi: 10.1128/JVI.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lambert SM, Langley DR, Garnett JA, Angell R, Hedgethorne K, Meanwell NA, Matthews SJ. 2014. The crystal structure of NS5A domain 1 from genotype 1a reveals new clues to the mechanism of action for dimeric HCV inhibitors. Protein Sci 23:723–734. doi: 10.1002/pro.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wold MS. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 64.Brill SJ, Stillman B. 1989. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature 342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 65.Kim C, Snyder RO, Wold MS. 1992. Binding properties of replication protein A from human and yeast cells. Mol Cell Biol 12:3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sali DL, Ingram R, Wendel M, Gupta D, McNemar C, Tsarbopoulos A, Chen JW, Hong Z, Chase R, Risano C, Zhang R, Yao N, Kwong AD, Ramanathan L, Le HV, Weber PC. 1998. Serine protease of hepatitis C virus expressed in insect cells as the NS3/4A complex. Biochemistry 37:3392–3401. doi: 10.1021/bi972010r. [DOI] [PubMed] [Google Scholar]
- 67.Lee J, Chastain PD II, Kusakabe T, Griffith JD, Richardson CC. 1998. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol Cell 1:1001–1010. doi: 10.1016/S1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 68.Marians KJ. 1992. Prokaryotic DNA replication. Annu Rev Biochem 61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 69.Newport JW, Lonberg N, Kowalczykowski SC, von Hippel PH. 1981. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. II. Specificity of binding to DNA and RNA. J Mol Biol 145:105–121. [DOI] [PubMed] [Google Scholar]
- 70.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. 1997. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 71.Quezada EM, Kane CM. 2009. The hepatitis C virus NS5A stimulates NS5B during in vitro RNA synthesis in a template specific manner. Open Biochem J 3:39–48. doi: 10.2174/1874091X00903010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shulla A, Randall G. 2012. Hepatitis C virus-host interactions, replication, and viral assembly. Curr Opin Virol 2:725–732. doi: 10.1016/j.coviro.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee HC, Aalto AP, Yang Q, Chang SS, Huang G, Fisher D, Cha J, Poranen MM, Bamford DH, Liu Y. 2010. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol 8(10):e1000496. doi: 10.1371/journal.pbio.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benson KH, Kreuzer KN. 1992. Plasmid models for bacteriophage T4 DNA replication: requirements for fork proteins. J Virol 66:6960–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu H, Wang Y, Bleuit JS, Morrical SW. 2001. Helicase assembly protein Gp59 of bacteriophage T4: fluorescence anisotropy and sedimentation studies of complexes formed with derivatives of Gp32, the phage ssDNA binding protein. Biochemistry 40:7651–7661. doi: 10.1021/bi010116n. [DOI] [PubMed] [Google Scholar]
- 76.Perumal SK, Nelson SW, Benkovic SJ. 2013. Interaction of T4 UvsW helicase and single-stranded DNA binding protein gp32 through its carboxy-terminal acidic tail. J Mol Biol 425:2823–2839. doi: 10.1016/j.jmb.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xi J, Zhang Z, Zhuang Z, Yang J, Spiering MM, Hammes GG, Benkovic SJ. 2005. Interaction between the T4 helicase loading protein (gp59) and the DNA polymerase (gp43): unlocking of the gp59-gp43-DNA complex to initiate assembly of a fully functional replisome. Biochemistry 44:7747–7756. doi: 10.1021/bi047296w. [DOI] [PubMed] [Google Scholar]
- 78.Yuzhakov A, Turner J, O'Donnell M. 1996. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell 86:877–886. doi: 10.1016/S0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 79.Yuzhakov A, Kelman Z, O'Donnell M. 1999. Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell 96:153–163. doi: 10.1016/S0092-8674(00)80968-X. [DOI] [PubMed] [Google Scholar]
- 80.Eki T, Matsumoto T, Murakami Y, Hurwitz J. 1992. The replication of DNA containing the simian virus 40 origin by the monopolymerase and dipolymerase systems. J Biol Chem 267:7284–7294. [PubMed] [Google Scholar]
- 81.Yuzhakov A, Kelman Z, Hurwitz J, O'Donnell M. 1999. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J 18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Levin MK, Patel SS. 2002. Helicase from hepatitis C virus, energetics of DNA binding. J Biol Chem 277:29377–29385. doi: 10.1074/jbc.M112315200. [DOI] [PubMed] [Google Scholar]
- 83.Lai MM. 1992. RNA recombination in animal and plant viruses. Microbiol Rev 56:61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem 280:36417–36428. doi: 10.1074/jbc.M508175200. [DOI] [PubMed] [Google Scholar]
- 85.Shirota Y, Luo H, Qin W, Kaneko S, Yamashita T, Kobayashi K, Murakami S. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J Biol Chem 277:11149–11155. doi: 10.1074/jbc.M111392200. [DOI] [PubMed] [Google Scholar]
- 86.Appel N, Herian U, Bartenschlager R. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J Virol 79:896–909. doi: 10.1128/JVI.79.2.896-909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liang Y, Ye H, Kang CB, Yoon HS. 2007. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 46:11550–11558. doi: 10.1021/bi700776e. [DOI] [PubMed] [Google Scholar]
- 88.Verdegem D, Badillo A, Wieruszeski JM, Landrieu I, Leroy A, Bartenschlager R, Penin F, Lippens G, Hanoulle X. 2011. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic alpha-helical propensity and is a substrate of cyclophilin A. J Biol Chem 286:20441–20454. doi: 10.1074/jbc.M110.182436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hanoulle X, Verdegem D, Badillo A, Wieruszeski JM, Penin F, Lippens G. 2009. Domain 3 of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem Biophys Res Commun 381:634–638. doi: 10.1016/j.bbrc.2009.02.108. [DOI] [PubMed] [Google Scholar]
- 90.Hanoulle X, Badillo A, Verdegem D, Penin F, Lippens G. 2010. The domain 2 of the HCV NS5A protein is intrinsically unstructured. Protein Pept Lett 17:1012–1018. doi: 10.2174/092986610791498920. [DOI] [PubMed] [Google Scholar]
- 91.Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol 82:7964–7976. doi: 10.1128/JVI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ross-Thriepland D, Amako Y, Harris M. 2013. The C terminus of NS5A domain II is a key determinant of hepatitis C virus genome replication, but is not required for virion assembly and release. J Gen Virol 94:1009–1018. doi: 10.1099/vir.0.050633-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hwang J, Huang L, Cordek DG, Vaughan R, Reynolds SL, Kihara G, Raney KD, Kao CC, Cameron CE. 2010. Hepatitis C virus nonstructural protein 5A: biochemical characterization of a novel structural class of RNA-binding proteins. J Virol 84:12480–12491. doi: 10.1128/JVI.01319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. 1995. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem 270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 95.Piccininni S, Varaklioti A, Nardelli M, Dave B, Raney KD, McCarthy JE. 2002. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J Biol Chem 277:45670–45679. doi: 10.1074/jbc.M204124200. [DOI] [PubMed] [Google Scholar]
- 96.Zuccola HJ, Filman DJ, Coen DM, Hogle JM. 2000. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol Cell 5:267–278. doi: 10.1016/S1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]
- 97.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. 1996. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87:297–306. doi: 10.1016/S0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 98.Appleton BA, Loregian A, Filman DJ, Coen DM, Hogle JM. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol Cell 15:233–244. doi: 10.1016/j.molcel.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 99.Indiani C, O'Donnell M. 2006. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol 7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]