ABSTRACT
Influenza A viruses (IAVs) rely on host factors to support their life cycle, as viral proteins hijack or interact with cellular proteins to execute their functions. Identification and understanding of these factors would increase our knowledge of the molecular mechanisms manipulated by the viruses. In this study, we searched for novel binding partners of the influenza A virus NS2 protein, the nuclear export protein responsible for overcoming host range restriction, by a yeast two-hybrid screening assay and glutathione S-transferase-pulldown and coimmunoprecipitation assays and identified AIMP2, a potent tumor suppressor that usually functions to regulate protein stability, as one of the major NS2-binding candidates. We found that the presence of NS2 protected AIMP2 from ubiquitin-mediated degradation in NS2-transfected cells and AIMP2 functioned as a positive regulator of IAV replication. Interestingly, AIMP2 had no significant effect on NS2 but enhanced the stability of the matrix protein M1. Further, we provide evidence that AIMP2 recruitment switches the modification of M1 from ubiquitination to SUMOylation, which occurs on the same attachment site (K242) on M1 and thereby promotes M1-mediated viral ribonucleoprotein complex nuclear export to increase viral replication. Collectively, our results reveal a new mechanism of AIMP2 mediation of influenza virus replication.
IMPORTANCE Although the ubiquitination of M1 during IAV infection has been observed, the precise modification site and the molecular consequences of this modification remain obscure. Here, we demonstrate for the first time that ubiquitin and SUMO compete for the same lysine (K242) on M1 and the interaction of NS2 with AIMP2 facilitates the switch of the M1 modification from ubiquitination to SUMOylation, thus increasing viral replication.
INTRODUCTION
Influenza A virus (IAV) is a significant cause of morbidity and mortality in both humans and animal species owing to its ability to cause yearly epidemics and occasional pandemics (1–3). Like other viruses, IAV hijacks the host cellular machinery to support its life cycle. Thus, identification and characterization of the interactions between viral components and specific host factors would help provide an understanding of the mechanisms by which the viruses acquire human pandemic potential.
IAV belongs to the Orthomyxoviridae family, and its genome consists of eight negative-sense RNA segments encoding up to 17 viral proteins (4–9). The viral RNA (vRNA) exists as a form of viral ribonucleoprotein complexes (vRNPs) containing vRNA, nucleoprotein (NP), and three viral polymerase proteins (PB1, PB2, and PA). Unlike most other RNA viruses, influenza virus transcribes and replicates its genome in the nucleus. Thus, after it enters a host cell, vRNPs enter the nucleus to complete transcription and replication (10). The newly synthesized vRNPs are exported from the nucleus for packaging into progeny virions (11). In this regard, efficient nuclear export of vRNPs is essential for productive infection.
NS2, also known as nuclear export protein (NEP), acts as an adaptor to mediate the nuclear export of vRNPs by forming the Crm1-NS2-M1-vRNP complex (12, 13). Recently, this adaptor protein has been shown to play an important role in overcoming host range restriction. Adaptive mutations in NS2 can increase viral RNA accumulation, which compensates for the reduced activity of avian viral polymerase in mammalian cells, thereby allowing highly pathogenic avian H5N1 influenza viruses to overcome host restriction (14). It has also been reported that NS2 promotes the efficient release of budding virions by recruiting F1Fo ATPase (15). Consequently, NS2 appears to perform different functions during the viral life cycle.
M1, another key regulator of vRNP nuclear export (11, 16), is a multifunctional protein that plays essential structural and functional roles in various steps of the influenza virus life cycle. The proper subcellular localization of M1 is necessary for its functions in the viral life cycle. Early in infection, newly synthesized M1 translocates to the nucleus, where it blocks the transcription of viral mRNA by binding to vRNPs and mediates vRNP nuclear export (16–19). Late in infection, M1 is exported from the nucleus to block the reentry of vRNPs into the nucleus, mediate viral assembly and budding, and control virus morphology (11, 20–22). Posttranslational modifications of M1 play important roles in the regulation of its cellular localization and function. Phosphorylation of M1 at Y132 mediates the nuclear import of M1 (23), whereas SUMOylation of M1 regulates the nuclear export of vRNPs and promotes virion assembly and budding (21). Ubiquitination of M1 has been implicated in IAV release from the endosomes (24). However, the precise modification site and the biological functions of M1 ubiquitination are not well understood.
Aminoacyl-tRNA synthetase interacting multifunctional protein 2 (AIMP2; also known as JTV-1 or p38) was first identified to be a component of the multi-aminoacyl-tRNA synthetase (ARS) complex (25, 26). Besides stabilizing the ARS complex to promote efficient protein synthesis, AIMP2 has recently been shown to dissociate from the ARS complex following DNA damage or oncogenic stimuli and work as a potent tumor suppressor through the regulation of ubiquitin-mediated degradation of target proteins. AIMP2 binds to and sequesters p53 from Mdm2-dependent ubiquitination in response to oxidative stress (27). Following transforming growth factor β treatment, AIMP2 interacts with and mediates the ubiquitination of FBP (28). AIMP2 also promotes tumor necrosis factor alpha (TNF-α)-induced ubiquitin-dependent degradation of TRAF2 (29). Moreover, AIMP2 itself is a substrate of the E3 ligase Parkin (30, 31).
In this study, we performed a yeast two-hybrid assay to screen NS2-interacting host proteins and identified AIMP2 to be its potential binding partner. We present data that NS2 interacts with AIMP2 and protects it from ubiquitin-mediated degradation. AIMP2 functions as a positive regulator of IAV replication by facilitating the switch from ubiquitination to SUMOylation of M1.
MATERIALS AND METHODS
Cell culture, viruses, and antibodies.
A549 (human type II alveolar epithelial), 293T (human embryonic kidney), HeLa (human epithelial carcinoma) cells, and MDCK (Madin-Darby canine kidney) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Paisley, United Kingdom).
Recombinant influenza virus A/WSN/33 (H1N1) (WSN) was generated using a 12-plasmid-based reverse genetics approach and propagated in MDCK cells (32). A/PR/8/34 (H1N1) virus was propagated in specific-pathogen-free chicken embryos. The viral titer was measured using a standard plaque assay or hemagglutinin (HA) assay (33, 34). The M1 K242R mutant virus was generated using reverse genetics in which segment 7 (wild-type [WT] M gene) was replaced with the mutant construct (AAA → AGA at residue 749).
Mouse anti-Flag antibody (M2; catalog number F3165) was purchased from Sigma (St. Louis, MO, USA). Mouse anti-HA (catalog number sc-7392), anti-Myc (catalog number sc-40), and anti-influenza A virus NP (catalog number sc-101352) antibodies, goat anti-influenza A virus NS2 (catalog number sc-17598) and anti-JTV1 (AIMP2) (catalog number sc-160454) antibodies, and horseradish peroxidase-conjugated donkey anti-goat IgG were provided by Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit anti-lamin B1 was purchased from Boster (Wuhan, China). Mouse antitubulin (catalog number ZS-80350) and anti-His antibodies, tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-goat IgG, and TRITC-conjugated anti-mouse IgG were purchased from Zhongshan Golden Bridge Biotechnology (Beijing, China). Fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG and FITC-conjugated anti-mouse IgG were from Jackson ImmunoResearch Laboratories. Rabbit polyclonal antibody against influenza A virus NP and mouse monoclonal anti-influenza A virus M1 antibody were kindly provided by Wenjun Liu (Institute of Microbiology, Chinese Academy of Sciences, Beijing, China).
Plasmids.
The NS2 gene was amplified from influenza A virus A/WSN/33 (H1N1) cDNA by PCR and cloned into the pGEX4T-1, pBridge, pXJ40-HA, pcDNA3.0-Flag, or pcDNA4.0-Myc/His vector. The NS2 gene of A/PR/8/34 (H1N1) or A/QH/12/05 (H5N1) was cloned into pXJ40-HA. M1 cDNA was derived from the A/WSN/33 strain and cloned into the pcDNA3.0-Flag or pcDNA4.0-Myc/His vector. Full-length M1 carrying a K102R, K187R, K230R, or K242R mutation was generated by PCR with specific primers and cloned into the pcDNA3.0-Flag or pcDNA4-Myc/His vector. AIMP2 was cloned into the pGADT7, pXJ40-HA, pcDNA3.0-Flag, or pcDNA4.0-Myc/His vector. Ubiquitin (Ub)-conjugated HA (HA-Ub), HA-K48-Ub, and HA-K63-Ub were kindly provided by Xin Ye (Institute of Microbiology, Chinese Academy of Sciences, Beijing, China). HA–SUMO-1 was cloned into the pXJ40-HA vector. The Ubc9 plasmid was kindly provided by Hong Tang (Institute of Biophysics, Chinese Academy of Sciences, Beijing, China). Plasmids for expressing nontagged PB1, PB2, PA, and NP were derived from an influenza A virus reverse genetics system. A polymerase I-expressing plasmid carrying an influenza virus-like RNA encoding firefly luciferase (vNS-Luc) was kindly provided by Xin Ye.
Yeast two-hybrid screening.
The GAL4 BD-NS2 fusion bait plasmid was transformed into Saccharomyces cerevisiae strain CG1945 using the lithium acetate protocol. The transformant was grown on synthetic dropout (SD) agar plates without Trp and spread on SD agar plates without His and Trp for self-activation estimation. The cDNA library derived from human fetal brain was screened following the Matchmaker two-hybrid system protocol (Clontech). Approximately 107 Trp+ Leu+ transformants were selected on SD agar plates without His, Leu, and Trp for the primary screening and then tested by the LacZ assay for the second screening. After rescue, the potential positive plasmids were isolated and retransformed into yeast strain CG1945 containing the NS2 bait plasmid. Only the clones that were positive for all the reporters and confirmed to be positive by at least two independent tests were selected for specific interactions and sequenced.
siRNA oligonucleotide sequences and transfection.
The sequences of small interfering RNA (siRNA) targeting AIMP2 (siAIMP2) were as follows: for siAIMP2 sense, 5′-ACACCAGAUGCAGACUUGAUGUAA-3′; for siAIMP2 antisense, 5′-UUACAUCAAGUCUGCAUCUGGUGU-3′ (35). The siRNA oligonucleotides were chemically synthesized (Guangzhou Ribobio, Guangzhou, China) and transfected into 293T or A549 cells using the Lipofectamine 2000 reagent according to the manufacturer's instructions (Invitrogen).
Quantitative real-time PCR analysis.
Total RNA was isolated from A/WSN/33 virus-infected cells using the TRIzol reagent (TianGen Biotech Co., Ltd., Beijing, China). Two-microgram RNA aliquots were used for first-strand cDNA synthesis using Moloney murine leukemia virus reverse transcriptase (Promega, USA). The primers used for reverse transcription of vRNA, cRNA, and mRNA were 5′-AGCGAAAGCAGG-3′, 5′-AGTAGAAACAAGG-3′, and oligo(dT), respectively. Real-time PCR was conducted using SYBR Ex Taq II premix (TaKaRa Co., Ltd., DaLian, China). The cycle conditions included an initial denaturation step at 95°C for 30 s, followed by 40 cycles of amplification for 3 s at 95°C and 1 min at 60°C. The amounts of viral RNA were normalized by the amount of 18S rRNA. The primer sequences used in the real-time PCR were as follows: 5′-GCCTGCCTGCCTGTGTGTATGGAT-3′ for the NP forward primer and 5′-GGCATGCCATCCACACCAGTTGAC-3′ for the NP reverse primer, 5′-GTAACCCGTTGAACCCCATT-3′ for the 18S rRNA forward primer and 5′-CCATCCAATCGGTAGTAGCG-3′ for the 18S rRNA reverse primer (36), 5′-CCCCAATGTCTCTGTTGTTGAC-3′ for the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) forward primer and 5′-CAGCCTTCACTACCCTCTTGAT-3′ for the GAPDH reverse primer; and 5′-ACCTGTCTCTGCAAGCTCTT-3′ for the AIMP2 forward primer and 5′-GTGGTTAAAGTCGTGGGCTC-3′ for the AIMP2 reverse primer.
Luciferase assay.
293T cells were transfected with the IAV PA, PB1, PB2, and NP expression vectors, along with a luciferase reporter (vNS-Luc) and the Renilla luciferase reporter vector pRL-TK (Promega, USA), which was used as an internal control. The cells were also cotransfected with Flag-AIMP2 and HA-NS2. The cell lysates were prepared at 30 h posttransfection, and the luciferase activity was measured using a dual-luciferase reporter assay system (Promega, USA) according to the manufacturer's recommendations. Each set of assays was performed in triplicate.
Western blot analysis.
Cell lysates were prepared by washing the transfected or virus-infected cells with cold phosphate-buffered saline (PBS) and extracting the total protein using cell lysis buffer (Cell Signaling Technology). To detect the cellular localization of influenza virus proteins, cytoplasmic or nuclear extracts were prepared using a nuclear and cytoplasmic protein extraction kit (Beyotime Institute of Biotechnology, Beijing, China). The extracted proteins were separated using SDS-PAGE and transferred to a polyvinylidene difluoride membrane. After blocking, the membrane was incubated with the primary antibody overnight at 4°C, followed by incubation with a horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. Bands were detected using enhanced chemiluminescence (Applygen, China).
Immunofluorescence assay.
The transfected or A/WSN/33 virus-infected 293T or A549 cells were washed three times with PBS, fixed in 4% paraformaldehyde for 20 min at room temperature, and permeabilized with 0.2% Triton X-100 for 4 min. After blocking in 5% bovine serum albumin for 30 min, the cells were incubated for 35 to 40 min with the primary antibodies at 37°C. After washing with PBS, the cells were incubated for 40 min with TRITC conjugates and fluorescein isothiocyanate conjugates at 37°C. Cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) according to the manufacturer's recommendations. The cells were observed with a Leica confocal microscope.
GST-pulldown and coimmunoprecipitation assays.
Glutathione S-transferase (GST) and GST fusion proteins were purified from Escherichia coli BL21(DE3) cells using glutathione-Sepharose 4B beads (Amersham Biosciences, Uppsala, Sweden). An equal amount of either GST or GST fusion protein (2 μg) bound to the beads was incubated with the lysates from transiently transfected 293T cells in NP-40 lysis buffer for 4 h at 4°C. The beads were then washed five times with PBS containing 0.1% Triton X-100. The bound proteins were eluted by boiling in 2× SDS loading buffer and analyzed by Western blotting with an anti-HA antibody.
For the coimmunoprecipitation experiments, transfected or infected cells were lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, 20 mM HEPES, pH 7.5, 10% glycerol, 1 mM EDTA, protease inhibitors), and the cell lysates were incubated with antibodies at 4°C for 2 h. Protein G agarose beads (Sigma, USA) were then added, and the mixture was incubated at 4°C overnight. The beads were washed three times with lysis buffer, boiled in 2× SDS loading buffer for 5 min, and then analyzed by Western blotting.
Statistical analyses.
The data are presented as the mean ± standard deviation (SD). Statistical comparisons were performed using SPSS (version 11.5) software (SPSS, Inc., Chicago, IL, USA). Parametric data were compared using Student's t test. One-way analysis of variance (ANOVA) was used to determine the difference between independent groups. The differences between the variants were considered statistically significant if P was <0.05.
RESULTS
NS2 interacts with AIMP2 in vitro and in IAV-infected cells.
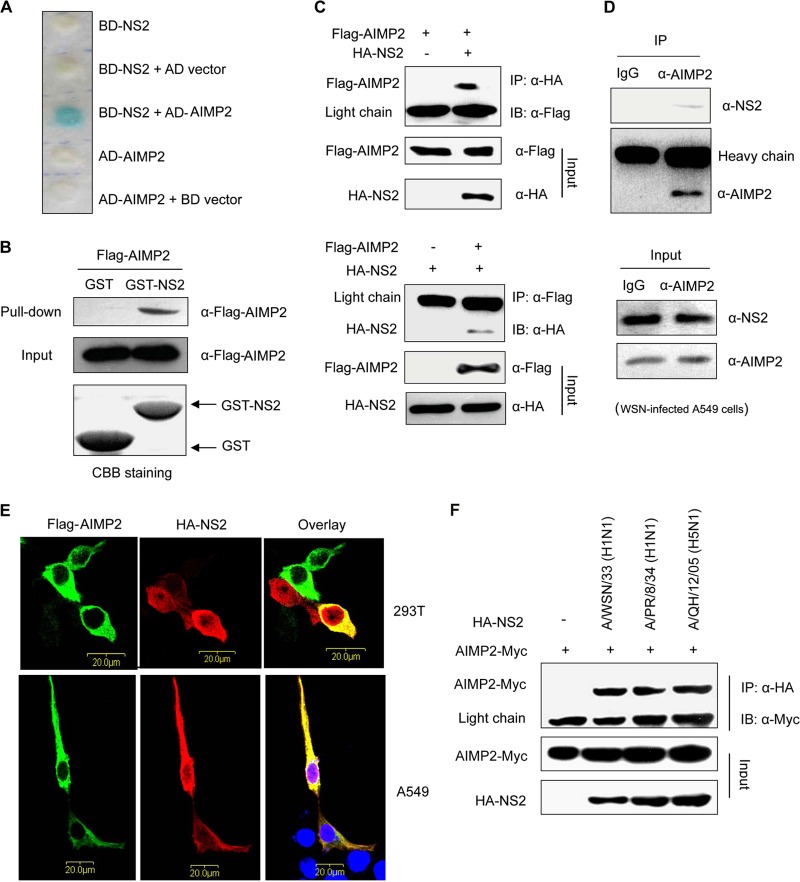
To better understand the roles of NS2 in the pathogenesis of influenza viruses, we performed yeast two-hybrid assays to identify NS2-binding proteins and obtained four proteins, including AIMP2, ATP5E, HINT2, and SMC3. Among them, we focused on AIMP2 (Fig. 1A). The interaction between NS2 and AIMP2 was further confirmed in the in vitro GST-pulldown assay (Fig. 1B) and an in vivo coimmunoprecipitation study in 293T cells cotransfected with vectors expressing Flag-tagged AIMP2 and HA-tagged NS2 (Fig. 1C). Next we examined whether endogenous AIMP2 could interact with NS2 in IAV-infected cells by another coimmunoprecipitation study. As shown in Fig. 1D, NS2 could be coimmunoprecipitated in WSN-infected A549 cell extracts when using a precipitation antibody to endogenous AIMP2, whereas NS2 was not detected in the immunoprecipitates by a control IgG. Finally, we performed immunofluorescence analyses to detect the cellular colocalization of AIMP2 with NS2 in 293T or A549 cells. AIMP2 colocalized specifically with NS2 in the cytoplasm (Fig. 1E) but not with NS1 (data not shown). Together, these data clearly demonstrate that NS2 is able to physically bind to AIMP2.
FIG 1.
NS2 interacts with AIMP2 in vitro and in IAV-infected cells. (A) Yeast two-hybrid assay analysis of the interaction between NS2 and AIMP2. Yeast strain CG1945 was cotransformed with the BD and AD plasmids, as indicated. The presence of a blue product indicates a positive interaction. (B) GST-pulldown assay showing the direct interaction between NS2 and AIMP2. Lysates of 293T cells transfected with Flag-AIMP2 were incubated with an equal amount of GST or GST-NS2 bound to glutathione-Sepharose 4B beads. The bound proteins were analyzed by Western blotting with an anti-Flag antibody. CBB, Coomassie brilliant blue. (C) Coimmunoprecipitation of HA-NS2 and Flag-AIMP2 in 293T cells. 293T cells were cotransfected with HA-NS2 and Flag-AIMP2. The cell lysates were immunoprecipitated (IP) and immunoblotted (IB) with specific antibodies, as indicated. (D) Coimmunoprecipitation showing the interaction of endogenous AIMP2 and NS2 in A/WSN/33 virus-infected A549 cells. A549 cells were infected with A/WSN/33 virus at an MOI of 2 for 16 h, and the cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies. (E) Colocalization of HA-NS2 and Flag-AIMP2 in 293T and A549 cells. 293T and A549 cells were cotransfected with HA-NS2 and Flag-AIMP2. At 28 h posttransfection, the cells were fixed, permeabilized, and stained for Flag-AIMP2 (green) and HA-NS2 (red). The area stained blue with DAPI represents the nucleus. (F) Coimmunoprecipitation of AIMP2-Myc and HA-NS2 from different IAV strains in 293T cells. 293T cells were cotransfected with AIMP2-Myc and HA-NS2 from the indicated IAV strains. At 28 h posttransfection, the cell lysates were prepared, immunoprecipitated, and immunoblotted with the indicated antibodies. The data in panels A to F are representative of those from at least three independent experiments.
To investigate whether the NS2 proteins of other IAV strains also interact with AIMP2, we performed coimmunoprecipitation assays with Myc-tagged AIMP2 and HA-tagged NS2 from a low-pathogenic strain, A/PR/8/34 (H1N1), the NS2 protein of which exhibited 95% identity with WSN NS2, and a highly pathogenic strain, A/QH/12/05 (H5N1), the NS2 protein of which had 93% homology with WSN NS2. As shown in Fig. 1F, AIMP2 interacted with the NS2 proteins from the two different IAV strains as well.
Identification of the NS2-AIMP2-binding region.
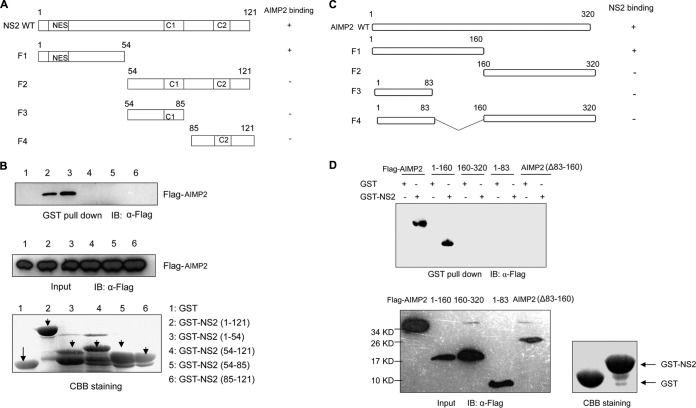
NS2 can be divided into an N-terminal protease-sensitive domain and a C-terminal protease-resistant domain (Fig. 2A). The N-terminal domain contains a highly conserved nuclear export signal which is thought to be required for binding with Crm1. The C-terminal domain consists of two α-helices, C1 and C2, and is essential for M1 binding (37). Thus, NS2 is believed to be an adaptor protein during Crm1-mediated nuclear export of vRNPs by forming the vRNP-M1-NS2-Crm1 complex (12). Here, we found that the N-terminal domain but not the C-terminal (M1-binding) domain of NS2 was responsible for the binding with AIMP2 by the GST-pulldown assay (Fig. 2B). Furthermore, we also identified the region of AIMP2 involved in its binding with NS2 by the GST-pulldown assay. The peptide spanning amino acids (aa) 1 to 160 of AIMP2 interacted with NS2, while the peptides spanning aa 160 to 320, 1 to 83, and 1 to 320 with deletion of aa 83 to 160 [AIMP2(Δ83–160)] did not interact with NS2, implying that AIMP2 amino acid residues 83 to 160 are essential for binding with NS2 (Fig. 2C and D).
FIG 2.
Mapping of the sites of interaction between NS2 and AIMP2. (A) Schematic representation of the deletion mutants of NS2. NES, nuclear export signal. (B) Lysates of 293T cells transfected with Flag-AIMP2 were incubated with an equal amount of GST, GST-NS2 from aa 1 to 121, GST-NS2 from aa 1 to 54, GST-NS2 from aa 54 to 121, GST-NS2 from 54 to 85, or GST-NS2 from aa 85 to 121. Bound proteins were analyzed by Western blotting with an anti-Flag antibody. (C) Diagram of the deletion mutants of AIMP2. (D) The regions of AIMP2 that interact with NS2 were identified using GST-pulldown assays. The data in panels B and D are representative of those from three independent experiments.
NS2 inhibits AIMP2 ubiquitination.
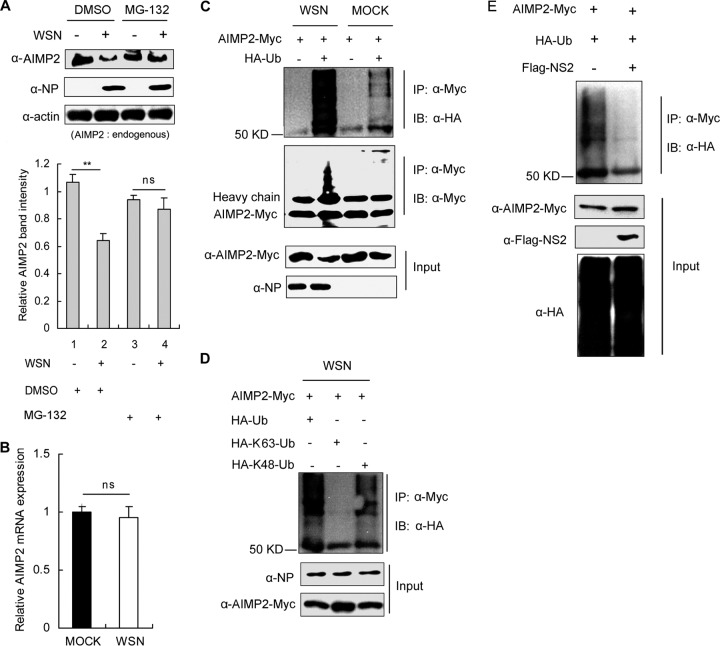
Under physiological conditions, AIMP2 is an integral component of the ARS complex. In response to signals, AIMP2 dissociates from the ARS complex to participate in a variety of cellular processes (28, 38). The released AIMP2 can be recognized by E3 ligases, such as Parkin, leading to its ubiquitination and degradation quickly through the proteasome pathway (39). Here, we observed that IAV infection significantly decreased the protein levels of AIMP2, while treatment with the proteasome inhibitor MG-132 attenuated the reduction (Fig. 3A). Furthermore, there was no significant difference in AIMP2 mRNA levels between the cells with or without WSN infection (Fig. 3B). These results suggest that IAV infection may trigger ubiquitin-mediated degradation of AIMP2. To further confirm the ubiquitination of AIMP2 in IAV-infected cells, 293T cells were cotransfected with AIMP2-Myc and HA-Ub, followed by infection with A/WSN/33 virus. Immunoprecipitation experiments clearly indicated that IAV infection enhanced the ubiquitination of AIMP2 (Fig. 3C). It is known that K48-linked polyubiquitin chains target proteins for proteasome-mediated degradation, whereas K63-linked polyubiquitin chains usually regulate a variety of nonproteolytic cellular functions. Here, we found that WSN virus infection induced the K48- but not K63-linked ubiquitination of AIMP2 (Fig. 3D). Given the clear evidence of the interaction between NS2 and AIMP2, we next investigated whether NS2 was involved in the regulation of AIMP2 ubiquitination. Since no recombinant WSN virus lacking the NS2 gene is available, we studied the effects of NS2 on AIMP2 ubiquitination in cells transfected with NS2. We observed that the presence of NS2 eliminated the ubiquitin chain from AIMP2 (Fig. 3E), supporting the negative regulatory roles of NS2 on AIMP2 ubiquitination. Taken together, our results indicate that IAV infection induces ubiquitin-dependent AIMP2 degradation and that binding with NS2 protects AIMP2 from ubiquitin modification.
FIG 3.
NS2 inhibits AIMP2 ubiquitination. (A) IAV accelerates the degradation of AIMP2 through the proteome pathway. 293T cells were infected with the A/WSN/33 virus at an MOI of 1 or left uninfected. At 12 h postinfection, the proteasome inhibitor MG-132 or a dimethyl sulfoxide (DMSO)-treated control was added to the cells for 4 h. Cell lysates were prepared and immunoblotted for AIMP2, NP, and β-actin. Relative AIMP2 band intensities were measured using ImageJ software (NIH). Data are shown as the mean ± SD from three independent experiments. Statistical significance was determined by ANOVA. (B) IAV does not alter mRNA levels of AIMP2. 293T cells were infected with the A/WSN/33 virus at an MOI of 1 or left uninfected. At 12 h postinfection, AIMP2 mRNA expression was evaluated by quantitative reverse transcription-PCR with GAPDH as an internal control. Data represent the mean ± SD from three independent experiments. Statistical significance was determined by Student's t test. (C) IAV promotes AIMP2 ubiquitination. 293T cells were cotransfected with AIMP2-Myc and HA-Ub or an HA control vector for 28 h and then infected with the A/WSN/33 virus at an MOI of 1 or left uninfected for an additional 12 h. The cell lysates were extracted and immunoprecipitated with anti-Myc antibody. Immunoblotting was performed with HA, Myc, or NP antibodies. (D) IAV induces K48-linked ubiquitination of AIMP2. 293T cells were transfected with the indicated plasmids for 28 h and then infected with the A/WSN/33 virus at an MOI of 1 for 12 h. Cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies. (E) NS2 suppresses AIMP2 ubiquitination. Lysates of 293T cells transfected with AIMP2-Myc, HA-Ub, and Flag-NS2 were immunoprecipitated and immunoblotted with the indicated antibodies. The data in panels C to E are representative of those from at least three independent experiments. **, P < 0.01; ns, no significant difference.
AIMP2 enhances IAV replication by promoting vRNP nuclear export.
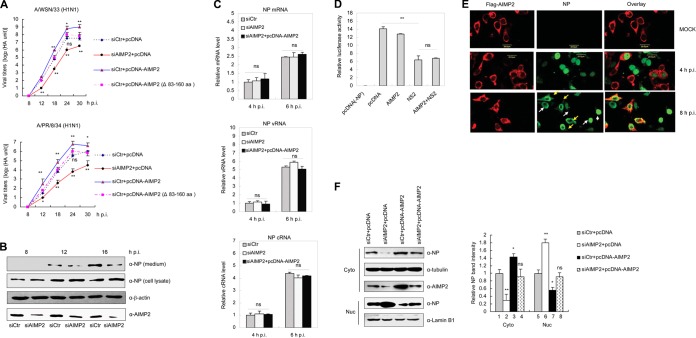
We next investigated whether AIMP2 affected IAV replication. To this end, 293T cells were transfected with plasmid pcDNA-AIMP2 with wild-type AIMP2, deletion mutant plasmid pcDNA-AIMP2(Δ83–160) lacking the NS2-binding site, or specific siRNA targeting AIMP2 (siAIMP2), followed by infection of A/WSN/33 or A/PR/8/34 virus at a multiplicity of infection (MOI) of 0.5. The supernatants of infected cells were harvested to determine virus titers by hemagglutinin assay. As shown in Fig. 4A, for both the A/WSN/33 and A/PR/8/34 strains, viral titers were increased in AIMP2-overexpressing cells and decreased in AIMP2-knockdown cells (P < 0.01), whereas no significant difference was observed in cells transfected with pcDNA-AIMP2(Δ83–160). Therefore, it is likely that AIMP2 functions as a positive regulator of IAV replication by binding to NS2. We then investigated how AIMP2 could influence IAV replication. 293T cells were transfected with control siRNA (siCtr) or siRNA targeting AIMP2 for 48 h, followed by infection with A/WSN/33 virus at an MOI of 0.5. At 8, 12, and 16 h postinfection (p.i.), both the culture supernatants and whole-cell lysates were assayed by Western blotting with anti-NP antibody. We observed that AIMP2 knockdown had no significant effect on NP levels in cellular lysates at 8 and 12 h p.i. but decreased the NP levels in the culture supernatants (virus particle) at 12 and 16 h p.i. (Fig. 4B). These results indicate that AIMP2 may not be involved in the early step of viral replication but most likely has an effect in the late stage, when viral protein synthesis has started to occur (24, 40). We also performed real-time PCR to examine whether AIMP2 had any effect on the transcription and replication of the viral genome in the early step of viral replication. 293T cells were transfected with siCtr, siAIMP2, or siAIMP2 and pcDNA-AIMP2, followed by infection with A/WSN/33 virus. The levels of mRNA, cRNA, and vRNA specific for the NP segment were measured by real-time PCR with specific primers. No significant differences in NP mRNA, cRNA, or vRNA levels were observed among the three groups at 4 and 6 h p.i. (Fig. 4C). Meanwhile, we applied a minireplicon reporter assay to confirm the negligible effect of AIMP2 on RNA-dependent RNA polymerase (RdRp) activity. As expected, overexpression of NS2, which has been shown to inhibit reporter gene expression in this system, did significantly inhibit RdRp activity (P < 0.01). In contrast, overexpression of AIMP2 did not alter RdRp activity or NS2-mediated RdRp inhibition (Fig. 4D). Thus, AIMP2 appeared not to regulate transcription or replication of the genome. Previously, NS2 has been shown to act as a key adaptor in vRNP nuclear export (12, 41). The interaction between NS2 and AIMP2 prompted us to investigate whether AIMP2 regulates vRNP nuclear export. To better track the subcellular localization of vRNPs, we performed an immunofluorescence assay with anti-NP antibody because NP is associated with viral genomic RNA. NP (Fig. 4E, green) was localized in the nucleus at 4 h p.i. and exported to the cytoplasm at 8 h p.i. in AIMP2-overexpressing cells (Fig. 4E, red), while NP was still retained in the nucleus at 8 h p.i. in AIMP2-nonoverexpressing cells (Fig. 4E), indicating that AIMP2 facilitates vRNP nuclear export. The enhancement of the nuclear export of vRNPs by AIMP2 was further confirmed by analysis of relative NP levels in the cytoplasm and nucleus when AIMP2 expression was up- or downregulated. As shown in Fig. 4F (right), AIMP2 knockdown leads to a decrease in NP levels in the cytoplasmic fractions (column 2; P < 0.01) and an increase in the nuclear fractions (column 6; P < 0.01). Conversely, AIMP2 overexpression increases the amounts of NP in the cytoplasmic fractions (column 3; P < 0.05) and decreases the nuclear retention of NP (column 7; P < 0.05). While the ectopic expression of AIMP2 was silenced by siAIMP2, there were no alterations in NP levels in the cytoplasmic fractions or the nuclear fractions (columns 4 and 8, respectively). Collectively, AIMP2 functions as a positive regulator for IAV replication, acting in the steps of vRNP nuclear export.
FIG 4.
AIMP2 promotes IAV replication. (A) Effect of AIMP2 on IAV replication. 293T cells were transfected with siCtr, siAIMP2, siCtr plus pcDNA-AIMP2, or siCtr plus pcDNA-AIMP2(Δ83–160) for 48 h, followed by infection with the A/WSN/33 or A/PR/8/34 virus at an MOI of 0.5. The supernatants of the cell culture were collected at different times postinfection, as indicated, and assayed for virus titers by hemagglutinin assay. The data represent the mean ± SD from three independent experiments. One-way ANOVA was used to determine the difference between independent groups. The differences between the variants were considered statistically significant if P was <0.05. (B) 293T cells were transfected with siAIMP2 or siCtr for 48 h, followed by infection with the A/WSN/33 virus at an MOI of 0.5. At 8, 12, and 16 h p.i., both the media and whole-cell lysates were assayed by Western blotting with anti-NP antibody. Data are representative of those from three independent experiments. (C) AIMP2 has no significant effect on the replication and transcription of the viral genome. 293T cells were transfected with siAIMP2, siCtr, or siAIMP2 and pcDNA-AIMP2. At 36 h posttransfection, the cells were infected with the A/WSN/33 virus at an MOI of 0.1. At 4 and 6 h p.i., total RNAs were extracted and subjected to quantitative reverse transcription-PCR for detection of the RNA of the NP segment, with 18S rRNA used as an internal control. Data represent the mean ± SD from three independent experiments. Statistical significance was determined by ANOVA. The differences between the variants were considered statistically significant if P was <0.05. (D) AIMP2 has no effect on RdRp activity. 293T cells were transfected with pPolI-Luc and plasmids for the expression of viral PB1, PB2, PA, and NP proteins. Renilla luciferase was used as an internal control. The cells were also cotransfected with AIMP2 or NS2, as indicated. At 30 h posttransfection, luciferase activity was measured. The data represent the mean ± SD from three independent experiments. Statistical significance was determined by ANOVA. (E) AIMP2 promotes vRNP nuclear export. 293T cells were transfected with Flag-AIMP2 and infected with the A/WSN/33 virus at an MOI of 1 or left uninfected. At 4 and 8 h p.i., cells were fixed, permeabilized, and stained for Flag-AIMP2 (red) and NP (green). Yellow arrows, AIMP2-overexpressing cells; white arrows, AIMP2-nonoverexpressing cells. Data are representative of those from three independent experiments. (F) AIMP2 promotes vRNP nuclear export. 293T cells were transfected with siCtr, siAIMP2, siCtr plus pcDNA-AIMP2, or siAIMP2 plus pcDNA-AIMP2 for 48 h, followed by infection with the A/WSN/33 virus at an MOI of 2 for an additional 12 h. Cytoplasmic (Cyto) and nuclear (Nuc) proteins were extracted and assayed by Western blotting with the indicated antibodies. Tubulin and lamin B1 were used as loading controls for the cytoplasmic and nuclear fractions, respectively. Relative NP band intensity was measured using ImageJ software (NIH). Data represent the mean ± SD from three independent experiments. Statistical significance was determined by ANOVA. **, P < 0.01; *, P < 0.05; ns, no significant difference.
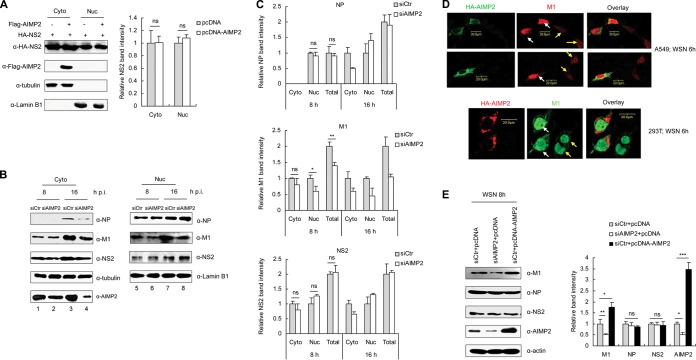
AIMP2 enhances the stability of M1.
Subsequently, we investigated how AIMP2 regulates vRNP nuclear export. Since AIMP2 directly binds NS2 and NS2 is an essential adaptor for vRNP nuclear export, we hypothesized that AIMP2 may facilitate vRNP nuclear export by regulating the cellular distribution or expression of NS2. To test this hypothesis, we transfected 293T cells with Flag-AIMP2 and HA-NS2 and analyzed the levels of NS2 in cytoplasmic and nuclear extracts. The results of Western blot analysis showed that AIMP2 had no significant effect on either the expression or the distribution of NS2 in the transfected cells (Fig. 5A). Thus, it is likely that AIMP2 regulates vRNP nuclear export by targeting other proteins rather than by directly regulating NS2. To confirm this, we then determined the effects of AIMP2 knockdown on viral NP and M1, two other important proteins involved in mediating the nuclear export of vRNPs in IAV-infected cells. 293T cells were transfected with control siRNA or siRNA targeting AIMP2 for 48 h, followed by infection with A/WSN/33 virus, and the cytoplasmic and nuclear proteins were subjected to Western blot analysis. As shown in Fig. 5B and C, knockdown of AIMP2 had no significant effect on either NS2 or NP protein levels or cellular localization at the early stage of infection (8 h p.i.). Interestingly, both the protein levels and the nuclear localization of M1 were reduced in AIMP2-knockdown cells at 8 h p.i. (P < 0.05), suggesting that the disruption of AIMP2 negatively regulated M1. These results were further confirmed by tracing the distribution of M1 in IAV-infected A549 and 293T cells with or without AIMP2 overexpression. The intensity of immunofluorescence for M1 in AIMP2-overexpressing cells was higher than that in AIMP2-nonoverexpressing cells, although M1 was mainly localized in the nucleus in both AIMP2-overexpressing and AIMP2-nonoverexpressing cells at 6 h p.i. (Fig. 5D). The positive regulation of M1 by AIMP2 was further confirmed by the increased amount of M1 in AIMP2-overexpressing cells (P < 0.05) and the decreased amount of M1 in AIMP2-knockdown cells (P < 0.01) (Fig. 5E). Together, these results suggest that AIMP2 increases M1 protein levels upon IAV infection.
FIG 5.
AIMP2 does not alter the subcellular distribution of NS2 but enhances the stability of M1. (A) AIMP2 does not alter the cellular localization of NS2. 293T cells were cotransfected with Flag-AIMP2 and HA-NS2. At 30 h posttransfection, cytoplasmic and nuclear proteins were extracted and assayed by Western blotting with the indicated antibodies. Tubulin and lamin B1 were used as loading controls for the cytoplasmic and nuclear fractions, respectively. Relative NS2 band intensities were measured using ImageJ software (NIH). Data represent the mean ± SD from three independent experiments. One-way ANOVA was used to determine the differences between independent groups. The differences between the variants were considered statistically significant if P was <0.05. (B) Knockdown of AIMP2 decreases the nuclear import and stability of M1. 293T cells were transfected with siCtr or siAIMP2 and infected with the A/WSN/33 virus at an MOI of 2. At 8 and 16 h p.i., cytoplasmic and nuclear proteins were extracted and assayed by Western blotting with the indicated antibodies. Data are representative of those from three independent experiments. (C) Quantification of relative NP, M1, and NS2 band intensities from panel B using ImageJ software (NIH). Data represent the mean ± SD from three independent experiments. Statistical significance was determined by ANOVA. (D) Enhancement of M1 nuclear localization in AIMP2-overexpressing cells. A549 and 293T cells were transfected with HA-AIMP2 and infected with A/WSN/33 at an MOI of 2. At 6 h p.i., the cells were fixed, permeabilized, and stained for HA-AIMP2 and M1. White arrows, AIMP2-overexpressing cells; yellow arrows, AIMP2-nonoverexpressing cells. Data are representative of those from three independent experiments. (E) 293T cells were transfected with siCtr, siAIMP2, or siCtr plus pcDNA-AIMP2 for 48 h, followed by infection with the A/WSN/33 virus at an MOI of 2 for 8 h. Whole-cell lysates were assayed by Western blotting with the indicated antibodies. Relative band intensities were measured using ImageJ software (NIH). Data represent the mean ± SD from three independent experiments. Statistical significance was determined by ANOVA. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, no significant difference.
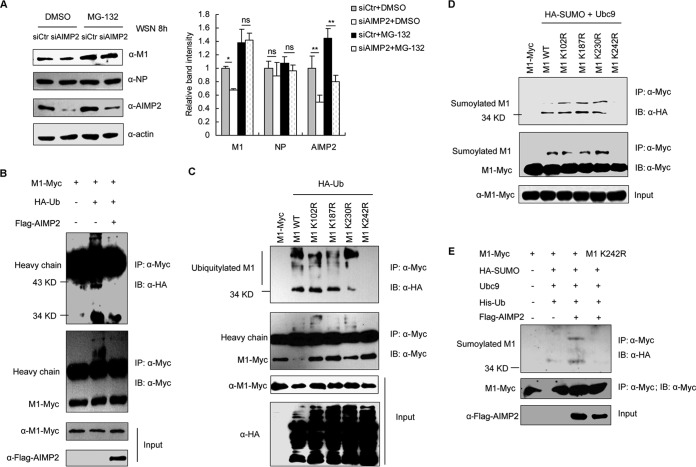
AIMP2 promotes the switch from ubiquitination to SUMOylation of M1 on K242.
AIMP2 increases M1 protein levels (Fig. 5B and E) but not mRNA levels (data not shown), suggesting a possible role of AIMP2 in the regulation of M1 stability. The ubiquitin-proteasome pathway is the most common method used by cells to degrade endogenous proteins. Therefore, we tested if it plays a role in the AIMP2-mediated regulation of M1 stability upon IAV infection. After treatment with the proteasome inhibitor MG-132, the degradation of M1 in AIMP2-knockdown cells was attenuated (P < 0.05) (Fig. 6A), suggesting that AIMP2 protects M1 from ubiquitin-mediated degradation. Furthermore, we observed that overexpression of AIMP2 reduced M1 ubiquitination (Fig. 6B). We next determined the potential ubiquitinated sites in M1. On the basis of the ubiquitination sites predicted by two online software tools (BDM-PUB and ubPred), K102, K187, K230, and K242 were selected for further study. To verify these predicted sites, we constructed M1 mutants including K102R, K187R, K230R, and K242R. When K242 was mutated to Arg (R), the mutant M1 protein in the cells could no longer be modified by ubiquitin (Fig. 6C), indicating that K242 is the major, if not the only, ubiquitin modification site of M1. Interestingly, K242 has been shown to be the SUMO modification site on M1 (21). This finding was also confirmed in our experiments (Fig. 6D). As both modifiers target the same site on M1, a positive action of AIMP2 on the SUMOylation of M1 was proposed. Accordingly, overexpression of AIMP2 increased the efficiency of SUMOylation of M1 (Fig. 6E). Collectively, these data strongly suggest that AIMP2 decreases the ubiquitination of K242 on M1 and enhances the SUMOylation of the same site.
FIG 6.
AIMP2 regulates ubiquitin/SUMO modification on K242 of M1. (A) Knockdown of AIMP2 accelerates proteasome-dependent degradation of M1. 293T cells were transfected with siCtr or siAIMP2 for 48 h, followed by infection with A/WSN/33 at an MOI of 2 along with DMSO or MG-132 treatment for 8 h. Cell lysates were extracted and assayed by Western blotting with the indicated antibodies. Relative M1, NP, and AIMP2 band intensities were measured using ImageJ software (NIH). Data represent the mean ± SD from three independent experiments. Statistical significance was determined by ANOVA. **, P < 0.01; *, P < 0.05; ns, no significant difference. (B) AIMP2 inhibits ubiquitination of M1. 293T cells were cotransfected with the indicated plasmids for 26 h. The cell lysates were extracted and immunoprecipitated with anti-Myc antibody. Immunoblotting was performed with HA, Myc, and Flag antibodies. (C) Ubiquitination of M1 is dependent on K242. 293T cells were cotransfected with the indicated plasmids for 26 h. The cell lysates were extracted and immunoprecipitated with an anti-Myc antibody. Immunoblotting was performed with the indicated antibodies. (D) K242 is the SUMOylation site of M1. 293T cells were cotransfected with the indicated plasmids for 26 h. The cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies. (E) AIMP2 enhances K242-mediated SUMOylation of M1. 293T cells were cotransfected with the indicated plasmids for 26 h. The cell lysates were extracted and immunoprecipitated with an anti-Myc antibody, followed by immunoblotting with the indicated antibodies. The data in panels B to E are representative of those from at least three independent experiments.
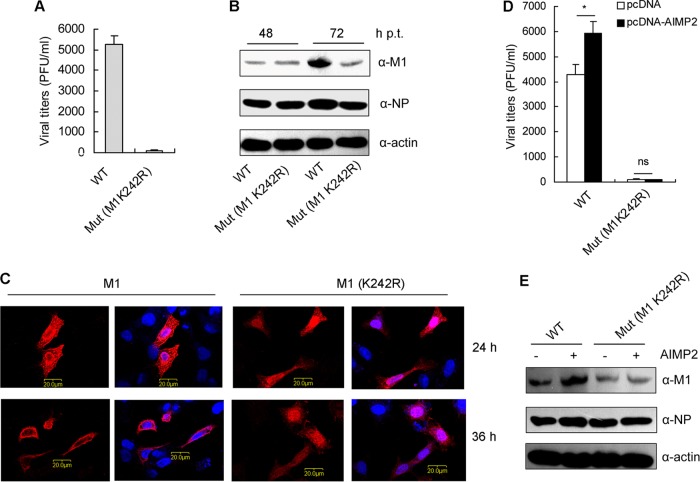
K242 of M1 is essential for IAV replication.
To assess the roles of K242 on M1 in IAV replication, we attempted to generate M1 mutant (K242R) virus by reverse genetics. The M1 mutant (K242R) virus could be rescued but replicated to significantly lower titers than WT virus (100-fold less) (Fig. 7A) and produced smaller plaques than WT virus (data not shown). To further characterize the impact of M1 K242 on the viral life cycle, we then detected the expression of M1 in 293T cells transfected with the 12 plasmids of the virus rescue system containing WT or mutant (K242R) M1. There was no significant difference in the protein levels between WT M1 and mutant (K242R) M1 at 48 h posttransfection (p.t.). At 72 h posttransfection, the levels of WT M1 were higher than those of mutant (K242R) M1, which can be explained by the more efficient WT virus replication at that time point (Fig. 7B). We also traced the intracellular distribution of M1 in 293T cells transfected with the 12 plasmids of the virus rescue system containing WT or mutant (K242R) M1. Wild-type M1 could be detected in both the cytoplasm and the nucleus at 24 h posttransfection (p.t.) and concentrated on the cell membrane at 36 h p.t. Surprisingly, the K242R mutant was still retained in the nucleus at 36 h p.t. (Fig. 7C), suggesting that the K242R mutation resulted in the nuclear retention of M1, which may lead to inefficient vRNP trafficking and virion assembly. To further clarify the effect of AIMP2 on WT or mutant virus rescue, 293T cells were cotransfected with pcDNA-AIMP2 along with the 12 plasmids containing WT or mutant M1. The plaque assays indicated that AIMP2 overexpression promoted WT but not mutant virus propagation (Fig. 7D). The increased efficiency of infectious virus particle production from AIMP2-overexpressing cells may benefit from the enhanced M1 protein levels (Fig. 7E). On the contrary, AIMP2 had no effect on the protein levels of K242R mutant M1. These data suggest that K242 of M1 is essential for IAV replication and AIMP2-mediated enhancement of IAV replication is dependent on K242 of M1.
FIG 7.
K242 of M1 is essential for IAV replication. (A) K242 of M1 is essential for virus rescue. 293T cells were transfected with the 12 plasmids of the IAV rescue system containing wild-type or mutant (Mut) (K242R) M1. At 72 h posttransfection, the media were collected and subjected to plaque assays on MDCK cells. Data represent the mean ± SD from three independent experiments. (B) 293T cells were transfected as described in the legend to panel A. At 48 and 72 h posttransfection, the cells were lysed for Western blot analysis with anti-M1 and anti-NP antibodies. Data are representative of those from three independent experiments. (C) 293T cells were transfected as described in the legend to panel A. At 24 and 36 h posttransfection, the cells were fixed, permeabilized, and stained for M1 (red) and with DAPI (blue). Data are representative of those from three independent experiments. (D) AIMP2 promotes wild-type but not M1 mutant (K242R) virus rescue. 293T cells were cotransfected with pcDNA-AIMP2 and the 12 plasmids containing wild-type or mutant (K242R) M1. At 72 h posttransfection, the media were collected and subjected to plaque assays on MDCK cells. Data represent the mean ± SD from three independent experiments. Statistical significance was determined by ANOVA. *, P < 0.05; ns, no significant difference. (E) 293T cells were transfected as described in the legend to panel D. At 48 h posttransfection, the cells were lysed for Western blot analysis with anti-M1 and anti-NP antibodies. Data are representative of those from three independent experiments.
DISCUSSION
Although ubiquitination of M1 during IAV infection has been observed (42), the precise modification site and the molecular consequences of this modification remain obscure. Here, we showed that ubiquitin and SUMO competed for the same lysine (K242) site on M1 to control the cellular localization of M1 and regulate its function in vRNP nuclear export and virus assembly during IAV infection. Interaction of NS2 with AIMP2 facilitated the switch from the ubiquitination to the SUMOylation of M1, thereby increasing virus replication.
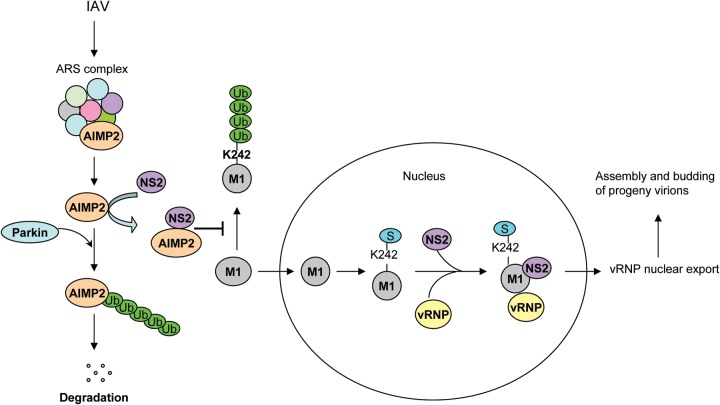
Our model for the regulatory roles of the AIMP2-NS2 interaction in M1 posttranslational modifications is presented in Fig. 8. AIMP2 is incorporated into the ARS complex under physiological conditions and dissociates from the complex upon IAV infection. The released AIMP2 can be recognized and ubiquitinated by E3 ubiquitin ligases, such as Parkin, and then degraded. To prevent ubiquitin-mediated degradation of AIMP2, NS2 quickly interacts with AIMP2 to keep it away from E3 ligase. Following binding with NS2, AIMP2 recruitment prevents the K242-mediated ubiquitination and degradation of M1 by an unknown mechanism. M1 then translocates into the nucleus, where it is SUMOylated at K242. The SUMOylation of M1 may induce a conformational change or an alteration of the protein-protein interaction, an effect likely attributable to vRNP nuclear export and the following viral assembly and budding.
FIG 8.
Proposed model for AIMP2 as a positive regulator of influenza virus replication. AIMP2 is incorporated into the ARS complex under physiological conditions. Following IAV infection, AIMP2 dissociates from the ARS complex and the released AIMP2 can be ubiquitinated by Parkin or another E3 ligase for degradation. To prevent ubiquitin-mediated degradation of AIMP2, NS2 is able to interact with AIMP2 to keep it away from ubiquitination. Following binding with NS2, AIMP2 recruitment prevents K242-mediated ubiquitination and degradation of M1. M1 then translocates into the nucleus, where it is SUMOylated at K242. The SUMOylation of M1 facilitates the formation of the NS2-M1-vRNP complex, modulates the nuclear export of vRNPs, and promotes the assembly and budding of progeny virions.
Ubiquitination of M1 was first reported by Liu and colleagues (42), who showed that overexpression of CypA increases the ubiquitination and degradation of M1 to restrict IAV replication. A recent report by Su et al. has demonstrated that Itch, an E3 ligase, interacts with M1 and mediates the ubiquitination of M1 during the virus entry process (24). They speculate that Itch-mediated ubiquitination of M1 facilitates the dissociation of viral RNA from M1 and the subsequent transport of vRNP to the nucleus (24). In this regard, ubiquitination of M1 during the virus entry process is beneficial to the viral life cycle. However, for the newly synthesized M1, Itch- or other E3 ligase-mediated ubiquitination and degradation of M1 are undoubtedly limiting factors for the production of progeny virus. Therefore, influenza virus develops strategies to evade recognition by host E3 ligase. Here we present evidence that the interaction between NS2 and AIMP2 prevents M1 from ubiquitin-mediated degradation. Moreover, we identify K242 to be the major, if not the only, ubiquitin modification site of M1. Interestingly, K242 has been shown to be the SUMO modification site on M1 (21). Different from ubiquitin-mediated degradation, SUMOylation usually alters protein-protein interactions, induces a conformational change, or promotes a relocalization of the target protein within the cell (43). K242-mediated M1 SUMOylation has been suggested to control the maturation and assembly of influenza virus particles by facilitating the interaction between M1 and vRNPs (21). As both modifiers target the same lysine on M1, the negative effect of ubiquitin on M1 may result not only from the degradation of M1 but also from the occupation of the SUMO attachment site lysine residue on M1. Therefore, AIMP2-mediated inhibition of M1 ubiquitination leads to increased M1 stability and the release of K242 for SUMO modification.
AIMP2 is primarily an integral component of the translational machinery, the ARS complex. In response to signals, such as DNA damage (27, 38) or TNF-α stimulation (29), AIMP2 dissociates from the complex to participate in a variety of cellular processes. AIMP2 has been shown to stabilize p53 via an apparently independent mechanism in response to oxidative stress. On the one hand, AIMP2 directly interacts with p53, preventing MDM2-mediated ubiquitination and the degradation of p53 (27); on the other hand, AIMP2 coactivates FBP to induce ubiquitin-specific peptidase 29 (USP29) expression. USP29 binds to p53, cleaves polyubiquitin chains from p53, and stabilizes p53 (38). Upon IAV infection, AIMP2 dissociates from the ARS complex in order to participate in the cellular antiviral response. We present data that the released AIMP2 could be readily hijacked by NS2 to prevent M1 from ubiquitin-mediated degradation. How AIMP2 inhibits M1 ubiquitination remains unclear. It is possible that AIMP2 disturbs the interaction between M1 and its E3 ligase or regulates the activity of E3 ligase or a deubiquitinating enzyme that acts on M1. These questions need to be further investigated.
Collectively, the findings from this study provide proof of principle that AIMP2 promotes IAV replication by interaction with NS2 to facilitate the switch from K242-mediated ubiquitination to SUMOylation of M1. The present work may point toward a newly identified regulatory mechanism used by IAV to ensure its life cycle.
ACKNOWLEDGMENTS
This work was supported by grants from the National Basic Research Program of China (973 Program; no. 2011CB504706, no. 2012CB518900, no. 2012CB519003, no. 2011CB504805, and no. 2010CB912201), the National Natural Science Foundation of China (no. 81171572), the National Major Scientific and Technological Special Project for Major New Drugs Innovation and Development (2014ZX09101041), and the Guangdong Innovative Research Team Program (no. 2009010058).
We thank Wenjun Liu and Xin Ye for providing materials.
REFERENCES
- 1.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, Qi X, Cui L, Zhou M, Wang H. 2013. Human co-infection with novel avian influenza A H7N9 and influenza A H3N2 viruses in Jiangsu province, China. Lancet 381:2134. doi: 10.1016/S0140-6736(13)61135-6. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. 2009. New influenza A(H1N1) virus infections: global surveillance summary, May 2009. Wkly Epidemiol Rec 84:173–179. [PubMed] [Google Scholar]
- 4.Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. 2012. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog 8:e1002998. doi: 10.1371/journal.ppat.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. 2013. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol 87:2455–2462. doi: 10.1128/JVI.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 8.Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P. 2009. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol 83:8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasin AV, Temkina OA, Egorov VV, Klotchenko SA, Plotnikova MA, Kiselev OI. 2014. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res 185:53–63. doi: 10.1016/j.virusres.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Stegmann T, White JM, Helenius A. 1990. Intermediates in influenza induced membrane fusion. EMBO J 9:4231–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin K, Helenius A. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117–130. doi: 10.1016/0092-8674(91)90576-K. [DOI] [PubMed] [Google Scholar]
- 12.Shimizu T, Takizawa N, Watanabe K, Nagata K, Kobayashi N. 2011. Crucial role of the influenza virus NS2 (NEP) C-terminal domain in M1 binding and nuclear export of vRNP. FEBS Lett 585:41–46. doi: 10.1016/j.febslet.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Akarsu H, Burmeister WP, Petosa C, Petit I, Muller CW, Ruigrok RW, Baudin F. 2003. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J 22:4646–4655. doi: 10.1093/emboj/cdg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manz B, Brunotte L, Reuther P, Schwemmle M. 2012. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat Commun 3:802. doi: 10.1038/ncomms1804. [DOI] [PubMed] [Google Scholar]
- 15.Gorai T, Goto H, Noda T, Watanabe T, Kozuka-Hata H, Oyama M, Takano R, Neumann G, Watanabe S, Kawaoka Y. 2012. F1Fo-ATPase, F-type proton-translocating ATPase, at the plasma membrane is critical for efficient influenza virus budding. Proc Natl Acad Sci U S A 109:4615–4620. doi: 10.1073/pnas.1114728109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bui M, Wills EG, Helenius A, Whittaker GR. 2000. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J Virol 74:1781–1786. doi: 10.1128/JVI.74.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sha B, Luo M. 1997. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat Struct Biol 4:239–244. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- 18.Baudin F, Petit I, Weissenhorn W, Ruigrok RW. 2001. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 281:102–108. doi: 10.1006/viro.2000.0804. [DOI] [PubMed] [Google Scholar]
- 19.Ye ZP, Pal R, Fox JW, Wagner RR. 1987. Functional and antigenic domains of the matrix (M1) protein of influenza A virus. J Virol 61:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burleigh LM, Calder LJ, Skehel JJ, Steinhauer DA. 2005. Influenza A viruses with mutations in the M1 helix six domain display a wide variety of morphological phenotypes. J Virol 79:1262–1270. doi: 10.1128/JVI.79.2.1262-1270.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu CY, Jeng KS, Lai MM. 2011. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J Virol 85:6618–6628. doi: 10.1128/JVI.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. 2010. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci U S A 107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Zhao Z, Bi Y, Sun L, Liu X, Liu W. 2013. Tyrosine 132 phosphorylation of influenza A virus M1 protein is crucial for virus replication by controlling the nuclear import of M1. J Virol 87:6182–6191. doi: 10.1128/JVI.03024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su WC, Chen YC, Tseng CH, Hsu PW, Tung KF, Jeng KS, Lai MM. 2013. Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc Natl Acad Sci U S A 110:17516–17521. doi: 10.1073/pnas.1312374110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nechushtan H, Kim S, Kay G, Razin E. 2009. Chapter 1: the physiological role of lysyl tRNA synthetase in the immune system. Adv Immunol 103:1–27. doi: 10.1016/S0065-2776(09)03001-6. [DOI] [PubMed] [Google Scholar]
- 26.Ofir-Birin Y, Fang P, Bennett SP, Zhang HM, Wang J, Rachmin I, Shapiro R, Song J, Dagan A, Pozo J, Kim S, Marshall AG, Schimmel P, Yang XL, Nechushtan H, Razin E, Guo M. 2013. Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol Cell 49:30–42. doi: 10.1016/j.molcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han JM, Park BJ, Park SG, Oh YS, Choi SJ, Lee SW, Hwang SK, Chang SH, Cho MH, Kim S. 2008. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc Natl Acad Sci U S A 105:11206–11211. doi: 10.1073/pnas.0800297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim MJ, Park BJ, Kang YS, Kim HJ, Park JH, Kang JW, Lee SW, Han JM, Lee HW, Kim S. 2003. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat Genet 34:330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- 29.Choi JW, Kim DG, Park MC, Um JY, Han JM, Park SG, Choi EC, Kim S. 2009. AIMP2 promotes TNFalpha-dependent apoptosis via ubiquitin-mediated degradation of TRAF2. J Cell Sci 122:2710–2715. doi: 10.1242/jcs.049767. [DOI] [PubMed] [Google Scholar]
- 30.Ko HS, Lee Y, Shin JH, Karuppagounder SS, Gadad BS, Koleske AJ, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM. 2010. Phosphorylation by the c-Abl protein tyrosine kinase inhibits Parkin's ubiquitination and protective function. Proc Natl Acad Sci U S A 107:16691–16696. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Karuppagounder SS, Shin JH, Lee YI, Ko HS, Swing D, Jiang H, Kang SU, Lee BD, Kang HC, Kim D, Tessarollo L, Dawson VL, Dawson TM. 2013. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat Neurosci 16:1392–1400. doi: 10.1038/nn.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song L, Liu H, Gao S, Jiang W, Huang W. 2010. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol 84:8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Chi X, Wei H, Chen Y, Chen Z, Huang S, Chen JL. 2014. Influenza A virus-induced degradation of eukaryotic translation initiation factor 4B contributes to viral replication by suppressing IFITM3 protein expression. J Virol 88:8375–8385. doi: 10.1128/JVI.00126-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han JM, Lee MJ, Park SG, Lee SH, Razin E, Choi EC, Kim S. 2006. Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J Biol Chem 281:38663–38667. doi: 10.1074/jbc.M605211200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Li G, Ye X. 2010. Cyclin T1/CDK9 interacts with influenza A virus polymerase and facilitates its association with cellular RNA polymerase II. J Virol 84:12619–12627. doi: 10.1128/JVI.01696-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paterson D, Fodor E. 2012. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog 8:e1003019. doi: 10.1371/journal.ppat.1003019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Chung HJ, Vogt M, Jin Y, Malide D, He L, Dundr M, Levens D. 2011. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J 30:846–858. doi: 10.1038/emboj.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko HS, von Coelln R, Sriram SR, Kim SW, Chung KK, Pletnikova O, Troncoso J, Johnson B, Saffary R, Goh EL, Song H, Park BJ, Kim MJ, Kim S, Dawson VL, Dawson TM. 2005. Accumulation of the authentic Parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci 25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demirov D, Gabriel G, Schneider C, Hohenberg H, Ludwig S. 2012. Interaction of influenza A virus matrix protein with RACK1 is required for virus release. Cell Microbiol 14:774–789. doi: 10.1111/j.1462-5822.2012.01759.x. [DOI] [PubMed] [Google Scholar]
- 41.Neumann G, Hughes MT, Kawaoka Y. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J 19:6751–6758. doi: 10.1093/emboj/19.24.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Sun L, Yu M, Wang Z, Xu C, Xue Q, Zhang K, Ye X, Kitamura Y, Liu W. 2009. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell Microbiol 11:730–741. doi: 10.1111/j.1462-5822.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 43.Everett RD, Boutell C, Hale BG. 2013. Interplay between viruses and host sumoylation pathways. Nat Rev 11:400–411. doi: 10.1038/nrmicro3015. [DOI] [PubMed] [Google Scholar]