ABSTRACT
HIV-1 incorporates various host membrane proteins during particle assembly at the plasma membrane; however, the mechanisms mediating this incorporation process remain poorly understood. We previously showed that the HIV-1 structural protein Gag localizes to the uropod, a rear-end structure of polarized T cells, and that assembling Gag copatches with a subset, but not all, of the uropod-directed proteins, i.e., PSGL-1, CD43, and CD44, in nonpolarized T cells. The latter observation suggests the presence of a mechanism promoting virion incorporation of these cellular proteins. To address this possibility and identify molecular determinants, in the present study we examined coclustering between Gag and the transmembrane proteins in T and HeLa cells using quantitative two-color superresolution localization microscopy. Consistent with the findings of the T-cell copatching study, we found that basic residues within the matrix domain of Gag are required for Gag–PSGL-1 coclustering. Notably, the presence of a polybasic sequence in the PSGL-1 cytoplasmic domain significantly enhanced this coclustering. We also found that polybasic motifs present in the cytoplasmic tails of CD43 and CD44 also promote their coclustering with Gag. ICAM-1 and ICAM-3, uropod-directed proteins that do not copatch with Gag in T cells, and CD46, a non-uropod-directed protein, showed no or little coclustering with Gag. However, replacing their cytoplasmic tails with the cytoplasmic tail of PSGL-1 significantly enhanced their coclustering with Gag. Altogether, these results identify a novel mechanism for host membrane protein association with assembling HIV-1 Gag in which polybasic sequences present in the cytoplasmic tails of the membrane proteins and in Gag are the major determinants.
IMPORTANCE Nascent HIV-1 particles incorporate many host plasma membrane proteins during assembly. However, it is largely unknown what mechanisms promote the association of these proteins with virus assembly sites within the plasma membrane. Notably, our previous study showed that HIV-1 structural protein Gag colocalizes with a group of uropod-directed transmembrane proteins, PSGL-1, CD43, and CD44, at the plasma membrane of T cells. The results obtained in the current study using superresolution localization microscopy suggest the presence of a novel molecular mechanism promoting the association of PSGL-1, CD43, and CD44 with assembling HIV-1 which relies on polybasic sequences in HIV-1 Gag and in cytoplasmic domains of the transmembrane proteins. This information advances our understanding of virion incorporation of host plasma membrane proteins, some of which modulate virus spread positively or negatively, and suggests a possible new strategy to enrich HIV-1-based lentiviral vectors with a desired transmembrane protein.
INTRODUCTION
HIV-1 assembles at the plasma membrane in most cell types. The viral structural protein Gag, synthesized as a precursor polyprotein (Pr55), initiates and coordinates viral assembly at the plasma membrane. Membrane binding and targeting of Gag require cotranslational, N-terminal myristoylation of Gag (1, 2) as well as the highly basic region (HBR) of the matrix (MA) domain, which specifically interacts with the plasma membrane phospholipid phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] (3–7). The MA HBR also binds RNA (8–11). The MA-bound RNA is able to inhibit interactions of Gag with other acidic phospholipids; however, this inhibition can be overcome by PI(4,5)P2 interaction (12–16). This mechanism is likely to suppress nonspecific membrane binding of Gag to other cellular membranes and ensure Gag localization to the plasma membrane. Recent in vitro studies also implicated acyl chains of other phospholipids and cholesterol in Gag binding to lipid membranes (16–18). While the role of the phospholipid acyl chains in cells remains to be determined, previous studies showed that cholesterol depletion reduces Gag membrane binding in cells (19).
While Gag membrane binding and multimerization drive particle assembly, infectious virion production also involves a coordinated and highly regulated process in which Gag recruits other viral and cellular components to assembly sites and to assembling virions. This process includes incorporation of the viral envelope glycoprotein Env, encapsidation of viral genomic RNA, and incorporation of other viral and cellular proteins (20). Proteomics studies showed that a large number (∼100) of cellular proteins associated with the plasma membrane are incorporated into HIV-1 particles released by infected macrophages (21). However, the specific molecular mechanisms that mediate incorporation of host membrane proteins into virions remain largely unexplored, except for those for BST-2/tetherin (22) and virus receptors (23–25). This is in contrast to the incorporation of viral glycoproteins, where several mechanisms have been investigated (26, 27). Indeed, previous studies suggested that most membrane proteins are incorporated into retrovirus particles passively, i.e., with no enrichment relative to the plasma membrane (28). During assembly, Gag has been observed to associate with plasma membrane microdomains, such as lipid rafts (29, 30) and tetraspanin-enriched microdomains (TEMs) (31–36), and reorganize these domains in ways that do not occur in uninfected cells (37–39). Therefore, at least a subset of membrane proteins (e.g., glycosylphosphatidylinositol-anchored proteins and tetraspanins) may be incorporated into virions actively (as opposed to by passive incorporation, which does not involve enrichment of membrane proteins at assembly sites) without specific interactions, other than association with the plasma membrane microdomains. However, it is unknown whether other virion-associated membrane proteins are incorporated passively or whether specific interactions (direct or indirect) with Gag contribute to their incorporation.
T cells, which are a natural host of HIV-1 infection in vivo, are highly motile and exhibit a polarized morphology in secondary lymphoid tissues (40–44), where cell-to-cell HIV-1 transmission is likely to occur (45, 46). In polarized T cells, Gag localizes to a rear-end protrusion known as the uropod (34, 47). In our previous studies, we observed that when clustering was induced by specific antibodies, a subset of uropod-directed proteins, including PSGL-1, CD43, and CD44 (here referred to as class I uropod-directed proteins), colocalized or copatched with Gag, even in nonpolarized T cells (34, 39). This colocalization was found to be dependent upon the charge, but not the specific amino acid sequence, of the MA HBR (39). Intriguingly, we also observed that Gag failed to copatch with a second class of uropod-directed proteins (class II); CD59, ICAM-1, and ICAM-3 were observed to localize to uropods but not to copatch with Gag in unpolarized T cells. Furthermore, ICAM-3, which copatched with the class I uropod-directed protein CD44 in uninfected cells, dissociated from CD44 in the presence of Gag through an unknown mechanism (39). Most of these membrane proteins described above are cell adhesion molecules, and some of them are implicated in modulation of immunological synapses (48–50). Therefore, the association of these cellular proteins with virus assembly sites may have important implications in the formation or stability of cell-cell contact structures that efficiently transmit HIV-1 particles, such as the virological synapse (VS) (51). However, while the high degree of copatching suggests that HIV-1 may have evolved to preferentially recruit uropod-directed proteins to virus assembly sites, the mechanisms remain to be determined.
The previous studies were performed using an antibody-induced copatching assay (38) in which proteins of interest are clustered to form patches that can be readily distinguished by conventional, diffraction-limited microscopy. However, the protein distribution determined by this technique does not necessarily correlate with the native distribution of proteins in live cells because clustering is induced by antibodies prior to fixation. A great deal has been learned about mechanisms for the specific incorporation of viral glycoproteins into assembling virus particles using scanning electron microscopy or superresolution fluorescence microscopy (52–55), which allow analyses at a resolution higher than that possible by conventional fluorescence microscopy. However, except for the molecular determinants for BST-2/tetherin incorporation (56, 57), the molecular determinants for the incorporation of host transmembrane proteins have not been examined at a high resolution. High-resolution microscopy methods not only allow assessment of whether the protein of interest is incorporated into assembling virions but also provide information regarding the degree of enrichment of the protein at assembly sites relative to that in surrounding areas, which is essential in determining whether the protein is actively recruited to assembly sites. Indeed, in our previous study (56), superresolution localization microscopy methods, but not confocal microscopy, allowed us to detect differences in the degree of BST-2/tetherin–Gag coclustering among multimerization-competent Gag mutants. In this study, we employed the same quantitative two-color superresolution localization microscopy techniques to elucidate the molecular mechanism by which a subset of cellular transmembrane proteins is specifically recruited to sites where viral assembly occurs and incorporated into HIV-1 particles.
MATERIALS AND METHODS
Plasmids.
pNL4-3/Gag-mEos3.2 was described previously (56). pNL4-3/Fyn(10)/Gag-mEos3.2, pNL4-3/Fyn(10)/6A2T/Gag-mEos3.2, pNL4-3/Fyn(10)/HBR/RKswitch/Gag-mEos3.2, and pNL4-3/Fyn(10)/ΔMA/Gag-mEos3.2 were constructed using MA-containing sequences from pNL4-3/Gag-Venus versions of these constructs, which were described previously (38, 39). pNL4-3/Kmyr/ΔMA/Gag-mEos3.2 was constructed by replacing the MA domain of Gag with the N-terminal, Ras-derived membrane-anchoring sequence MGSSKSKDGKKKKKKSKTK (58, 59).
A plasmid encoding a full-length cDNA clone of human PSGL-1 (GenBank accession number NM_003006.3), pCMV6-AC/PSGL-1, was obtained from OriGene (Rockville, MD). pCMV6/empty was created by digesting pCMV6-AC/PSGL-1 with AgeI to remove the entire multiple-cloning site, followed by religation of the vector. The pCMV6-AC/PSGL-1/C310A and ΔCT (with a stop codon in place of L331) constructs were generated by standard PCR mutagenesis techniques. The PSGL-1 expression plasmid that we obtained encodes transcript variant 2, which lacks 1 of 12 10-amino-acid tandem repeats (corresponding to residues 132 to 141 of transcript variant 1 used in another study [60]) in the extracellular region. Accordingly, the C320A mutation that inhibits PSGL-1 dimerization reported previously (60) corresponds to the C310A mutation in our study. pCMV6-AC/PSGL-1/3A and pCMV6-AC/PSGL-1/6A constructs were generated by making the following substitutions (mutations are in bold) using a single-stranded PCR mutagenesis approach: RLSRK → ALSAA (residues 334 to 338) in pCMV6-AC/PSGL-1/3A and EPREDREGDD → APRAARAGAA (residues 385 to 394) in pCMV6-AC/PSGL-1/6A. Briefly, phosphorylated forward primers spanning mutated regions were used for PCR amplification with Pfu Ultra II high-fidelity DNA polymerase (Agilent, Santa Clara, CA). Next, methylated parental DNA was digested with the DpnI enzyme (NEB, Ipswich, MA), followed by heat inactivation of the enzyme. PCR products were then transformed directly into chemically competent Escherichia coli cells, and the resulting clones were sequenced to verify the correct sequence of the entire open reading frame.
Plasmids carrying full-length cDNA clones of the following genes were obtained from OriGene (Rockville, MD) (GenBank accession numbers are given in parentheses): human CD43 (NM_003123.2), pCMV6-XL5/CD43; CD44 (NM_000610.3), pCMV6-XL5/CD44; ICAM-1 (NM_000201.1), pCMV6-XL5/ICAM-1; ICAM-3 (NM_002162.2), pCMV6-AC/ICAM-3; and CD46 (NM_002389.3), pCMV6-XL4/CD46. Basic motif mutants pCMV6-XL5/CD43/6A and pCMV6-XL5/CD44/6A were generated by single-stranded PCR mutagenesis as described above for the pCMV6-AC/PSGL-1/3A and pCMV6-AC/PSGL-1/6A constructs. The chimeric constructs pCMV6-AC/ICAM-1/PCT, pCMV6-AC/ICAM-3/PCT, and pCMV6-AC/CD46/PCT were constructed by inserting the sequences encoding the extracellular and transmembrane domains of ICAM-1, ICAM-3, and CD46, respectively, into pCMV6-AC/PSGL-1 in place of the PSGL-1 extracellular and transmembrane domains by standard molecular cloning techniques. The boundary sequences of the chimeras are as follows (the sequences of ICAM-1, ICAM-3, and CD46 are shown in bold): for ICAM-1 with the PSGL-1 cytoplasmic tail (ICAM-1/PCT), MAPSSP … LSTYLYAVRLSR; for ICAM-3/PCT, MATMVP … ALMYVFAVRLSR; and for CD46/PCT, MEPPGR … ICVVPYAVRLSR. Each construct was verified by sequencing the entire open reading frame.
Antibodies.
Monoclonal mouse antibodies directed against human PSGL-1 (clone KPL-1), CD43 (1G10), CD44 (515), ICAM-1 (LB-2), ICAM-3 (TU41), and CD46 (E4.3) were obtained from BD Biosciences (San Jose, CA). Rabbit monoclonal antibody against human ICAM-1 (EP1442Y) was obtained from Abcam (Cambridge, MA). For microscopy experiments, antibodies were directly labeled with Alexa Fluor 647 using an Alexa Fluor 647 antibody labeling kit obtained from Life Technologies (Carlsbad, CA). For immunoblotting, unlabeled primary antibodies were used along with either Alexa Fluor 488- or Alexa Fluor 594-conjugated antihuman or antirabbit secondary antibodies (Life Technologies, Carlsbad, CA). Anti-HIV Ig was obtained from the NIH AIDS Research and Reference Reagent Program (NABI and NHLBI).
Cells, transfection, infection, immunostaining, and analyses for cell surface expression, virus-like particle (VLP) release, and incorporation of membrane proteins.
HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 5% fetal bovine serum, penicillin-streptomycin, and l-glutamine. For superresolution localization microscopy analysis, cells were plated at a density of 4.2 × 104 cells per well in number 1.5, 4-chamber coverslips from Fisher Scientific (Pittsburgh, PA). After overnight incubation, cells were transfected with 1 μg pNL4-3/Gag-mEos3.2 plasmids and 50 ng pCMV6 plasmids per well, using the Lipofectamine 2000 reagent, according to the manufacturer's instructions (Life Technologies, Carlsbad, CA). Cells were then incubated for 16 h prior to fixation with phosphate-buffered saline (PBS) containing 4% paraformaldehyde and 0.1% glutaraldehyde for 10 min, followed by immunostaining with Alexa Fluor 647-conjugated antibodies.
For flow cytometry analysis of cellular transmembrane proteins expressed on the transfected HeLa cell surface, HeLa cells were plated at a density of 5.6 × 105 cells per well in 6-well plates and 1 day later transfected with 100 ng pCMV6 plasmids per well. Cells were incubated for 16 h prior to collection of cells as suspensions and fixation with PBS containing 4% paraformaldehyde for 30 min. Cells were immunostained with Alexa Fluor 647-conjugated antibodies and analyzed using a BD FACSCanto flow cytometer.
The VLP release assay used for the Gag-mEos3.2 constructs was described previously (56). For analysis of virion incorporation of CD43, CD43/6A, ICAM-1, or ICAM-1/PCT, HeLa cells cotransfected with pNL4-3 and an expression plasmid carrying one of the membrane proteins were cultured for 16 h or 2 days, and cell and virus lysates were prepared as previously described (61). Immunoblotting was performed using anti-CD43 (1G10) or anti-ICAM-1 (EP1442Y) and anti-HIV Ig in combination with appropriate secondary antibodies, and fluorescence signals were detected using a Typhoon scanner (GE Healthcare).
Maintenance, fixation, and immunostaining of P2 cells and their parental A3.01 cells were performed as described previously (34, 39). For superresolution localization microscopy analysis of T cells, A3.01 cells were infected with vesicular stomatitis virus G glycoprotein (VSV-G)-pseudotyped viruses encoding Gag derivatives as described previously (34, 39). Two days later, cells (∼0.5 × 106) were resuspended in RPMI medium containing no serum and plated onto the coverslip area of MatTek number 1.5 glass-bottom dishes. The coverslip area was precoated with 5 μg/ml fibronectin in PBS. After a 50-min incubation at 37°C, cells were fixed with PBS containing 4% paraformaldehyde and 0.1% glutaraldehyde for 10 min, followed by immunostaining with Alexa Fluor 647-conjugated antibodies.
Superresolution localization microscopy and image processing.
Sample preparation, conditions for image acquisition, reconstruction of superresolution images, and data analysis methods were described previously (56). Briefly, single fluorophores of Alexa Fluor 647- and mEos3.2-labeled proteins were imaged simultaneously under total internal reflection (TIRF) using an Olympus IX81-XDC inverted microscope with a cellTIRF module, a ×100 UAPO TIRF objective (numerical aperture = 1.49), and active Z-drift correction (ZDC) (Olympus America). Two emission channels were projected onto the same electron-multiplying charge-coupled-device camera (iXon-897; Andor) using a DV2 splitter (Photometrics). Single molecules captured in each emission channel were identified and localized in individual image frames and culled to remove outliers in intensity, width, or localization error, and then high-resolution images were reconstructed from localized centers acquired from at least 5,000 acquired raw images. Reconstructed images from the two emission channels were aligned by sampling space with fiducial particles both before and after imaging. Pair cross-correlation [C(r)] functions were tabulated from masked images of reconstructed localized centers as described previously (62). Cross-correlation curves are normalized to be 1 at long distances, which corresponds to a random distribution. Additionally, cross-correlation curves from individual cells were set to 1 at 1 μm in order to avoid potential artifacts arising from uneven illumination of the sample or inaccurate masking of individual cells. To compare total coclustering in cross-correlation analyses, the area (A) under the cross-correlation curves was summed using the following equation for radii between 0 and 200 nm: , where n is the number of radii included in the sum, i is an index describing the radius, ri, and r is radius. The average radii of the Gag puncta were determined by fitting the Gag autocorrelation function to a Gaussian shape, with the average radius being the standard deviation of this curve (62).
RESULTS
Two-color superresolution microscopy analysis detects MA HBR-dependent coclustering of Gag with PSGL-1 in T cells.
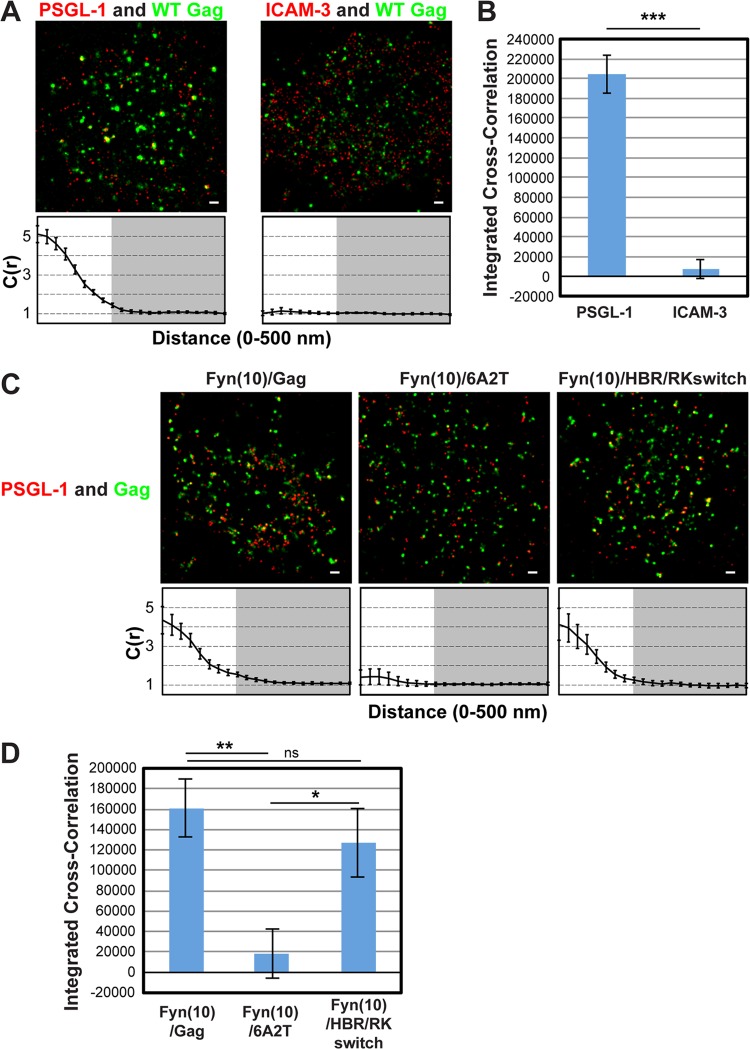
To examine whether class I uropod-directed proteins associate with Gag in the absence of pretreatment with cross-linking antibodies, we analyzed coclustering between PSGL-1 and wild-type (WT) Gag-mEos3.2, a Gag construct fused to the photoswitchable, monomeric fluorescent protein mEos3.2 (63), in T cells using two-color superresolution localization microscopy techniques (56). T cells infected with VSV-G-pseudotyped HIV-1 encoding WT Gag-mEos3.2 were fixed with PBS containing 4% paraformaldehyde and 0.1% glutaraldehyde prior to immunostaining with an anti-PSGL-1 monoclonal antibody directly conjugated to Alexa Fluor 647 and examined by TIRF illumination. In superresolution localization microscopy, signals for two colocalizing proteins do not necessarily appear to localize at exactly the same spots due to the high-resolution nature of the method (25 nm for Gag-mEos3.2 and 15 nm for PSGL-1); rather, colocalizing proteins are often detected adjacent to each other. To assess the coclustering (or lack thereof) between Gag-mEos3.2 and PSGL-1 quantitatively, we performed cross-correlation analysis (Fig. 1A, bottom), as in our previous studies (56). This analysis measures the probability of finding one protein (e.g., PSGL-1) at a given distance from another (e.g., Gag) or vice versa normalized by the probability that would be detected from a random distribution of the same average density (62). A normalized cross correlation of 1 is observed when the distributions of the two proteins are independent from each other, whereas a value greater than 1 indicates coclustering of the two proteins. To facilitate comparison of total coclustering between different conditions, the area between cross-correlation curves and the baseline [C(r) = 1; no cross-correlation] was summed for radii between 0 and 200 nm on a cell-by-cell basis, as described in Materials and Methods, and the average values were compared (Fig. 1B). This value, presented as an integrated cross-correlation, is 0 if two proteins distribute completely randomly. In this analysis, we found that Gag coclusters with PSGL-1 but not with ICAM-3 (Fig. 1A and B), an observation that correlates well with our previous results obtained in the copatching assays (39).
FIG 1.
Two-color superresolution microscopy analysis detects MA HBR-dependent coclustering of Gag with PSGL-1 in T cells. (A) A3.01 cells infected with VSV-G-pseudotyped HIV-1 whose genome encodes the Gag-mEos3.2 fusion protein were plated onto fibronectin-coated coverslips, fixed, immunostained for PSGL-1 or ICAM-3 with Alexa Fluor 647-labeled monoclonal antibodies, and imaged by TIRF microscopy in a reducing buffer (at least 5,000 image frames per cell). Representative reconstructed images were produced as described in Materials and Methods. Images show Gag-mEos3.2 in green and PSGL-1 and ICAM-3 in red. Cross-correlation measurements were performed using images of a total of at least 15 cells per condition. Curves below the images represent mean cross-correlation [C(r)] values at distances ranging from 0 to 500 nm. The range that is not shaded corresponds to 0 to 200 nm, which was used for the calculation of total coclustering in panel B. (B) Total coclustering was calculated by integrating the area under the cross-correlation curves from 0 to 200 nm, as described in Materials and Methods. The values shown indicate means ± SEMs. ***, P < 0.0005. (C) A3.01 cells were infected with VSV-G-pseudotyped viruses whose genomes encode the indicated Fyn(10)/Gag-mEos3.2 derivatives and analyzed as described in the legend to panel A. (D) The analysis of total coclustering was performed as described in the legend to panel B using cross-correlation curves for images of a total of 9 to 15 cells. **, P < 0.005; *, P < 0.05; ns, not significant. The fields shown in panels A and C are 10 μm by 10 μm. Bars = 500 nm.
To further evaluate the copatching results analyzed in our previous study, we next examined the effect of amino acid substitutions in MA HBR on Gag–PSGL-1 coclustering (Fig. 1C and D). To compensate for the membrane-binding defect caused by MA alterations, we examined Gag derivatives that have an N-terminal triple acylation sequence derived from Fyn (Fig. 2A). In Fyn(10)/6A2T/Gag, the 8 basic residues of the HBR, which facilitate interactions between Gag and PI(4,5)P2, have been replaced with neutral amino acids (39), whereas in Fyn(10)/HBR/RKswitch Gag, the identity of the 8 basic residues has been exchanged (R → K and K → R) (RKswitch). We previously observed that when fused to yellow fluorescent protein and examined by the antibody-induced copatching assay, the 6A2T substitutions but not the RKswitch changes in the context of Fyn(10)/Gag abolished colocalization between Gag and PSGL-1 (39). Consistent with the copatching results, we found in superresolution localization microscopy analysis that in T cells, Fyn(10)/6A2T Gag-mEos3.2 failed to cocluster with PSGL-1, whereas Fyn(10)/HBR/RKswitch Gag-mEos3.2 showed a level of coclustering with PSGL-1 similar to that shown by Fyn(10)/Gag, which retains the intact MA sequence (Fig. 1C and D). Altogether, the two-color superresolution localization microscopy analysis confirmed the MA-dependent association between Gag and PSGL-1 in T cells, as previously suggested on the basis of the copatching studies (39).
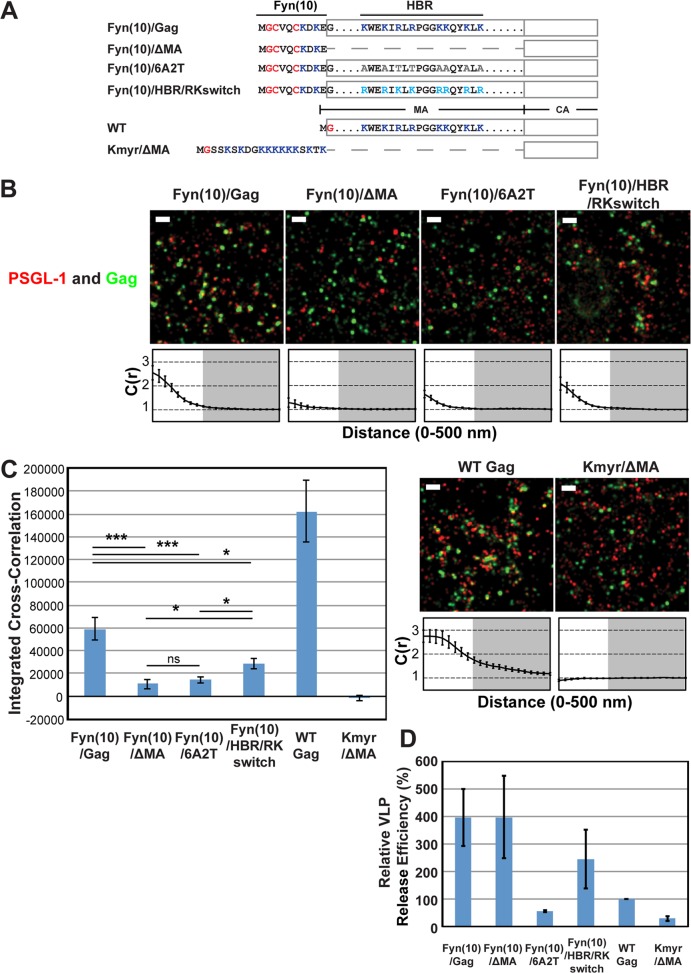
FIG 2.
Coclustering of Gag and PSGL-1 in HeLa cells is dependent on the intact MA HBR sequence but does not correlate with Gag membrane binding or VLP production. (A) Schematic representation of the MA mutants examined in this study. The amino acid residues of the Fyn(10), HBR, and Kmyr sequences are shown. Blue, basic residues; gray, uncharged amino acids; turquoise, switched residues; red, lipid-modified residues. (B) HeLa cells were transfected with plasmids carrying the indicated Gag-mEos3.2 fusion constructs and wild-type PSGL-1. Cells were fixed and immunostained for PSGL-1 with a monoclonal antibody against PSGL-1, which was directly labeled with Alexa Fluor 647, and imaged by TIRF microscopy in a reducing buffer (7,500 image frames per cell). Representative reconstructed images were produced as described in Materials and Methods. Images show Gag-mEos3.2 in green and PSGL-1 in red. Bars = 500 nm. Cross-correlation curves are shown below their corresponding representative images. Cross-correlation measurements were performed as described in the legend to Fig. 1A using images of a total of at least 10 cells per condition from 2 independent experiments. (C) Total coclustering was calculated as described in the legend to Fig. 1B. Values shown indicate means ± SEMs. ***, P < 0.0005; *, P < 0.05; ns, not significant. (D) A VLP release assay was performed on Gag-mEos3.2 constructs in HeLa cells. Results represent the averages of three independent experiments normalized to the value for WT Gag-mEos3.2.
Coclustering of Gag and PSGL-1 in HeLa cells is dependent on the intact MA HBR sequence but does not correlate with Gag membrane binding per se or VLP production efficiency.
We conducted subsequent experiments in this study using HeLa cells because HeLa cells do not express endogenous uropod proteins PSGL-1, CD43, and ICAM-3 (see below) and yet are very amenable to transfection by plasmid DNA, properties that are essential for examining molecular determinants of potential interactions between Gag and uropod proteins. Nonetheless, it is possible that the Gag-uropod-protein associations are dependent on T-cell-specific factors that are absent in adherent cell lines. Therefore, we first sought to determine whether the MA-dependent nature of the association between Gag and uropod-directed proteins in T cells is also observed in HeLa cells. To this end, we expressed PSGL-1 along with Gag-mEos3.2 constructs (63) in HeLa cells and compared Gag derivatives with MA deletion or amino acid substitutions (Fig. 2A). As described above, PSGL-1 was detected with a specific monoclonal antibody conjugated directly to Alexa Fluor 647. In addition to the Fyn(10)/Gag derivatives examined in the experiments presented in Fig. 1, Fyn(10)/ΔMA/Gag, which lacks the entire MA sequence (5), was examined for coclustering with PSGL-1 (Fig. 2A).
When expressed in HeLa cells, all of these constructs were readily detected at the plasma membrane, as was PSGL-1 (Fig. 2B). We found that deletion of MA or basic-to-neutral substitutions in MA HBR reduced the level of coclustering of Fyn(10)/Gag-mEos3.2 with PSGL-1 4- to 6-fold (Fig. 2B and C), consistent with our findings in T cells (39) (Fig. 1). In contrast, coclustering was reduced only 2-fold by the HBR/RKswitch mutation (Fig. 2B and C). These results indicate that Gag is able to associate with PSGL-1, a class I uropod-directed protein, when expressed in HeLa cells. They also confirm that the HBR positive charge in MA is an important determinant of this association, as observed in T cells, although we note that the HBR/RKswitch mutant may associate more efficiently with PSGL-1 in T cells than in HeLa cells.
Using the same experimental settings described above, we further sought to determine whether myristoylation and a polybasic sequence are sufficient to recruit PSGL-1 or whether other features of the MA domain contribute to PSGL-1 recruitment. To test these possibilities, we replaced the MA domain of Gag with a synthetic membrane anchor, Kmyr, which binds to the plasma membrane through myristoylation and basic residues (Fig. 2A) (59), and compared this construct (Kmyr/ΔMA/Gag) with Fyn(10)/Gag derivatives and WT Gag. We found that WT Gag-mEos3.2 coclustered with PSGL-1 even more efficiently than Fyn(10)/Gag-mEos3.2, indicating that dual palmitoylation is not necessary for coclustering of Gag with PSGL-1 in the presence of the MA sequence. We detected signals for Kmyr/ΔMA/Gag-mEos3.2 at the plasma membrane by TIRF illumination, indicating that the Kmyr peptide supports membrane binding of Gag in the absence of the MA domain (Fig. 2B). However, Kmyr/ΔMA/Gag-mEos3.2 failed to show any coclustering with PSGL-1 (Fig. 2B and C). These results indicate that plasma membrane binding via a combination of myristoylation and basic charge is insufficient to facilitate PSGL-1 coclustering with HIV-1 assembly sites and that an additional property of the MA domain is necessary.
We next sought to determine whether efficient VLP production correlates with the recruitment of PSGL-1. We compared the VLP release efficiencies of the constructs used in the above-described experiments in HeLa cells (Fig. 2D). The VLP release efficiency of Fyn(10)/Gag was higher than that of WT Gag, as observed previously for untagged versions of these constructs (5, 19). The higher membrane binding ability conferred by the triple acylation signal of Fyn attached to Gag is likely to explain at least partly the increased VLP release efficiency of Fyn(10)/Gag (5, 19). Likewise, Fyn(10)/ΔMA/Gag and Fyn(10)/HBR/RKswitch Gag released VLPs more efficiently than WT Gag. In contrast, Fyn(10)/6A2T/Gag and Kmyr/ΔMA/Gag were less efficient in VLP release than WT Gag, although the defective step in VLP assembly/release of Fyn(10)/6A2T/Gag and Kmyr/ΔMA/Gag is currently unknown. Of note, comparison of Fyn(10)/Gag derivatives and WT Gag indicates that VLP release efficiencies do not correlate with the Gag–PSGL-1 coclustering shown in Fig. 2C. Likewise, we did not find any correlation between Gag–PSGL-1 coclustering and the size of Gag puncta, which are within 44 to 52 nm (in radius) for all Gag derivatives. Altogether, PSGL-1 coclustering with HIV-1 Gag is correlated with the presence of the MA domain but not with strong Gag membrane binding per se or efficient VLP production.
Efficient coclustering of Gag and PSGL-1 in HeLa cells is dependent on a basic stretch in the cytoplasmic domain of PSGL-1.
In order to better understand how Gag coclusters with class I uropod-directed proteins, we sought to identify which parts of PSGL-1 are necessary for this localization. To this end, we constructed PSGL-1 derivatives (Fig. 3A), transfected them into HeLa cells, and compared the cells with cells transfected with wild-type PSGL-1 by immunoblotting, flow cytometry, and superresolution localization microscopy analyses (Fig. 3B to D). While all constructs were readily detected in cell lysates (Fig. 3B) and on the cell surface (Fig. 3C), we observed that there were severalfold differences in total and cell surface expression levels among PSGL-1 derivatives. It is important to note, however, that the cross-correlation method used in this study has been shown to be insensitive to relative protein expression levels (62). Moreover, in superresolution microscopy, cells with readily detectable expression levels were examined, and this was likely to minimize, if any, the effects of the low expression levels of some PSGL-1 mutants.
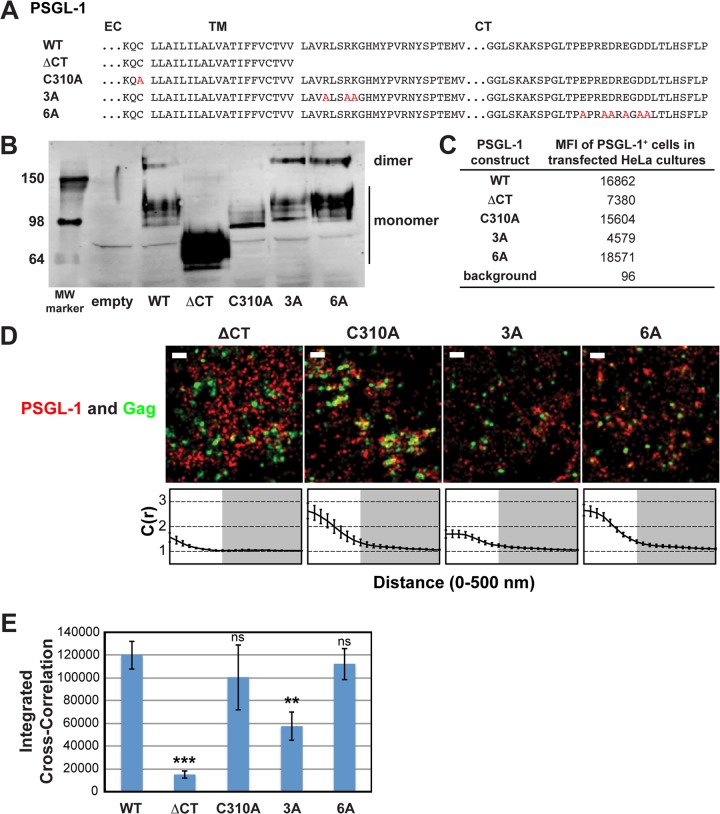
FIG 3.
A basic sequence in the cytoplasmic tail of PSGL-1 enhances coclustering with HIV-1 Gag. (A) Partial amino acid sequences of WT and mutant PSGL-1 constructs. EC, extracellular domain; TM, transmembrane domain; CT, cytoplasmic tail. Amino acid substitutions are shown in red. (B) WT PSGL-1 and PSGL-1 mutants were expressed in HeLa cells, and cell lysates were analyzed by SDS-PAGE (nonreducing conditions), followed by immunoblotting using an anti-PSGL-1 antibody. Numbers on the left are molecular weights (MWs), in kilodaltons. (C) HeLa cells transfected with plasmids carrying the indicated PSGL-1 constructs or an empty vector were recovered as cell suspensions at 16 h posttransfection, fixed, immunostained with an anti-PSGL-1 antibody labeled with Alexa Fluor 647, and analyzed by flow cytometry. The background mean fluorescence intensity was determined using cells transfected with the empty vector. Cells expressing transfected PSGL-1 derivatives, which were readily distinguished on the basis of the much higher fluorescence intensity, were gated, and their mean fluorescence intensities (MFIs) were determined. (D) HeLa cells were transfected with plasmids carrying WT Gag-mEos3.2 and the indicated PSGL-1 constructs and imaged as described in the legend to Fig. 2B. Representative reconstructed images are shown. Bars = 500 nm. Gag-mEos3.2 is shown in green, and PSGL-1 is shown in red. Cross-correlation curves, prepared as described in the legend to Fig. 1A, are shown. (E) The analysis of total coclustering was performed as described in the legend to Fig. 1B using cross-correlation curves for images of a total of 10 cells per condition from 2 independent experiments. Values shown indicate means ± SEMs. P values were calculated for each construct relative to WT PSGL-1. ***, P < 0.0005; **, P < 0.005; ns, not significant.
We first asked whether dimerization is necessary for Gag–PSGL-1 coclustering, since PSGL-1 is always present as a dimer in cells. For some proteins, multimerization enhances microdomain association (64–66), which in turn may promote association with assembly sites. To this end, we introduced into our expression construct a previously characterized mutation (C310A) that abolishes the dimerization of PSGL-1 (60) (Fig. 3A and B). We found that this mutation did not significantly decrease Gag–PSGL-1 coclustering, ruling out a role for dimerization (Fig. 3D and E).
We also hypothesized that the PSGL-1 cytoplasmic tail (PCT) contributes to localization of PSGL-1 to virus assembly sites via direct or indirect interactions with Gag or other viral components. Indeed, we found that deletion of the cytoplasmic tail of PSGL-1 (truncated after residue 331) substantially reduced Gag–PSGL-1 coclustering (Fig. 3D and E). While the cytoplasmic tail deletion also appeared to abolish dimerization, dimerization per se is not necessary for Gag–PSGL-1 coclustering, as shown above. Altogether, this experiment identifies the cytoplasmic tail of PSGL-1 to be the major determinant of Gag–PSGL-1 coclustering.
With regard to the features of the cytoplasmic tail of PSGL-1 that promote its localization to HIV-1 assembly sites, we speculated that an acidic stretch near the C terminus of the cytoplasmic tail of PSGL-1 may interact directly with the HBR of the MA domain. To test this hypothesis, we generated a mutant, PSGL-1/6A, in which the 6 acidic residues in this stretch have been replaced with an uncharged amino acid, alanine (Fig. 3A). When expressed in HeLa cells, we found that PSGL-1/6A coclusters with Gag-mEos3.2 to a similar extent as wild-type PSGL-1 (Fig. 3D and E). These results rule out a direct interaction between the highly acidic C-terminal region of PSGL-1 and the basic residues of the MA HBR.
Our alternative hypothesis was based on the observation that PSGL-1, CD43, and CD44 all possess basic amino acids immediately C terminal to their transmembrane domains. Basic sequences in the juxtamembrane domains of several transmembrane proteins are shown to interact with acidic phospholipids (67–73). Given that Gag can bind to acidic lipids, such as PI(4,5)P2 and phosphatidylserine (PS), through the HBR (5, 12, 39), we speculated that clustering of acidic lipids at assembly sites by multimerizing Gag molecules may attract PSGL-1 and possibly also CD43 and CD44. As the first step toward testing this hypothesis, we generated a mutant, PSGL-1/3A, in which three juxtamembrane basic residues have been replaced with alanine (Fig. 3A). We found that unlike the 6A mutation, the 3A mutation significantly reduced the coclustering of Gag and PSGL-1 (Fig. 3D and E). These results indicate that a basic motif near the plasma membrane significantly enhances localization of PSGL-1 to HIV-1 assembly sites. While the residual coclustering suggests the presence of additional determinants within the cytoplasmic tail of PSGL-1, the membrane-proximal basic motif RLSRK is likely a major determinant of the association between Gag and PSGL-1.
Basic sequences in cytoplasmic domains are required for efficient coclustering of CD43 and CD44 with Gag in HeLa cells.
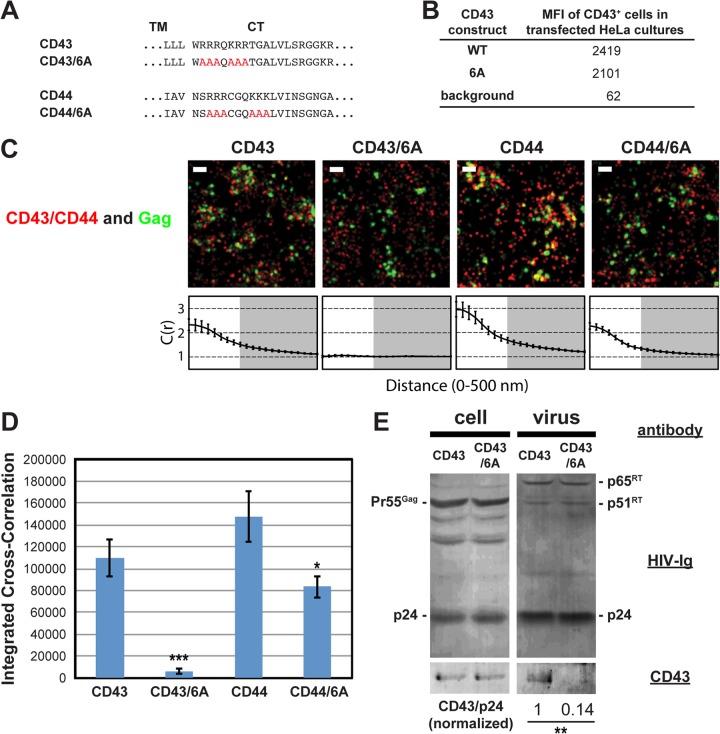
We next investigated whether juxtamembrane basic motifs in the cytoplasmic tails of CD43 and CD44 promote recruitment of these proteins to plasma membrane sites where HIV-1 assembles, as observed with PSGL-1. To do this, we generated mutants in which 6 basic residues, present in two clusters in both proteins, were mutated to alanine (CD43/6A and CD44/6A, respectively) (Fig. 4A). Flow cytometry analysis confirmed the cell surface expression of CD43 and CD43/6A (Fig. 4B). Due to the endogenous expression of CD44, the cell surface expression of transfected CD44 constructs could not be measured (data not shown). When expressed in HeLa cells, we found that wild-type CD43 and CD44, detected with Alexa Fluor 647-labeled antibodies, significantly coclustered with Gag-mEos3.2 (Fig. 4C and D). As was observed with the 3A mutation of PSGL-1, we found that mutation of the basic residue stretches in the CD43 and CD44 cytoplasmic tails significantly decreased their coclustering with Gag-mEos3.2 (Fig. 4C and D). We note that coclustering between Gag and the CD44/6A mutant may be overestimated due to the presence of endogenous wild-type CD44. Consistent with the superresolution localization microscopy results, incorporation of CD43/6A into virions was reduced approximately 7-fold relative to that of wild-type CD43 (Fig. 4E). These results support a model in which a group of uropod-directed proteins, including PSGL-1, CD43, and CD44, is recruited to plasma membrane sites where HIV-1 assembly occurs in a manner dependent on juxtamembrane basic residues in the cytoplasmic tails of these proteins.
FIG 4.
Coclustering of CD43 and CD44 with HIV-1 Gag is enhanced by juxtamembrane polybasic sequences. (A) Partial amino acid sequences of CD43 and CD44 mutants. (B) The mean fluorescence intensities of HeLa cells expressing transfected CD43 constructs were determined as described in the legend to Fig. 3C. Note that the mean fluorescence intensities of HeLa cells transfected with CD44 expression plasmids were not determined due to a high endogenous expression of CD44. (C) HeLa cells were transfected with plasmids carrying WT Gag-mEos3.2 and the indicated constructs and imaged as described in the legend to Fig. 2. Representative reconstructed images and cross-correlation curves are shown. Bars = 500 nm. Gag-mEos3.2 is shown in green, and the indicated uropod-directed proteins or their derivatives are shown in red. (D) The analysis of total coclustering was performed as described in the legend to Fig. 1B using cross-correlation curves for images of a total of 10 cells per condition from 2 independent experiments. Values shown indicate means ± SEMs. P values were calculated for each mutant construct relative to its wild-type counterpart. ***, P < 0.0005; *, P < 0.05. (E) HeLa cells were transfected with a WT HIV-1 molecular clone (pNL4-3) and expression plasmids carrying wild-type CD43 or CD43/6A and cultured for 16 h. Cell and virus lysates were collected and subjected to immunoblotting analysis using HIV-Ig and anti-CD43. Note that no obvious difference in virus release efficiency was observed between CD43- and CD43/6A-expressing cells. The results shown are representative of the results from three independent experiments. The normalized CD43/p24 ratio shown at the bottom was calculated and averaged from the results of the three experiments. **, P < 0.005. RT, reverse transcriptase.
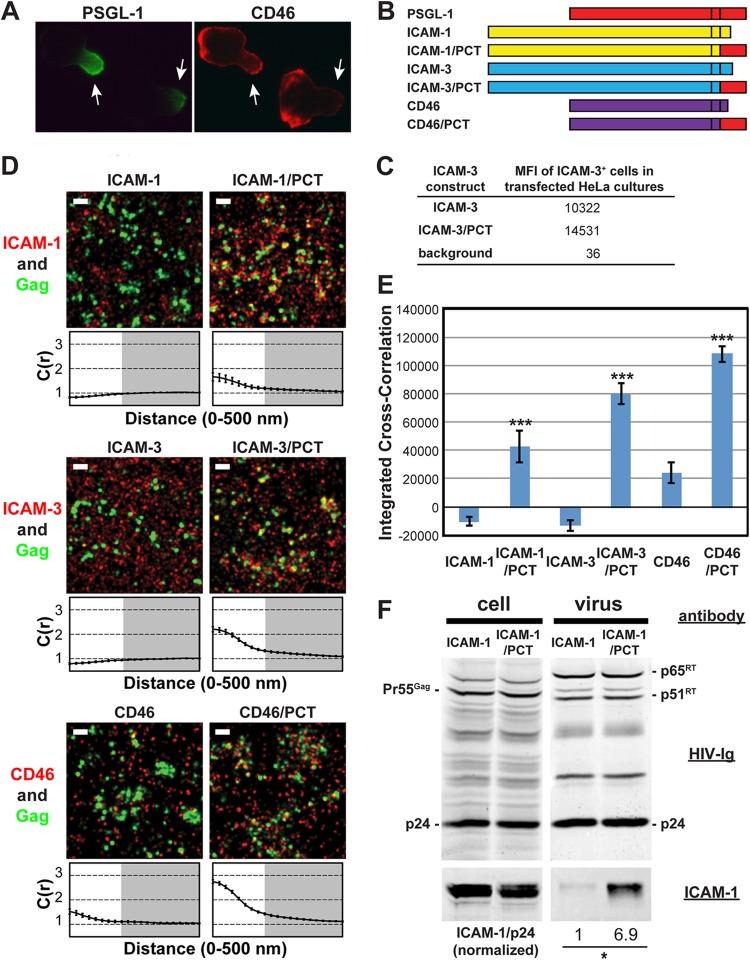
Chimeric transmembrane proteins containing the PSGL-1 cytoplasmic tail cocluster with Gag more efficiently than the native counterparts.
We further examined whether the PSGL-1 cytoplasmic domain can promote coclustering of other host cell proteins with assembling HIV-1. To do this, we examined ICAM-1 and ICAM-3, which localize to uropods but do not copatch with Gag in T cells (34, 39), and CD46, which does not specifically localize to uropods (Fig. 5A). Transfection of these proteins as well as their chimeric derivatives (Fig. 5B) led to an increase in mean fluorescence intensity in flow cytometry (data not shown), confirming their ectopic expression in HeLa cells; however, only ICAM-3 and its chimeric construct were measured accurately for cell surface expression levels due to the endogenous expression of ICAM-1 and CD46 (Fig. 5C). Consistent with previous observations obtained using T cells (39) (Fig. 1), we found that these proteins, detected with Alexa Fluor 647-labeled antibodies, do not associate strongly with HIV-1 Gag when expressed in HeLa cells (Fig. 5D and E). Notably, we found that both ICAM-1 and ICAM-3 give integrated cross-correlation values of less than 0, indicating that there is a tendency for these proteins to be excluded from HIV-1 assembly sites, rather than simply being randomly distributed on the cell surface (giving an integrated cross-correlation value of ≈0).
FIG 5.
Chimeric transmembrane proteins containing the PSGL-1 cytoplasmic tail cocluster with HIV-1 Gag more efficiently than their native counterparts. (A) T cells with a polarized morphology (P2 cells) were dually immunostained for PSGL-1 and CD46. As indicated by arrows, PSGL-1 localized to uropods, whereas CD46 localized to the entire cell surface. (B) Schematic representation of the chimeric constructs. (C) The mean fluorescence intensities of HeLa cells expressing transfected ICAM-3 constructs were determined as described in the legend to Fig. 3C. Note that the mean fluorescence intensities of HeLa cells transfected with plasmids carrying ICAM-1 or CD46 were not determined due to endogenous expression of these proteins. (D) HeLa cells were transfected with plasmids carrying WT Gag-mEos3.2 and the indicated constructs and imaged as described in the legend to Fig. 1. Representative reconstructed images and cross-correlation curves are shown. Bars = 500 nm. Gag-mEos3.2 is shown in green, and the indicated proteins are shown in red. (E) The analysis of total coclustering was performed as described in the legend to Fig. 1B using cross-correlation curves for images of a total of 10 cells per condition from 2 independent experiments. Values shown indicate means ± SEMs. P values were calculated for each PCT construct relative to its wild-type counterpart. ***, P < 0.0005. (F) HeLa cells were transfected with a WT HIV-1 molecular clone (pNL4-3) and expression plasmids carrying wild-type ICAM-1 or ICAM-1/PCT and cultured for 2 days. Cell and virus lysates were collected and subjected to immunoblotting analysis using HIV-Ig and anti-ICAM-1. Note that wild-type ICAM-1 was detected in virus lysates but at a much smaller amount than ICAM-1/PCT. The results shown are representative of the results from three independent experiments. The normalized ICAM-1/p24 ratio shown at the bottom was calculated and averaged from the results of the three experiments. *, P < 0.05.
We hypothesized that replacement of the cytoplasmic tails of these proteins with the PCT could enhance their recruitment to plasma membrane sites where assembling HIV-1 localizes (Fig. 5B). When expressed in HeLa cells, we found by cross-correlation analysis that the PCT-containing chimeric proteins were more strongly associated with HIV-1 Gag than the corresponding wild-type proteins (Fig. 5D and E). Of note, due to the presence of endogenous wild-type ICAM-1 in HeLa cells, the cross-correlation between Gag and ICAM-1/PCT may be underestimated. Although wild-type CD46 was weakly associated with Gag-mEos3.2, this coclustering was enhanced approximately 4- to 5-fold by replacement of its native cytoplasmic tail with PCT (Fig. 5D and E). These results demonstrate that the cytoplasmic tail of PSGL-1, which contains the major determinants for coclustering of this protein with HIV-1 assembly sites (Fig. 3), is sufficient to promote efficient recruitment of other transmembrane proteins to plasma membrane sites where HIV-1 assembly takes place.
ICAM-1/PCT is incorporated into virions more efficiently than WT ICAM-1.
It is likely that transmembrane proteins that cocluster with Gag at the plasma membrane are more efficiently incorporated into virions than those that distribute randomly relative to Gag. To examine whether this is the case with chimeric proteins that contain the PSGL-1 cytoplasmic domain, we transfected HeLa cells with a WT HIV-1 molecular clone (pNL4-3) and an expression plasmid for WT ICAM-1 or ICAM-1/PCT and examined cell and virus lysates for the presence of the transmembrane proteins by immunoblotting (Fig. 5F). The ICAM-1 pair was chosen for this analysis because an antibody usable for immunoblotting was available. A much higher virion-associated ICAM-1 signal was observed when HeLa cells were transfected with ICAM-1/PCT than WT ICAM-1, suggesting that ICAM-1/PCT was more efficiently incorporated into virions than wild-type ICAM-1. When coexpressed with a mutant HIV-1 molecular clone that encodes nonmyristoylated Gag and, hence, is unable to produce virions, ICAM-1/PCT was not detected in the virus lysate preparation, suggesting that the proteins detected in WT virus lysates indeed associated with virus particles but not cell-derived microvesicles (data not shown). Altogether, these results suggest that coclustering between host transmembrane proteins and Gag at the plasma membrane leads to enhanced incorporation of the transmembrane proteins into released HIV-1 particles.
DISCUSSION
Virus particles of HIV-1 and other retroviruses incorporate various heterologous transmembrane proteins, some of which drastically affect the fate of nascent virions. These proteins may be passively incorporated into assembling particles due to their presence at the plasma membrane or specifically recruited to assembly sites via direct or indirect molecular interactions with Gag. To determine the specificity of the plasma membrane protein association with assembly sites and molecular determinants for this association, high-resolution and quantitative microscopy analysis is essential. However, such a mechanistic analysis for the association of heterologous cellular transmembrane proteins with assembly sites has not been performed, except for ones performed in studies on BST-2/tetherin (56, 57). In the present study, we employed superresolution localization microscopy methods capable of resolving single virus assembly sites as well as single uropod-directed proteins. These studies were carried out in cells that had been fixed prior to immunostaining, allowing observation of the native distribution of these proteins with respect to HIV-1 assembly sites.
Previous studies showed that copatching of lipid raft- and TEM-associated proteins with Gag occurs even when the MA sequence is replaced with heterologous membrane binding sequences (35). In contrast and consistent with previous findings from a copatching study performed in T cells (39), we found that coclustering of Gag with PSGL-1 is dependent upon the HBR of the MA domain in both T and HeLa cells. Therefore, the mechanism for Gag–PSGL-1 coclustering is likely to be different from that mediated by lipid rafts or TEMs. Replacing 8 basic amino acids within the HBR with neutral amino acids substantially reduced Gag–PSGL-1 coclustering. Exchanging the basic residues of the HBR from lysine to arginine and vice versa still allowed coclustering of PSGL-1 with Gag at high (in T cells) or moderate (in HeLa cells) levels (Fig. 1 and 2). Although the Gag construct containing this change (HBR/RKswitch) is incapable of binding to PI(4,5)P2, it is still able to bind liposomes containing PS after RNase treatment (39). Therefore, the coclustering between Fyn(10)/HBR/RKswitch Gag and PSGL-1 observed in this study may reflect the involvement of acidic phospholipids (discussed below). Of note, Fyn(10)/HBR/RKswitch Gag expressed in T cells showed copatching and coclustering with PSGL-1 to the same extent as Fyn(10)/Gag, which has an intact MA sequence, whereas the level of coclustering of Fyn(10)/HBR/RKswitch Gag with PSGL-1 was lower than that of Fyn(10)/Gag in HeLa cells. It is unclear at this point whether this difference arose from the difference in lipid compositions or other components of the plasma membrane of different cell types. It is also unclear why WT Gag showed higher coclustering with PSGL-1 than Fyn(10)/Gag, particularly in HeLa cells (Fig. 2). However, as observed in cross-correlation curves, WT Gag, but not Fyn(10)/Gag, tended to have a substantial cross-correlation even at a distance of 200 nm in HeLa cells. This longer-range association between WT Gag and PSGL-1 may be due to the tendency of WT Gag puncta, but not Fyn(10)/Gag puncta, to form clusters of puncta (data not shown), since, in such a case, PSGL-1 associated with one Gag assembly site contributes to a cross-correlation with Gag molecules in neighboring assembly sites. Regardless, both WT Gag and Fyn(10)/Gag showed similar levels of coclustering with PSGL-1 at short distances, suggesting that the extent of PSGL-1 coclustering with single assembly sites is similar for both Gag proteins.
We have observed that basic residues are required on both sides of the association between Gag and PSGL-1 (Fig. 3) and that juxtamembrane polybasic residues also facilitate the association of CD43 and CD44 with assembly sites (Fig. 4). Based on these findings, we hypothesize the existence of an acidic intermediate which links Gag and class I uropod-directed proteins. This factor could potentially be a lipid, protein, nucleic acid, or other negatively charged molecule. Given the likely proximity of the required basic residues on MA and PSGL-1 to the plasma membrane, we further hypothesize that this unknown factor may be an acidic lipid present in the inner leaflet of the plasma membrane. Consistent with this hypothesis, both PS and PI(4,5)P2 have been shown to form clusters on the cytoplasmic leaflet of the plasma membrane (74–78). Moreover, a recent study also showed the formation of an acidic interface, composed of plasma membrane acidic phospholipids, at the virological synapses between murine leukemia virus-infected and virus receptor-expressing cells (79). Altogether, our current findings and previous studies are consistent with a model in which basic residues within the HBR of the MA domain result in local clustering of acidic lipids (35, 80), potentially driven by the multimerization of Gag, which in turn selectively recruits some host cell plasma membrane proteins on the basis of the presence of polybasic motifs in their cytoplasmic tails. It is also possible that a preexisting microdomain enriched in acidic phospholipids attracts both Gag and cellular transmembrane proteins with juxtamembrane polybasic sequences. At this point, we also do not rule out the possibility that nonlipid factors, such as proteins involved in membrane-cytoskeleton interactions (e.g., ezrin), directly or indirectly promote the basic sequence-dependent coclustering. Further analyses using lipid and cytoskeleton probes and Gag multimerization mutants in superresolution localization microscopy may distinguish these possibilities.
Interestingly, while the manuscript was in revision, a study using a scanning electron microscopy method demonstrated that CD4, another transmembrane protein with a juxtamembrane polybasic sequence, is efficiently recruited to assembling HIV-1 (81). However, not all transmembrane proteins with polybasic sequences associate with HIV-1 assembly sites. While ICAM-1, ICAM-3, and CD46 do contain basic amino acid-rich sequences in their cytoplasmic domains, we observed that these proteins did not specifically cocluster with assembling HIV-1. Therefore, the availability of the basic sequences and/or additional regions present in the cytoplasmic domains of type I uropod-directed proteins may also play important roles.
Of note, while we observed that ICAM-1 neither copatches with Gag in T cells that express endogenous ICAM-1 (39) nor coclusters with Gag in transfected HeLa cells (this study), previous studies showed that ICAM-1 is nonetheless incorporated into HIV-1 particles when virions were produced by 293T or Cos7 cells transfected with ICAM-1 expression plasmids (82–84). We speculate that these previous observations likely represent a passive mode of incorporation. Alternatively, in the overexpression context used in these studies, a small population of ICAM-1 molecules may escape from a binding partner that would prevent specific recruitment to assembly sites and, as a result, engage in direct or indirect interactions with Gag (85). Consistent with these interpretations, a previous study showed that the amounts of ICAM-1 present on the virions correlate with the levels of ICAM-1 expression on virus-producing 293T cells (86). We also observed that HeLa cells transfected with an expression plasmid for ICAM-1 produced virions that contain a faint but detectable level of ICAM-1 (Fig. 5F). However, the amount of ICAM-1 incorporated into virions in these cells was much less than that incorporated into virions in cells transfected with ICAM-1/PCT, despite the similar total expression levels in cells.
In summary, this study illuminated the presence of a novel molecular mechanism that promotes the specific recruitment of a class of cellular transmembrane proteins to HIV-1 assembly sites. This mechanism relies on basic sequences in HIV-1 MA and in cytoplasmic domains of the transmembrane proteins. Coclustering with HIV-1 assembly sites via this mechanism leads to the enhanced incorporation of transmembrane proteins into virions compared to the level of incorporation achieved by random colocalization. Therefore, further analyses of the roles played by the polybasic sequences will likely help with the development of a new strategy to improve the targeting specificity of lentiviral vectors via pseudotyping. It is known that PSGL-1, CD43, and CD44 participate in adherence, motility, cell-cell contact, and cell signaling events in lymphocytes (48–50, 87–90). However, although a mechanism promoting the specific recruitment of these proteins to assembly sites does exist, as revealed in this study, it is unknown whether these proteins play important roles in HIV-1 replication. Our current efforts are focused on determining what role these proteins play in the replication and dissemination of HIV-1 and whether specific plasma membrane lipids are involved in the recruitment of these proteins to HIV-1 assembly sites.
ACKNOWLEDGMENTS
We thank members of the A. Ono and S. L. Veatch labs for helpful discussions. We thank G. Nick Llewellyn, a former Ono lab member, for help with plasmid construction. We thank Matthew Stone for help with image processing and Brian DeVree for help with sample preparation for microscopy. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: catalog no. 3957, HIV-IG from NABI and NHLBI.
This work was supported by NIH grants R21 AI095022 and R01 AI071727 (to A.O.) and R00 GM087810 and R01 GM110052 (to S.L.V.). J.R.G. is supported by a Clayton Willison- and Emma Elizabeth Willison-endowed graduate fellowship.
REFERENCES
- 1.Bryant M, Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A 87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottlinger HG, Sodroski JG, Haseltine WA. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A 101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A 103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag membrane binding. J Virol 82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saad JS, Ablan SD, Ghanam RH, Kim A, Andrews K, Nagashima K, Soheilian F, Freed EO, Summers MF. 2008. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol 382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono A. 2010. HIV-1 assembly at the plasma membrane. Vaccine 28(Suppl 2):B55–B59. doi: 10.1016/j.vaccine.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang CY, Chang YF, Wang SM, Tseng YT, Huang KJ, Wang CT. 2008. HIV-1 matrix protein repositioning in nucleocapsid region fails to confer virus-like particle assembly. Virology 378:97–104. doi: 10.1016/j.virol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimarelli A, Luban J. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol 73:5388–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lochrie MA, Waugh S, Pratt DG Jr, Clever J, Parslow TG, Polisky B. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 Gag polyprotein. Nucleic Acids Res 25:2902–2910. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purohit P, Dupont S, Stevenson M, Green MR. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7:576–584. doi: 10.1017/S1355838201002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chukkapalli V, Oh SJ, Ono A. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A 107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chukkapalli V, Inlora J, Todd GC, Ono A. 2013. Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. J Virol 87:7155–7159. doi: 10.1128/JVI.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfadhli A, McNett H, Tsagli S, Bachinger HP, Peyton DH, Barklis E. 2011. HIV-1 matrix protein binding to RNA. J Mol Biol 410:653–666. doi: 10.1016/j.jmb.2011.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dick RA, Kamynina E, Vogt VM. 2013. Effect of multimerization on membrane association of Rous sarcoma virus and HIV-1 matrix domain proteins. J Virol 87:13598–13608. doi: 10.1128/JVI.01659-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alfadhli A, Still A, Barklis E. 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol 83:12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dick RA, Goh SL, Feigenson GW, Vogt VM. 2012. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc Natl Acad Sci U S A 109:18761–18766. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vlach J, Saad JS. 2013. Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc Natl Acad Sci U S A 110:3525–3530. doi: 10.1073/pnas.1216655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono A, Waheed AA, Freed EO. 2007. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology 360:27–35. doi: 10.1016/j.virol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW Jr, Sowder RC II, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol 80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neil SJ. 2013. The antiviral activities of tetherin. Curr Top Microbiol Immunol 371:67–104. doi: 10.1007/978-3-642-37765-5_3. [DOI] [PubMed] [Google Scholar]
- 23.Henriksson P, Bosch V. 1998. Inhibition of cellular glycoprotein incorporation into human immunodeficiency virus-like particles by coexpression of additional cellular interaction partner. Virology 251:16–21. doi: 10.1006/viro.1998.9403. [DOI] [PubMed] [Google Scholar]
- 24.Monde K, Maeda Y, Tanaka Y, Harada S, Yusa K. 2007. Gp120 V3-dependent impairment of R5 HIV-1 infectivity due to virion-incorporated CCR5. J Biol Chem 282:36923–36932. doi: 10.1074/jbc.M705298200. [DOI] [PubMed] [Google Scholar]
- 25.Young JA, Bates P, Willert K, Varmus HE. 1990. Efficient incorporation of human CD4 protein into avian leukosis virus particles. Science 250:1421–1423. doi: 10.1126/science.2175047. [DOI] [PubMed] [Google Scholar]
- 26.Checkley MA, Luttge BG, Freed EO. 2011. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson MC. 2011. Mechanisms for Env glycoprotein acquisition by retroviruses. AIDS Res Hum Retroviruses 27:239–247. doi: 10.1089/aid.2010.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammarstedt M, Wallengren K, Pedersen KW, Roos N, Garoff H. 2000. Minimal exclusion of plasma membrane proteins during retroviral envelope formation. Proc Natl Acad Sci U S A 97:7527–7532. doi: 10.1073/pnas.120051597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen DH, Hildreth JE. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol 74:3264–3272. doi: 10.1128/JVI.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono A, Freed EO. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A 98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. 2006. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol 173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. 2006. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol 172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jolly C, Sattentau QJ. 2007. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol 81:7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llewellyn GN, Hogue IB, Grover JR, Ono A. 2010. Nucleocapsid promotes localization of HIV-1 Gag to uropods that participate in virological synapses between T cells. PLoS Pathog 6:e1001167. doi: 10.1371/journal.ppat.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hogue IB, Llewellyn GN, Ono A. 2012. Dynamic association between HIV-1 Gag and membrane domains. Mol Biol Int 2012:979765. doi: 10.1155/2012/979765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grigorov B, Attuil-Audenis V, Perugi F, Nedelec M, Watson S, Pique C, Darlix JL, Conjeaud H, Muriaux D. 2009. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology 6:28. doi: 10.1186/1742-4690-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krementsov DN, Rassam P, Margeat E, Roy NH, Schneider-Schaulies J, Milhiet PE, Thali M. 2010. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic 11:1401–1414. doi: 10.1111/j.1600-0854.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hogue IB, Grover JR, Soheilian F, Nagashima K, Ono A. 2011. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J Virol 85:9749–9766. doi: 10.1128/JVI.00743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llewellyn GN, Grover JR, Olety B, Ono A. 2013. HIV-1 Gag associates with specific uropod-directed microdomains in a manner dependent on its MA highly basic region. J Virol 87:6441–6454. doi: 10.1128/JVI.00040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller MJ, Wei SH, Parker I, Cahalan MD. 2002. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science 296:1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 41.Bajenoff M, Egen JG, Koo LY, Laugier JP, Brau F, Glaichenhaus N, Germain RN. 2006. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity 25:989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hugues S, Fetler L, Bonifaz L, Helft J, Amblard F, Amigorena S. 2004. Distinct T cell dynamics in lymph nodes during the induction of tolerance and immunity. Nat Immunol 5:1235–1242. doi: 10.1038/ni1134. [DOI] [PubMed] [Google Scholar]
- 43.Miller MJ, Wei SH, Cahalan MD, Parker I. 2003. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proc Natl Acad Sci U S A 100:2604–2609. doi: 10.1073/pnas.2628040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mrass P, Takano H, Ng LG, Daxini S, Lasaro MO, Iparraguirre A, Cavanagh LL, von Andrian UH, Ertl HC, Haydon PG, Weninger W. 2006. Random migration precedes stable target cell interactions of tumor-infiltrating T cells. J Exp Med 203:2749–2761. doi: 10.1084/jem.20060710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, Tager AM, Luster AD, Mempel TR. 2012. HIV-infected T cells are migratory vehicles for viral dissemination. Nature 490:283–287. doi: 10.1038/nature11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sewald X, Gonzalez DG, Haberman AM, Mothes W. 2012. In vivo imaging of virological synapses. Nat Commun 3:1320. doi: 10.1038/ncomms2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FY, Li XD, Asmuth DM, Huser T, Chen BK. 2009. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science 323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dustin ML, Chakraborty AK, Shaw AS. 2010. Understanding the structure and function of the immunological synapse. Cold Spring Harb Perspect Biol 2:a002311. doi: 10.1101/cshperspect.a002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-Madrid F, Serrador JM. 2009. Bringing up the rear: defining the roles of the uropod. Nat Rev Mol Cell Biol 10:353–359. doi: 10.1038/nrm2680. [DOI] [PubMed] [Google Scholar]
- 50.Cullinan P, Sperling AI, Burkhardt JK. 2002. The distal pole complex: a novel membrane domain distal to the immunological synapse. Immunol Rev 189:111–122. doi: 10.1034/j.1600-065X.2002.18910.x. [DOI] [PubMed] [Google Scholar]
- 51.Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med 199:283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgenson RL, Vogt VM, Johnson MC. 2009. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J Virol 83:4060–4067. doi: 10.1128/JVI.02425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lucas TM, Lyddon TD, Grosse SA, Johnson MC. 2010. Two distinct mechanisms regulate recruitment of murine leukemia virus envelope protein to retroviral assembly sites. Virology 405:548–555. doi: 10.1016/j.virol.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muranyi W, Malkusch S, Muller B, Heilemann M, Krausslich HG. 2013. Super-resolution microscopy reveals specific recruitment of HIV-1 envelope proteins to viral assembly sites dependent on the envelope C-terminal tail. PLoS Pathog 9:e1003198. doi: 10.1371/journal.ppat.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy NH, Chan J, Lambele M, Thali M. 2013. Clustering and mobility of HIV-1 Env at viral assembly sites predict its propensity to induce cell-cell fusion. J Virol 87:7516–7525. doi: 10.1128/JVI.00790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grover JR, Llewellyn GN, Soheilian F, Nagashima K, Veatch SL, Ono A. 2013. Roles played by capsid-dependent induction of membrane curvature and Gag-ESCRT interactions in tetherin recruitment to HIV-1 assembly sites. J Virol 87:4650–4664. doi: 10.1128/JVI.03526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lehmann M, Rocha S, Mangeat B, Blanchet F, Uji IH, Hofkens J, Piguet V. 2011. Quantitative multicolor super-resolution microscopy reveals tetherin HIV-1 interaction. PLoS Pathog 7:e1002456. doi: 10.1371/journal.ppat.1002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy MO, Leventis R, Silvius JR. 2000. Mutational and biochemical analysis of plasma membrane targeting mediated by the farnesylated, polybasic carboxy terminus of K-ras4B. Biochemistry 39:8298–8307. doi: 10.1021/bi000512q. [DOI] [PubMed] [Google Scholar]
- 59.Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. 2006. Receptor activation alters inner surface potential during phagocytosis. Science 313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- 60.Epperson TK, Patel KD, McEver RP, Cummings RD. 2000. Noncovalent association of P-selectin glycoprotein ligand-1 and minimal determinants for binding to P-selectin. J Biol Chem 275:7839–7853. doi: 10.1074/jbc.275.11.7839. [DOI] [PubMed] [Google Scholar]
- 61.Hogue IB, Hoppe A, Ono A. 2009. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding. J Virol 83:7322–7336. doi: 10.1128/JVI.02545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Veatch SL, Machta BB, Shelby SA, Chiang EN, Holowka DA, Baird BA. 2012. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS One 7:e31457. doi: 10.1371/journal.pone.0031457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, Chen J, Liu B, Lu J, Liu Y, Zhang J, Xu P, Xu T. 2012. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods 9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 64.Harder T, Scheiffele P, Verkade P, Simons K. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol 141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langhorst MF, Reuter A, Stuermer CA. 2005. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol Life Sci 62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parton RG, Simons K. 2007. The multiple faces of caveolae. Nat Rev Mol Cell Biol 8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 67.Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. 2013. Architecture and membrane interactions of the EGF receptor. Cell 152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deng W, Cho S, Li R. 2013. FERM domain of moesin desorbs the basic-rich cytoplasmic domain of L-selectin from the anionic membrane surface. J Mol Biol 425:3549–3562. doi: 10.1016/j.jmb.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lam AD, Tryoen-Toth P, Tsai B, Vitale N, Stuenkel EL. 2008. SNARE-catalyzed fusion events are regulated by syntaxin1A-lipid interactions. Mol Biol Cell 19:485–497. doi: 10.1091/mbc.E07-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsushita C, Tamagaki H, Miyazawa Y, Aimoto S, Smith SO, Sato T. 2013. Transmembrane helix orientation influences membrane binding of the intracellular juxtamembrane domain in Neu receptor peptides. Proc Natl Acad Sci U S A 110:1646–1651. doi: 10.1073/pnas.1215207110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Michailidis IE, Rusinova R, Georgakopoulos A, Chen Y, Iyengar R, Robakis NK, Logothetis DE, Baki L. 2011. Phosphatidylinositol-4,5-bisphosphate regulates epidermal growth factor receptor activation. Pflugers Arch 461:387–397. doi: 10.1007/s00424-010-0904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sengupta P, Bosis E, Nachliel E, Gutman M, Smith SO, Mihalyne G, Zaitseva I, McLaughlin S. 2009. EGFR juxtamembrane domain, membranes, and calmodulin: kinetics of their interaction. Biophys J 96:4887–4895. doi: 10.1016/j.bpj.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams D, Vicogne J, Zaitseva I, McLaughlin S, Pessin JE. 2009. Evidence that electrostatic interactions between vesicle-associated membrane protein 2 and acidic phospholipids may modulate the fusion of transport vesicles with the plasma membrane. Mol Biol Cell 20:4910–4919. doi: 10.1091/mbc.E09-04-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. 2011. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol 194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spira F, Mueller NS, Beck G, von Olshausen P, Beig J, Wedlich-Soldner R. 2012. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat Cell Biol 14:640–648. doi: 10.1038/ncb2487. [DOI] [PubMed] [Google Scholar]
- 76.James DJ, Khodthong C, Kowalchyk JA, Martin TF. 2008. Phosphatidylinositol 4,5-bisphosphate regulates SNARE-dependent membrane fusion. J Cell Biol 182:355–366. doi: 10.1083/jcb.200801056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McLaughlin S, Murray D. 2005. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 78.van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, Willig KI, Hubrich BE, Dier M, Hell SW, Grubmuller H, Diederichsen U, Jahn R. 2011. Membrane protein sequestering by ionic protein-lipid interactions. Nature 479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li F, Jin J, Herrmann C, Mothes W. 2013. Basic residues in the matrix domain and multimerization target murine leukemia virus Gag to the virological synapse. J Virol 87:7113–7126. doi: 10.1128/JVI.03263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kerviel A, Thomas A, Chaloin L, Favard C, Muriaux D. 2013. Virus assembly and plasma membrane domains: which came first? Virus Res 171:332–340. doi: 10.1016/j.virusres.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 81.Gregory DA, Olinger GY, Lucas TM, Johnson MC. 2014. Diverse viral glycoproteins as well as CD4 co-package into the same human immunodeficiency virus (HIV-1) particles. Retrovirology 11:28. doi: 10.1186/1742-4690-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fortin JF, Cantin R, Lamontagne G, Tremblay M. 1997. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J Virol 71:3588–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fortin JF, Cantin R, Tremblay MJ. 1998. T cells expressing activated LFA-1 are more susceptible to infection with human immunodeficiency virus type 1 particles bearing host-encoded ICAM-1. J Virol 72:2105–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rizzuto CD, Sodroski JG. 1997. Contribution of virion ICAM-1 to human immunodeficiency virus infectivity and sensitivity to neutralization. J Virol 71:4847–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beausejour Y, Tremblay MJ. 2004. Interaction between the cytoplasmic domain of ICAM-1 and Pr55Gag leads to acquisition of host ICAM-1 by human immunodeficiency virus type 1. J Virol 78:11916–11925. doi: 10.1128/JVI.78.21.11916-11925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paquette JS, Fortin JF, Blanchard L, Tremblay MJ. 1998. Level of ICAM-1 surface expression on virus producer cells influences both the amount of virion-bound host ICAM-1 and human immunodeficiency virus type 1 infectivity. J Virol 72:9329–9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Clark MC, Baum LG. 2012. T cells modulate glycans on CD43 and CD45 during development and activation, signal regulation, and survival. Ann N Y Acad Sci 1253:58–67. doi: 10.1111/j.1749-6632.2011.06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kawashima H, Fukuda M. 2012. Sulfated glycans control lymphocyte homing. Ann N Y Acad Sci 1253:112–121. doi: 10.1111/j.1749-6632.2011.06356.x. [DOI] [PubMed] [Google Scholar]
- 89.Zarbock A, Ley K, McEver RP, Hidalgo A. 2011. Leukocyte ligands for endothelial selectins: specialized glycoconjugates that mediate rolling and signaling under flow. Blood 118:6743–6751. doi: 10.1182/blood-2011-07-343566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jacobelli J, Bennett FC, Pandurangi P, Tooley AJ, Krummel MF. 2009. Myosin-IIA and ICAM-1 regulate the interchange between two distinct modes of T cell migration. J Immunol 182:2041–2050. doi: 10.4049/jimmunol.0803267. [DOI] [PubMed] [Google Scholar]