ABSTRACT
Hepatitis C virus (HCV) glycoprotein E2 is considered a major target for generating neutralizing antibodies against HCV, primarily due to its role of engaging host entry factors, such as CD81, a key cell surface protein associated with HCV entry. Based on a series of biochemical analyses in combination with molecular docking, we present a description of a potential binding interface formed between the E2 protein and CD81. The virus side of this interface includes a hydrophobic helix motif comprised of residues W437LAGLF442, which encompasses the binding site of a neutralizing monoclonal antibody, mAb41. The helical conformation of this motif provides a structural framework for the positioning of residues F442 and Y443, serving as contact points for the interaction with CD81. The cell side of this interface likewise involves a surface-exposed hydrophobic helix, namely, the D-helix of CD81, which coincides with the binding site of 1D6, a monoclonal anti-CD81 antibody known to block HCV entry. Our illustration of this virus-host interface suggests an important role played by the W437LAGLF442 helix of the E2 protein in the hydrophobic interaction with the D-helix of CD81, thereby facilitating our understanding of the mechanism for antibody-mediated neutralization of HCV.
IMPORTANCE Characterization of the interface established between a virus and host cells can provide important information that may be used for the control of virus infections. The interface that enables hepatitis C virus (HCV) to infect human liver cells has not been well understood because of the number of cell surface proteins, factors, and conditions found to be associated with the infection process. Based on a series of biochemical analyses in combination with molecular docking, we present such an interface, consisting of two hydrophobic helical structures, from the HCV E2 surface glycoprotein and the CD81 protein, a major host cell receptor recognized by all HCV strains. Our study reveals the critical role played by hydrophobic interactions in the formation of this virus-host interface, thereby contributing to our understanding of the mechanism for antibody-mediated neutralization of HCV.
INTRODUCTION
Hepatitis C virus (HCV) infects more than 170 million people worldwide. Approximately 70% of infected people fail to clear the virus during the acute phase of the disease and become chronic carriers. Liver cirrhosis, which develops in about 10 to 20% of chronically infected patients, is linked with a high risk for hepatocellular carcinoma in later life (1, 2). Although the FDA recently approved a number of highly effective antiviral drugs for treatment of HCV infections, prophylaxis is still an unmet medical need. Disease prevention by use of virus-specific neutralizing antibodies remains the most cost-effective and realistic way to control HCV infection (and reinfection) and significantly reduces the burden of HCV-related diseases (3, 4).
Protective immunity against HCV has been difficult to establish in humans, as the antibodies generated during natural HCV infection are incapable of resolving chronic infections, for unknown reasons (5). Nevertheless, strong evidence exists for antibodies to play a significant part in clearance of HCV infections. For example, a longitudinal follow-up of patients after acute HCV infections revealed that neutralizing antibodies elicited early correlated with viral clearance (6–8).
The involvement of the E2 protein in HCV entry into liver cells makes this viral surface protein a major target for eliciting neutralizing antibodies. The majority of neutralizing antibodies reported to date have been shown to block the interaction of E2 with CD81, the major cellular receptor for all HCV strains. Antibodies that block the E2-CD81 interaction recognize both linear and conformational epitopes, mostly within conserved segments that are discontinuous in the E2 primary sequence, thus reflecting the complexity of the formation of the E2-CD81 interface. Numerous studies on neutralizing antibody specificities have shown that there are three dominant binding regions on E2, which include residues 412 to 423, 436 to 447, and 523 to 540 (9–25). Several extensive mutagenesis studies have further confirmed the importance of most of these regions by showing that the specific residues critical for E2 binding to CD81 include W420, Y527, W529, G530, and D535 (16) and the G436WLAGLFY443 motif (17).
Two recent publications reported crystal structures of the E2 core, including the E2 core in complex with a neutralizing antibody, AR3C (26), and the E2 core in complex with a nonneutralizing antibody (27). In the E2 core-AR3C complex, the E2 core is described as having a β-sheet central core that is sandwiched between two additional protein layers. These layers are composed largely of loops, with the front layer having a short stretch of α-helical structure which includes a portion of the epitope II region of E2. The flanking protein layers observed in the E2 core have residues from two of the three dominant regions of the E2 protein, including residues 436 to 447 (front layer) and 523 to 540 (CD81 binding loop), purported to be involved in CD81 binding. The E2 region comprised of residues 412 to 423 (i.e., the epitope I region) was found to be disordered. The structural determination of the E2 core has greatly facilitated an overall understanding of how these different regions of E2 might be involved in CD81 binding. However, a description of how these regions of E2 interact specifically with CD81 remains elusive.
Previously, we generated a murine monoclonal antibody, mAb41, which could neutralize HCV in vitro by binding to a linear region of E2 that includes residues 436 to 447 and that resides within epitope II of the E2 protein (23). The same region of E2 was originally described by Drummer et al. to be involved in CD81 interaction and viral entry (17). We thus focused our efforts on the region of E2 from residues 436 to 447 to investigate its potential involvement as an interface for CD81 interaction. In our recent structural studies of a panel of monoclonal antibodies that are similar to mAb41 and recognize the same set of residues within the epitope II region of E2, we revealed that the G436WLAGLFY443 motif, as observed in the E2 core structure (26), adopts an α-helical conformation (28, 29). The helical structure of the G436WLAGLFY443 motif was also confirmed by Krey et al. by use of two different antibodies (30). These observations suggest that the helical formation is not an induced fit conferred by the antibody.
Complementary mutagenesis and antibody blocking studies have also been well documented for CD81, in addition to the determination of the crystal structure of its large extracellular loop (LEL) (12, 31–33). Residues of CD81 that were reported to contribute to E2 interaction include F186, I182, L185, and N184, which comprise the D-helix, and L162, T163, and T166, which reside at the N-terminal end of the C-helix. Interestingly, a study by Bertaux and Dragic showed that the anti-CD81 antibodies 1D6 and JS-81 inhibited the interactions between CD81 and E2, but by distinct mechanisms (34). This conclusion was drawn based on the difference observed in the ratios of the 50% effective concentration (EC50) to 50% inhibitory concentration (IC50) of ID6 and JS-81, in which complete inhibition by 1D6 involved occupying all available CD81 binding sites, whereas JS-81 required binding to only one of the four CD81 molecules to achieve complete inhibition of viral entry. From this, Bertaux and Dragic proposed that 1D6, by recognizing a linear epitope, can block viral entry by directly inhibiting the E2-CD81 interaction, whereas JS-81, by recognizing a conformational epitope, may be involved in the blocking of postbinding steps in addition to E2-CD81 binding.
In this study, the identification of linear binding sites for both antibodies (mAb41 and 1D6) was used as a basis for molecular docking, which allowed us to explore both surfaces of a potential E2-CD81 interface. Our results suggest that an interface can be formed between epitope II of E2 and the large extracellular loop of CD81. The description of this interface provides a first glimpse of how epitope II engages in CD81 interaction, thus highlighting the relevance of this region of the E2 protein in antibody-mediated neutralization of HCV.
MATERIALS AND METHODS
Antibodies and Fab fragment generation.
MAb41 belongs to a panel of antibodies that were generated by immunizing mice with a synthesized peptide encompassing residues 412 to 447 of the E2 protein (Harlan Bioproducts for Science, Indianapolis, IN). The neutralizing activity of mAb41 was previously demonstrated in a cell-based HCV neutralization assay (23). The Fab fragment of mAb41 was generated by digestion of purified mAb41 with ficin according to the manufacturer's procedures (Thermo Scientific, Rockford, IL), followed by gel filtration and ion-exchange chromatography. 1D6, an anti-CD81 monoclonal antibody known to block HCV entry in vitro (9, 34, 35), was purchased from AbD Serotec (Raleigh, NC).
Peptide synthesis.
All peptides were synthesized in the Core Laboratory of the Center of Biologics Evaluation and Research, FDA, with an Applied Biosystems (Foster City, CA) model 433A peptide synthesizer, using standard FastMoc chemistry. Biotinylated peptides were synthesized with an Fmoc-Lys(biotin-LC)-Wang resin (AnaSpec, San Jose, CA).
Plasmid constructs and protein expression.
A cDNA fragment of HCV strain H77 corresponding to residues 384 to 661 of the E2 protein was cloned into the mammalian cell expression vector pFUSE-Fc2-IL2ss (Invivogen, San Diego, CA) to produce an IgG1 Fc-fused E2 protein, referred to as E2-16Fc. Similarly, the CD81-Fc construct was produced by cloning a cDNA fragment corresponding to the large extracellular domain of human CD81 (CD81-LEL; residues 113 to 201) into the same expression vector. Huh7 cells constitutively secreting these Fc fusion proteins were established by selecting Zeocin-resistant clones. The fusion proteins were purified from the serum-free culture medium by use of a Hi-Trap protein G column according to the manufacturer's protocols (GE Healthcare, Uppsala, Sweden). In addition, the E2 protein (residues 384 to 661), fused with an N-terminal FLAG tag containing the sequence DYKDDDDK and named E2-FLAG, was transiently expressed in Huh7 cells. This FLAG-tagged E2 protein was also secreted into the serum-free culture medium and purified by use of anti-FLAG M2 magnetic beads (Sigma-Aldrich, St. Louis, MO).
For an alternative assay to evaluate the binding of CD81 to E2, human CD81-LEL was produced as an N-terminal fusion protein with Gaussia luciferase (New England BioLabs, Beverly, MA) and labeled CD81-Luc. Since luciferase is a naturally secreted protein, CD81-Luc was collected from the cell culture medium after transient transfections of an expression construct (pFUSE-Luc-CD81) into Huh7 cells. The presence of CD81-Luc in the culture medium was demonstrated by the detection of luciferase activity. The luciferase activity was quantified using a BioLUX Gaussia luciferase flex kit (New England BioLabs) followed by measurement of the luminescence intensity using a microtiter plate reader equipped with luminescence optics (Infinite F500 model; Tecan, Männedorf, Switzerland).
Site-directed mutagenesis and expression of E2-16Fc mutants.
Mutations that included the E431A, E431A/N434E, W437A, L438A, L441A, F442A, and P525A mutations were introduced into the backbone of E2-16Fc by using a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) and were confirmed by DNA sequencing.
Pulldown assays to evaluate the interaction of E2 with CD81-Fc.
Protein G magnetic beads (GE Healthcare) were incubated with either purified CD81-Fc or human immunoglobulin, intravenous (IGIV), as a nonspecific, negative control. After extensive washing with phosphate-buffered saline containing 0.05% Tween 20 (PBS-T), the beads bearing CD81-Fc or IGIV were mixed with E2-FLAG. For antibody blocking experiments, E2-FLAG was preincubated with mAb41, Fab41, or Fab41 complexed with either an epitope II peptide (N430ESLNTGWLAGLFYQHK446) or an epitope II mimic peptide (EWLSTLS) to block Fab41. All binding experiments were performed at room temperature with gentle rocking for 1 h. After washing of the beads with PBS-T, the bound proteins were eluted with 0.1 M glycine-HCl buffer (pH 3.0), neutralized with 1 M Tris-HCl (pH 8.0), and analyzed by Western blotting, probing with either 1D6 or mAb41.
Similarly, another set of pulldown assays was performed to examine the binding of mutant forms of E2 to CD81. In this case, E2-16Fc and its mutants were captured, individually, on protein A magnetic beads. After washing with PBS-T, the beads bearing E2-16Fc or individual E2-16Fc mutants were then mixed with CD81-Luc and left to bind at room temperature for 1 h. After extensive washing, bound proteins on the beads were eluted with 0.1 M glycine-HCl buffer (pH 3.0) and neutralized with 1 M Tris-HCl (pH 8.0). Aliquots of the eluted proteins were dispensed in duplicate into a 96-well Nunc white luminescence plate for luciferase activity measurement on a Tecan microplate reader. The observed luciferase activity was normalized with the amount of E2-16Fc present in the same eluate, as determined by Western blotting band analysis using AlphaView Q software (Alpha Innotech Corporation, San Leandro, CA).
Blocking of the interaction between E2-16Fc and CD81-Luc by specific peptides.
E2-16Fc was immobilized on a 96-well plate precoated with protein A (Thermo Scientific). As negative and positive controls for the assay, wells were coated with the mAb41 and 1D6 antibodies, respectively. After washing, CD81-Luc, which had been preincubated with various amounts of peptides (epitope II, epitope II mimic, or a nonspecific peptide), was added to the wells. After extensive washing, the captured luciferase activity was measured using a Tecan microplate reader. All assays were performed in duplicate, and results were expressed as means ± standard deviations (SD). The IC50s were determined using GraphPad Prism software (36).
Mapping of 1D6 binding site by screening of random-peptide phage display libraries.
Selection of peptides representing the 1D6 binding site was performed by screening random-peptide phage display libraries as described previously (18). The binding site was determined by comparison of the consensus sequence of the selected peptides with the human CD81-LEL protein sequence.
Molecular docking of CD81-LEL with E2 peptides.
Docking studies were performed using the AutoDock Vina program (37). The crystal structure of human CD81-LEL (PDB code 1G8Q) (31) was used as the target receptor for the docking of the two ligands, i.e., the epitope II peptide (N430ESLTNTGWLAGLFYQHK446) and the epitope II mimic peptide (EWLSTLS). The conformations that the peptide ligands most likely adopted were determined by a combinatorial approach of secondary structure prediction using the PEP-FOLD server (38; http://bioserv.rpbs.univ-paris-diderot.fr/PEP-FOLD/) and comparisons of predicted models to the structure of the epitope II peptide in complex with a neutralizing antibody (28) and to the recently published E2 core structure (26).
Docking was performed using a flexible ligand-rigid receptor approach in which flexible peptide ligands were docked to a rigid monomer of CD81-LEL. The AutoDock Tools utility was used to prepare both target receptor and ligands (37). For the receptor, polar hydrogens were added. For each peptide ligand, hydrogens were added, Gasteiger charges were calculated, and nonpolar hydrogens were merged. Additionally, the peptide backbone torsions for each peptide ligand were restricted to maintain a helical conformation among residues W437LAGLF442 and W437LAGL441 within the epitope II peptide. For the remaining residues of each peptide ligand, the backbone torsions with exclusion of the amide bonds were designated as being rotatable. In addition, the side chains for all residues in these peptides were also assigned as being rotatable. Each ligand was docked to a CD81-LEL monomer, using identical docking parameters. The dimensions of the search space used for all peptide ligand docking had grid point dimensions of 36 Å, 36 Å, and 36 Å, with grid-point spacing of 1 Å. The grid search space of the receptor was oriented in each case to include residues I182SNLF186 of CD81-LEL (the D-helix). At least two rounds of docking were performed to generate 9 binding modes per round for each peptide ligand. The binding modes of peptide ligands obtained after each round of docking were analyzed with PyMOL (39).
For comparison to the epitope II peptide, fexofenadine, a small molecule reported to weakly inhibit the E2-CD81 interaction (40, 41), was also used as a ligand for the docking of CD81-LEL. Fexofenadine was prepared for docking by using the same procedures as those for the peptide ligands, including the addition of hydrogens and charges and the merging of nonpolar hydrogens. All allowable bonds were assigned as being rotatable. The same docking approach and parameters used for the peptide ligands were used for fexofenadine. The binding modes for fexofenadine were analyzed with PyMOL and compared to those of the peptide ligands.
RESULTS
Characterization of key residues within epitope II required for binding of neutralizing antibody mAb41.
mAb41 was previously shown to neutralize HCV infection in a cell-based assay (23); however, the underlying mechanism has not been fully elucidated. We initially examined the key residues on the E2 protein that are involved in mAb41 binding by the screening of random-peptide phage display libraries (23). From 30 independent phage-displayed peptide sequences, we identified the consensus sequence WLXXL as the core binding motif (Fig. 1A). There was also a trend in the presence of an E or D residue in close proximity to the consensus sequence; however, the relevance of these residues to mAb41 recognition is presently unknown. We further examined selected phage clones by Western blotting under denaturing conditions, using mAb41 as a probe, and found that the peptide (EWLSTLS) displayed on phage 51 could react with mAb41 (Fig. 1A), resulting in an intense signal at an approximate molecular mass of 51 kDa (Fig. 1B). This result was confirmed by using a chemically synthesized version of this short peptide which was indistinguishably detected by either mAb41 or Fab41 in comparison to the epitope II peptide by enzyme-linked immunosorbent assay (ELISA) (data not shown). These results allowed us to conclude that the EWLSTLS peptide represents a linear mimic of epitope II on the E2 protein (referred as epitope II mimic) which can bind to mAb41.
FIG 1.
mAb41 recognizes the consensus sequence WLXXL. (A) Results of phage display screening with mAb41. Bold letters denote the consensus sequence. (B) Western blot analysis under reducing conditions of phage 51 probed by mAb41.
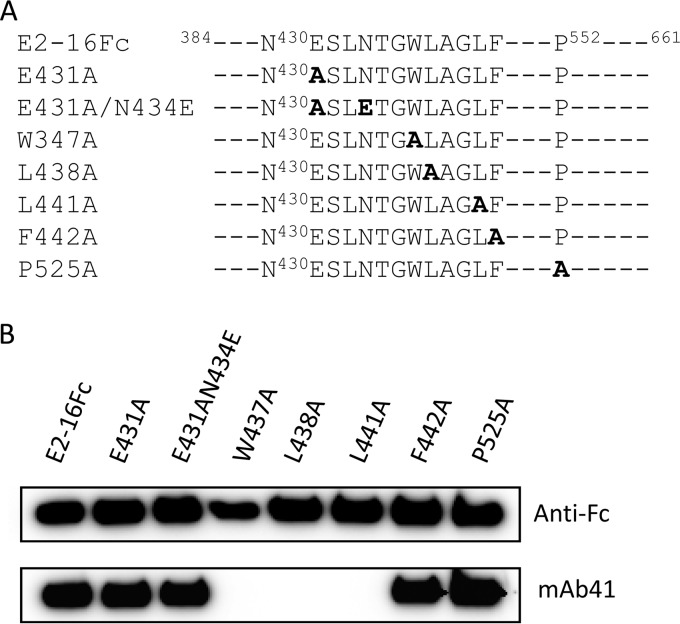
To further characterize the binding site of mAb41, we produced soluble forms of E2 spanning residues 384 to 661, along with a series of its mutants, in Huh7 cells, as human IgG1 Fc fusion proteins (Fig. 2A). We found that the E2-16Fc W437A, L438A, and L441A mutants could not be recognized by mAb41 on Western blots (Fig. 2B). In contrast, the F442A substitution did not affect mAb41 recognition. Similarly, other E2-16Fc mutants, including the E431A, E431A/N434E, and P525A mutants, could still be recognized by mAb41 (Fig. 2B). These results confirm that the key contact residues in the E2 protein for the binding of mAb41 include W437, L438, and L441.
FIG 2.
Residue-specific binding of mAb41 to E2-16Fc and its mutants. (A) Schematic representation of E2-16Fc and its mutants used in Western blot analysis. (B) E2-16Fc and its mutants were purified from cell culture medium by use of protein G magnetic beads. Eluates were separated by SDS-PAGE and probed with either anti-human IgG1 Fc (anti-Fc) or mAb41.
Neutralizing antibody mAb41 blocks E2-CD81 interaction.
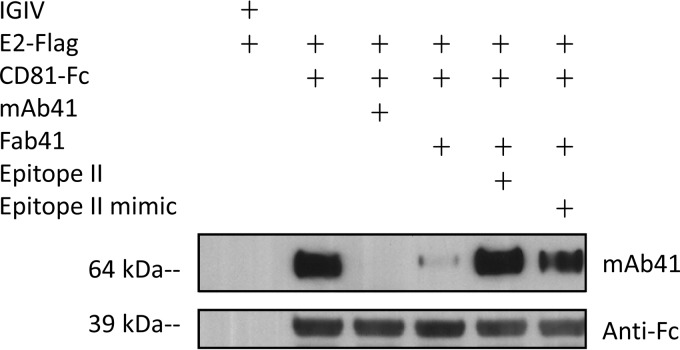
We tested whether mAb41 could block the interaction of E2 with CD81 in a pulldown assay in which E2-FLAG was incubated with CD81-Fc captured on protein G beads in the presence or absence of mAb41 or Fab41. As expected, E2-FLAG was pulled down with CD81-Fc, in contrast to the IGIV preparation used as a negative control. However, the interaction between E2 and CD81 was significantly reduced in the presence of either mAb41 or Fab41 (Fig. 3). Furthermore, the blocking effect of Fab41 could be reversed completely by preincubation of the antibody with either the epitope II peptide or the epitope II mimic peptide, suggesting that mAb41 may occupy a region on the E2 protein to which CD81 can bind.
FIG 3.
mAb41 pages the E2-CD81 interaction. CD81-Fc was captured on protein G magnetic beads and then incubated with E2-FLAG, with or without mAb41, Fab41, or Fab41 complexed with either epitope II or the epitope II mimic peptide. An IGIV which lacks any detectable anti-HCV activity was used as a negative control. Proteins bound on the beads were eluted, separated by SDS-PAGE under reducing conditions, and probed with either anti-Fc or mAb41.
Characterization of the E2-CD81 interface.
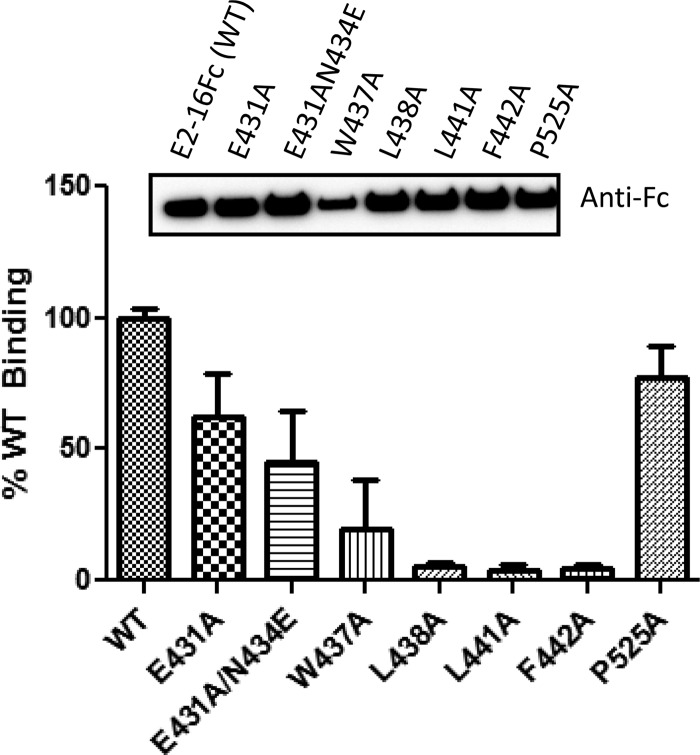
To examine the effects of specific E2 residues on the E2-CD81 interaction, we created CD81-Luc as a reporter for quantitative measurements of its ability to bind to E2-16Fc or E2-16Fc mutants in a luciferase-based binding assay. Compared with E2-16Fc, the W437A, L438A, and L441A mutants showed significant reductions in CD81 binding (Fig. 4). In contrast, the P525A mutation had no effect on CD81 binding. Similarly, the E431A or E431A/N434E mutations led to a detectable reduction of binding, but the effect was not as pronounced as that observed with the W437A, L438A, and L441A mutants. The F442A mutation did not affect mAb41 binding, as shown in Fig. 2B; however, this mutant exhibited a significant decrease in CD81 interaction (Fig. 4). Therefore, it appears that the binding site of mAb41 overlaps with that of CD81 on the E2 protein; however, these two binding sites are distinguishable from each other with respect to residue F442 within epitope II.
FIG 4.
Interaction of CD81-Luc with various mutant forms of E2-16Fc. The binding of CD81-Luc to individual mutants of E2-16Fc is indicated by percentages relative to the binding to E2-16Fc (WT). The inset shows the amounts of E2 protein bound to protein G magnetic beads, which were quantitated after Western blot analysis with anti-Fc, using AlphaView Q software (Alpha Innotech Corp.) to normalize the protein input. The data from three independent experiments are presented.
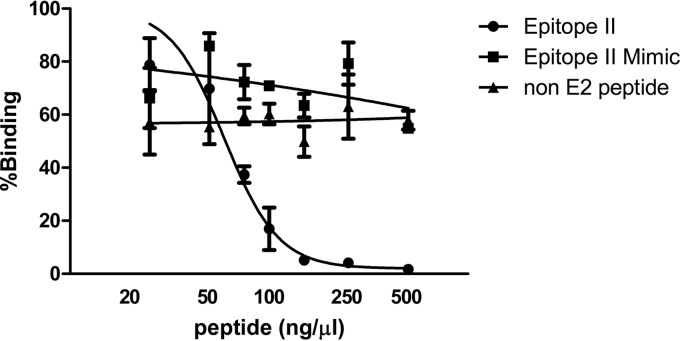
To further determine the association of the mAb41 binding site with the E2-CD81 interacting surface, we tested whether the epitope II peptide and the linear mimic peptide of epitope II could act as competitors of the E2 protein for binding to CD81. E2-16Fc was immobilized to a 96-well luminescence plate that was precoated with protein A and then incubated with CD81-Luc in the presence of various amounts of epitope II peptide, epitope II mimic peptide, or an unrelated peptide as a negative control. We observed that the epitope II peptide was able to compete with the E2 protein for CD81 binding in a dose-dependent manner. An approximate IC50 of 60 ng/μl (95% confidence interval [CI] = 48.2 to 73.5 ng/μl; R2 = 0.92) was obtained by nonlinear regression analysis (Fig. 5). In contrast, the addition of the epitope II mimic peptide, in which F442 was replaced by a serine, did not affect the interaction, nor did the addition of an unrelated peptide control (Fig. 5). These observations confirm that epitope II, especially residue F442, is a key part of the binding surface on the E2 protein for CD81 interaction, although we cannot exclude the possibility that the other flanking residues may influence the stability of local E2 conformations for the interaction.
FIG 5.
Epitope II peptide interferes with E2-CD81 interaction. E2-16Fc was immobilized on a protein A-coated luminescence plate and then incubated with CD81-Luc in the presence of different amounts of epitope II peptide, epitope II mimic peptide, or a non-E2 peptide as a negative control. Luciferase activity representing CD81 binding, shown by the percentage relative to the level of a control CD81-Luc sample in the absence of a peptide competitor, was measured. The data from two independent experiments are presented. A nonlinear regression analysis of these data was performed.
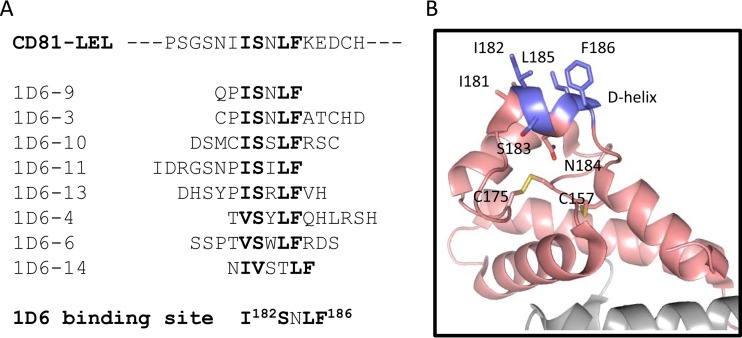
Mapping of the 1D6 binding site on CD81.
The anti-CD81 antibody 1D6 is known to block the interaction of CD81 with E2 (9, 34, 35). We set out to map the binding site of 1D6 by screening random-peptide phage display libraries. The consensus sequence ISXLF emerged after the selection, which allowed us to precisely map the 1D6 binding site to the I182SNLF186 sequence on CD81-LEL (Fig. 6A). As shown in Fig. 6B, the binding site adopts a helical conformation, previously referred to as the D-helix (31), which is supported by two flanking loops and two conserved disulfide bonds. The other residues of the binding site appear to be exposed on the D-helix, despite their hydrophobic nature. Residue F186, which is positioned in the last turn of the helix, has been reported to be the most critical residue for the interaction with E2, although other residues (I182 and N184) on this hydrophobic helix have also been implicated in the binding to E2 (9, 12, 32). These results suggest that the D-helix, as the target site of neutralizing antibody 1D6, may represent a crucial hydrophobic surface for CD81 that participates in the interaction with epitope II of the E2 protein.
FIG 6.
Mapping of the 1D6 binding site to the D-helix of CD81. (A) Peptide sequences isolated from phage display screening with 1D6. Residues in bold represent the consensus located on CD81. (B) Cartoon representation of CD81-LEL structure (31). Residues I182, S183, L185, and F186, which compose the 1D6 binding site as presented in panel A, are highlighted, with carbons in metallic blue, oxygens in red, and nitrogens in blue. Other residues of CD81-LEL are also presented, with carbons in pale pink, oxygens in red, nitrogens in blue, and sulfurs in yellow. Conserved disulfide bond pairs are shown between cysteine residues at positions 175-157 and 156-190. The figure was generated using PyMOL.
Molecular docking of the interaction of epitope II and fexofenadine with CD81.
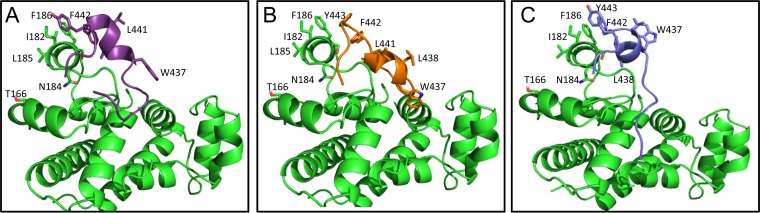
The identification of the binding sites for 1D6 on CD81 and mAb41 on the E2 protein provided the basis for investigating the potential involvement of the CD81 D-helix and epitope II in forming an interface. To visualize this interface, molecular docking was performed using AutoDock Vina software (37). The CD81 crystal structure (PDB code 1G89) (31) was selected as the target receptor for docking. The epitope II peptide was used as the ligand, with the backbone of the W437LAGL441 (or W437LAGLF442) motif constrained as a helix (26, 28–30). After each round of docking, binding modes of epitope II were generated and ranked by the docking software according to an estimated binding energy (affinity) in terms of kcal/mol, and the most dominant binding modes were analyzed (Fig. 7).
FIG 7.
Molecular docking of epitope II peptide to CD81-LEL. (A) Cartoon representation of one of three predominant conformations of epitope II peptide docked to CD81-LEL, with the C-α of residues W437LAGLF442 restrained as a helix. Residues of the epitope II peptide and CD81-LEL are shown in stick representation. Backbone and side chain atoms of epitope II are indicated, with carbons shown in purple, oxygens in red, and nitrogens in blue. Atoms of backbone and side chain residues of CD81-LEL are also shown, with carbons in green, oxygens in red, and nitrogens in blue. (B) Cartoon representation of a predominant epitope II conformer obtained from docking to CD81-LEL with the C-α of residues W437LAGL441 restrained as a helix, with less restriction than that in the conformer obtained for panel A. Residues in the epitope II peptide are shown in stick representation, with carbon atoms shown in metallic blue, oxygens in red, and nitrogens in blue. The color scheme for CD81-LEL atoms is the same as that for panel A. (C) Third representative conformer of epitope II docked to CD81-LEL, shown in cartoon representation. Docking was performed with W437LAGLF442 restrained as a helix. The same color schemes as those denoted for panels A and B were used for all atoms but the carbons of epitope II, which are shown in metallic blue.
When the W437LAGL441 or W437LAGLF442 motif in the epitope II peptide was set as a helix, we obtained two dominant conformers after several rounds of docking to the D-helix of CD81. Among all conformers, residue F442 was positioned in close proximity for potential hydrophobic interactions with both F186 and I182 of the D-helix. Additionally, residue Y443 was also positioned near F186 and I182 of the D-helix. These observations indicate that residues of these two hydrophobic helices may provide an interacting surface that is optimal for stabilizing the CD81-E2 interface (Fig. 7A and B). When less constraint was imposed on the W437LAGLF442 helical motif, both F442 and Y443 were similarly positioned in proximity to F186 and I182 of the D-helix (Fig. 7C), reinforcing the notion that an interaction between these two hydrophobic helices occurs. However, in the majority of the conformers obtained from docking, residues W437, L438, and L441 were not positioned to make direct contact with the CD81 D-helix (Fig. 7A to C); instead, these residues were more likely to maintain the helical conformation of the W437LAGL441 motif of epitope II, which supports the positioning of residues F442 and Y443 on a more flexible region of this helix motif.
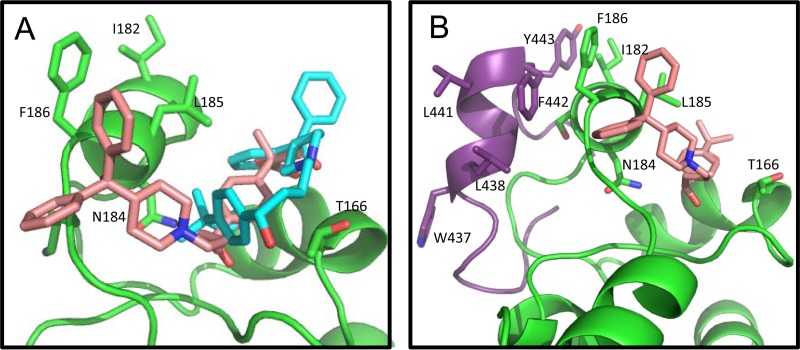
The small molecule fexofenadine was also docked to the CD81-LEL, utilizing the same methods as those used for the peptide ligands. Fexofenadine is an active, nontoxic form of terfenadine, which was reported to moderately inhibit (approximately 27% reduction at 50 μM) the interaction between E2 and CD81-LEL (40). More importantly, fexofenadine was reported to interact with the residues of the D-helix of CD81 (41); therefore, we included this small molecule in our analysis for comparison. In the majority of the docked models of fexofenadine, at least two of the three phenyl moieties were positioned to enable hydrophobic contacts with the residues of the D-helix of CD81 (Fig. 8A). A comparison between the docked models of fexofenadine and the epitope II peptide showed that the two dominant fexofenadine conformers were all docked within the crevice between the D- and C-helices of CD81 (Fig. 8A), whereas the epitope II conformers were docked on the other side of the D-helix (Fig. 8B). In this docked orientation, fexofenadine was situated to make contacts not only with F186 and L185 on the D-helix but also with residue T163, located on the C-helix, and residue N184, located on a downturn of the D-helix which orients underneath the D-helix (Fig. 8B). Interestingly, the C- and D-helices of CD81 were recently described as forming a significant E2-interacting surface based on the results of an electron microscopic (EM) reconstruction of a ternary complex of E2 core, CD81-LEL, and antibody Fab AR2A, an E2-specific antibody (26).
FIG 8.
Fexofenadine docked to CD81-LEL D-helix. (A) Cartoon representation of comparison between two dominant conformers of fexofenadine docked to CD81-LEL. Atoms of fexofenadine are colored with the following scheme: carbons are in cyan (conformer 1) or pale pink (conformer 2), nitrogens in blue, and oxygens in red. (B) Comparison of the dominant conformers of fexofenadine and epitope II peptide docked to CD81-LEL. Fexofenadine is shown in pale pink (same conformer as in panel A). The epitope II peptide is shown in purple (same conformer as that shown in Fig. 7A). The color scheme for atoms of CD81-LEL is the same as that denoted in the legend to Fig. 7.
When the conformers of the epitope II peptide obtained from docking were superimposed with the E2 core structure, we observed that residues F442 and Y443, as a part of the helix motif W437LAGLFY443, were located within a surface-exposed cavity and were accessible to residues F186 and I182 on the D-helix of CD81 (Fig. 9), thus suggesting that these residues may serve as major hydrophobic contact points for CD81 interaction. This suggestion is consistent with the observation that the epitope II mimic peptide, due to the replacement of F442 by a much less hydrophobic serine residue, is unable to compete with the interaction between the E2 protein and CD81. Furthermore, residues W437, L438, and L441 were shown to be buried in the E2 core, which may limit their surface accessibility. Therefore, these residues might not be involved in direct contact with the CD81 D-helix. Instead, these residues may contribute to stabilization of the helical conformation of epitope II to support interactions with CD81 and subsequent conformational changes of the E2 protein upon binding.
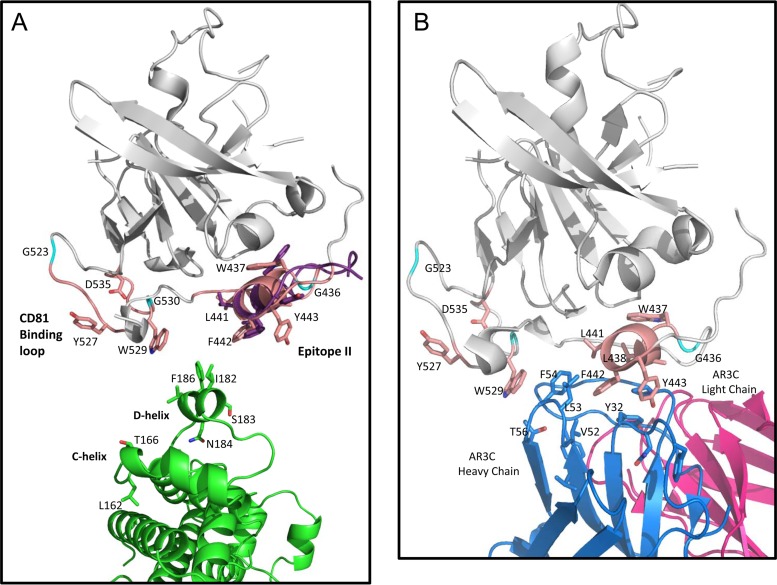
FIG 9.
(A) Cartoon representation of the docked conformer of epitope II peptide superimposed with the E2 core structure (PDB ID 4MWF). The E2 core is shown in gray, with regions involved in CD81 binding highlighted pale pink and specific side chains associated with CD81 binding displayed as follows: carbons in pale pink, oxygens in red, and nitrogens in blue, with the exception of glycines, which are shown in cyan. The docked epitope II peptide conformer superimposed on the same sequence in E2 is shown with side chains displayed in stick representation, with carbons shown in purple, oxygens in red, and nitrogens in blue. The CD81-LEL dimer is shown with the same color scheme as that denoted in the legends to Fig. 7 and 8, with the side chains of the D-helix shown in stick representation and positioned as a reference to how this region may interact with the E2 helix motif, W437LAGLFY443. (B) Cartoon representation of the published E2 core structure in complex with antibody AR3C (PDB ID 4MWF), with residues of the HC CDR loop regions of AR3C that show similarity to the D-helix of CD81 displayed. The color scheme for the E2 core is the same as that described for panel A. The light chain of AR3C is shown in magenta, the heavy chain is shown in blue, and residues of the HC CDR2 loop (including residues V52, L53, and F54) and the CDR1 loop (including residue Y32) are shown in stick representation, with carbons in blue and oxygens in red.
Interestingly, upon close examination of the E2 core-AR3C structure complex, we found that the interactions between the HC CDR2 loop of AR3C and the W437LAGLFY443 helix motif of E2 core have a notable similarity to the interactions indicated in the epitope II-CD81 docking models. As shown in Fig. 9B, the HC CDR2 loop, consisting of residues V52, L53, and F54, closely resembles the D-helix of CD81 in terms of hydrophobicity and shape. Furthermore, residues F54 and L53 on the HC CDR2 loop, which are comparable to F186 and I182 on the D-helix of CD81, participate significantly in the interactions with residues F442 and Y443 of the E2 core.
DISCUSSION
In this study, we described a binding interface formed between the HCV E2 protein and the host entry factor CD81. On the virus side, the interacting surface includes a hydrophobic stretch of epitope II within E2, comprised of residues W437LAGLF442, previously characterized as an α-helix that is recognized by the neutralizing antibody mAb41 (26, 28–30). On the cell side, the interface has a surface-exposed hydrophobic helix, composed of residues I182SNLF186 within the CD81-LEL, and coincides with the binding site of 1D6, a monoclonal antibody known to block HCV entry in vitro (9, 34, 35).
Our observation of the involvement of W437LAGLF442 in CD81 binding is in agreement with the previous findings of Drummer et al. (17), whose study showed that replacement of residues W437, L438, L441, and F442 by either an alanine or a valine resulted in significant reductions in viral entry that were correlated with a substantial decrease in CD81 binding. A recent study by Keck et al. (42) also showed that alanine substitutions of residues W437, L441, and F442 led to significant reductions in CD81 binding.
One of the distinctive features discerned from our study is the identification of the critical role of residues F442 and Y443 within epitope II in CD81 binding, although these residues appear to be nonessential for the binding of the neutralizing antibody mAb41. This observation is consistent with our previous studies which showed that the binding of residues W437, L438, and L441 of epitope II is necessary, but may not in itself be sufficient, for antibody-mediated neutralization (28, 29). The mAb41-mediated neutralization may be attributable simply to steric hindrance by its binding to a region that is crucial for the E2-CD81 interface. However, the hydrophobicity of F442 within the W437LAGLF442 motif appears to be essential in establishing the interface with CD81, as indicated by the inability of the epitope II mimic peptide, which has a serine in place of F442, to compete with the E2 protein for binding to CD81.
In our search for an analogous hydrophobic surface of CD81 that can potentially interact with the W437LAGLF442 helix of epitope II on the E2 protein, we focused our attention on 1D6, an antibody shown to inhibit the CD81-E2 interaction. As opposed to JS-81, which binds a conformational epitope on CD81, we found from results of phage display studies that 1D6 recognizes a linear epitope which corresponds to the I182SNLF186 sequence (i.e., the D-helix) located on the surface of CD81 (31). Since the D-helix has been shown to participate in E2 interaction, specifically involving the highly conserved residues F186, I182, and L185, the binding of 1D6 to the D-helix demonstrates that the D-helix is an important interface for interacting with the E2 protein.
Based on the results of molecular docking and comparative analysis of the epitope II conformers with the E2 core structure, we propose that the W437LAGLF442 motif of E2 forms a stable hydrophobic helix that serves in two capacities, which include supporting the position of the major contact residues F442 and Y443 and providing stabilization during binding and subsequent conformational changes.
By examining the EM reconstruction of a complex consisting of E2, CD81-LEL, and the anti-E2 antibody AR2A, Kong et al. recently proposed that various regions of the E2 core, including the W437LAGLFY443 motif, make contacts with both the D- and C-helices of CD81 (26). With our results, we further suggest that the hydrophobic helix motif W437LAGL441 on the surface of E2 is able to support the positioning of residue F442, and also Y443, to serve as the major contact points with the highly conserved residues F186, I182, and L185 on the CD81 D-helix. According to the epitope II-CD81 docking models, both faces of the D-helix are accessible to the W437LAGLFY443 motif of the E2 protein. Interestingly, the epitope II docking models are supported in part by the E2 core-AR3C structure, in which the HC CDR2 loop shows striking similarity to the D-helix of CD81 and makes interactions with residues F442 and Y443 similar to those observed in the epitope II-CD81 docking models.
In the E2 core structure, the W437LAGLF442 helix is situated in close proximity to other, more flexible regions of E2 that have also been implicated in E2 binding, specifically residues 523 to 540. Several glycine residues reside in these regions and may provide the flexibility that can accommodate conformational changes subsequent to binding. In fact, this concept is supported by a study from Drummer et al. showing that a G436P mutation in E2, as either virion-incorporated or cell lysate-derived E1-E2 pseudoparticles, negatively affected HCV entry; however, it did not abrogate CD81 binding (17). This finding indicates, as concluded by Drummer et al., that the conserved residue G436 is likely involved in facilitating postbinding conformational changes essential for viral entry. Additionally, our previous structural studies showed the significance of G436 in guiding the spatial arrangement of essential residues of epitope II (i.e., the N-terminal loop and the C-terminal helix), thus providing a degree of flexibility of epitope II in adopting distinct conformations in antibody binding (28, 29). Taken together, the glycines residing between these regions of E2 reflect an inherent flexibility between different parts of E2 that are involved in protein interactions.
Further analysis of the E2 core crystal structure indicates that both residues W437 and L438, as the key residues for binding of mAb41, are buried. This configuration precludes the possibility of direct contact of these residues with either CD81 or mAb41. Recently we demonstrated that discrete conformations exist within epitope II, which supports the possibility of dynamic structural transitions occurring that may expose these residues (29). In addition, residues L438 and L441 on the W437LAGLF442 helix in the E2 core structure protrude into the shallow cavity formed by two regions of E2 that include residues 436 to 443 (epitope II) and 523 to 535 (CD81 binding loop). The unique location of these two residues may provide, upon the conformational transitions of the helix, an opportunity for the W437LAGLF442 helix to indirectly affect the interaction of the E2 protein with CD81. Given the flexibility of the surrounding regions, it is likely that the W437LAGLF442 helix in the E2 core-AR3C structure complex represents one bound form of several possible conformations of the E2 structure. Therefore, depending on the binding partner, e.g., different antibodies, CD81, or other receptors, different surfaces of E2 may become accessible for interaction.
The interaction between E2 and CD81, which is a significant early step in virus entry, is anticipated to be a dynamic process most likely involving a series of conformational changes. Moreover, we and others have shown that other highly conserved residues of the E2 protein, such as W420, Y527, W529, G530, and D535, are also involved in the E2-CD81 interaction (16, 25). The question that remains to be answered is how the residues within these different regions of the E2 protein cooperatively interact with CD81. Further characterization of this dynamic process on both sides of the E2-CD81 interface should yield useful insights into how to develop neutralizing antibodies against specific conformations that can effectively disrupt virus entry.
ACKNOWLEDGMENTS
This study was funded by a Modernizing Science grant from FDA CBER.
We thank Stephen Feinstone and Hongying Duan for providing critical reagents and John Finlayson for comments on the study.
REFERENCES
- 1.Lauer GM, Walker BD. 2001. Hepatitis C virus infection. N Engl J Med 345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2012. CDC health information for international travel. Oxford University Press, New York, NY. [Google Scholar]
- 3.Alter HJ, Liang TJ. 2012. Hepatitis C: the end of the beginning and possibly the beginning of the end. Ann Intern Med 156:317–318. doi: 10.7326/0003-4819-156-4-201202210-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honegger JR, Zhou Y, Walker CM. 2014. Will there be a vaccine to prevent HCV infection? Semin Liver Dis 34:79–88. doi: 10.1055/s-0034-1371081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logvinoff C, Major ME, Oldach D, Heyward S, Talal A, Balfe P, Feinstone SM, Alter H, Rice CM, McKeating JA. 2004. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci U S A 101:10149–10154. doi: 10.1073/pnas.0403519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavillette D, Morice Y, Germanidis G, Donot P, Soulier A, Pagkalos E, Sakellariou G, Intrator L, Bartosch B, Pawlotsky JM, Cosset FL. 2005. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol 79:6023–6034. doi: 10.1128/JVI.79.10.6023-6034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, Patel AH, Meisel H, Baumert J, Viazov S, Rispeter K, Blum HE, Roggendorf M, Baumert TF. 2007. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A 104:6025–6030. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowd KA, Netski DM, Wang XH, Cox AL, Ray SC. 2009. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology 136:2377–2386. doi: 10.1053/j.gastro.2009.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating JA. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol 73:6235–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allander T, Drakenberg K, Beyene A, Rosa D, Abrignani S, Houghton M, Widell A, Grillner L, Persson MA. 2000. Recombinant human monoclonal antibodies against different conformational epitopes of the E2 envelope glycoprotein of hepatitis C virus that inhibit its interaction with CD81. J Gen Virol 81:2451–2459. [DOI] [PubMed] [Google Scholar]
- 11.Triyatni M, Vergalla J, Davis AR, Hadlock KG, Foung SKH, Liang TJ. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124–132. doi: 10.1006/viro.2002.1463. [DOI] [PubMed] [Google Scholar]
- 12.Drummer HE, Wilson KA, Poumbourios P. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J Virol 76:11143–11147. doi: 10.1128/JVI.76.21.11143-11147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci U S A 100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roccasecca R, Ansuini H, Vitelli A, Meola A, Scarselli E, Acali S, Pezzanera M, Ercole BB, McKeating J, Yagnik A, Lahm A, Tramontano A, Cortese R, Nicosia A. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J Virol 77:1856–1867. doi: 10.1128/JVI.77.3.1856-1867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owsianka A, Tarr AW, Juttla VS, Lavillette D, Bartosch B, Cosset FL, Ball JK, Patel AH. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol 79:11095–11104. doi: 10.1128/JVI.79.17.11095-11104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owsianka AM, Timms JM, Tarr AW, Brown RJ, Hickling TP, Szwejk A, Bienkowska-Szewczyk K, Thomson BJ, Patel AH, Ball JK. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol 80:8695–8704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drummer HE, Boo I, Maerz AL, Poumbourios P. 2006. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J Virol 80:7844–7853. doi: 10.1128/JVI.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P, Wu CG, Mihalik K, Virata-Theimer ML, Yu MY, Alter HJ, Feinstone SM. 2007. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc Natl Acad Sci U S A 104:8449–8454. doi: 10.1073/pnas.0703039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson DX, Voisset C, Tarr AW, Aung M, Ball JK, Dubuisson J, Persson MA. 2007. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc Natl Acad Sci U S A 104:16269–16274. doi: 10.1073/pnas.0705522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perotti M, Mancini N, Diotti RA, Tarr AW, Ball JK, Owsianka A, Adair R, Patel AH, Clementi M, Burioni R. 2008. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the hepatitis C virus E2 protein. J Virol 82:1047–1052. doi: 10.1128/JVI.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med 14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 22.Mancini N, Diotti RA, Perotti M, Sautto G, Clementi N, Nitti G, Patel AH, Ball JK, Clementi M, Burioni R. 2009. Hepatitis C virus (HCV) infection may elicit neutralizing antibodies targeting epitopes conserved in all viral genotypes. PLoS One 4:e8254. doi: 10.1371/journal.pone.0008254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan H, Kachko A, Zhong L, Struble E, Pandey S, Yan H, Harman C, Virata-Theimer ML, Deng L, Zhao Z, Major M, Feinstone S, Zhang P. 2012. Amino acid residue-specific neutralization and nonneutralization of hepatitis C virus by monoclonal antibodies to the E2 protein. J Virol 86:12686–12694. doi: 10.1128/JVI.00994-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. 2012. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A 109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Z, Zhong L, Elrod E, Struble E, Ma L, Yan H, Harman C, Deng L, Virata-Theimer ML, Liu P, Alter H, Grakoui A, Zhang P. 2014. A neutralization epitope in the hepatitis C virus E2 glycoprotein interacts with host entry factor CD81. PLoS One 9:e84346. doi: 10.1371/journal.pone.0084346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. 2013. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. 2014. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng L, Zhong L, Struble E, Duan H, Ma L, Harman C, Yan H, Virata-Theimer ML, Zhao Z, Feinstone S, Alter H, Zhang P. 2013. Structural evidence for a bifurcated mode of action in the antibody-mediated neutralization of hepatitis C virus. Proc Natl Acad Sci U S A 110:7418–7422. doi: 10.1073/pnas.1305306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng L, Ma L, Virata-Theimer ML, Zhong L, Yan H, Zhao Z, Struble E, Feinstone S, Alter H, Zhang P. 2014. Discrete conformations of epitope II on the hepatitis C virus E2 protein for antibody-mediated neutralization and nonneutralization. Proc Natl Acad Sci U S A 111:10690–10695. doi: 10.1073/pnas.1411317111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krey T, Meola A, Keck ZY, Damier-Piolle L, Foung SKH, Rey FA. 2013. Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog 9:e1003364. doi: 10.1371/journal.ppat.1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitadokoro K, Bordo D, Galli G, Petracca R, Falugi F, Abrignani S, Grandi G, Bolognesi M. 2001. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J 20:12–18. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higginbottom A, Quinn ER, Kuo CC, Flint M, Wilson LH, Bianchi E, Nicosia A, Monk PN, McKeating JA, Levy S. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J Virol 74:3642–3649. doi: 10.1128/JVI.74.8.3642-3649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol 78:1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertaux C, Dragic T. 2006. Different domains of CD81 mediate distinct stages of hepatitis C pseudoparticle entry. J Virol 80:4940–4948. doi: 10.1128/JVI.80.10.4940-4948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meuleman P, Hesselgesser J, Paulson M, Vanwolleghem T, Desombere I, Reiser H, Leroux-Roels G. 2008. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology 48:1761–1768. doi: 10.1002/hep.22547. [DOI] [PubMed] [Google Scholar]
- 36.GraphPad Software. 2007. GraphPad Prism, version 5 for Windows. GraphPad Software, La Jolla, CA. [Google Scholar]
- 37.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maupetit J, Derreumaux P, Tuffery P. 2009. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res 37:W498–W503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLano WL. 2002. The PyMOL molecular graphics system. http://www.pymol.org.
- 40.Holzer M, Ziegler S, Albrecht B, Kronenberger B, Kaul A, Bartenschlager R, Kattner L, Klein CD, Hartmann RW. 2008. Identification of terfenadine as an inhibitor of human CD81-receptor HCV-E2 interaction: synthesis and structure optimization. Molecules 13:1081–1110. doi: 10.3390/molecules13051081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajesh S, Sridhar P, Tews BA, Fénéant L, Cocquerel L, Ward DG, Berditchevski F, Overduin M. 2012. Structural basis of ligand interactions of the large extracellular domain of tetraspanin CD81. J Virol 86:9606–9616. doi: 10.1128/JVI.00559-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, Li AY, Patel AH, Lemon SM, Bukh J, Rey FA, Foung SKH. 2012. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization in a genotype 2a isolate. PLoS Pathog. 8:e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]