ABSTRACT
Bone marrow stromal cell antigen 2 (BST2) is a cellular restriction factor with a broad antiviral activity. In sheep, the BST2 gene is duplicated into two paralogs termed oBST2A and oBST2B. oBST2A impedes viral exit of the Jaagsiekte sheep retroviruses (JSRV), most probably by retaining virions at the cell membrane, similar to the “tethering” mechanism exerted by human BST2. In this study, we provide evidence that unlike oBST2A, oBST2B is limited to the Golgi apparatus and disrupts JSRV envelope (Env) trafficking by sequestering it. In turn, oBST2B leads to a reduction in Env incorporation into viral particles, which ultimately results in the release of virions that are less infectious. Furthermore, the activity of oBST2B does not seem to be restricted to retroviruses, as it also acts on vesicular stomatitis virus glycoproteins. Therefore, we suggest that oBST2B exerts antiviral activity using a mechanism distinct from the classical tethering restriction observed for oBST2A.
IMPORTANCE BST2 is a powerful cellular restriction factor against a wide range of enveloped viruses. Sheep possess two paralogs of the BST2 gene called oBST2A and oBST2B. JSRV, the causative agent of a transmissible lung cancer of sheep, is known to be restricted by oBST2A. In this study, we show that unlike oBST2A, oBST2B impairs the normal cellular trafficking of JSRV envelope glycoproteins by sequestering them within the Golgi apparatus. We also show that oBST2B decreases the incorporation of envelope glycoprotein into JSRV viral particles, which in turn reduces virion infectivity. In conclusion, oBST2B exerts a novel antiviral activity that is distinct from those of BST2 proteins of other species.
INTRODUCTION
BST2/tetherin is an interferon (IFN)-stimulated gene with potent antiviral properties against a variety of enveloped viruses (1–4). Human bone marrow stromal cell antigen 2 (hBST2) restricts viral-particle release of human immunodeficiency virus type 1 (HIV-1) lacking the viral accessory protein Vpu by tethering nascent virions, which are subsequently endocytosed, to the cell membrane (2, 5). Moreover, hBST2 has been shown to be an innate sensor of viral-particle release since, upon HIV-1 virion retention, it activates the NF-κB signaling pathway and induces expression of proinflammatory genes (6–8). BST2 is a type II transmembrane protein with a unique topology characterized by a short amino-terminal cytoplasmic tail followed by a transmembrane region, an ectodomain, and a carboxy-terminal glycosylphosphatidylinosotol (GPI) anchor. hBST2 is present in different intracellular compartments, including the trans-Golgi network (TGN) and early and recycling endosomes, and within lipid rafts (9). It is the unique topology of BST2, rather than the primary sequence, that determines the ability of this protein to restrict virus release, since an artificial “art tetherin” with no sequence similarity to BST2, but which maintains its structural features, is still capable of restricting the release of enveloped viral particles (5). At least for HIV, hBST2 restriction involves direct interaction between BST2 and nascent virions. BST2 acquires an axial conformation with pairs of either the amino- or carboxy-terminal transmembrane anchors inserted into the virions and the cell membrane, resulting in the tethering of viral particles (10). hBST2 exhibits a broad antiviral spectrum, since it is able to block not only HIV but also several other retroviruses (11) and, most importantly, many other enveloped viruses (12). However, viruses have evolved a variety of countermeasures to overcome BST2 restriction that involve the physical separation of BST2 from the site of virus assembly (12). Primate lentiviruses use different viral proteins, such as Vpu, Nef, and Env, to antagonize BST2 antiviral activity in different ways. For example, HIV-1 Vpu blocks hBST2 transit to the cell surface and promotes its endosomal degradation (13), whereas HIV-2 Env sequesters hBST2 within the TGN (12, 14). Importantly, the host-specific adaptation of HIV-1 to overcome hBST2 restriction through the adaptation of Vpu played a major role in the transmission of pandemic HIV-1 group M from chimpanzees to humans, highlighting the importance of BST2 in determining host range (15).
Jaagsiekte sheep retrovirus (JSRV) is an exogenous betaretrovirus that causes ovine pulmonary adenocarcinoma (OPA), a naturally occurring lung cancer of sheep (16–19). The sheep genome harbors at least 27 copies of endogenous retroviruses highly related to JSRV (enJSRVs) (20, 21). enJSRV transcripts and proteins are particularly abundant in the genital tract of the ewe (22, 23), are essential for the reproductive biology of this species (23), and can interfere with the replication of their exogenous counterparts (20, 23–25). To date, one copy of the BST2 gene has been found in most vertebrates. In ruminants, the BST2 gene was duplicated prior to the speciation of sheep and cows (26, 27). Thus, sheep possess two BST2 paralogs, referred to as oBST2A and oBST2B (26). We previously showed that interferon tau (IFNT), the pregnancy recognition protein in ruminants, induces the expression of the two ovine BST2 genes both in vitro and in vivo and that oBST2A blocks cell exit of JSRV and enJSRVs (26). Curiously, oBST2A appeared to be much more efficient than oBST2B at restricting JSRV particle release (26). Importantly, a recent study showed that the bovine BST2B (bBST2B) protein displays Golgi localization and reduces in vitro production of bovine leukemia virus, albeit at a lower level than that of the two bBST2As analyzed (27). In this study, we further investigated the restriction mechanism of oBST2B on JSRV. We highlight several biological properties that distinguish oBST2B from oBST2A and provide data suggesting that oBST2B possesses a distinct antiviral activity which complements the classical tethering restriction provided by oBST2A.
MATERIALS AND METHODS
Plasmids.
pCMV2JS21 expressing the full-length JSRV21 molecular clone has been described previously (19, 28). pCMV3JS21ΔGP is an expression plasmid for JSRV Env (29). pCMV5a-JS21Flag is an expression plasmid for JSRV Env with a Flag epitope fused at its carboxy terminus (Env-CFlag) (30). Plasmids expressing the hemagglutinin (HA)-tagged version of ovine BST2A and BST2B, pCIoBST2A-HA and pCIoBST2B-HA, have also been described previously (26). pcDNA3.1 and pDsRed-Golgi (Ds-Red-Monomer coding sequence fused to the N-terminal 81 amino acids of human beta 1,4-galactosyltransferase) plasmids were purchased from Invitrogen and Clontech, respectively.
Protein alignment and prediction analysis.
oBST2A and oBST2B proteins were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Predictions of putative oBST2A/2B domains and posttranslational modifications were made using the Expasy Proteomics server (http://expasy.org/tools). TMHMM, version 2, and NetNGly1.0 were used to predict potential transmembrane regions and N-linked glycosylation sites, respectively. GPI-SOM was used to predict putative glycosylphosphatidylinositol (GPI) anchor sites.
Cells.
HEK-293T cells were cultured in Dulbecco's modified Eagle medium. Sheep choroid plexus (CPT-Tert) cells were cultured in Iscove's modified Dulbecco's medium (26). All cell lines were supplemented with 10% fetal bovine serum and grown at 37°C, 5% CO2, and 95% humidity.
Transfections, cell lysates, and Western blotting.
To assess the glycosylation status of oBST2, cell lysates were treated with the peptide N-glycosidase F (PNGase F; New England BioLabs). Briefly, 293T cells were transfected in 10-cm plates with 1 μg of pCIoBST2A-HA or pCIoBST2B-HA using a calcium phosphate protocol. At 24 h posttransfection, cells were harvested in 1% NP-40. The cell lysates were then diluted to a final volume of 35 μl containing 200 μg of total proteins before the addition of 10 μl of 5× reaction buffer (250 mM sodium phosphate, pH 7.5) and 2.5 μl of denaturation solution (2% sodium dodecyl sulfate and 1 M 2-mercaptoethanol). Samples were then boiled at 100°C for 5 min, and 2.5 μl of 15% Triton X-100 in phosphate-buffered saline (PBS) (vol/vol) was added, followed by 2 μl of PNGase F (0.01 U) or PBS as a control. All samples were then incubated at 37°C for 6 h. The reaction was stopped by the addition of 4 μl of 4× SDS-PAGE reducing buffer (250 mM Tris-HCl [pH 6.8], 8% SDS, 40% glycerol [vol/vol], 0.008% bromophenol blue, 20% 2-mercaptoethanol) and boiling for 5 min prior to SDS-PAGE/Western blot analysis.
CPT-Tert cells were transfected with increasing amounts (0.25 to 2 μg) of an empty vector (pcDNA3.1) or expression plasmids for oBST2B (with or without an HA epitope tag) in 12-well plates, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A second transfection was performed 24 h later with 5 μg of pCMV2JS21. At 48 h posttransfection, Western blotting was performed on concentrated viral particles and cell lysates as previously described (31). Gag proteins were detected with a rabbit polyclonal serum against the JSRV major capsid protein (CA) (32). JSRV Env was detected using a monoclonal antibody against the surface (SU) domain (33). Ovine BST2B proteins tagged with HA were detected using a mouse monoclonal anti-HA antibody (Covance). Membranes were exposed to the appropriate peroxidase-conjugated secondary antibodies and then developed by chemiluminescence using ECL Plus (Amersham) or SuperSignal West Pico substrate (Thermo Scientific). The Western blot signal for the expression of CA and SU was quantified in the viral pellets using Image Studio Lite software. The values of CA and SU in the absence of oBST2B were arbitrarily assigned a value of 100, and the ratios between SU and CA were calculated. Each experiment (from transfection to Western blotting) was performed independently at least three times. Statistical analyses were performed with two-way analysis of variance (ANOVA).
Immunofluorescence and confocal microscopy.
Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. CPT-Tert cells were plated onto two-well chambered glass slides (Lab-Tek; Nalge Nunc International) and cotransfected with pCMV2JS21 or pCMV5a-JS21Flag (1 μg) and either pDsRed-Golgi, pCIoBST2A-HA or -2B-HA, or pcDNA3.1 as a control plasmid (0.25 μg of each plasmid). In some experiments, as indicated in Results, cells were treated with brefeldin A (Sigma) at a concentration of 200 ng/ml for 90 min before fixation with 5% formaldehyde in PBS at 4°C for 20 min and were immunolabeled for confocal microscopy. Immunofluorescence was performed as described previously (32, 34) using the following antibodies: JSRV SU (1/200), JSRV Gag p23 (1/200), anti-FLAG (1/750; Sigma), TGN46 (1/1,000; Abcam), giantin (1/100; Abcam), p115 (1/100; Abcam), anti-HA (1/100; Covance) or pan-cadherin (1/500; Abcam). Secondary antibodies included Alexa Fluor 488- or Alexa Fluor 546-conjugated goat anti-rabbit and Alexa Fluor 546 donkey anti-mouse antibodies (Life Technologies). Cells were examined using a Leica TCS SP2 confocal microscope. Image analysis was performed using Image-Pro Analyzer 7.0 (Media Cybernetics). Statistical analyses were performed with a χ2 test to compare the intracellular distribution patterns of each protein. Protein colocalization signals were measured using Image-Pro Plus software (Media Cybernetics), and Pearson's correlation coefficient was estimated. Values ranged from −1 to 1, with 0.5 representing the value of the significance threshold. Values are presented as the average number of Pearson's correlation coefficients measured from random fields of at least 50 cells in two independent experiments (± standard error).
Entry assays.
Murine leukemia virus (MLV)-derived pseudoparticles were designed and generated as previously described (35). Briefly, HEK-293T cells stably expressing oBST2B-HA (293T/B-HA) or an empty vector as a control (293T-cont) were cultured in 10-cm plates and transfected by calcium phosphate with three expression plasmids encoding (i) MLV core-packaging components (8 μg); (ii) an MLV retroviral transfer vector harboring the green fluorescent protein (GFP) marker (8 μg); and (iii) the envelope glycoprotein of JSRV (pCMV3JS21ΔGP) (29) (5 μg, 2 μg, or 0.8 μg) or vesicular stomatitis virus (VSV-G) (2 μg) to generate pseudoparticles referred to as JSRVpp-5 μg, JSRVpp-2 μg, JSRVpp-0.8 μg, or VSVpp, respectively. Pseudoparticles with no envelope glycoprotein, termed NoEnvpp, were generated by transfecting only expression plasmids for GFP and MLV core-packaging components. At 36 h posttransfection, cell supernatants were recovered, filtered through a 0.45-μm-pore-size filter and used to infect CPT-Tert cells for 5 h. At 72 h postinfection, cells were analyzed by a FACSCalibur system (BD Biosciences) to quantify the percentage of GFP-positive cells, using the values obtained with NoEnvpp preparations as the threshold. The percentage of cells infected by JSRVpp-5 μg, JSRVpp-2 μg, JSRVpp-0.8 μg, or VSVpp produced in 293T-cont cells was arbitrarily assigned a value of 100% and compared to the percentage of cells infected by the corresponding JSRVpp-5 μg, JSRVpp-2 μg, JSRVpp-0.8 μg, or VSVpp pseudoparticles produced in 293T/B-HA cells. Experiments were performed independently at least three times. Statistical analysis was performed with two-way ANOVA using R software (Comprehensive R Archive Network; http://www.R-project.org). In parallel, pseudoparticles were purified and concentrated by ultracentrifugation at 120,000 × g for 2 h through a 20% sucrose cushion. Viral pellets were then subjected to Western blot analysis using mouse anti-SU (1/10) or anti-VSV-G (P5D4; 1/1,000) antibodies and a rat anti-MLV-CA (R187; 1/1,000) antibody, as described above. The Western blot signals and SU/CA ratios of JSRVpp-2 μg and JSRVpp-0.8 μg were determined as described above. Statistical analyses were carried out using one-way ANOVA to compare the median ratios of JSRVpp-2 μg and JSRVpp-0.8 μg produced in 293T/B-HA cells.
RESULTS
oBST2A and oBST2B have distinct cellular localizations.
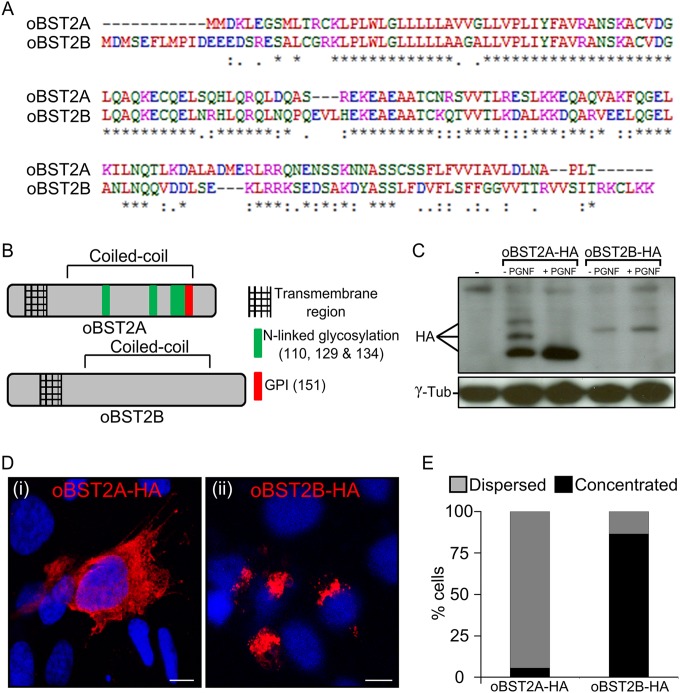
oBST2A and oBST2B share 77.5% identity at the nucleotide level and 48.9% at the amino acid level (Fig. 1A). oBST2A has the typical BST2 topology, including a predicted amino-terminal transmembrane region, an ectodomain with three N-linked glycosylation sites, and a GPI anchor (Fig. 1B). However, oBST2B appears to lack both the predicted GPI anchors and the N-linked glycosylation sites (Fig. 1B). Previously, we generated and characterized oBST2 HA-tagged versions and showed that they are fully functional and comparable to the wild-type oBST2 proteins (26). Thus, to assess the glycosylation status of both oBST2 paralogs, HEK-293T cells were first transfected with plasmids expressing HA-tagged versions of oBST2A (oBST2A-HA) and oBST2B (oBST2B-HA) and were then subjected to endoglycosidase treatment (PNGase F) to remove the carbohydrate chains from N-linked glycosylated proteins. In the absence of PNGase F treatment, oBST2A-HA was detectable as three bands corresponding to the unglycosylated and glycosylated forms of the protein (Fig. 1C). However, we found that PNGase F treatment altered the migration pattern of oBST2A, reducing it to a single band. In contrast, we observed that oBST2B migrated as a single band in either the presence or absence of PNGase F treatment (Fig. 1C). A lack of glycosylation may have a profound impact on oBST2B structure, intracellular localization, and function. Thus, we next investigated whether oBST2A/2B shared a similar intracellular localization. Sheep choroid plexus cells (CPT-Tert) were transfected with oBST2A-HA or oBST2B-HA and subjected to immunofluorescence followed by confocal microscopy. We observed that unlike oBST2A-HA, oBST2B-HA displayed a predominantly concentrated distribution surrounding the nucleus that could be detected throughout the cytoplasm (Fig. 1D and E) (χ2 test, P < 10−3). oBST2B-HA showed a significant degree of colocalization with several Golgi markers, including p115, giantin, and TGN46 (Fig. 2A). Protein colocalization signals were measured using Image-Pro Plus software (Media Cybernetics), and Pearson's correlation coefficient, with values ranging from −1 to 1, where 0.5 represents the value of the significance threshold, were estimated. Pearson's correlation values ranged between 0.6 and 0.7 for Golgi markers (Fig. 2B). In contrast, we did not observe this localization pattern for oBST2A-HA, and Pearson's correlation values were never above 0.3 with any of the Golgi markers (Fig. 2B). In addition, upon treatment with brefeldin A, a drug that fragments and redistributes Golgi-based proteins (36), oBST2B-HA redistributes within the cytoplasm similarly to what we observed for giantin (Fig. 3A and B) (χ2 test, P < 10−3). Altogether, these data show that unlike oBST2A, oBST2B is localized primarily to the Golgi.
FIG 1.
oBST2A and -2B have different biological features. (A) Alignment of the amino acid sequences of the ovine BST2 proteins (oBST2A and -2B). An asterisk (*) indicates an identical amino acid, a period (.) indicates weak similarity, and a colon (:) indicates strong similarity. (B) Schematic diagram illustrating the main structural features and predicted posttranslational modifications of oBST2A and -2B. (C) HEK-293T cells were transiently transfected with expression plasmids for oBST2A-HA or -2B-HA. Cell lysates were harvested and incubated in the presence and absence of PNGase F overnight. Proteins were then separated by SDS-PAGE and visualized by Western blotting using an HA antibody. γ-Tubulin was used as a sample-loading control. (D) Confocal microscopy images of oBST2A-HA and oBST2B-HA in transiently transfected CPT-Tert cells display two different patterns: (i) dispersed within the cytoplasm and cell membrane and (ii) concentrated in a perinuclear region. Scale bars in both panels represent 10 μm. (E) Graph representing the number (%) of cells in which oBST2A-HA- and oBST2B-HA-staining patterns were counted as dispersed or concentrated. At least 75 cells in random fields from two independent experiments were scored.
FIG 2.
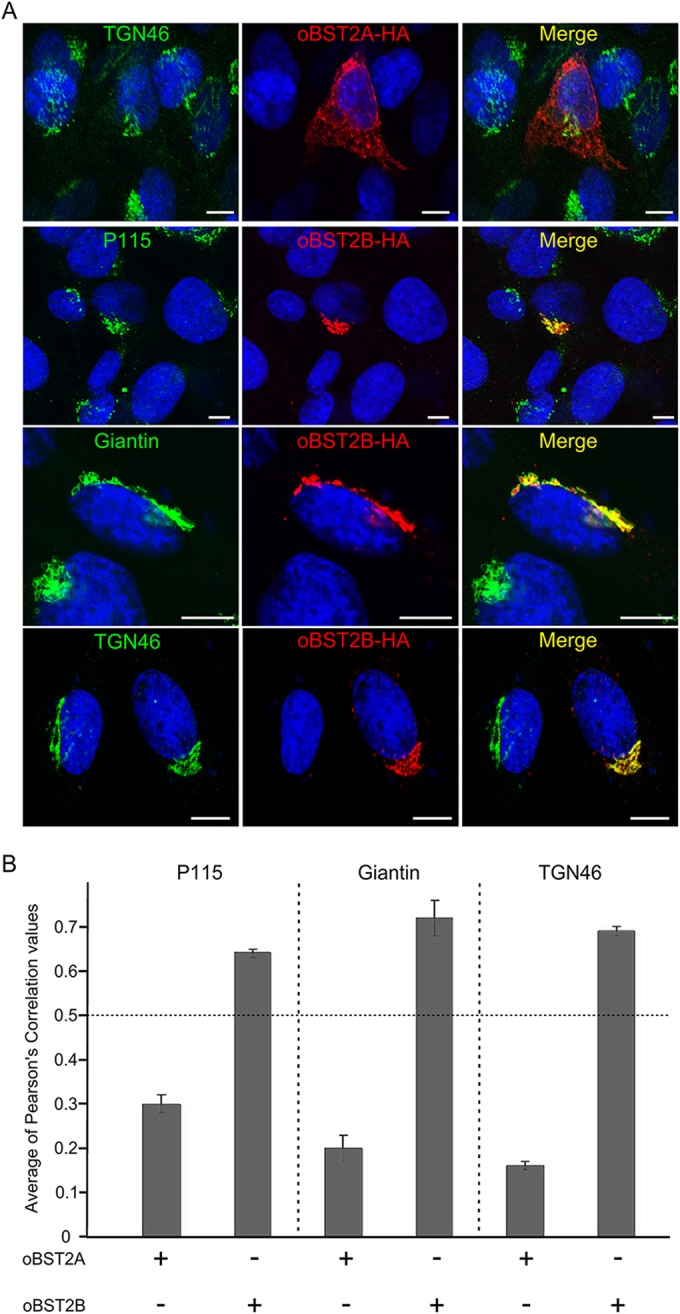
oBST2B localizes to the Golgi apparatus. (A) CPT-Tert cells were transfected with oBST2A-HA or oBST2B-HA and analyzed 18 h after transfection by confocal microscopy using, respectively, antibodies against the HA epitope (to detect oBST2A-HA or oBST2B-HA protein) and the Golgi markers: p115, giantin, TGN46, and appropriate secondary conjugated antibodies. Scale bars in all panels represent 10 μm. (B) Colocalization of oBST2A-HA and oBST2B-HA with Golgi markers was measured in at least 50 cells from two independent experiments with Image-Pro Plus software using Pearson's correlation coefficient. Any values above 0.5 were regarded as representing significant colocalization.
FIG 3.
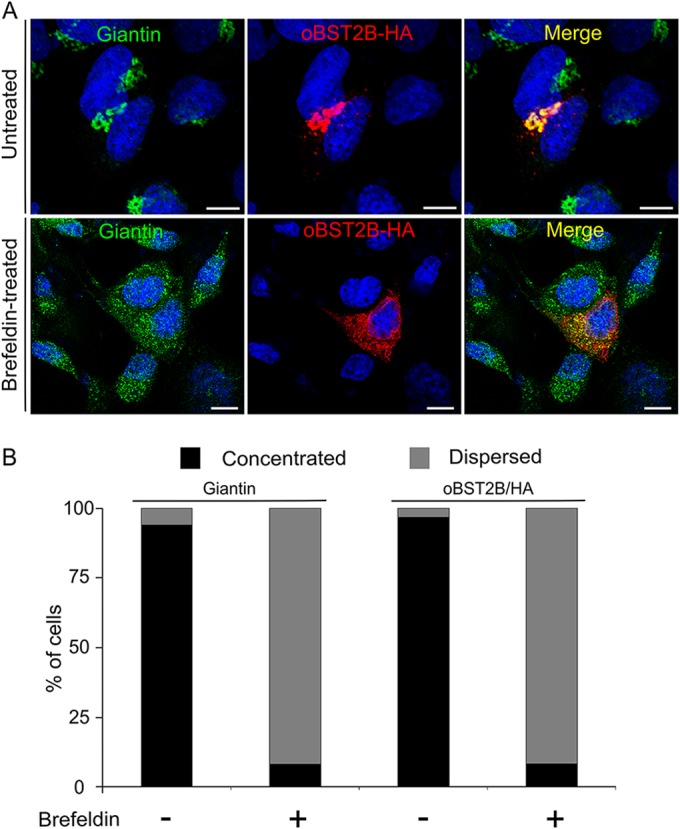
oBST2B localization is altered by treatment with brefeldin A. (A) CPT-Tert cells were transfected with oBST2B-HA expression plasmids. Eighteen hours after transfection, cells were treated or not treated with 200 ng/ml of brefeldin A for 90 min, fixed, and analyzed by confocal microscopy using antibodies to the giantin Golgi marker and the HA epitope as indicated. Two different staining patterns were observed: (i) dispersed within the cytoplasm and (ii) concentrated in a perinuclear region. Scale bars in all panels represent 10 μm. (B) Graph representing the number (%) of cells in which oBST2B-HA and giantin staining were observed to be concentrated as opposed to dispersed. At least 100 cells from two independent experiments were evaluated randomly.
oBST2B redistributes intracellular JSRV Env and Gag proteins.
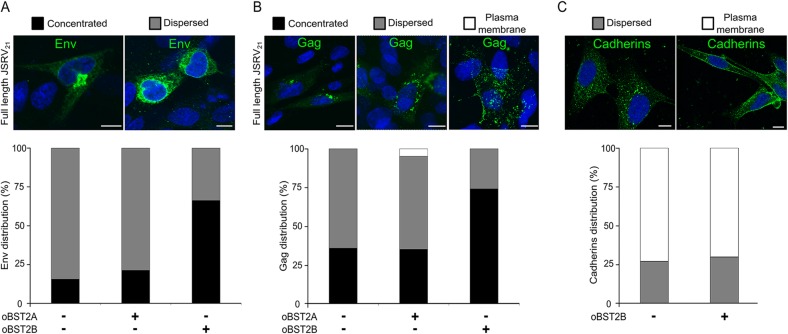
Next, we sought to determine the effect of both oBST2A and oBST2B on the intracellular localization of Env and Gag, the two major structural proteins of JSRV. To this end, the full-length molecular clone JSRV21 was cotransfected with oBST2A-HA or oBST2B-HA expression plasmids in CPT-Tert cells. By confocal microscopy, the distribution of JSRV Env was quantified as either concentrated close to the nucleus or dispersed throughout the cytoplasm (Fig. 4A). While oBST2A had no significant effect on the intracellular Env localization of JSRV (χ2 test, P = 0.1387), a significant increase in the concentrated pattern (from 15.6% up to 66.3%) was observed in the presence of oBST2B (Fig. 4A) (χ2 test, P < 10−3). We next investigated the effects that oBST2A and oBST2B exert on the intracellular distribution of JSRV Gag proteins. We divided the viral Gag localization into three patterns: concentrated, dispersed, and spotted at the plasma membrane (Fig. 4B). As shown in Fig. 4B, oBST2A increased, albeit at a low level (4.7%), the concentration of JSRV Gag at the plasma membrane (χ2 test, P = 0.8521). On the other hand, oBST2B redistributed Gag to a concentrated perinuclear localization (from 35.9% to 74.3%) (χ2 test, P < 10−3) (Fig. 4B). Furthermore, we assessed whether oBST2B had an effect on other cellular proteins, such as the cadherins, that normally traffic through the Golgi before reaching the plasma membrane (37). As shown in Fig. 4C, oBST2B has no major influence on the intracellular localization of cadherins, suggesting that oBST2B does not disrupt the overall Golgi-plasma membrane transport pathways.
FIG 4.
oBST2A and oBST2B both redistribute intracellular Env and Gag. CPT-Tert cells were transfected with the full-length JSRV21 expression plasmid with either an empty plasmid or expression plasmids for oBST2A-HA or oBST2B-HA. CPT-Tert cells were also transfected with an expression plasmid for oBST2B-HA in order to assess the influence of this protein on the intracellular distribution of cellular cadherins. Upon immunostaining with the appropriate antibody, the intracellular distributions of Env (A), Gag (B), and cadherins (C) were scored and counted as concentrated or dispersed or at the plasma membrane using confocal microscopy. Scale bars in all panels represent 10 μm. Graphs represent the number (%) of cells in which the intracellular distribution of Env (A), Gag (B), or cadherins (C) displays a concentrated, dispersed, or plasma membrane staining pattern. At least 100 cells in random fields from two independent experiments were counted.
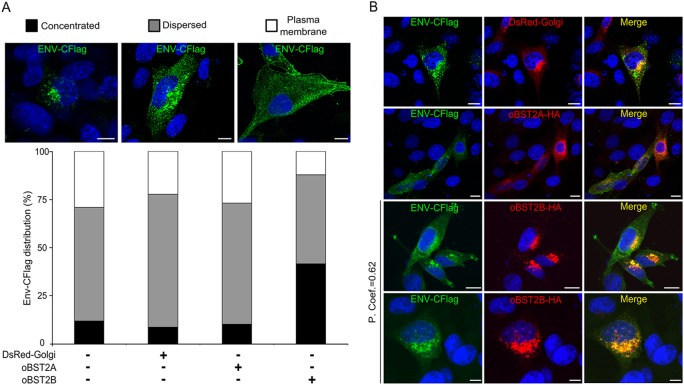
CPT-Tert cells were then transfected with an expression plasmid for JSRV Env tagged with a Flag epitope at the C terminus (30) in the presence or absence of pDsRed-Golgi used as a control or the oBST2A-HA or oBST2B-HA expression plasmid. After immunostaining with a Flag antibody, expression of oBST2B, but not of oBST2A or DsRed-Golgi, resulted in an increased concentrated phenotype of JSRV Env/Flag (χ2 test, P < 10−3) (Fig. 5A). In addition, JSRV Env/Flag colocalizes in a perinuclear region with oBST2B, while it only partially overlaps with oBST2A or DsRed-Golgi proteins (Fig. 5B). Overall, these experiments show that oBST2B specifically impairs the normal trafficking of JSRV Env, which remains in a perinuclear region, most probably in the Golgi apparatus. Likely as a result of altered trafficking of the JSRV Env, the Gag protein also appears to concentrate in the vicinity of the nucleus.
FIG 5.
oBST2B colocalizes with JSRV Env. (A) CPT-Tert cells transfected with the JSRV Env-CFlag-expressing plasmid with either an empty pcDNA3.1 vector, pDsRed-Golgi, pCIoBST2A-HA, or -2B-HA were analyzed by confocal microscopy. The intracellular distribution of Env-CFlag was scored as concentrated, dispersed, or at the plasma membrane using confocal microscopy. Scale bars in all panels represent 10 μm. The graph represents the number (%) of cells in which the intracellular distribution of Env-CFlag displays a concentrated, dispersed, or plasma membrane staining pattern. At least 100 cells in random fields from two independent experiments were counted. (B) Representative pictures of CPT-Tert cells coexpressing Env-CFlag with DsRed-Golgi, oBST2A-HA, or oBST2B-HA. Colocalization of Env-CFlag with oBST2B-HA in a perinuclear region was measured in at least 50 cells from two independent experiments with Image-Pro Plus software using Pearson's correlation coefficient (P. Coef.). Any value above 0.5 was regarded as significant. Scale bars in all panels represent 10 μm.
oBST2B reduces the incorporation of Env into JSRV viral particles.
Given that oBST2B redistributes or sequesters JSRV Env in a perinuclear compartment; we reasoned that oBST2B could hamper Env trafficking and restrict its incorporation into nascent viral particles. Thus, CPT-Tert cells were transiently transfected with increasing amounts of wild-type oBST2B or the oBST2B-HA-tagged expression plasmid and the full-length JSRV21 expression plasmid. Subsequently, Gag (anti-CA) and Env (anti-SU) proteins were analyzed by Western blotting on both total cell lysates and viral pellets. The intensities of the CA and SU staining in the viral pellets were quantified, and the ratio between SU and CA was also calculated for each sample. We found no significant difference (two-way ANOVA, P = 0.74) in the amounts of Gag (CA) released in the supernatants of cells transfected with JSRV21 alone or in the presence of oBST2B or oBST2B-HA (Fig. 6A). However, we observed a decrease in the amount of Env associated with viral particles when JSRV21 was expressed in the presence of oBST2B or its HA-tagged version (Fig. 6B and C). The SU/CA ratio decreased significantly with increasing amounts of oBST2B (or oBST2B-HA) (Fig. 6B and D) (two-way ANOVA, P < 0.05). These data show that oBST2B does not tether JSRV particles to the cell membrane but rather reduces the incorporation of Env.
FIG 6.
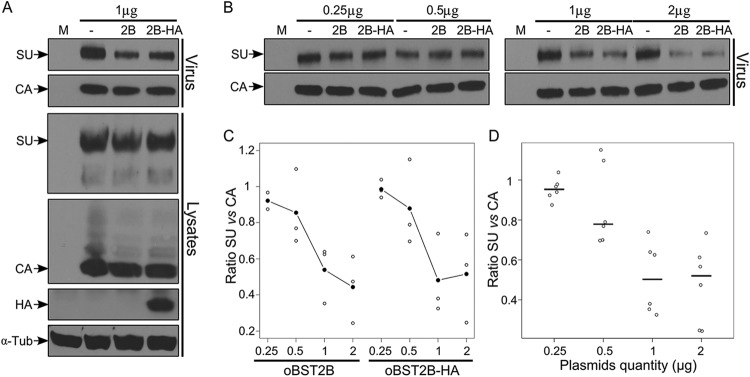
oBST2B reduces the incorporation of JSRV Env into virions. (A) Representative Western blots of concentrated viral particles from supernatants (virus) and cellular extracts (lysates) of mock-CPT-Tert-transfected cells (M) or cells transfected with expression plasmids for full-length JSRV21 and 1 μg of expression plasmids for oBST2B (2B) or oBST2B-HA (2B-HA) or an empty pcDNA3.1 control (−). Blots were incubated with the appropriate antisera as indicated in each panel. (B) Viral-particle release and Env incorporation in the presence of an increasing amount of oBST2B or oBST2B-HA (0.25 μg to 2 μg) were detected with an anti-Gag (CA) and anti-Env (SU) antibodies and quantified by chemifluorescence. (C and D) Values for CA and SU expression were related to values obtained for cells cotransfected with an expression plasmid for JSRV21 and the empty pcDNA3.1 plasmid (taken as 100). Blots represent the ratios of SU/CA expression calculated from three independent experiments. (C) A two-way statistical ANOVA shows that there is no difference in the relative SU/CA ratios between cells transfected with oBST2B- and oBST2B-HA-expressing plasmids (P = 0.74). (D) These ratios are negatively correlated with the quantity of transfected plasmids (P = 0.006). Open circles are ratio values; black circles (C) or horizontal bars (D) represent the mean ratios of the data obtained.
oBST2B reduces the infectivity of MLV-derived pseudotypes.
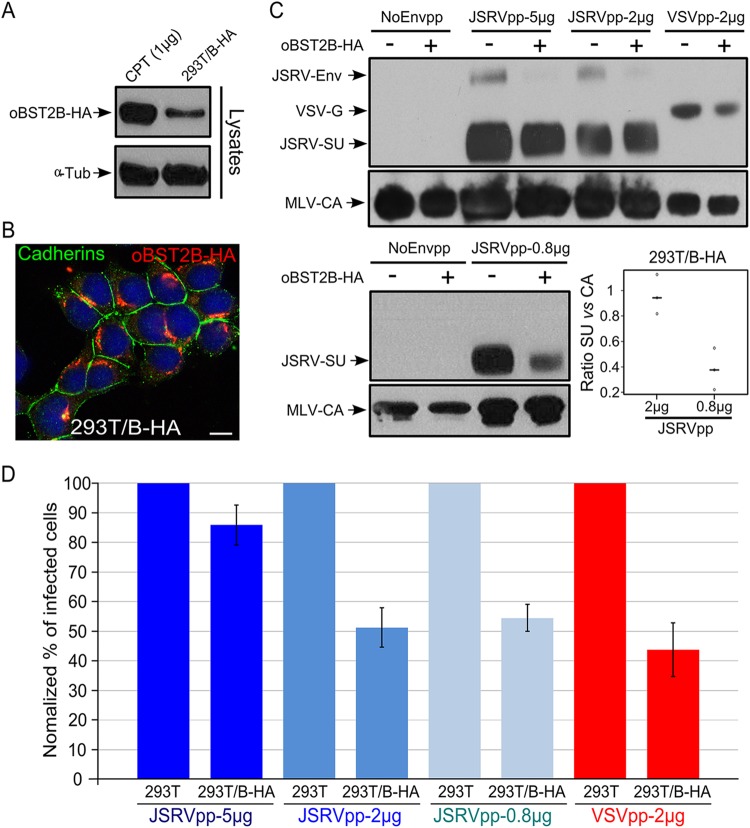
Next, we performed entry assays to determine whether oBST2B was affecting viral infectivity in addition to Env incorporation. Unfortunately, there is no cell culture system supporting abundant replication of JSRV; therefore, we used murine leukemia virus-based retroviral vectors to assess the role of oBST2B in the infectivity of viral particles pseudotyped by the JSRV Env or VSV glycoprotein (VSV-G). To this end, we generated HEK-293T cells stably expressing oBST2B-HA (293T/B-HA) or an empty vector as a control (293T-cont) in order to avoid overexpression of transient transfection systems. As shown in Fig. 7A, 293T/B-HA cells expressed fewer oBST2B-HA proteins than CPT-Tert cells transfected with 1 μg of oBST2B-HA expression plasmid. Moreover, 293T/B-HA cells expressed oBST2B-HA with a concentrated and perinuclear cellular distribution, as was previously observed in CPT-Tert cells (Fig. 7B). Both cell lines were then transfected with plasmids to generate JSRV Env pseudotypes (JSRVpp) and VSV-G pseudotypes (VSVpp). The supernatants of transfected cells were then analyzed by Western blotting and entry assays. As shown in Fig. 7C, JSRVpp-5 μg, JSRVpp-2 μg, and VSVpp-2 μg produced in 293T/B-HA cells harbored less JSRV Env (70 kDa) and VSV-G (57.5 kDa) than those produced in 293T-cont cells, while a less obvious difference was observed for JSRV SU (48 kDa). However, JSRVpp-0.8 μg displayed a clear decrease in SU levels when produced in 293T/B-HA cells (Fig. 7C). Indeed, the SU/CA ratio was significantly reduced in JSRVpp-0.8 μg compared to that in JSRVpp-2 μg (Fig. 7C) (one-way ANOVA, P < 0.05). Importantly, similar amounts of MLV CA (30 kDa) were produced from 293T-cont and 293T/B-HA cell lines in Western blot analyses (Fig. 7C). Notably, both JSRVpp and VSVpp, produced in 293T/B-HA, had reduced infectivity (up to 50%) compared to those produced in 293T-cont cells (Fig. 7D). As expected, the inhibitory effect of oBST2B-HA was stronger on JSRVpp-2 μg and JSRVpp-0.8 μg than on JSRVpp-5 μg, likely due to a dose-dependent effect of oBST2B-HA on Env. In particular, statistical analysis shows that the effect of oBST2B-HA on JSRVpp-2 μg, JSRVpp-0.8 μg, and VSVpp-2 μg was significant (two-way ANOVA, P < 0.05).
FIG 7.
oBST2B reduces infectivity of MLV-based vectors pseudotyped by JSRV Env or VSV-G glycoproteins. (A) Representative Western blots of cellular extracts (lysates) of CPT-Tert transfected with 1 μg of the expression plasmid for oBST2B-HA [CPT (1 μg)] or 293T cells stably expressing oBST2B-HA (293T/B-HA). oBST2B-HA was detected using an anti-HA antibody. α-Tubulin (α-Tub) was used as a sample-loading control. (B) 293T/B-HA cells were analyzed by confocal microscopy using antibodies to the HA epitope and cadherins, as indicated in the panel. The scale bar represents 10 μm. (C) Western blots of concentrated viral particles recovered from the supernatants of 293T or 293T/B-HA cells (indicated by a minus or a plus sign, respectively) transfected with the expression plasmids in the amounts (5 μg, 2 μg, or 0.8 μg) reported above the blots. Blots were incubated with anti-MLV-CA and anti-JSRV-SU or anti-VSV-G antibodies, as indicated. Each experiment was repeated independently three times, and representative Western blots are shown. Viral-particle release and Env incorporation of JSRVpp-2 μg and JSRVpp-0.8 μg produced in 293T and 293T/B-HA cells were assessed by chemifluorescence. Values for CA and SU expression in 293T/B-HA cells were related to values obtained in 293T control cells (arbitrarily set as 100%). The graph represents the SU/CA ratios of JSRVpp-2 μg and JSRVpp-0.8 μg produced in 293T/B-HA cells. Note that there is a statistically significant difference (P = 0.01, calculated by one-way ANOVA) in 293T/B-HA cells that depends on the amount of JSRV Env expression plasmid transfected. Open circles indicate ratio values, while black horizontal bars represent the mean ratio values of the data obtained. (D) NoEnvpp-, JSRVpp-, and VSVpp-infected CPT-Tert cells were analyzed by fluorescence-activated cell sorter (FACS) at 72 h postinfection to quantify the percentage of GFP-positive cells. The percentage of cells infected with JSRVpp and VSVpp produced in control 293T cells was designated 100% of infectivity. Experiments were performed independently at least three times with two biological replicates for each CPT-Tert infection. Bars indicate standard errors.
DISCUSSION
The BST2 gene was duplicated during the ruminant evolution more than 25 million years ago before speciation within the Bovinae subfamily (26). The fixation and maintenance of oBST2B in the ruminant genome strongly suggest a biological function for this gene. In this study, we showed that oBST2B restricts sheep betaretroviruses with a unique mechanism that is distinct from the paralog oBST2A and the much studied human ortholog. In a previous study, we found that oBST2B is less efficient than oBST2A at blocking viral exit, as measured by the release of JSRV Gag proteins in transfected cells (26). Here, we found that unlike oBST2A, oBST2B is not glycosylated and localizes in the Golgi apparatus. N-linked glycosylation is reported to be involved in a number of processes, including protein orientation relative to membranes, protein turnover and regulation of stabilization against denaturation/proteolysis, enhanced solubility, and modulation of immune responses (38). Thus, a lack of glycosylation may have a profound impact on oBST2B structure, intracellular localization, and, in turn, function. Importantly, we found that oBST2B impairs the normal trafficking of Env to the cell membrane and sequesters Env proteins within the Golgi apparatus. Additionally, oBST2B does not seem to impair the global Golgi-plasma membrane trafficking, as it has no influence on the cellular distribution of cadherins. Future experiments will determine whether oBST2B disturbs one particular flow of protein cargo or interacts (directly or indirectly) with JSRV Env. oBST2A slightly increases the presence of Gag proteins at the plasma membrane, similar to what is expected for the classic tethering model with other BST2 orthologs that retain virions at the cell surface. In contrast, oBST2B induces Gag accumulation in a perinuclear region. Previous studies have shown that the Gag proteins of betaretroviruses such as Mason-Pfizer monkey retrovirus (M-PMV) and JSRV target the pericentriolar region in a dynein- and microtubule-dependent fashion (34, 39, 40). The assembled viral particles then traffic to the cell membrane by a mechanism that is influenced by the recycling endosomes (34) and, at least for M-PMV, by the viral Env protein (39). Therefore, the absence or strong depletion of Env at the plasma membrane and, subsequently, in recycling endosomes could influence the intracellular trafficking of the newly formed virions and lead to the accumulation of Gag proteins in a pericentrosomal area. Overall, these data suggest that the oBST2 paralogs exert different effects over the intracellular distribution of the two JSRV structural proteins, Gag and Env, suggesting that they may have distinct antiviral mechanisms. We previously demonstrated that oBST2B blocks viral exit less efficiently than oBST2A and only following transient transfections with proportionally more than twice the maximal amount of oBST2B expression plasmid used in this study (26). Betaretroviral particles can still exit from the cells in the absence of viral Env, although it is expected that this would occur at a lower rate than in its presence (39). Therefore, the sequestration of Env within the Golgi could explain the weak effect of oBST2B on JSRV particle release (detected as released Gag in supernatant of transfected cells) that we previously observed in vitro (26). However, it is important to mention that oBST2s are IFN-induced proteins (26) that could be highly expressed in vivo, leading to a strong viral restriction potentially able to reduce JSRV particle release. Indeed, sequestration of the Env glycoproteins within the Golgi by oBST2B could be a very efficient way to prevent exit of infectious virions from cells. In addition, our data show that incorporation of JSRV Env into viral particles is hampered in cells expressing oBST2B. Thus, while the restriction of oBST2B on viral particle release appears to be only moderate and evident only when large amounts of oBST2B are expressed (26), the lack of Env incorporation from the viral surface imposed by oBST2B will reduce viral infectivity. Indeed, using a murine leukemia virus (MLV)-based pseudotyping assay, we observed a significant decrease in the infectivity of viruses pseudotyped with JSRV Env (JSRVpp-2 μg and JSRVpp-0.8 μg) or VSV envelope glycoproteins (VSVpp-2 μg) and produced in 293T cells constitutively expressing oBST2B-HA. In Western blots, significant differences in the amounts of JSRV SU incorporated into viral particles produced in 293T-cont and 293T/B-HA cells were observed when a small amount (0.8 μg) of JSRV Env expression plasmid was transfected, whereas the differences were less evident with a large amount (5 μg or 2 μg) of viral glycoprotein expression plasmid. Of note, the entry assays presented in this study were performed in 293T cells stably expressing oBST2B-HA protein whose level is lower than the one produced in cotransfection experiments. Therefore, in 293T/B-HA cells, a strong intracellular expression level of JSRV ENV or VSV-G may partially overcome the restriction of oBST2B. Our data demonstrate that oBST2B activity is not restricted to JSRV (Retroviridae family) but can also function on the glycoproteins of VSV, which belongs to the Rhabdoviridae family. Recently, Takeda et al. (2012) showed that the antiviral activity of bBST2B against VSV was weaker than that of bBST2A (27); however, VSV may have evolved specific countermeasures to overcome bBST2B antiviral activity which is likely absent in the pseudotyping assays used here or that is not effective against ovine BST2. Similarly, even though oBST2A and -2B exert antiviral activity against JSRV, we cannot completely rule out the presence of a BST2 antagonist within the JSRV genome, and it could be interesting to assess oBST2's antiviral activities on a betaretrovirus unrelated to sheep, such as M-PMV. hBST2 has broad antiviral activity, and several studies have found this protein to block not only HIV but also several other retroviruses belonging to different genera, including Alpharetrovirus, Betaretrovirus, Deltaretrovirus, and Spumavirus (11). Importantly, hBST2 has been found to restrict replication of many enveloped viruses, including filoviruses and arenaviruses (11, 41–43). Therefore, it will also be interesting to determine if oBST2B and/or oBST2A specifically displays broad antiviral activity against other pathogenic viruses.
JSRV causes lung cancer in sheep by a unique mechanism where its Env acts as a dominant oncogene both in vitro and in vivo (17, 44). Indeed, JSRV Env alone can transform various cell lines in vitro, and it is sufficient to induce lung tumors in lambs and mice (16, 19, 33, 45, 46). Given that oBST2B sequesters JSRV Env within the TGN, we can speculate that it could also modulate Env oncogenic activity. On the other hand, expression of enJSRV Env is absolutely necessary for conceptus development and placental morphogenesis in sheep (23). Interestingly, there is a physical separation of BST2 and enJSRV Env expression within the uterus: BST2 is expressed in the stroma of the uterus, while enJSRV Env is expressed in the luminal and glandular epithelia (26). Taken together, these observations further support the role of BST2 in modulating JSRV Env trafficking within the cell, given that BST2 expression is removed from sites where enJSRV activity is fundamental for sheep reproductive biology. Nonetheless, in vivo, oBST2B may have a major protective effect against JSRV and other viruses of ruminants by reducing their infectivity.
In summary, our study highlights that oBST2B displays a novel antiviral activity acting on the viral envelope glycoprotein, reducing its incorporation into virions and, as a consequence, compromising its infectivity, a mechanism that is distinct but complementary to classical virion tethering.
ACKNOWLEDGMENTS
This study was funded by the Ecole Pratique des Hautes Etudes, the Institut National de la Recherche Agronomique, the University of Lyon 1, the Rhône-Alpes region, and the Wellcome Trust.
We thank Marie-Pierre Confort for her technical help, and we acknowledge the contribution of the Confocal Microscopy and Flow Cytometry Platforms of SFR BioSciences Gerland Lyon Sud (UMS3444/US8). We also thank Alessia Armezzani for revising the manuscript and members of our laboratories for useful suggestions.
REFERENCES
- 1.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 3.Neil SJ. 2013. The antiviral activities of tetherin. Curr Top Microbiol Immunol 371:67–104. doi: 10.1007/978-3-642-37765-5_3. [DOI] [PubMed] [Google Scholar]
- 4.Weinelt J, Neil SJ. 2014. Differential sensitivities of tetherin isoforms to counteraction by primate lentiviruses. J Virol 88:5845–5858. doi: 10.1128/JVI.03818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocka LJ, Bates P. 2012. Identification of alternatively translated Tetherin isoforms with differing antiviral and signaling activities. PLoS Pathog 8:e1002931. doi: 10.1371/journal.ppat.1002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ. 2012. Innate sensing of HIV-1 assembly by Tetherin induces NFkappaB-dependent proinflammatory responses. Cell Host Microbe 12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tokarev A, Suarez M, Kwan W, Fitzpatrick K, Singh R, Guatelli J. 2013. Stimulation of NF-kappaB activity by the HIV restriction factor BST2. J Virol 87:2046–2057. doi: 10.1128/JVI.02272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. 2003. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 10.Venkatesh S, Bieniasz PD. 2013. Mechanism of HIV-1 virion entrapment by tetherin. PLoS Pathog 9:e1003483. doi: 10.1371/journal.ppat.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol 83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Tortorec A, Willey S, Neil SJ. 2011. Antiviral inhibition of enveloped virus release by tetherin/BST-2: action and counteraction. Viruses 3:520–540. doi: 10.3390/v3050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arias JF, Iwabu Y, Tokunaga K. 2012. Sites of action of HIV-1 Vpu in BST-2/tetherin downregulation. Curr HIV Res 10:283–291. doi: 10.2174/157016212800792423. [DOI] [PubMed] [Google Scholar]
- 14.Gupta RK, Mlcochova P, Pelchen-Matthews A, Petit SJ, Mattiuzzo G, Pillay D, Takeuchi Y, Marsh M, Towers GJ. 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc Natl Acad Sci U S A 106:20889–20894. doi: 10.1073/pnas.0907075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murgia C, Caporale M, Ceesay O, Di Francesco G, Ferri N, Varasano V, de las Heras M, Palmarini M. 2011. Lung adenocarcinoma originates from retrovirus infection of proliferating type 2 pneumocytes during pulmonary post-natal development or tissue repair. PLoS Pathog 7:e1002014. doi: 10.1371/journal.ppat.1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmarini M, Cousens C, Dalziel RG, Bai J, Stedman K, DeMartini JC, Sharp JM. 1996. The exogenous form of Jaagsiekte retrovirus is specifically associated with a contagious lung cancer of sheep. J Virol 70:1618–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmarini M, Fan H. 2001. Retrovirus-induced ovine pulmonary adenocarcinoma, an animal model for lung cancer. J Natl Cancer Inst 93:1603–1614. doi: 10.1093/jnci/93.21.1603. [DOI] [PubMed] [Google Scholar]
- 19.Palmarini M, Sharp JM, de las Heras M, Fan H. 1999. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J Virol 73:6964–6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnaud F, Caporale M, Varela M, Biek R, Chessa B, Alberti A, Golder M, Mura M, Zhang YP, Yu L, Pereira F, Demartini JC, Leymaster K, Spencer TE, Palmarini M. 2007. A paradigm for virus-host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog 3:e170. doi: 10.1371/journal.ppat.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chessa B, Pereira F, Arnaud F, Amorim A, Goyache F, Mainland I, Kao RR, Pemberton JM, Beraldi D, Stear MJ, Alberti A, Pittau M, Iannuzzi L, Banabazi MH, Kazwala RR, Zhang YP, Arranz JJ, Ali BA, Wang Z, Uzun M, Dione MM, Olsaker I, Holm LE, Saarma U, Ahmad S, Marzanov N, Eythorsdottir E, Holland MJ, Ajmone-Marsan P, Bruford MW, Kantanen J, Spencer TE, Palmarini M. 2009. Revealing the history of sheep domestication using retrovirus integrations. Science 324:532–536. doi: 10.1126/science.1170587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black SG, Arnaud F, Burghardt RC, Satterfield MC, Fleming JA, Long CR, Hanna C, Murphy L, Biek R, Palmarini M, Spencer TE. 2010. Viral particles of endogenous betaretroviruses are released in the sheep uterus and infect the conceptus trophectoderm in a transspecies embryo transfer model. J Virol 84:9078–9085. doi: 10.1128/JVI.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE. 2006. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci U S A 103:14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmarini M, Hallwirth C, York D, Murgia C, de Oliveira T, Spencer T, Fan H. 2000. Molecular cloning and functional analysis of three type D endogenous retroviruses of sheep reveal a different cell tropism from that of the highly related exogenous jaagsiekte sheep retrovirus. J Virol 74:8065–8076. doi: 10.1128/JVI.74.17.8065-8076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mura M, Murcia P, Caporale M, Spencer TE, Nagashima K, Rein A, Palmarini M. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci U S A 101:11117–11122. doi: 10.1073/pnas.0402877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnaud F, Black SG, Murphy L, Griffiths DJ, Neil SJ, Spencer TE, Palmarini M. 2010. Interplay between ovine bone marrow stromal cell antigen 2/tetherin and endogenous retroviruses. J Virol 84:4415–4425. doi: 10.1128/JVI.00029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeda E, Nakagawa S, Nakaya Y, Tanaka A, Miyazawa T, Yasuda J. 2012. Identification and functional analysis of three isoforms of bovine BST-2. PLoS One 7:e41483. doi: 10.1371/journal.pone.0041483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmarini M, Sharp JM, Lee C, Fan H. 1999. In vitro infection of ovine cell lines by Jaagsiekte sheep retrovirus. J Virol 73:10070–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda N, Palmarini M, Murgia C, Fan H. 2001. Direct transformation of rodent fibroblasts by jaagsiekte sheep retrovirus DNA. Proc Natl Acad Sci U S A 98:4449–4454. doi: 10.1073/pnas.071547598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zavala G, Pretto C, Chow YH, Jones L, Alberti A, Grego E, De las Heras M, Palmarini M. 2003. Relevance of Akt phosphorylation in cell transformation induced by Jaagsiekte sheep retrovirus. Virology 312:95–105. doi: 10.1016/S0042-6822(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 31.Varela M, Chow YH, Sturkie C, Murcia P, Palmarini M. 2006. Association of RON tyrosine kinase with the Jaagsiekte sheep retrovirus envelope glycoprotein. Virology 350:347–357. doi: 10.1016/j.virol.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 32.Murcia PR, Arnaud F, Palmarini M. 2007. The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. J Virol 81:1762–1772. doi: 10.1128/JVI.01859-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wootton SK, Metzger MJ, Hudkins KL, Alpers CE, York D, DeMartini JC, Miller AD. 2006. Lung cancer induced in mice by the envelope protein of jaagsiekte sheep retrovirus (JSRV) closely resembles lung cancer in sheep infected with JSRV. Retrovirology 3:94. doi: 10.1186/1742-4690-3-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnaud F, Murcia PR, Palmarini M. 2007. Mechanisms of late restriction induced by an endogenous retrovirus. J Virol 81:11441–11451. doi: 10.1128/JVI.01214-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douam F, Dao Thi VL, Maurin G, Fresquet J, Mompelat D, Zeisel MB, Baumert TF, Cosset FL, Lavillette D. 2014. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology 59:776–788. doi: 10.1002/hep.26733. [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. 1988. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem 263:18545–18552. [PubMed] [Google Scholar]
- 37.Yap AS, Crampton MS, Hardin J. 2007. Making and breaking contacts: the cellular biology of cadherin regulation. Curr Opin Cell Biol 19:508–514. doi: 10.1016/j.ceb.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helenius A, Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 39.Sfakianos JN, Hunter E. 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4:671–680. doi: 10.1034/j.1600-0854.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- 40.Sfakianos JN, LaCasse RA, Hunter E. 2003. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic 4:660–670. doi: 10.1034/j.1600-0854.2003.00125.x. [DOI] [PubMed] [Google Scholar]
- 41.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. 2009. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A 106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. 2009. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J Virol 83:9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakuma T, Noda T, Urata S, Kawaoka Y, Yasuda J. 2009. Inhibition of Lassa and Marburg virus production by tetherin. J Virol 83:2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmarini M, Fan H. 2003. Molecular biology of jaagsiekte sheep retrovirus. Curr Top Microbiol Immunol 275:81–115. doi: 10.1007/978-3-642-55638-8_4. [DOI] [PubMed] [Google Scholar]
- 45.Caporale M, Cousens C, Centorame P, Pinoni C, De las Heras M, Palmarini M. 2006. Expression of the jaagsiekte sheep retrovirus envelope glycoprotein is sufficient to induce lung tumors in sheep. J Virol 80:8030–8037. doi: 10.1128/JVI.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wootton SK, Halbert CL, Miller AD. 2005. Sheep retrovirus structural protein induces lung tumours. Nature 434:904–907. doi: 10.1038/nature03492. [DOI] [PMC free article] [PubMed] [Google Scholar]