ABSTRACT
Flaviviruses undergo large conformational changes during their life cycle. Under acidic pH conditions, the mature virus forms transient fusogenic trimers of E glycoproteins that engage the lipid membrane in host cells to initiate viral fusion and nucleocapsid penetration into the cytoplasm. However, the dynamic nature of the fusogenic trimer has made the determination of its structure a challenge. Here we have used Fab fragments of the neutralizing antibody DV2-E104 to stop the conformational change of dengue virus at an intermediate stage of the fusion process. Using cryo-electron microscopy, we show that in this intermediate stage, the E glycoproteins form 60 trimers that are similar to the predicted “open” fusogenic trimer.
IMPORTANCE The structure of a dengue virus has been captured during the formation of fusogenic trimers. This was accomplished by binding Fab fragments of the neutralizing antibody DV2-E104 to the virus at neutral pH and then decreasing the pH to 5.5. These trimers had an “open” conformation, which is distinct from the “closed” conformation of postfusion trimers. Only two of the three E proteins within each spike are bound by a Fab molecule at domain III. Steric hindrance around the icosahedral 3-fold axes prevents binding of a Fab to the third domain III of each E protein spike. Binding of the DV2-E104 Fab fragments prevents domain III from rotating by about 130° to the postfusion orientation and thus precludes the stem region from “zipping” together the three E proteins along the domain II boundaries into the “closed” postfusion conformation, thus inhibiting fusion.
INTRODUCTION
Enveloped viruses enter cells by fusing their lipid membrane with specific membranes of the host cell. In the endosome, this process is often triggered by the acidic environment, which promotes virion glycoproteins to form oligomeric (usually trimeric) structures in which each monomer within the oligomer has a hydrophobic peptide at its extremity. Trimeric fusogenic structures have been identified in numerous viruses that have been classified into at least three types based on the nature of the fusion peptide (1, 2). In many cases, the structure of the mature virus and the postfusion structure are known. Based on these results, different fusion mechanisms have been proposed (3–8). Although the prefusion fusogenic structures of some viruses can be created by low-pH or particular lipid environments, it has been difficult to define the prefusion fusogenic state of flaviviruses because of their instability and propensity to fuse with neighboring virions.
Dengue virus (DENV) is a member of the Flaviviridae family of positive-stranded RNA viruses, which include arthropod-borne human pathogens such as West Nile, Japanese encephalitis, and yellow fever viruses. Each year, approximately 390 million people become infected by DENV, resulting in about 20,000 deaths (9). DENV infections cause a spectrum of clinical diseases ranging from acute dengue fever to severe, potentially fatal, dengue hemorrhagic fever and shock syndrome (10). Currently, there are no approved antiviral drugs or licensed vaccines available to reduce the disease burden of DENV infections (11).
DENV has an 11-kb genome that encodes an envelope glycoprotein (E), a precursor membrane protein (prM), a capsid protein, and seven nonstructural proteins (12). The E protein has three ectodomains (DI, DII, and DIII), a stem region, and a transmembrane region. DI and DIII have β-barrel structures, whereas DII has a long, finger-like domain that contains a highly conserved fusion peptide at its distal end (13, 14). The hinge angle between DI and DII varies, depending on whether the virus is immature, mature, or in a postfusion state (15). The stem region lies flat on the viral membrane and connects the ectodomains and transmembrane anchor (16). In the postfusion “closed” trimer, the stem region relocates from the viral membrane surface to the groove between adjacent DII domains (17, 18). This conformational change has been likened to a “zipper” stabilizing the “closed” form.
Under acidic pH conditions, the ectodomain of DENV E protein forms a postfusion “closed” trimer after inserting its fusion loop into a lipid membrane (5). In this postfusion conformation, the DII and DIII domains are rotated by about 30° and 70°, respectively, relative to DI compared with the E protein monomer in the dimer of the mature, smooth-surfaced virus at neutral pH (4, 13, 15). The stem region of E intercalates into the intermonomer groove in the postfusion trimer (18). An “open” fusion intermediate structure was predicted (18) in which the hinge angle between the DII and DI domains is similar to that of the dimer in the mature virus. This would result in a greater separation of the DII domains compared to the “closed” trimer.
In this study, we utilize Fab fragments of a neutralizing antibody (DV2-104) (19) against DENV serotype 2 (DENV-2) E protein, a class II fusion protein, to trap the protein in a structure that was previously predicted to be a fusogenic trimer.
MATERIALS AND METHODS
DENV propagation and purification.
DENV-2 strain 16681 was propagated in C6/36 Aedes albopictus mosquito cells at 28°C and purified by gradient ultracentrifugation as described previously (4).
Antibody production and Fab fragmentation.
DV2-104 is an anti-DENV-2 monoclonal antibody (MAb) that recognizes an epitope on the C-C′ loop in DIII of the E protein (19). Purified IgG was generated after protein A Sepharose affinity purification of hybridoma supernatants. Fab fragments were generated after papain digestion and purified by sequential protein A Sepharose and Superdex 75 16/60 size exclusion chromatography.
Plaque assay.
DENV-2 in NTE buffer (120 mM NaCl, 20 mM Tris-HCl, and 1 mM EDTA [pH 8.0]) was incubated with Fab fragments of DV2-E104 (100 μg/ml) for 40 min at 4 and 35°C. Two control samples with DENV-2 in the absence of Fab fragments were incubated in parallel. The samples were then buffer exchanged and centrifuged five times in 100-kDa-cutoff centrifuge tubes to remove unbound Fab fragments. The samples were then added to BHK21 cells, and a standard plaque assay was performed as described previously (19).
Cryo-electron microscopy data collection and three-dimensional reconstruction.
DENV-2 and Fab fragments of DV2-E104 were incubated together in NTE buffer for 1 h at 35°C. Acidic buffer (0.1 M morpholineethanesulfonic acid [MES]-hydrogen acetate [HAc] [pH 4.5]) was then added to a final pH of 5.5. After incubating for 5 min, the samples were placed on grids and frozen. Cryo-electron microscopy (cryo-EM) images of virions were acquired using the FEI Titan Krios operated at 300 kV with a dose of ∼24 electrons per Å2 on each image. The spiky particles were boxed using the program e2boxer.py. The contrast transfer function on the images was corrected by flipping the phase. A reference free classification and class averages were calculated using the program refine2d.py. Ten initial models were created by the program e2initialmodel.py, assuming icosahedral symmetry (20). The three models with the best scores were used as a starting model and then refined with the whole data set using the program refine. After ∼16 cycles, the three reconstructions converged to the same structure as shown by visual comparison (Fig. 1A). In addition, a different structure, the immature dengue virus structure low pass filtered to 60-Å resolution, was used as a starting model. The correlation coefficient between this immature structure and the final structure of the low-pH Fab-DENV complex was 0.5 at a resolution of about 180 Å. This reconstruction also converged to the same structure after 30 cycles of refinement as judged by a Fourier shell coefficient of 0.5 at a resolution of about 30 Å (Fig. 1A). The original data, consisting of 1,048 particles, was randomly divided into two parts, and each was refined independently starting with the acidic pH structure low pass filtered to 80-Å resolution. After 16 iterations, the final Fourier shell coefficient between these two independent reconstructions was 0.143 at 26-Å resolution (Fig. 1B). Finally, the two half data sets were recombined, and a total of 528 particles were used to calculate the reconstruction shown in Fig. 2A.
FIG 1.
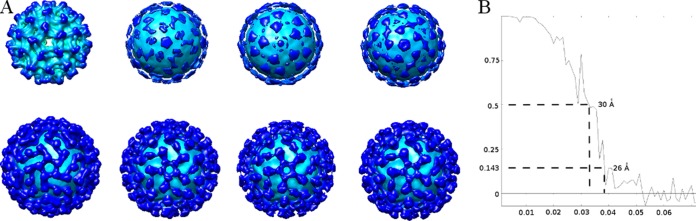
Cryo-EM reconstruction. (A) Initial model and the corresponding results after refinement. The top row from left to right shows immature DENV and three initial models created by EMAN2. The bottom row from left to right shows the refined results corresponding to the initial models in the top row. (B) Fourier shell coefficient (FSC) between two reconstructions refined independently from two independent data sets. Based on the 0.143 standard, the resolution of the reconstruction was about 26 Å. The x axis shows 1/resolution in Å−1; the y axis shows the FSC value.
FIG 2.
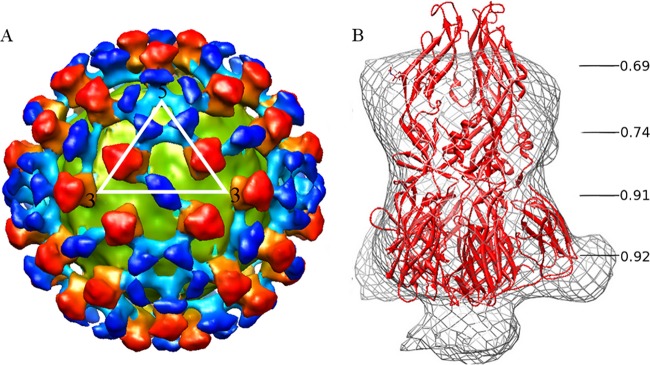
Three-dimensional cryo-EM reconstruction of DENV-2 and Fab fragments of DV2-E104 complex at pH 5.5. (A) The virus membrane is colored yellow-green, the densities corresponding to E trimers are colored red, and the densities corresponding to the Fab molecules are colored blue. Each color has greater intensity at larger radial distances from the viral center. (B) Postfusion crystallographic trimer coordinates of the E protein (5) were fitted into the spike densities of the cryo-EM complex. The correlation coefficients between the parts of the structure and the corresponding cryo-EM spike density are shown for different radial distances along the 3-fold axis of the spike.
Fitting of the complex structure.
The program EMfit (21) was used to fit the crystallographic structures into the cryo-EM electron density map. The structure of the postfusion trimer, represented by C-α atoms only, was fitted into the spike density. A “difference” map was then calculated by setting to zero all density within 6 Å of every fitted C-α atom in DI and DII. The Fab-DIII complex structure (22) (PDB accession no. 4FFZ) was then fitted into the “difference” map.
Cells.
Baby hamster kidney cells (BHK21-15) were cultured at 37°C and 5% CO2 in 1× high-glucose, l-glutamine-enriched Dulbecco modified Eagle medium (DMEM) (Gibco) with 10% fetal bovine serum (FBS), penicillin (100 U/ml), streptomycin (100 μg/ml), 10 mM HEPES, and 100 μM nonessential amino acids. C6/36 cells were cultured at 30°C and 5% CO2 in minimal essential medium (MEM) (Gibco) with 10% FBS, 7.5% sodium bicarbonate, penicillin (100 U/ml), streptomycin (100 μg/ml), 2 mM glutamine, and 100 μM nonessential amino acids. For virus production, HEPES (25 mM, pH 7.3) was added to the cell culture medium to maintain a neutral pH.
Virus growth and DiD labeling.
DENV-2 strain 16681 was propagated on C6/36 cells and purified by ultracentrifugation as described previously (23). Purified DENV particles were labeled with the lipophilic fluorescent probe 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD) (Invitrogen), according to a published procedure (23).
Binding and fusion assay.
BHK21-15 cells (20,000 cells/well) were seeded in chambered LabTek II slides (Nunc) 1 day before infection. Prior to the experiment, the cells were washed three times with cold serum-free, phenol red-free MEM (Gibco). Then, cold phenol red-free MEM containing 1% glucose was added, and incubation was continued for 10 min at 4°C. Subsequently, DiD-labeled DENV particles were added to the cells, and viral adsorption was allowed for 20 min at 4°C. Unbound virus particles were removed by washing, and increasing concentrations of MAb DV2-E104 dissolved in MEM containing 1% glucose were then added to the wells. Incubation was continued for 20 min at 4°C. Next, the medium was removed and replaced with fresh MEM containing 1% glucose at 4°C for the binding assay and 37°C for the fusion assay. For the binding assay, cells were directly used for microscopy. For the fusion assay, cells were further incubated for 30 min at 37°C to allow virus entry. In both assays, 20 snapshots of randomly selected fields were taken using differential interference contrast (DIC) optics and a 635-nm laser to visualize the DiD signal in a Leica Biosystems 6000B microscope. Images were analyzed with ImageJ using an in-house macro, as described previously (23). Anti-keyhole limpet hemocyanin (anti-KLH) antibody MAb0031 (R&D Systems) was used as an isotype control at a concentration of 4 μg/ml. As a negative control in the fusion assay, 50 mM NH4Cl was added to all steps of the assay to prevent endosomal membrane fusion (23). The binding and fusion assay data were normalized to 100% on the basis of the value for the no-antibody control. Moreover, in the analysis of the fusion assay, the background fluorescence level found for the NH4Cl control was set at 0% of fusion.
Low-pH native DENV virus.
A drop of concentrated (∼1 mg/ml, ∼1011 PFU per ml), purified DENV particles at pH 8.0 were placed on an ∼3-nm-thick carbon film on a transmission electron microscopy grid. A filter paper was used to blot excess buffer after keeping the drop on the grid for 1 min. A drop of an acidic buffer maintained at a different pH was immediately added to the grid after blotting. The samples were incubated for 5 min, and then a second filter paper was used to blot the excess buffer. The EM grid was then frozen immediately by plunging into liquid ethane. The frozen virions were examined using an FEI Titan Krios transmission electron microscope. Cryo-EM images were taken with a total dose of ∼40 electrons per Å2 on each image with a defocus of ∼5 μm. The individual spike densities were boxed and classified by EMAN program (24) refine2d.py.
Protein structure accession numbers.
The electron density map of the DENV-Fab DV2-E104 complex structure has been deposited in the Electron Microscopy Data Bank (EMDB accession no. EMD-6146), and the corresponding fitted atomic coordinates have been deposited in the Protein Data Bank (PDB accession no. 3J8D).
RESULTS AND DISCUSSION
The structure of DENV complexed with Fab E104 at pH 5.5.
Using yeast surface display and site-directed mutagenesis, the epitope of the neutralizing DV2-E104 antibody was localized to the C-C′ loop on DIII of the E protein (19). However, this epitope is not exposed on the surface of the smooth DENV structure. Consistent with this observation, cryo-EM studies showed that Fab fragments of DV2-E104 did not bind to DENV-2 at 4°C, although they could bind when incubated with the virus at 35°C and pH 7.8 for 1 h, conditions that are known to result in significant conformational changes of the mature virus (25, 26). The size of this complex was larger than the size of the high-temperature “bumpy” virus alone, confirming the presence of bound Fab. An attempt to perform a three-dimensional reconstruction from cryo-EM data was unsuccessful, possibly because of the lack of homogeneity of the Fab-virus complex. However, a 26-Å resolution, cryo-EM reconstruction of the complex of DENV-2 and DV2-E104 Fab fragments at 35°C was accomplished when the pH was lowered to 5.5. These temperature-dependent results were confirmed by plaque assays. Preincubation of Fab fragments of DV2-E104 with DENV at 35°C reduced the number of plaques by about 90%, whereas preincubation at 4°C reduced infection by only about 5%.
The structure of the low-pH virus-Fab fragment complex (Fig. 2A) consisted of 60 icosahedrally related spike densities representing trimeric E protein complexes on the surface of a spherical, 410-Å-diameter shell that represented the viral lipid membrane. The corresponding lipid bilayer in the immature and mature viruses had an outer diameter of 400 Å (27) and 410 Å (8, 16), respectively. The spike has an approximately triangular cross-section and extends about 110 Å beyond the lipid envelope, making the external diameter of the Fab-virus complex about 630 Å. This is larger than the 600-Å surface of the immature virus and the 500-Å surface of the mature virus. The spike is 40 Å wide on average and connects to the spherical lipid bilayer density by an approximately 25-Å-long neck density.
In addition to the spike densities, there is additional density near the icosahedral 5-fold and 2-fold axes (Fig. 2A). Given the placement of the E protein trimer, the additional densities represent the Fab molecules and have dimensions consistent with Fab fragments connected to the base of the spike. These Fab densities cover most of the area where the viral lipid bilayer is not covered by spikes, probably preventing aggregation of virus particles.
The E protein trimer in the low-pH DV2-E104–DENV complex has a conformation that appears similar to that of the predicted “open” state.
Although the DENV-2–Fab complex formed at pH 5.5 has not yet encountered host cell lipid membranes, the trimeric spikes of this complex are reminiscent of the postfusion trimers that occur when the virus encounters a lipid membrane at low pH. Hence, an attempt was made to fit the atomic structure of the postfusion E trimer (5) into the spike density (Fig. 2B). Visual inspection showed that the triangle-shaped part of the postfusion trimer structure, formed by domains DI and DIII, fits well into the bottom part of the spike density. The thin waist in the middle of the spike density corresponds to the narrow part of the postfusion trimer structure between domains DI and DII. However, the top part of the postfusion trimer structure is longer and thinner than the spike density. The mean correlation coefficient between the bottom part of the observed spike density and the fitted postfusion structure was 0.91. In contrast, the correlation coefficient of the top part of the spike was only 0.71. The poorer fitting of the closed trimer into the top part of the density might be because the usually flexible hinge angle (15) between DI and DII has changed. Accordingly, five different E trimer structures with different hinge angles between DI and DII (15) were used to fit into the spike density (Table 1). The best correlation coefficient was obtained when the angle between DI and DII was flexible, suggesting that all five structures might be present on the virus surface. Thus, most of the spike structures in the low-pH DENV-Fab complex have a conformation with a large hinge angle as predicted for a fusion intermediate (18) in an “open” state.
TABLE 1.
Correlation coefficient scores between spike density and trimer coordinates with different hinge angles
| Stage or coordinate | Correlation coefficienta | Distanceb (°) |
|---|---|---|
| Mature | 0.76 | 63 |
| Immature | 0.77 | 40 |
| sEhc | 0.73 | 51 |
| sEpc | 0.75 | 56 |
| Postfusion | 0.71 | 22 |
| Mixture | 0.88 |
Correlation coefficient scores between spike density and trimer coordinates.
The nearest distance between any two fusion loops in the trimer.
sEh and sEp are the atomic coordinates of the atoms in DENV E dimer crystal structure as found at Harvard and Purdue, respectively, with PDB accession nos. 1OAM and 1TGB, respectively.
Structure of DENV when complexed with Fab fragments of DV2-E104 at pH 5.5.
Fab fragments are bound to only two of the three DIII domains in each spike of the Fab DV2-E104-virus complex at pH 5.5 (Fig. 2A). This occurs because there is insufficient space near the icosahedral 3-fold axes for a third Fab to bind to the remaining E protein. As the atomic structure of the DV2-E104 Fab fragment was not known, the crystal structure of DENV DIII complexed with E111 Fab fragment (22), derived from another mouse antibody that binds to an analogous epitope on DIII of DENV-1, was used to interpret the cryo-EM density of the complex. However, upon superimposing DIII of the crystal structure onto DIII of the postfusion E trimer, the Fab clashed with the viral membrane and did not correspond in any possible way to the Fab density in the reconstruction (Fig. 3). Thus, the E111 Fab-DIII crystal structure was fitted by superimposing the Fab structure onto the Fab density in the reconstruction of the low-pH complex. In this fit, DIII of the crystal structure was located in a similar position but in an orientation that differed by about 130° to that of DIII in the postfusion trimer (Fig. 3). In this orientation of DIII, the carboxy end points down toward the viral membrane to which it is connected via the helical stem region of E (Fig. 3). The quality of the fit of the E111 Fab-DIII complex into the DENV-2–E104 cryo-EM density was verified with the EMfit program (Table 2). Given that the stem regions of the flavivirus E proteins dissociate from the viral membrane at acidic pH (28), the 25-Å-long neck density (Fig. 2B) between the spike densities and the viral lipid bilayer is formed by the stem regions of the three E proteins. However, if the E proteins were positioned in the known postfusion structure, then DIII would be oriented with its carboxy end pointing up, away from the viral membrane. Given that DIII is not in this orientation, we conclude that the movement of DIII is blocked by DV2-E104 during the prefusion-to-postfusion transition. This rearrangement of DIII prevents the stem region from “zipping up” the neighboring DII domains into a postfusion structure, effectively inhibiting fusion between the viral and cellular membranes.
FIG 3.
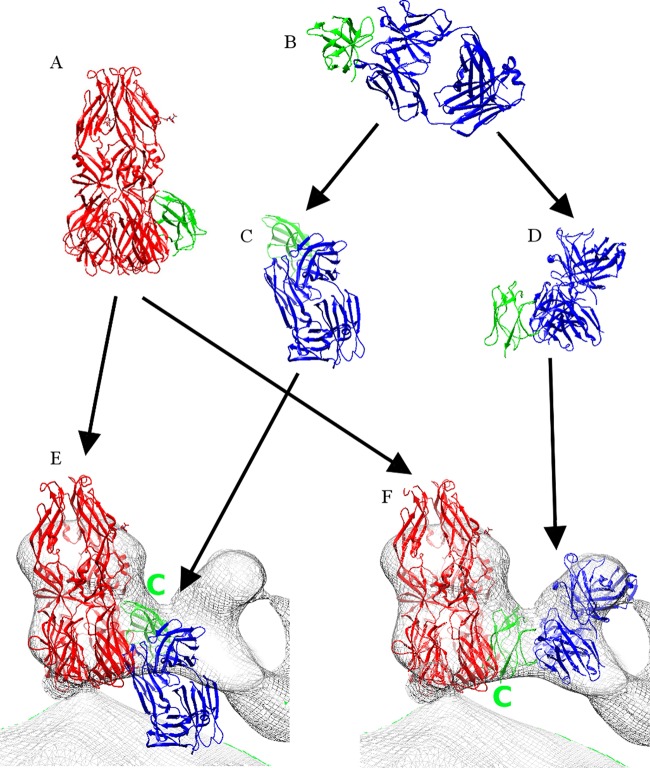
Fitting of the E111 Fab-DIII complex crystal structure into cryo-EM density of DENV-2 complexed with DV2-E104 Fab fragments at pH 5.5. (A) Crystal structure of the postfusion trimer shown as a ribbon diagram in red, except that DIII of one of the E monomers is shown in green. (B) Ribbon diagram of the E111 Fab-DIII complex crystal structure with the Fab molecule in blue and DIII in green. (C) The complex shown in panel B rotated to align DIII (green) with DIII (green) in panel A. (D) The complex shown in panel B rotated to align the blue Fab structure to fit into the cryo-EM density (gray) shown in panel F. (E) The cryo-EM density of the DENV complexed with DV2-E104 Fab fragments at pH 5.5 superimposed with the E trimer structure shown in panel A and the DIII-Fab complex shown in panel C. Note that the Fab molecule (blue) is far outside the cryo-EM density. (F) The cryo-EM density of the DENV complexed with DV2-E104 Fab fragments at pH 5.5 superimposed with the E trimer structure shown in panel A and the DIII-Fab complex shown in panel D. Note the good fit of the Fab molecule (blue) into the cryo-EM density. The green letter C in panels E and F shows the position of the DIII C terminus. The C terminus is pointing to the outside of the virus in panel E and to the inside of the virus in panel F.
TABLE 2.
Fitting the crystal structure of the Fab E111-DIII complex into the low-pH cryo-EM density using the EMfit program (21)
| Object being fitted | Fit order | Rcrita | Sumfb | Clashc | −dend |
|---|---|---|---|---|---|
| E trimere | 1 | 3.59 | 38.7 | 0 | 6.3 |
| 2 | 2.19 | 37.2 | 0 | 9.4 | |
| 3 | 1.09 | 36.0 | 0 | 10.3 | |
| 4 | 1.03 | 35.8 | 0 | 8.6 | |
| Fab near the 5-fold axesf | 1 | 1.94 | 22.5 | 0 | 0.4 |
| Fab near the 2-fold axesf | 1 | 9.53 | 21.3 | 3.2 | 3.5 |
Rcrit is the combined score (21) of the three different parameters (Sumf, Clash, and −den) used to establish the quality of the fit. The combination is the sum of the number of standard deviations each parameter is above the mean.
Sumf is the average height of the densities at the atomic positions.
Clash is the percentage of atoms that sterically interfere between neighboring molecules.
−den is the percentage of atoms in negative density.
E trimer represents the fit of the open E trimer structure. The best four fitting results are provided in rows 1, 2, 3, and 4. Based on the Rcrit criterion, the best fit is much better than the second best fit.
The “Fab near the 5-fold axes” and “Fab near the 2-fold axes” rows correspond to the fit of the Fab E111-DIII complex structure near the icosahedral 5-fold and 2-fold axes, respectively. After a complete three-dimensional angular search at 10° intervals, the best 25 fit results all converged.
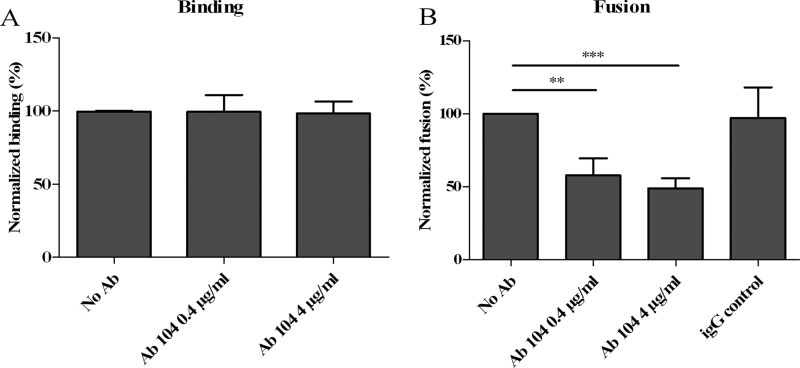
This neutralizing mechanism was confirmed by binding and fusion assays (23). The addition of excess MAb DV2-E104 to DiD-labeled virus particles (23) did not interfere with virus-cell binding (Fig. 4A). Nevertheless, in the presence of MAb DV2-E104, a significant reduction in DENV membrane fusion activity was observed (Fig. 4B). Together with the structural studies, this shows that MAb DV2-E104 interferes with membrane fusion activity.
FIG 4.
MAb DV2-E104 (Ab 104) does not affect virus cell binding but reduces DENV membrane fusion. (A) Virus-cell binding was quantified after adding increasing concentrations of antibody (Ab) to BHK21-15 cells that had been exposed to DENV particles at 4°C. The number of bound particles was normalized with respect to the value for control that lacked antibody. (B) Membrane fusion activity of DENV particles was determined in the presence of increasing concentrations of antibody. The level of membrane fusion was normalized to the control that lacked antibody (100%) and to the control that had added NH4Cl (0%). At least three independent experiments were carried out for each measurement, except for the IgG control, for which two independent experiments were performed. The values that were significantly different from the value for the no-antibody control are indicated by bars and asterisks as follows: **, P < 0.01; ***, P < 0.0001.
Capturing the low-pH E trimer structure in the absence of DV2-E104 Fab fragments.
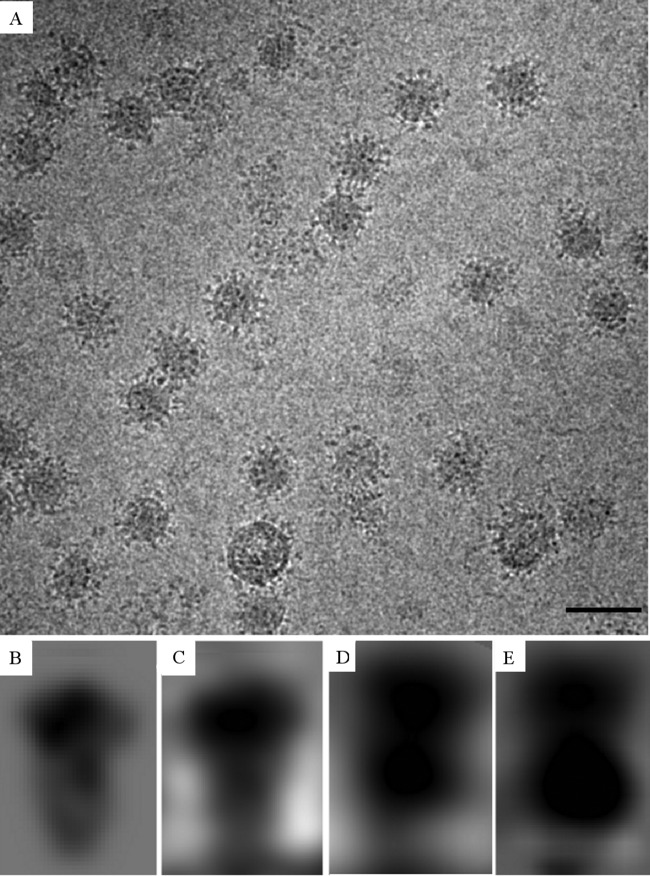
To determine the impact of acidic pH on DENV in the absence of Fab molecules, first the virus was fixed onto a carbon film to avoid particle aggregation. Electron microscopy grids were prepared by absorbing DENV particles at pH 8.0. The grids were then incubated at pH 7.0, 6.5, 6.0, or 5.5 for 5 min before freezing. At pH 7.0, most of the DENV particles had a smooth surface. When the pH was shifted to 6.0 or lower, most of the DENV particles acquired spikes (Fig. 5A).
FIG 5.
Cryo-EM image of spiky particles of DENV-2 at pH 5.5. (A) At pH 5.5, the DENV-2 particles have spikes on the surface. Bar, 100 nm. (B) Two-dimensional 25-Å-resolution electron density map created from postfusion trimer coordinates; (C) averaged individual trimer densities taken from surface spikes on the pH 5.5 DENV (“class one” representing a postfusion trimeric state); (D) averaged individual trimer densities taken from surface spikes on the pH 5.5 DENV (“class two” representing a prefusion trimeric state); (E) a projection of the trimer density from the DENV-Fab E104 low-pH complex. The black densities at the bottom of panels C, D, and E correspond to the density of viral lipid bilayer.
Side views of individual spikes were boxed in a set of highly defocused (5-μm) images taken with a dose rate of about 40 electrons per Å2. The boxed spikes were classified by a nonreference classification method (see Materials and Methods). Most of the spike side views belonged to one of two classes (Fig. 5B to E). The first class, containing 87 spikes, consisted of a globular head and longer stalk. The second class of 107 spikes consisted of two consecutive radially separated globes. The profiles belonging to the first class probably correspond to the known postfusion trimer with the elongated DII domain inserted into the viral membrane, whereas the profiles belonging to the second class likely are prefusion trimeric assemblies. Support for this profile being a prefusion trimer is that DENV retains the ability to fuse with liposomes for 10 min after exposure to pH 5.5 (29). Thus, at least some of the spikes on DENV particles at low pH must be in a prefusion state. This profile resembles the shape of the spike density in the low-pH DENV-Fab complex, which was interpreted above as being in an “open” state. These results show that the spikes in the DENV-Fab complex are constrained by the DV2-E104 Fab fragments to remain in an “open” prefusion structure.
The limited resolution of these results may be due of the lack of homogeneity when the pH is lowered and the temperature is increased (26). However, the spike formed by the E glycoproteins is clear and corresponds to a transient fusogenic trimer structure that has not been captured previously. Furthermore, our results show that during the transition from the prefusion state to the postfusion state, the E proteins became trapped in an intermediate trimeric state (Fig. 6). When Fab fragments of DV2-E104 were bound to DIII, this domain was unable to move to its postfusion position that normally would allow the stem region to zip together the DIIs of neighboring E proteins into a postfusion trimer. In the trimeric intermediate state (Fig. 6Aii), trapped by DV2-E104, the DIIs were observed in an open-like conformation with the fusion loop peptides exposed at the distal end and poised to insert into a lipid membrane. Thus, the major difference between the observed intermediate state and a prefusion fusogenic state would be the rotation of DIII by about 130°.
FIG 6.
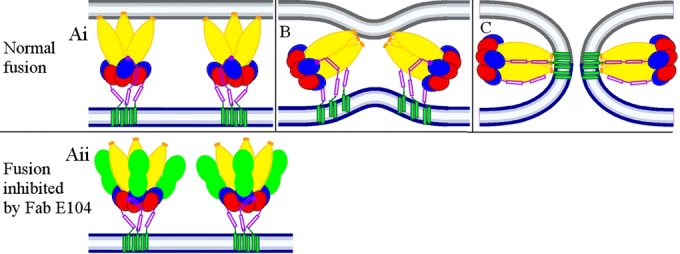
Proposed E protein trimers fusing with a cellular membrane at low pH. (Ai) At pH 5.5, the E proteins form fusogenic “open” trimers that have exposed fusion loops inserted into a cellular membrane (gray). DIII of the E protein (blue) has rotated to a position as is observed in postfusion structures. The C terminus of DIII (dark red dot) connects with the stem region (dark red) and is positioned close to domain DII of E (yellow). (B) The stem region has started to bind into the grove between neighboring DIIs, stabilizing the “closed” DII conformation. This brings the viral membrane (blue) closer to the cellular membrane. (C) The viral and cellular membranes have fused and created a channel for the release of the viral genome. (Aii) When Fab fragments of DV2-E104 (green) were bound to DIII, DIII was unable to rotate to a position as in panel Ai. Thus, DIII has been forced to rotate, causing its C terminus (dark red dot) to point away from DII. Thus, the stem region was unable to bind into the grove between neighboring DIIs, which is required to pull the trimer sideways (as in panels B and C) and, therefore, stops the fusion process.
The large conformational changes that occur when the assembled particles mature to become infectious (30, 31) and subsequently when a few components of the virus actually fuse with the host cell are made possible by the fluidity of the viral membrane. Because these movements are large, it has been challenging to determine the paths taken by the different proteins during their gyrations around each other. Our studies confirm prior predictions (18) and define an intermediate structure that occurs after the dimers of the mature virus have dissolved and shortly before the newly formed trimeric E spikes insert into the endosomal membranes. The anticipated change from an open-like trimeric state before fusion to a “closed” trimeric state after fusion represents a bridge in the steps that relate to membrane fusion. Although the present results are concerned with the fusion of a virus to a cell, the same mechanism may be valid for the fusion of cells (32).
ACKNOWLEDGMENTS
We thank Agustin Avila-Sakar and Valorie Bowman for maintaining the electron microscope facility at Hockmeyer Hall of Structural Biology. We are grateful to Sheryl Kelly for help in the preparation of the manuscript.
This work was supported by NIH grant R01 AI076331 to M.G.R. and NIH grant R01 AI077955 to M.S.D.
X.Z., M.G.R., T.E.H., and J.M.S. designed the experiments. X.Z., J.S., S.K.A., and T.E.H. performed the experiments. X.Z., T.E.H., J.M.S., M.S.D., R.J.K., and M.G.R. analyzed the data. X.Z., J.M.S., R.J.K., M.S.D., and M.G.R. wrote the paper.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Rey FA. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137–148. doi: 10.1016/S0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Modis Y. 2014. A novel membrane fusion protein family in Flaviviridae? Trends Microbiol 22:176–182. doi: 10.1016/j.tim.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlenghi I, Clarke M, Ruttan T, Allison SL, Schalich J, Heinz FX, Harrison SC, Rey FA, Fuller SD. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol Cell 7:593–602. doi: 10.1016/S1097-2765(01)00206-4. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725. doi: 10.1016/S0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modis Y, Ogata S, Clements D, Harrison SC. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 6.Stiasny K, Heinz FX. 2006. Flavivirus membrane fusion. J Gen Virol 87:2755–2766. doi: 10.1099/vir.0.82210-0. [DOI] [PubMed] [Google Scholar]
- 7.Harrison SC. 2008. Viral membrane fusion. Nat Struct Mol Biol. 15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. 2013. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol. 20:105–110. http://dx.doi.org/10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halstead SB, Chow JS, Marchette NJ. 1973. Immunological enhancement of dengue virus replication. Nat New Biol 243:24–25. [PubMed] [Google Scholar]
- 11.Halstead SB. 2007. Dengue. Lancet 370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 12.Lindenbach BD, Rice CM. 2001. Flaviviridae: the viruses and their replication, p 991-1041. In Knipe DM, Howley PM (ed), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Modis Y, Ogata S, Clements D, Harrison SC. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A 100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol 10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J 23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein DE, Choi JL, Harrison SC. 2013. Structure of a dengue virus envelope protein late-stage fusion intermediate. J Virol 87:2287–2293. doi: 10.1128/JVI.02957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS. 2010. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol 84:9227–9239. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Rossmann MG, Bernal R, Pletnev SV. 2001. Combining electron microscopic with X-ray crystallographic structures. J Struct Biol 136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 22.Austin SK, Dowd KA, Shrestha B, Nelson CA, Edeling MA, Johnson S, Pierson TC, Diamond MS, Fremont DH. 2012. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog 8:e1002930. doi: 10.1371/journal.ppat.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayala-Núñez NV, Wilschut J, Smit JM. 2011. Monitoring virus entry into living cells using DiD-labeled dengue virus particles. Methods 55:137–143. doi: 10.1016/j.ymeth.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 25.Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok S-M. 2013. Structural changes in dengue virus when exposed to a temperature of 37°C. J Virol 87:7585–7592. doi: 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. 2013. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc Natl Acad Sci U S A 110:6795–6799. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. 2007. Structure of immature West Nile virus. J Virol 81:6141–6145. doi: 10.1128/JVI.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Fremont DH, Kuhn RJ, Diamond MS, Rossmann MG. 2009. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog 5:e1000672. doi: 10.1371/journal.ppat.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaitseva E, Yang S-T, Melikov K, Pourmal S, Chernomordik LV. 2010. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog 6:e1001131. doi: 10.1371/journal.ppat.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. 2008. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 31.Yu I-M, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 32.DuBois RM, Vaney MC, Tortorici MA, Kurdi RA, Barba-Spaeth G, Krey T, Rey FA. 2013. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature 493:552–556. doi: 10.1038/nature11741. [DOI] [PubMed] [Google Scholar]