Abstract
Malawi polyomavirus (MWPyV) is a recently identified human polyomavirus. Serology for MWPyV VP1 indicates that infection frequently occurs in childhood and reaches a prevalence of 75% in adults. The MWPyV small T antigen (ST) binds protein phosphatase 2A (PP2A), and the large T antigen (LT) binds pRb, p107, p130, and p53. However, the MWPyV LT was less stable than the simian virus 40 (SV40) LT and was unable to promote the growth of normal cells. This report confirms that MWPyV is a widespread human virus expressing T antigens with low transforming potential.
TEXT
Polyomaviruses are small, nonenveloped, circular double-stranded DNA viruses that maintain persistent lifelong infections in humans and animals (1). Human polyomaviruses (HPyV), including JC polyomavirus (JCPyV) and BK polyomavirus (BKPyV), can cause significant disease, primarily in immunocompromised individuals, with increased virus levels contributing to the progression of a disease state (1, 2). Since 2007, the identification of 11 new HPyV has brought renewed attention to this family of DNA tumor viruses and their potential role in human disease. For example, trichodysplasia spinulosa-associated polyomavirus (TSPyV) was isolated from severely immunocompromised patients with trichodysplasia spinulosa (3). The Merkel cell polyomavirus (MCPyV) was found to be clonally integrated into a large percentage of Merkel cell carcinomas (MCC), a cancer with risk factors that include advanced age, prolonged UV exposure, and immunosuppression due to HIV-1/AIDS, hematologic malignancy, or solid-organ transplantation (4, 5). However, many new HPyV have not yet been associated with disease. For example, although WU polyomavirus (WUPyV) (6) and KI polyomavirus (KIPyV) (7) were isolated from nasopharyngeal secretions of children, they do not appear to contribute to pneumonia or other pulmonary disorders. HPyV6 (8) and HPyV7 (8) were found to be chronically shed from the skin; HPyV7 was recently associated with a pruritic rash in certain immunocompromised individuals (9). HPyV9 (10) was found in blood and urine and has sequence similarity to the B-lymphotropic polyomavirus previously isolated from African green monkey cells (11).
Malawi polyomavirus (MWPyV) (12), Saint Louis polyomavirus (STLPyV) (13), HPyV12 (14), and New Jersey polyomavirus (NJPyV) (15) are the most recently described HPyV. HPyV12 was isolated from resected human liver tissue, while NJPyV was found in endothelial cells of a pancreatic transplant patient. HPyV10 (16) and Mexico polyomavirus (MXPyV) (17) represent different isolates of MWPyV, while STLPyV, HPyV12, and NJPyV are genetically distinct HPyV species. MWPyV/MXPyV and STLPyV were isolated from human stool samples in children presenting with diarrhea. MWPyV and HPyV10 were also isolated from a wart in a patient with WHIM (warts, hypogammaglobulinemia, infections, and myelokathexis) syndrome. WHIM syndrome increases patient susceptibility to human papillomavirus (HPV) infection (18). It is possible that MWPyV/HPyV10 contributed to the development of the excised warts and that WHIM syndrome makes patients susceptible to infection with and disease caused by MWPyV and other HPyV, although this association has not been demonstrated.
We independently identified an HPyV genome, closely related to those of MWPyV, HPyV10, and MXPyV, from two serial stool samples from a child at 9 and 11 months of age living in Denver, CO, USA. These samples were part of the collections for the Environmental Determinants of Diabetes in the Young (TEDDY) study (19), although the child had no signs of diabetes at the time of sample collection. Total DNA was isolated from cleared stool samples using a QIAamp DNA Blood minikit (catalog no. 51104; Qiagen) following the manufacturer's suggested protocol. The resulting total DNA was subjected to whole-genome amplification (WGA) by the use of multiple displacement amplification (MDA) and a REPLI-g minikit (Qiagen) (20). WGA DNA was fragmented by sonication and then subjected to massively parallel sequencing by the use of a Illumina HiSeq 2000 sequencer as previously reported (21). DNA sequences matching human genome sequences were subtracted using PathSeq (22), and the remaining unmapped reads were de novo assembled into long contiguous sequences (contigs) using Inchworm software (23). The accuracy of the viral genome assembly and DNA base accuracy were validated by PCR amplification of two DNA fragments, a 2,954-bp DNA fragment and a 2,349-bp DNA fragment, from the original batches of total DNA without WGA, followed by Sanger sequencing. After combining contigs from the two samples, we were able to assemble a 4,939-bp-long circular virus genome. This strain of MWPyV is 94.9% identical at the nucleotide level to the previously reported strains of MWPyV and 99.7% identical to HPyV10 and MXPyV. In contrast, STLPyV, HPyV12, and NJPyV are quite divergent from this group, with less than 81% similarity at the nucleotide level, supporting their designation as distinct HPyV species (24).
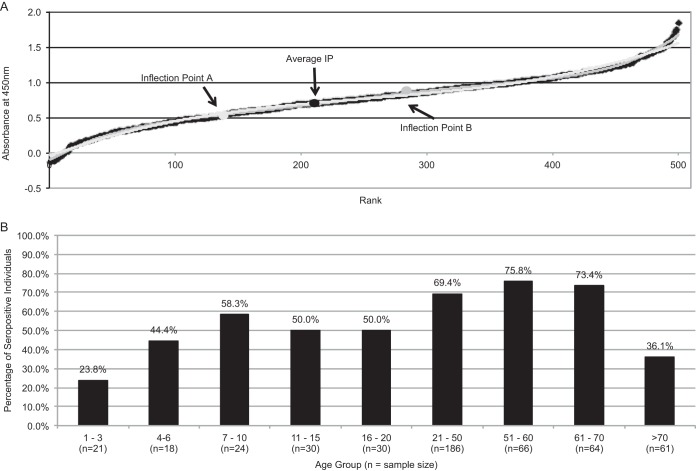
Serological studies have indicated that JCPyV and BKPyV infections, as well as KIPyV, WUPyV, and MCPyV infections, often occur during childhood (25). To determine the prevalence of MWPyV infection, the predicted MWPyV VP1 gene sequence was modified for optimal expression in Escherichia coli (GenScript) and used to study the serological prevalence of MWPyV infection in healthy individuals. We performed serological analysis using glutathione S-transferase (GST)-conjugated MWPyV-VP1 in a capsomere-based enzyme-linked immunosorbent assay (ELISA) as previously described (25). Serum samples from 500 individuals (a subset of a larger Colorado Multiple Institutional Review Board-approved collection) (25) were tested in triplicate, and values corresponding to their absorbance at 450 nm were plotted in ascending order to determine an inflection point and a cutoff value (Fig. 1A). By the use of this cutoff value, MWPyV-VP1 seroreactivity was detected in up to 75.8% of patient sample groups, with an age-related increase in seropositivity (Fig. 1B). Similar findings for MWPyV were recently reported using a virus-like particle-based ELISA, with seroprevalence reaching 42% in an adult population (26). The moderately high levels of seroprevalence in children from age 3 to age 21 suggest that primary infection with MWPyV occurs in childhood, and the increasing frequency of elevated antibody titers that occurs with age is consistent with persistent lifelong infections.
FIG 1.
Age-specific seroprevalence determined in a Denver, CO, USA, study population (n = 500) for MWPyV. (A) Determination of inflection points for MWPyV VP1 antigens assayed using the VP1-GST ELISA. Two best-fit functions, one sixth-order (A) and one third-order (B) polynomial, were derived from the data using Microsoft Excel. The inflection points were calculated by setting the second derivative of the corresponding function to zero. The average of the two inflection point (Average IP) values (average = 0.677) was the final assigned cutoff value. All absorbance values above that were considered to represent seropositivity. (B) MWPyV age-specific seroprevalence from 500 serum samples tested in triplicate, indicating an age-related increase in seropositivity.
Virus protein interactions can act as surrogates for human genetic variations, inducing disease states by influencing local and global properties of cellular networks (27). Classical studies have demonstrated that the model polyomavirus simian virus 40 (SV40) large T antigen (LT) promotes cellular proliferation at least in part by binding to pRb and p53 (28, 29). In addition, the SV40 small T antigen (ST) plays a critical role in cellular transformation by binding to the A and C subunits of protein phosphatase 2A (PP2A), thereby affecting multiple signaling pathways (30–33). A recent study indicated that LT from WUPyV, HPyV6, and HPyV7 could coprecipitate p53 and pRb (27).
To determine whether MWPyV LT could bind to p53 and pRb and whether MWPyV ST could bind to PP2A, MWPyV LT and ST open reading frame (ORF) cDNAs were generated by PCR-based Gateway cloning (Invitrogen) into destination vector MSCV-C-term-Flag-HA-IRES-Puro (CTAP) as previously described using the primer pairs listed in Table 1 (27, 34). The 2,954-bp viral DNA fragment, which included the large T antigen (LT) and small T antigen (ST) genes, was used to clone the T antigen cDNA constructs. The full-length MWPyV LT cDNA was generated by overlap extension PCR (27) using the individual LT exon PCR products. Recombinant retroviruses were generated in 293T cells and transduced into U-2OS human osteosarcoma cells followed by selection with puromycin (Sigma) (27) (2 μg/ml). Cells were then transfected with a control enhanced green fluorescent protein (EGFP)-expressing vector or V5-tagged pRb, p53, or PP2A-Aα (PPP2R1A) constructs using Lipofectamine 2000 (Invitrogen). After 48 h, cells were treated with 10 μM MG132 (Millipore) for 8 h before harvesting. Cell lysates were obtained using lysis buffer containing 150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 0.5% NP-40, 10% glycerol, and protease and phosphatase inhibitor cocktail sets (Calbiochem). Reciprocal immunoprecipitations from cell lysates were performed with anti-hemagglutinin (anti-HA) beads (Roche) or anti-V5 beads (Bethyl) as previously described (27). Beads were washed with lysis buffer, boiled in SDS sample buffer (Boston BioProducts), resolved by SDS-PAGE (Criterion TGX precast gels; Bio-Rad), and immunoblotted with antibodies to HA (C29F4; Cell Signaling), V5 (Invitrogen), and vinculin (H-10; Santa Cruz).
TABLE 1.
PCR primers for generating Gateway-compatible MWPyV ST and LT constructs and MWPyV LT LxCxE mutant
| Construct | Forward primer | Reverse primer |
|---|---|---|
| MWPyV ST | GGGGACAAGTTTGTACAAAAAAGCAGGCATGATAGAGTTCTTTCTAGAGAT | GGGGACCACTTTGTACAAGAAAGCTGGGTCGGAGTCCCATAAGTGGGA |
| MWPyV-LT-exon 1 | GGGGACAAGTTTGTACAAAAAAGCAGGCATGATAGAGTTCTTTCTAGAGAT | CCCATAAGTGGGATTTCCCTTTGCAGGAAAATAAACTT |
| MWPyV-LT-exon 2 | GGAAATCCCACTTATGGG | GGGGACCACTTTGTACAAGAAAGCTGGGTCTTGTGAATTAATTCCAGAGTCT |
| MWPyV-LT-E109K | GGATGGGATGAAGATTTAAGTTGTAATAAATCTTTTGCTCCCAGTGAT | ATCACTGGGAGCAAAAGATTTATTACAACTTAAATCTTCATCCCATCC |
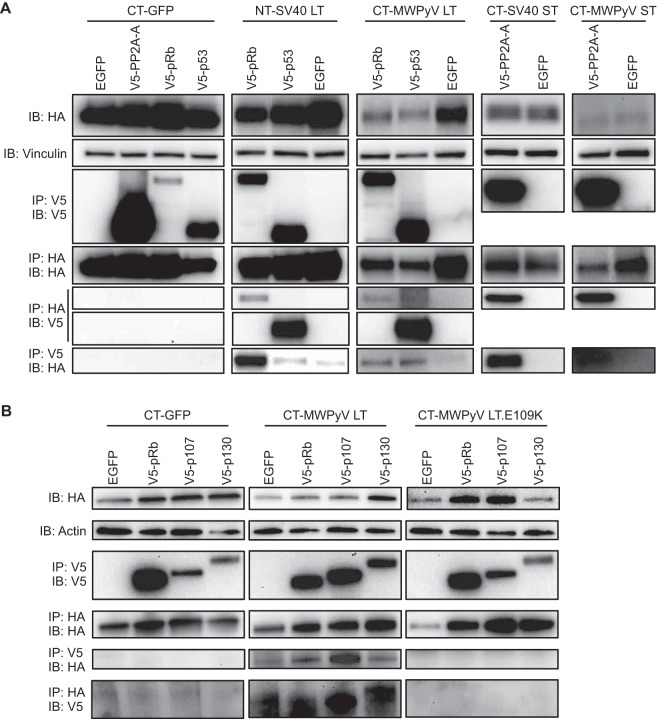
CTAP-MWPyV LT was able to coimmunoprecipitate both V5-tagged p53 and pRb, while V5-tagged p53 and pRb could pull down CTAP-MWPyV LT (Fig. 2A). An immunoprecipitation of CTAP-MWPyV ST revealed coimmunoprecipitation of the V5-tagged PP2A-Aα subunit, and, conversely, an immunoprecipitation of V5-PP2A-Aα coprecipitated CTAP-MWPyV ST (Fig. 2A). To determine if MWPyV LT could bind to other Rb-family proteins, we performed a similar coimmunoprecipitation experiment by transfecting U-2OS stably expressing CTAP-GFP, CTAP-MWPyV LT, and CTAP-MWPyV LT.E109K, which contains a mutated LXCXE (LXCXK) motif (1), with V5-tagged pRb, p107, p130, or a control EGFP-expressing vector. The E109K mutation was introduced into LT using the primer pairs listed in Table 1 and a QuikChange XL II site-directed mutagenesis kit (Agilent). An immunoprecipitation of CTAP-MWPyV LT, but not CTAP-MWPyV LT.E109K or NTAP-GFP, revealed weak binding of V5-tagged pRb, p107, and p130, and, conversely, immunoprecipitation of V5-tagged pRb, p107, and p130 was able to coprecipitate CTAP-MWPyV LT but not CTAP-MWPyV LT.E109K or NTAP-GFP (Fig. 2B). Actin-specific antibody (D6A8; Cell Signaling) was used as a loading control. These results confirm that MWPyV T antigens are able to bind specifically to cellular tumor suppressors in a manner similar to that seen with T antigens from SV40, although not as efficiently (1, 35).
FIG 2.
MWPyV T antigens bind to human tumor suppressors. (A) U-2OS cells stably expressing CTAP-GFP (CT-GFP), NTAP-SV40 LT (NT-SV40 LT), CT-MWPyV LT, CT-SV40 ST, and CT-MWPyV ST were transfected with the indicated V5-tagged constructs of PP2A-Aα, pRb, and p53 or with a control EGFP-expressing vector. After an 8-h treatment with 10 μM MG-132, cells were lysed and immunoprecipitated with either HA antibody-conjugated beads (IP: HA) or V5 antibody-conjugated beads (IP: V5). Blotting with HA (IB: HA) or V5 (IB: V5) was performed to detect IP of the CTAP/NTAP constructs or coimmunoprecipitation of the PP2A-Aα, pRb, and p53 constructs, respectively. Vinculin-specific antibody was used as a loading control. (B) U-2OS cells stably expressing CT-GFP, CT-MWPyV LT, and CT-MWPyV LT.E109K were transfected with the indicated V5-tagged constructs of pRb, p107, and p130 or with a control EGFP-expressing vector. After MG-132 treatment, cells were lysed and immunoprecipitated as described for panel A. Actin-specific antibody was used as a loading control.
Given the ability of MWPyV LT to bind to p53 and Rb-family proteins, we proceeded to test if it could regulate cell cycle-dependent gene expression in a fashion similar to that seen with SV40 LT. To determine the relative levels of induction of several E2F-target genes, U-2OS cells stably expressing CTAP-GFP, CTAP-MWPyV LT, or NTAP-SV40 LT were synchronized in late G1/S phase using l-mimosine, released with fresh media, and harvested every 3 h for 12 h as previously described (36). Although cells expressing SV40 LT had increased levels of cyclin E and B-Myb relative to cells expressing GFP, cells with MWPyV LT did not show increased levels of cyclin E, cyclin A, and B-Myb compared to cells expressing GFP (data not shown). pRb levels were also found to have remained unchanged in the three cell lines (data not shown).
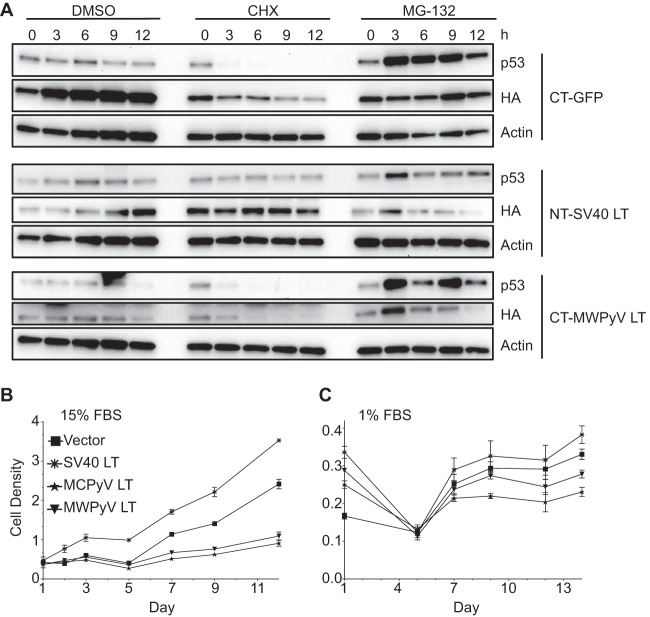
We observed that the levels of MWPyV LT appeared lower than those of SV40 LT in the stably transduced U-2OS cells. We sought to determine if MWPyV LT was being turned over in cells at a rate that was similar to or different from the rate seen with SV40 LT. U-2OS cells stably expressing CTAP-GFP, CTAP-MWPyV LT, or NTAP-SV40 LT were treated with cycloheximide (Sigma) (50 μg/ml), MG-132 (10 μM), or dimethyl sulfoxide (DMSO). Cells were harvested every 3 h for 12 h, lysed, and immunoblotted with HA and p53 (FL-293; Santa Cruz) antibodies (Fig. 3A). While NTAP-SV40 LT was quite stable under conditions of cycloheximide treatment, CTAP-MWPyV LT levels quickly diminished within 6 h. In cells expressing SV40 LT, p53 levels were increased, with a reduced turnover rate (37). We found that MWPyV LT failed to stabilize p53 compared to SV40 LT results following cycloheximide treatment (Fig. 3A).
FIG 3.
MWPyV LT is quickly degraded after cycloheximide treatment and fails to promote growth of IMR-90 human fibroblasts. (A) U-2OS cells stably expressing CTAP-GFP (CT-GFP), CT-MWPyV LT, or NTAP-SV40 LT (NT-SV40 LT) were treated with DMSO, 50 μg/ml cycloheximide (CHX), or 10 μM MG-132 and harvested at the indicated hours posttreatment. Cells were lysed and immunoblotted with antibodies against p53, HA, or actin. (B) IMR-90 cells expressing vector, SV40, MCPyV, and MWPyV LT in 15% FBS were seeded and grown for 12 days. (C) IMR-90 cells expressing vector, SV40, MCPyV, and MWPyV LT in 1% FBS were seeded and grown for 14 days. Cell density was determined by crystal violet staining followed by optical density measurement at 590 nm.
Expression of T antigens typically promotes cellular proliferation (38). To determine if MWPyV LT can promote cell growth, IMR-90 cells stably expressing NTAP-tagged MWPyV LT, SV40 LT, MCPyV LT, or vector, between passage 9 and passage 12, were seeded in triplicate in 24-well plates (day 0; 5 × 103 cells per well) and cultured in Dulbecco's modified Eagle medium (DMEM) (Cellgro) supplemented with 1% penicillin-streptomycin (Pen Strep; Gibco), 1% GlutaMAX (Gibco), and 1% nonessential amino acids (Gibco) and either 15% fetal bovine serum (FBS) (Atlanta Biologicals) (Fig. 3B) or 1% FBS (Fig. 3C). Cell density was measured by crystal violet assay at intervals after plating as previously described (27). Although the presence of SV40 LT led to an increase in the growth rate relative to the rate seen with the vector control cells, the presence of MWPyV LT led to a slight decrease in the growth rate of IMR-90 cells under both conditions (27, 32). It has been recently reported that MCPyV LT may have growth-inhibitory effects in human fibroblasts (39–41). We observed similar results and found that MWPyV LT-expressing cells grew only slightly faster than cells containing MCPyV LT but less well than the control cells cultured under identical conditions. These results indicate that MWPyV LT lacks transforming potential in vitro.
We have isolated a new strain of MWPyV and provided serological evidence that it is a virus infecting humans that is prevalent across a broad age range. The initial reports of MWPyV and the closely related MXPyV contained cautionary notes regarding their potential as environmental contaminants in stool rather than as human pathogens (12, 17). However, repeated isolation of identical viruses from temporally distinct stool samples, both in the original study (12) and in this study, suggests chronic infection with MWPyV. Furthermore, MWPyV/HPyV10 was found in high abundance in samples from a surgically excised wart (16), suggesting that human cells can support infection. Serological evidence for specific antibodies to HPyV VP1 provides additional strong support for its designation as a human polyomavirus.
We have determined that MWPyV LT cocomplexes with Rb-family proteins and p53 and that MWPyV ST cocomplexes with PP2A-Aα. Despite the ability of MWPyV T antigens to bind to known tumor suppressors, MWPyV T antigens did not promote cellular proliferation compared to control cell results. We found that MWPyV LT is rapidly turned over, which may explain its inability to promote cell growth. It remains to be determined whether MWPyV can contribute to human disease.
Nucleotide sequence accession number.
Sequence data for the 4,939-bp-long circular virus genome determined in this work was deposited in GenBank under accession no. KC690147.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants P01CA050661, RO1CA93804, and R01CA63113 to J.A.D. and R0137667 to R.L.G. and training grants 5T32AI007245 and F31CA177274 to C.B.
We thank Anna Korkhin for technical assistance.
The work described in this article was funded by DK 63829, 63861, 63821, 63865, 63863, 63836, 63790, and UC4DK095300 and contract no. HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the National Institute of Allergy and Infectious Diseases (NIAID), the National Institute of Child Health and Human Development (NICHD), the National Institute of Environmental Health Sciences (NIEHS), the Juvenile Diabetes Research Foundation (JDRF), and the Centers for Disease Control and Prevention (CDC).
The members of the TEDDY Study Group are available in Text S1 in the supplemental material.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02328-14.
REFERENCES
- 1.DeCaprio JA, Garcea RL. 2013. A cornucopia of human polyomaviruses. Nat Rev Microbiol 11:264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang M, Abend JR, Johnson SF, Imperiale MJ. 2009. The role of polyomaviruses in human disease. Virology 384:266–273. doi: 10.1016/j.virol.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog 6:e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodig SJ, Cheng J, Wardzala J, DoRosario A, Scanlon JJ, Laga AC, Martinez-Fernandez A, Barletta JA, Bellizzi AM, Sadasivam S, Holloway DT, Cooper DJ, Kupper TS, Wang LC, DeCaprio JA. 2012. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J Clin Invest 122:4645–4653. doi: 10.1172/JCI64116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog 3:e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. 2007. Identification of a third human polyomavirus. J Virol 81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho J, Jedrych JJ, Feng H, Natalie AA, Grandinetti L, Mirvish E, Crespo MM, Yadav D, Fasanella KE, Proksell S, Kuan S-FF, Pastrana DV, Buck CB, Shuda Y, Moore PS, Chang Y. 17 September 2014, posting date Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients. J Infect Dis doi: 10.1093/infdis/jiu524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauvage V, Foulongne V, Cheval J, Ar Gouilh M, Pariente K, Dereure O, Manuguerra JC, Richardson J, Lecuit M, Burguiere A, Caro V, Eloit M. 2011. Human polyomavirus related to African green monkey lymphotropic polyomavirus. Emerg Infect Dis 17:1364–1370. doi: 10.3201/eid1708.110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.zur Hausen H, Gissmann L. 1979. Lymphotropic papovaviruses isolated from African green monkey and human cells. Med Microbiol Immunol 167:137–153. doi: 10.1007/BF02121180. [DOI] [PubMed] [Google Scholar]
- 12.Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2012. Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol 86:10321–10326. doi: 10.1128/JVI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2013. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 436:295–303. doi: 10.1016/j.virol.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B. 2013. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS One 8:e58021. doi: 10.1371/journal.pone.0058021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra N, Pereira M, Rhodes RH, An P, Pipas JM, Jain K, Kapoor A, Briese T, Faust PL, Lipkin WI. 1 May 2014, posting date Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J Infect Dis doi: 10.1093/infdis/jiu250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA. 2012. Complete genome sequence of a tenth human polyomavirus. J Virol 86:10887. doi: 10.1128/JVI.01690-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G, Greninger AL, Isa P, Phan TG, Martinez MA, de la Luz Sanchez M, Contreras JF, Santos-Preciado JI, Parsonnet J, Miller S, DeRisi JL, Delwart E, Arias CF, Chiu CY. 2012. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS One 7:e49449. doi: 10.1371/journal.pone.0049449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai T, Malech HL. 2009. WHIM syndrome: congenital immune deficiency disease. Curr Opin Hematol 16:20–26. doi: 10.1097/MOH.0b013e32831ac557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagopian WA, Lernmark A, Rewers MJ, Simell OG, She JX, Ziegler AG, Krischer JP, Akolkar B. 2006. TEDDY–The Environmental Determinants of Diabetes in the Young: an observational clinical trial. Ann N Y Acad Sci 1079:320–326. doi: 10.1196/annals.1375.049. [DOI] [PubMed] [Google Scholar]
- 20.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci U S A 99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostic AD, Ojesina AI, Pedamallu CS, Jung J, Verhaak RG, Getz G, Meyerson M. 2011. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol 29:393–396. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johne R, Buck CB, Allander T, Atwood WJ, Garcea RL, Imperiale MJ, Major EO, Ramqvist T, Norkin LC. 2011. Taxonomical developments in the family Polyomaviridae. Arch Virol 156:1627–1634. doi: 10.1007/s00705-011-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kean JM, Rao S, Wang M, Garcea RL. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog 5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicol JT, Leblond V, Arnold F, Guerra G, Mazzoni E, Tognon M, Coursaget P, Touzé A. 30 October 2013. Seroprevalence of human Malawi polyomavirus. J Clin Microbiol doi: 10.1128/JCM.02730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, et al. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeCaprio JA. 2009. How the Rb tumor suppressor structure and function was revealed by the study of adenovirus and SV40. Virology 384:274–284. doi: 10.1016/j.virol.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Lane D, Levine A. 2010. p53 research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol 2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. 1990. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell 60:167–176. doi: 10.1016/0092-8674(90)90726-U. [DOI] [PubMed] [Google Scholar]
- 31.Yang SI, Lickteig RL, Estes R, Rundell K, Walter G, Mumby MC. 1991. Control of protein phosphatase 2A by simian virus 40 small-t antigen. Mol Cell Biol 11:1988–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol 22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sablina AA, Hector M, Colpaert N, Hahn WC. 2010. Identification of PP2A complexes and pathways involved in cell transformation. Cancer Res 70:10474–10484. doi: 10.1158/0008-5472.CAN-10-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. 2011. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest 121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krek W, DeCaprio JA. 1995. Cell synchronization. Methods Enzymol 254:114–124. [DOI] [PubMed] [Google Scholar]
- 37.Pipas JM, Levine AJ. 2001. Role of T antigen interactions with p53 in tumorigenesis. Semin Cancer Biol 11:23–30. doi: 10.1006/scbi.2000.0343. [DOI] [PubMed] [Google Scholar]
- 38.Howley PM, Livingston DM. 2009. Small DNA tumor viruses: large contributors to biomedical sciences. Virology 384:256–259. doi: 10.1016/j.virol.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng J, Rozenblatt-Rosen O, Paulson KG, Nghiem P, Decaprio JA. 20 March 2013. Merkel cell polyomavirus large T antigen has growth promoting and inhibitory activities. J Virol doi: 10.1128/JVI.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borchert S, Czech-Sioli M, Neumann F, Schmidt C, Wimmer P, Dobner T, Grundhoff A, Fischer N. 2014. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J Virol 88:3144–3160. doi: 10.1128/JVI.02916-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Wang X, Diaz J, Tsang SH, Buck CB, You J. 2013. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J Virol 87:9173–9188. doi: 10.1128/JVI.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.