ABSTRACT
Infectious spleen and kidney necrosis virus (ISKNV) is the type species of the Megalocytivirus genus, Iridoviridae family, causing a severe systemic disease with high mortality in mandarin fish (Siniperca chuatsi) in China and Southeast Asia. At present, the pathogenesis of ISKNV infection is still not fully understood. Based on a genome-wide bioinformatics analysis of ISKNV-encoded proteins, we found that ISKNV open reading frame 119L (ORF119L) is predicted to encode a three-ankyrin-repeat (3ANK)-domain-containing protein, which shows high similarity to the dominant negative form of integrin-linked kinase (ILK); i.e., viral ORF119L lacks the ILK kinase domain. Thus, we speculated that viral ORF119L might affect the host ILK complex. Here, we demonstrated that viral ORF119L directly interacts with particularly interesting Cys-His-rich protein (PINCH) and affects the host ILK-PINCH interaction in vitro in fathead minnow (FHM) cells. In vivo ORF119L overexpression in zebrafish (Danio rerio) embryos resulted in myocardial dysfunctions with disintegration of the sarcomeric Z disk. Importantly, ORF119L overexpression in zebrafish highly resembles the phenotype of endogenous ILK inhibition, either by overexpressing a dominant negative form of ILK or by injecting an ILK antisense morpholino oligonucleotide. Intriguingly, ISKNV-infected mandarin fish develop disorganized sarcomeric Z disks in cardiomyocytes. Furthermore, phosphorylation of AKT, a downstream effector of ILK, was remarkably decreased in ORF119L-overexpressing zebrafish embryos. With these results, we show that ISKNV ORF119L acts as a domain-negative inhibitor of the host ILK, providing a novel mechanism for the megalocytivirus pathogenesis.
IMPORTANCE Our work is the first to show the role of a dominant negative inhibitor of the host ILK from ISKNV (an iridovirus). Mechanistically, the viral ORF119L directly binds to the host PINCH, attenuates the host PINCH-ILK interaction, and thus impairs ILK signaling. Intriguingly, ORF119L-overexpressing zebrafish embryos and ISKNV-infected mandarin fish develop similar disordered sarcomeric Z disks in cardiomyocytes. These findings provide a novel mechanism for megalocytivirus pathogenesis.
INTRODUCTION
Infectious spleen and kidney necrosis virus (ISKNV) is the type species of the genus Megalocytivirus, family Iridoviridae. ISKNV and closely related isolates infect a wide range of marine and freshwater fish species, including mandarin fish (Siniperca chuatsi) (1), orange-spotted grouper (Epinephelus coioides) (2), large yellow croaker (Larimichthys crocea) (3), Aplocheilichthys normani (4), turbot (Scophthalmus maximus), zebrafish (Danio rerio) (5), and more than 50 species of marine fish (6). In China, ISKNV causes a serious disease with high mortality in mandarin fish, leading to economic loss and ecological problems. To control the virus, we started this viral pathogenesis study by sequencing the ISKNV genome.
In this study, we found that ISKNV open reading frame 119L (ORF119L) is predicted to encode an ankyrin (ANK) repeat-containing protein but without the kinase domain. The NH2-terminal three-ankyrin-repeat (3ANK)-containing domain of ORF119L is highly similar to that of integrin-linked kinase (ILK). The myxoma virus M-T5 (7, 8), myxoma nuclear factor (MNF) (9), poxvirus ankyrin repeat proteins (10, 11), vaccinia virus K1L protein (12), and ISKNV ORF124L (13) have this ankyrin repeat-containing domain. All of these proteins have been reported to be involved in viral pathogenesis.
In the host ILK signaling, ILK complexes with particularly interesting Cys-His-rich protein (PINCH) and parvin and plays a crucial role in cell adhesion and intracellular signal transduction (14, 15). Structurally, the NH2 terminus of ILK is a 3ANK-containing domain capable of binding to the LIM1 domain of PINCH (16–19), while the COOH terminus of ILK contains a kinase domain responsible for interacting with parvin. Functionally, ILK phosphorylates protein kinase B (PKB; also known as AKT) and glycogen synthase kinase 3 (GSK-3) for intracellular signal transduction (18). Mice with the germ line-mutated ILK in which Val386 and Thr387 in the kinase domain are replaced with glycine residues (ILK-VT/GG) die of vasculogenesis defects at embryonic day 12.5 due to the fact that VT/GG substitutions decrease ILK protein stability, leading to decreased ILK levels and reduced binding to paxillin and α-parvin (20). Heart failure and pericardial edema phenotypes are found in zebrafish expressing ILKL308P (kinase-dead mutant) during early embryogenesis (21). In mammalian cells, overexpression of the 3ANK domain of ILK without its kinase domain disrupts the assembly of the PINCH-ILK-parvin complex, exerting its dominant negative inhibitory (DNI) effect on ILK-mediated signaling (22, 23).
All of these prompted us to hypothesize that ORF119L may play a role in host ILK signaling. We studied the function of ORF119L in vivo by using the zebrafish model because we could observe the rapid embryonic development of zebrafish and manipulate its embryos outside the parent animal (24–26). Furthermore, zebrafish have been used to study the viral gene function and pathogenesis of ISKNV, as previously described (27–31). Here, we demonstrated that ORF119L interacts with PINCH and affects the binding of ILK to PINCH, which leads to cardiovascular defects in zebrafish, likely because of its ability to reduce ILK-mediated AKT phosphorylation. Consistent with this, inhibiting the endogenous zebrafish ILK effectively mimics the ORF119L-induced abnormal phenotype. Taken together, our data suggest that ISKNV ORF119L may function as a novel DNI-like factor of ILK.
MATERIALS AND METHODS
Collection of ISKNV-infected fish and isolation of viral DNA.
Moribund mandarin fish showing common symptoms of ISKNV infection were collected and kept at −80°C. ISKNV infection in mandarin fish, virus purification, and viral DNA extraction (universal genomic DNA extraction kit, v.3.0; TaKaRa, Dalian, China) were performed as described previously (5, 27).
Zebrafish maintenance, plasmid construction and microinjection.
A zebrafish transgenic line, Tg(flk1:GFP), expressing a green fluorescent protein (GFP) gene whose expression is driven by the promoter of the gene encoding zebrafish flk1 (also known as vascular endothelial growth factor receptor) was used, with wild-type zebrafish as the control group. All zebrafish were maintained at 28°C as previously described (31). Zebrafish embryos were kept in E3 zebrafish water containing 5.0 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4 (pH 7.4) at 28.5°C, and their developmental stages were defined as hours postfertilization (hpf) or days postfertilization (dpf) (27, 32). To construct the plasmids for microinjection in zebrafish embryos, the full-length ISKNV ORF119L was PCR amplified (primers in Table 1) using the ISKNV genomic DNA. The PCR products were subcloned into pDsRed2-C1 (TaKaRa Bio Company; Clontech, Mountain View, CA) to generate the RFP-ORF119L-expressing plasmid pRFP-ORF119L. Similarly, the ORF119L PCR products were subcloned into the pEGFP-N3 vector (Clontech, Mountain View, CA) to generate the ORF119L-EGFP-expressing plasmid pORF119L-EGFP. The RFP-ORF119L- and ORF119L-EGFP-overexpressing embryos are referred to as ORF119L embryos here. To overexpress the ORF119L mutant lacking the 3ANK-containing domain (119LΔ3ANK), the 119LΔ3ANK gene sequence was PCR amplified and subcloned into the pEGFP-N3 plasmid to generate the 119LΔ3ANK-EGFP-expressing plasmid p119LΔ3ANK-EGFP. The zebrafish first-strand cDNA was synthesized as previously described (31). To overexpress the 3ANK domain of ILK (ILK3ANK), the zebrafish ILK3ANK gene sequence (without the portion encoding the ILK kinase domain) was PCR amplified from zebrafish cDNA and then subcloned into the pEGFP-N3 vector to generate the ILK3ANK-EGFP-expressing plasmid pILK3ANK-EGFP. All plasmids and inserts were confirmed by bidirectional sequencing. The protocol for plasmid microinjection and image capture was described previously (31). Briefly, the plasmids were linearized and purified (QIAquick PCR purification kit), and resuspended in water at 100 ng/μl. Plasmids were microinjected (IM300 microinjector; Narishige, Japan) into one-cell-stage zebrafish embryos at 1 nl per embryo. Embryos were photographed at different developmental stages using an OlympusDP71 digital camera mounted on an Olympus MVX10 fluorescence stereomicroscope.
TABLE 1.
Summary of primers used in this study
| Purpose and primer | Sequence (5′-3′)a |
|---|---|
| Plasmid microinjection | |
| pORF119L-EGFP-F | CGGAATTCATGCCTGTACATGGGTGTGT |
| pORF119L-EGFP-R | CGGGATCCTCGCCTTGTGTTCTGTTTT |
| pRFP-ORF119L-F | CGGGATCCATGCCTGTACATGGGTGTGT |
| pRFP-ORF119L-R | CGGAATTCTCGCCTTGTGTTCTGTTTT |
| pILK3ANK-EGFP-F | CGGAATTCATGGATGACATCTTCACTCAG |
| pILK3ANK-EGFP-R | CGGGATCCAGGGACTTTTGACAGG |
| p119LΔ3ANK-EGFP-F | CGGAATTCTATGGACCTGCGGGCAGT |
| p119LΔ3ANK-EGFP-R | CGGGATCCTCGCCTTGTGTTCTGTTTT |
| Mammalian cell expression vector | |
| pMYC-ORF119L-F | CGGAATTCAATGCCTGTACATGGGTGT |
| pMYC-ORF119L-R | CGGGATCCTCATCGCCTTGTGTTCT |
| pMYC-ILK-F | CGGAATTCAATGGATGACATCTTCACTCAG |
| pMYC-ILK-R | CGGGATCCTTATTTGTCTTGCATCTTCTC |
| pFLAG-PINCH-F | CGGAATTCAATGCTGGGGGTGTCAG |
| pFLAG-PINCH-R | CGGGATCCTTACTTGCGGCCCA |
| pMYC-119LΔ3ANK-F | CGGAATTCAATGGACCTGCGGGCAGT |
| pMYC-119LΔ3ANK-R | CGGGATCCTTATCGCCTTGTGTTCTGTTTT |
| GST fusion constructs | |
| pGST-119L-F | CGGGATCCATGCCTGTACATGG |
| pGST-119L-R | CGGAATTCTCGCCTTGTGTTCTGTT |
| pGST-ILK-F | CGGGATCCATGGATGACATCTTCACT |
| pGST-ILK-R | CGGAATTCTTTGTCTTGCATCTTCTCCAG |
| Whole mount in situ hybridization of anf | |
| pGEM-T-anf-F | GACAGTCTTAATCAGGGGGCCGGTA |
| pGEM-T-anf-R | TGGGAGCCAACGTTGAGATTTTTTCCAATC |
| qRT-PCR | |
| anf-QF | GACGGATGTACAAGCGCACACGTTGAG |
| anf-QR | CGGTGTTGCTGTCTTCATAATCTACGGCTC |
| nkx2.5-QF | TTCACCTACAACACCTACCCTGCGTTTAGT |
| nkx2.5-QR | TGGATGCTGGACATGCTCGACGGATAG |
| cmlc2-QF | GCAGCATATCTCAAGAGCCAAGGACCAG |
| cmlc2-QR | CTCAGCACCCATCACTGTTCCGTTTCC |
Underlining indicates restriction endonuclease cleavage sites.
Bioinformatics analysis.
A BLAST (Basic Local Alignment Search Tool) (33) search was performed to compare the ISKNV ORF119L protein sequence to the NCBI (National Center for Biotechnology Information) zebrafish protein database (reference sequence 39495). Phylogenetic tree analysis was performed using the fast minimum evolution method (34). Domain features from different proteins were analyzed by the SMART program (http://smart.embl-heidelberg.de/) (35). The model structure of the proteins were generated using the SWISS-MODEL workspace (http://swissmodel.expasy.org/workspace/) (36). Multiple sequence alignments were performed as described previously (http://www.ebi.ac.uk/Tools/clustalw) (37).
Immunofluorescence staining.
To examine the intracellular distribution of PINCH in the presence or absence of ORF119L, MYC-ORF119L, MYC-ILK, and FLAG-PINCH fusion protein expressing plasmids were constructed. Full-length ORF119L and 119LΔ3ANK sequences were PCR amplified (primers in Table 1) by using the ISKNV genomic DNA and subcloned into pc-Myc-CMV-2 vector (Sigma, Ronkonkoma, NY, USA) to generate the MYC-ORF119L- and MYC-119LΔ3ANK-expressing plasmid pMYC-ORF119L and pMYC-119LΔ3ANK, respectively. The full-length zebrafish ILK and PINCH gene sequences were PCR amplified (primers in Table 1) by using the zebrafish cDNA, subcloned into pc-Myc-CMV-2 and pFLAG-CMV-2 vector (Sigma, Ronkonkoma, NY, USA), to generate the MYC-ILK-expressing plasmid pMYC-ILK and the FLAG-PINCH-expressing plasmid pFLAG-PINCH, respectively. Fathead minnow (FHM) cells were cultured on coverslips and transfected with pMYC-ORF119L, pMYC-ILK, and pFLAG-PINCH (as specified for each experiment). At 1 day posttransfection, the samples were fixed with methanol for 30 min at 4°C. After being washed in three changes of phosphate-buffered saline (PBS) and blocked (PBS with 10% normal blocking serum), samples were incubated with 1:500-diluted primary antibodies (rabbit anti-FLAG antibody and mouse anti-MYC antibody; Sigma) overnight at 4°C. After three washes, the cells were incubated in 1:500-diluted secondary antibodies (Alexa Fluor 488-conjugated goat anti-mouse IgG, heavy plus light chain [H+L], antibody and Alexa Fluor 555-conjugated goat anti-rabbit IgG [H+L] antibody; Life Technologies) for 1 h at room temperature in the dark. Hoechst 33342 was then applied for nucleus staining. After five rinses in PBS, cells were examined under a Zeiss LSM7 Duo NLO confocal microscope.
GST pulldown assay.
The full-length zebrafish ILK gene and ISKNV ORF119L were PCR amplified (primers in Table 1) and subcloned into pGEX-4T-1 vector (GE Healthcare) to generate the glutathione S-transferase (GST)-ILK-expressing plasmid pGST-ILK and the GST-ORF119L-expressing plasmid pGST-ORF119L, respectively. Plasmids pGEX-4T-1, pGST-ILK, and pGST-ORF119L were transformed into Escherichia coli BL21 cells to express GST, GST-ILK, and GST-ORF119L, respectively. Human embryonic kidney 293T (HEK293T) cells were cultured in Dulbecco's modified Eagle medium with 10% fetal bovine serum at 5% CO2. Plasmid pFLAG-PINCH was transfected (Lipofectamine 2000; Life Technologies) into HEK293T cells (cultured in 10-cm plates) to express the FLAG-PINCH fusion proteins. GST pulldown assays were performed as described previously (31), according to the manufacturer's instructions (MagneGST pull-down system; Promega, Madison, WI, USA). Briefly, 1 ml GST-, GST-ILK-, and GST-ORF119L-expressing BL21 bacterial cells were harvested, lysed with 200 μl of MagneGST cell lysis reagent, and incubated for 30 min on a rotating platform, and then precleared lysates were added to the tube containing the pre-equilibrated MagneGST glutathione particles (20 μl for each sample). After incubation for 30 min at room temperature, the GST control, GST-ILK, and GST-ORF119L immobilized particles were captured by a magnet stand; the particles were then washed with MagneGST binding/wash buffer three times for 5 min each and resuspended in 20 μl MagneGST binding/wash buffer. Aliquots of 5 μl of particles bound to the GST control and the GST-ILK and GST-ORF119L fusion proteins were saved for analysis of the specificity and efficiency of immobilization by Coomassie staining of SDS-PAGE gels. The FLAG-PINCH-expressing HEK293T cells were lysed with 1 ml cell lysis buffer (Beyotime, Jiangsu, China) containing a phosphatase/protease inhibitor cocktail. A 100-μl portion of cell lysate was stored at −20°C as a loading control, and the other 800 μl was added to the GST control, GST-ILK, and GST-ORF119L preimmobilized particles and incubated for 1 h at room temperature on a rotating platform. Nonspecific binding was removed by washing in 400 μl MagneGST binding/wash buffer five times for 5 min each. Finally, the FLAG-PINCH bound to particles was released by boiling in 20 μl 1× SDS loading buffer for 5 min. The samples were analyzed by Western blotting (rabbit anti-FLAG antibody; Life technologies).
Coimmunoprecipitation (co-IP) assay.
HEK293T cells were cultured in complete medium in 10-cm culture plates. Transfected cells (as specified for each experiment) were rinsed twice with cold PBS and lysed directly on the plate with 1 ml cell lysis buffer (Beyotime, Jiangsu, China) containing the phosphatase/protease inhibitor cocktail. Co-IP was performed according to the manufacturer's instructions (Dynabeads protein G immunoprecipitation kit; Life Technologies). Briefly, aliquots of 100 μl cell lysate were stored at −20°C as a loading control, and the other 800 μl lysate was added to mouse anti-MYC antibody (Sigma) preimmobilized on protein G beads and incubated for 1 h at room temperature. The beads were washed three times with PBS containing 0.1% Tween 20. Proteins bound to the beads were released by boiling in 20 μl 1× SDS loading buffer for 5 min. The samples were analyzed by Western blotting (rabbit anti-FLAG antibody; Life Technologies). For co-IP analysis with ORF119L-GFP coexpression, the 800 μl cell lysate was added to the prewashed anti-FLAG M2 affinity gel (FLAG-tagged protein immunoprecipitation kit; Sigma, Ronkonkoma, NY, USA). The mixture of cell lysate and anti-FLAG resin was incubated for 4 h at 4°C on a rotating platform. The beads were washed three times with 0.5 ml of 1× wash buffer (0.5 M Tris HCl, pH 7.4, with 1.5 M NaCl). Proteins bound to the beads were released by boiling for 5 min in 20 μl 1× SDS loading buffer. The samples were analyzed by Western blotting (mouse anti-MYC antibody; Sigma).
Whole-mount alkaline phosphatase (AP) staining.
Zebrafish embryos at 3 dpf were fixed in 4% paraformaldehyde (PFA) in PBS for 2 h at room temperature. Fixed embryos were dehydrated in methanol and stored overnight at −20°C. After permeabilization in acetone at −20°C for 30 min, embryos were washed in PBS and were incubated in staining buffer for 45 min as described previously (38, 39). Briefly, the staining reaction was started by adding 5 μl nitro blue tetrazolium–5-bromo-4-chloro-3-indolyl phosphate (NBT-BCIP; Roche: Basel, Switzerland) per milliliter of staining buffer and stopped by washing in PBS-Tween (PBST) buffer three times for 5 min each. The stained embryos were mounted in 70% glycerol, and all images were captured using an OlympusDP71 digital camera mounted on an Olympus MVX10 fluorescence stereomicroscope (Olympus, Tokyo, Japan).
Hematoxylin-eosin (H&E) and transmission electron microscopy (TEM) analysis.
For H&E staining, samples were collected and treated as described previously (40). Samples were paraffin sectioned at 5 μm using a Leica RM2145 microtome, and then H&E staining was performed using a standard protocol. For TEM, samples were dechorionated and fixed in Karnofsky's fixative (2% paraformaldehyde, 2.5% glutaraldehyde, 5% sucrose, 0.1% CaCl2 in 0.2 M cacodylate buffer [pH 7.2]) overnight at 4°C. Samples were then washed three times with 0.1 M phosphate buffer for 1 h at 4°C, dehydrated in a graduated ethanol series, and embedded in Spurr's resin. The blocks were sectioned and double-stained with uranyl acetate and lead citrate (41). The samples were examined under a Philips CM10 electron microscope (Philips, Eindhoven, Netherlands).
Whole-mount RNA in situ hybridization.
Atrial natriuretic factor (anf) is a cardiac stretch-responsive gene and used as a marker for development of zebrafish ventricle and atrium during early embryogenesis (21). Partial cDNA sequences of zebrafish anf were amplified by PCR and cloned into pGEM-T Easy vector (Promega, Madison, WI, USA) as templates to generate an antisense riboprobe (digoxigenin [DIG] RNA labeling kit; Roche Applied Science, Germany) for in situ hybridization in the embryos (31). Briefly, embryos were incubated in 0.003% 1-phenyl-2-thiourea (PTU; Sigma) to block pigmentation. Embryos were fixed at 4% PFA at room temperature for 4 h and dehydrated in methanol at −20°C overnight. After prehybridization at 65°C for 6 h, embryos were incubated with the anf antisense RNA probe (0.25 ng/μl) buffer at 65°C overnight. After washing and blocking, the embryos were incubated in alkaline phosphatase-conjugated sheep anti-digoxigenin Fab antibody at 4°C overnight. After 4 washes for 30 min each time in PBS with 0.1% Tween 20, the anf expression signal was detected by incubation in the NBT-BCIP substrate.
Antisense morpholino oligonucleotide-mediated gene knockdown.
Morpholino oligonucleotides (MO) (Gene Tools, LLC), an antisense technology used as a research tool for reverse genetics to knock down gene expression, have been successfully applied in the zebrafish model (42, 43). An antisense MO with a sequence targeting the translation initiation site (ATG) of the zebrafish ILK gene (ILK-MO; 3′-TACCTACTGTAGAAGTGAGTCACGG-5′) was injected into one-cell-stage embryos (21, 44, 45). A standard control MO (Ctrl-MO) antisense oligonucleotide was injected at the same concentrations (27). Embryos were maintained in E3 medium at 28°C until analyzed.
Quantitative real-time PCR (qRT-PCR) assay.
Total RNA was extracted from 30 embryos that had received different treatments and reverse transcribed into first-strand cDNA as described previously (31, 46). The anf-, nkx2.5-, and cmlc2-specific primers (Table 1) were used to analyze their transcription quantitatively in different groups of embryos by a LightCycler480 System (Roche, Germany). The transcription of anf, nkx2.5, and cmlc2 was assayed in triplicate. The zebrafish GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene was used as housekeeping gene to normalize the starting RNA quantity. The changes in gene transcription levels were calculated using the 2−ΔΔCT relative quantification method.
Statistical analysis.
The data are presented as means ± standard errors of the means (SEM). Student's t test was used to calculate the comparisons between groups of numerical data.
RESULTS
The sequence of ORF119L resembles the dominant negative form of ILK.
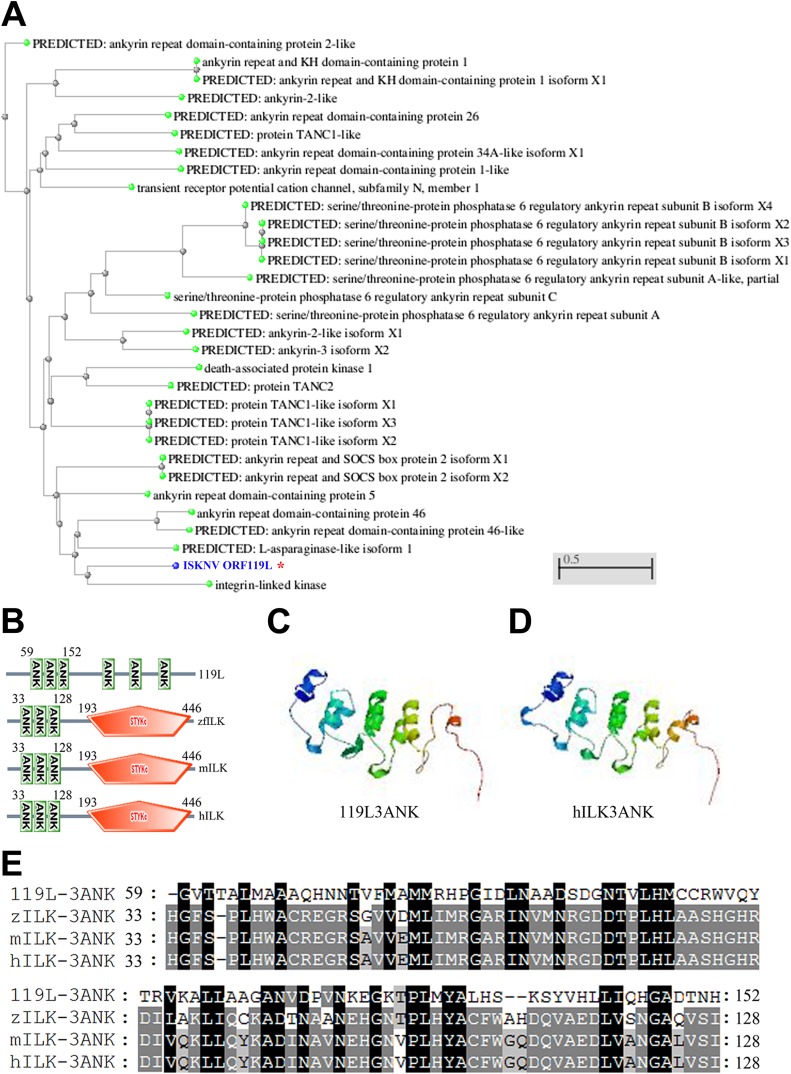
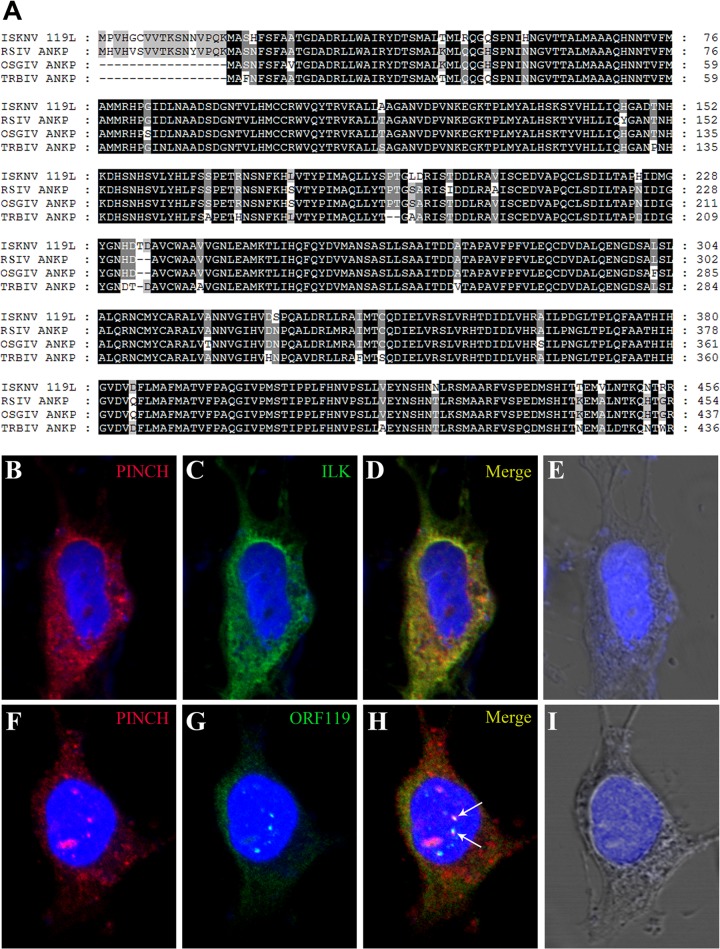
Through a genome-wide search for viral genes responsible for virus-host interactions that are critical for viral pathogenesis, we found several genes encoding ankyrin repeat-containing proteins in ISKNV. One of these genes, ISKNV ORF119L, 1,371 bp long, is predicted to encode a protein of 456 amino acid (aa) residues with a predicted molecular mass of 50.1 kDa. The BLAST analysis of ISKNV ORF119L revealed 30 highly similar sequences that are clustered with the gene for ILK (Fig. 1A). ISKNV ORF119L contains a sequence encoding a three-ankyrin-repeat domain (3ANK; 59 to 152 aa) which is aligned with ILK of zebrafish, mouse, and human (Fig. 1B). In zebrafish, mouse, and human ILK, a kinase domain is localized at the COOH-terminal end. However, in the COOH terminus of ORF119L, only three separate ankyrin motifs were found without a kinase domain (Fig. 1B). The model structure of the 3ANK domain from ISKNV ORF119L (Fig. 1C) shows high similarity to the 3ANK domain from human ILK (Fig. 1D). The 3ANK of ISKNV ORF119L is 42% identical to those of ILK from zebrafish, 43% identical to those of ILK from mouse, and 43% identical to those of ILK from human (Fig. 1E). In addition, ISKNV ORF119L shares an overall identity of 92%, as determined by multiple-sequence alignment analysis, with an orthologue found in red sea bream iridovirus (RSIV), 93% identity with a homolog in orange-spotted grouper iridovirus (OSGIV), and 92% identity with a homolog in turbot reddish body iridovirus (TRBIV) (Fig. 2A). Thus, ORF119L is evolutionarily conserved among megalocytiviruses, including RSIV, OSGIV, and TRBIV. All of these sequence analyses suggest that the viral 3ANK homologues might be unique among the megalocytiviruses, implying that they play a specific role in megalocytivirus pathogenesis. Specifically, we wanted to determine the functional consequence of ISKNV ORF119L expression in virus-host interactions.
FIG 1.
Sequence similarity alignment of ORF119L with the dominant negative form of ILK. (A) Phylogenetic tree analysis of ORF119L orthologues from NCBI zebrafish protein database. These orthologues score highest in levels of identity to ORF119L in the BLAST analysis. The GenBank accession numbers of proteins (in order) are as follows: XP_005157118.1, NP_001186697.1, XP_005161412.1, XP_697378.6, XP_001920876.2, XP_003199303.2, XP_005158347.1, XP_001920092.1, NP_899192.1, XP_689875.3, XP_005167841.1, XP_005167842.1, XP_005167840.1, XP_003200555.2, NP_001018164.1, XP_689244.2, XP_005166638.1, XP_005156096.1, NP_001093460.1, XP_002663935.3, XP_001920231.2, XP_005165903.1, XP_005165902.1, XP_005160666.1, XP_696390.3, NP_001020714.1, NP_991159.1, XP_002666119.1, XP_001923751.2, AAL98843, and NP_956865.1. (B) Analysis of the three-ankyrin-repeat-containing domain of ISKNV ORF119L and ILK proteins from zebrafish (zILK; GenBank accession no. AAH56593), mouse (mILK; GenBank accession no. NP_001155196), and human (hILK; GenBank accession no. CAG28601) with the SMART program (http://smart.embl-heidelberg.de). Compared to zILK, mILK, and hILK, the COOH terminus of ISKNV ORF119L lacks a kinase domain. The numbers indicate the positions of amino acid residues. (C and D) Model structures of the 119L 3ANK (C) and hILK 3ANK (D) domains were generated using the SWISS-MODEL Workspace. (E) Multiple-sequence alignment of the three-ankyrin-repeat-containing domains of ISKNV ORF119L and zebrafish, mice, and humans was performed by using the ClustalW program with default settings. 3ANK, three-ankyrin-repeat domain.
FIG 2.
Conservation of ORF119L among megalocytiviruses and alteration of cellular distribution of PINCH by coexpression of ORF119L. (A) ISKNV ORF119L (GenBank accession no. AAL98843) and the three-ankyrin-repeat-domain-containing proteins from orange-spotted grouper iridovirus (OSGIV) (GenBank accession no. AAX82423), turbot reddish body iridovirus (TRBIV) (GenBank accession no. ADE34453), red sea bream iridovirus (RSIV) (GenBank accession no. BAK14289) and rock bream iridovirus (RBIV) (GenBank accession no. AFR68193) were used for a multiple-sequence alignment analysis with default settings (http://www.ebi.ac.uk/Tools/msa/clustalw2/). (B to I) Immunofluorescence microscopy for cotransfection of MYC-ILK + FLAG-PINCH (B to E) and MYC-ORF119L + FLAG-PINCH (F to I) plasmids in FHM cells for immunofluorescence analysis. Alexa Fluor 555-conjugated (ILK and ORF119L, in red) or Alexa Fluor 488-conjugated (PINCH, in green) secondary antibodies were used for detection. ANKP, predicted three-ankyrin-repeat-domain-containing protein.
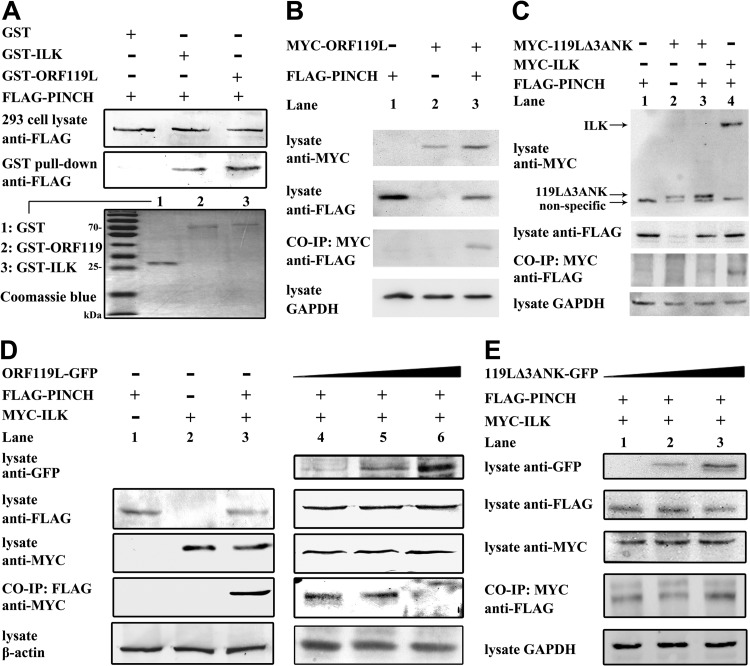
ORF119L interacts directly with PINCH to affect the PINCH-ILK interaction.
The NH2-terminal 3ANK domain of ILK is critical for its binding to the LIM domain of PINCH and the formation of PINCH-ILK-parvin complex in a host (22). Since we observed a high similarity between the 3ANK structure of the viral ORF119L and the host ILK, we hypothesized that ORF119L might directly bind to PINCH in virus-host interactions. An immunofluorescence microscopy analysis showed that zebrafish ILK and PINCH were colocalized in the cytoplasm in the absence of ORF119L in FHM cells (Fig. 2B to E). However, coexpression of viral ORF119L shifted PINCH to be colocalized with ORF119L in both the cytoplasm and nucleus (Fig. 2F to I), suggesting that ORF119L might directly interact with PINCH.
We performed a GST pulldown assay to explore the binding of ORF119L with PINCH (Fig. 3A). Coomassie blue staining of SDS-PAGE showed the homogeneity of GST, GST-ILK, and GST-ORF119L proteins obtained by GST tag affinity purification (Fig. 3A, panel 3 [panel numbers for this figure refer to the gels numbered from top to bottom]). When GST-, GST-ILK-, or GST-ORF119L-bound magnetic beads were incubated with a cellular lysate containing FLAG-tagged PINCH proteins, we found that PINCH specifically bound to GST-ILK fusion protein (positive control) (Fig. 3A, lane 2, panel 2) or GST-ORF119L fusion protein (Fig. 3A, lane 3, panel 2) but not to GST alone (negative control) (Fig. 3A, lane 1, panel 2), demonstrating that viral ORF119L directly bound to the zebrafish PINCH.
FIG 3.
ORF119 interacts with PINCH to affect the PINCH-ILK interaction. (A) In GST pulldown assays, the expression of FLAG-PINCH fusion proteins from transfected HEK293T cells was detected by anti-FLAG antibody Western blotting. After incubation with the GST-, GST-ILK-, and GST-ORF119L-bound beads, the associated FLAG-PINCH fusion proteins were detected by anti-FLAG antibody Western blotting. The purity of GST, GST-ILK, and GST-ORF119L was examined by Coomassie blue staining of SDS-PAGE gels. (B and C) In coimmunoprecipitation assays, plasmids pFLAG-PINCH + pc-Myc-CMV-2, pFLAG-CMV-2 + pMYC-ORF119L, and pFLAG-PINCH + pMYC-ORF119L (B) and plasmids pFLAG-PINCH + pc-Myc-CMV-2, pFLAG-CMV-2 + pMYC-119LΔ3ANK, pFLAG-PINCH + pMYC-119LΔ3ANK, and pFLAG-PINCH + pMYC-ILK (C) were cotransfected into HEK293T cells. The expression of MYC-ORF119L, MYC-ILK and FLAG-PINCH fusion proteins was examined by anti-MYC and anti-FLAG antibodies Western blotting respectively. After incubating with the anti-MYC antibody-bound protein G beads, the associated FLAG-PINCH fusion proteins were examined by anti-FLAG antibody Western blotting. GAPDH was used as the loading control. (D and E) In the coimmunoprecipitation assay, plasmids (pFLAG-PINCH + pc-Myc-CMV-2, pFLAG-CMV-2 + pMYC-ILK, and pFLAG-PINCH + pMYC-ILK) were cotransfected with pORF119L-EGFP (D, right panel, lanes 4 to 6) or p119LΔ3ANK-EGFP (E, lanes 1 to 3), respectively. The expression of ORF119L-GFP, 119LΔ3ANK-GFP, FLAG-PINCH, and MYC-ILK was detected by anti-GFP, anti-PINCH, and anti-MYC antibodies. After incubating with an anti-FLAG M2 affinity gel (D) or anti-MYC antibody-bound protein G beads (E), the associated MYC-ILK (D) or FLAG-PINCH (E) fusion proteins were examined by anti-MYC (D) or anti-FLAG (E) antibody Western blotting, respectively. β-Actin (D) and GAPDH (E) was used as the loading control, respectively.
We further confirmed the above results with a mammalian expression system. We coexpressed MYC-ORF119L and FLAG-PINCH in HEK293T cells and then performed a co-IP assay (Fig. 3B). When incubating the cell lysates with the anti-MYC antibody preimmobilized on protein G beads, we found that MYC-ORF119L specifically coimmunoprecipitated with FLAG-PINCH (Fig. 3B, lane 3, panel 3) but not with other controls (Fig. 3B, lane 1 and lane 2, panel 3), confirming the direct binding of ORF119L with PINCH. Subsequently, we generated an ORF119L mutant lacking the 3ANK-containing domain (designated 119LΔ3ANK) to investigate the binding domain of ORF119L to PINCH by the co-IP assay. Cell lysates containing FLAG-PINCH (Fig. 3C, lane 1), MYC-119LΔ3ANK (Fig. 3C, lane 2), FLAG-PINCH + MYC-119LΔ3ANK (Fig. 3C, lane 3), or FLAG-PINCH + MYC-ILK (Fig. 3C, lane 4) fusion proteins were separately incubated with the anti-MYC antibody-preimmobilized protein G beads. We found that FLAG-PINCH specifically coimmunoprecipitated with MYC-ILK (Fig. 3C, lane 4, panel 3, as a positive control) but not with the 119LΔ3ANK (Fig. 3C, lane 3, panel 3), demonstrating that the deletion of the 3ANK-containing domain abolished the ORF119L-PINCH interaction.
We then tried to test whether ORF119L could affect the PINCH-ILK interaction in cells. A co-IP assay was performed with coexpression of PINCH and ILK in cells in the absence or presence of ORF119L (Fig. 3D). In the absence of ORF119L, cell lysates containing FLAG-PINCH, MYC-ILK, or FLAG-PINCH + MYC-ILK fusion proteins were separately incubated with anti-FLAG (M2) monoclonal antibody-conjugated agarose beads. After three washes to remove nonspecific binding and then followed with anti-MYC antibody detection, we found that MYC-ILK was coimmunoprecipitated with FLAG-PINCH in the sample containing FLAG-PINCH and MYC-ILK (Fig. 3D, lane 3, left panel 3), but not in the sample containing FLAG-PINCH alone (Fig. 3D, lane1, left panel 3), or MYC-ILK alone (Fig. 3D, lane 2, left panel 3), demonstrating that PINCH interacts with ILK. However, in the presence of ORF119L in the same system as above, when ORF119L expression was increased (Fig. 3D, lanes 4 to 6, right panel 1), the bound MYC-ILK fusion proteins were decreased (Fig. 3D, lanes 4 and 5, right panel 4) and diminished (Fig. 3D, lane 6, right panel 4). This suggests that FLAG-PINCH binding to MYC-ILK was attenuated with the increasing levels of ORF119L expression, implying that ISKNV ORF119L competed with ILK for binding PINCH in a dose-dependent manner. To test the specificity of the competitive binding effect of ISKKV ORF119L, we performed the PINCH-ILK co-IP assay in the presence of the 119LΔ3ANK mutant. Importantly, when coexpression of 119LΔ3ANK mutant was increased (Fig. 3E, panel 1), the bound FLAG-PINCH fusion proteins was not affected (Fig. 3E, panel 4), demonstrating that the 3ANK-containing domain is critical for the competitive binding activity of ORF119L to PINCH.
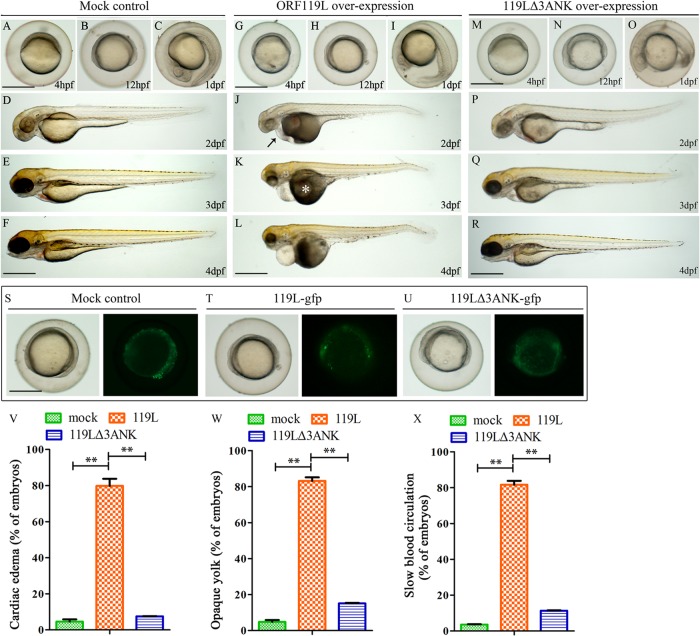
ORF119L overexpression affects zebrafish embryogenesis.
To explore the functions of ORF119L in vivo, we microinjected a recombinant plasmid expressing ORF119L-EGFP into wild-type zebrafish embryos while using an empty vector as a control. While the mock vector-injected embryos did not show any phenotypic abnormality from 4 hpf to 4 dpf (Fig. 4A to F), the ORF119L-EGFP-injected embryos developed normally at 4 hpf (Fig. 4G), 12 hpf (Fig. 4H), and 1 dpf (Fig. 4I). However, pericardial edema (Fig. 4J, arrow) and opaque yolk sac phenotype (Fig. 4K, asterisk) were evident at 2 dpf and 3 dpf, respectively, in the ORF119L-EGFP-injected embryos. These embryos became less active at 4 dpf (Fig. 4L), and most of them were dead at 5 dpf (data not shown). In contrast, embryos overexpressing the 119LΔ3ANK mutant were relatively normal from 4 hpf to 4 dpf (Fig. 4M to R). We observed GFP expression among the mock vector-injected control and ORF119L-EGFP- and 119LΔ3ANK-EGFP-expressing embryos at 12 hpf (Fig. 4S to U), indicating that ORF119L expression contributed to the abnormal embryogenesis. In ORF119L-expressing embryos at 3 dpf, we found statistically significant developmental defects, including cardiac edema (Fig. 4V) (mock vector-injected control, 4.58% ± 2.18%; ORF119L, 79.79% ± 6.93%; P = 0.005), opaque yolk (Fig. 4W) (mock vector-injected control, 4.80% ± 1.79%; ORF119L, 83.12% ± 3.51%; P = 7E-04), and slow blood cell circulation (Fig. 4X) (mock vector-injected control, 3.56% ± 0.37%; ORF119L, 81.65% ± 3.76%; P = 9E-04). In contrast, these defects were significantly reduced in 119LΔ3ANK-expressing embryos (Fig. 4V to X).
FIG 4.
ORF119L overexpression perturbs zebrafish embryogenesis. Morphology of mock vector-injected (A to F), ORF119L-overexpressing (G to L), and 119LΔ3ANK-overexpressing (M to R) embryos from 4 hpf to 4 dpf. Abnormal phenotypes were found in ISKNV ORF119L-overexpressing embryos (G to L). A black arrow indicates pericardial edema, and a white asterisk indicates opaque yolk sac abnormality. (S to U) GFP expression in mock vector-, pORF119L-EGFP-, or p119LΔ3ANK-EGFP-injected embryos at 12 hpf. (V to X). Percentage of embryos showing cardiac edema (V), opaque yolk sac (W), and slow blood circulation (X) phenotype at 3 dpf. Scale bars = 500 μm. **, P < 0.01.
ORF119L overexpression disturbs embryonic angiogenesis in zebrafish.
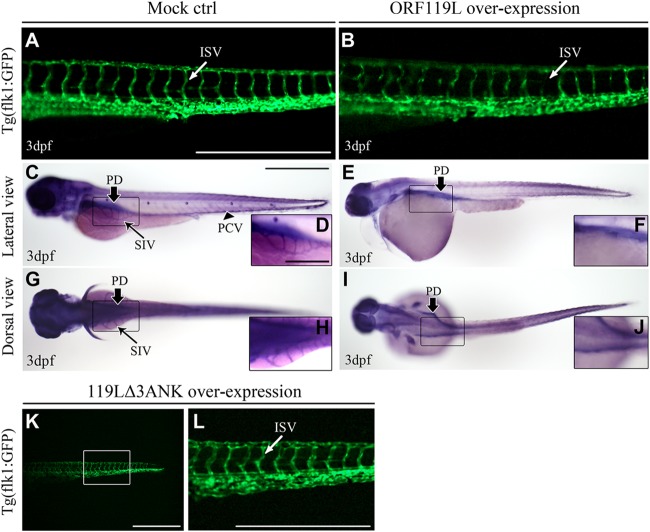
Previous reports showed that ILK is crucial for the vascular basement membrane development during some angiogenic sprouting (47, 48). Thus, we tested whether ORF119L could affect embryonic angiogenesis. An RFP-ORF119L (red-fluorescent-protein-tagged ORF119L)-expressing plasmid was microinjected into Tg(flk1:GFP) transgenic zebrafish embryos (GFP expression is driven by the vascular endothelium cell-specific promoter flk1). Compared to the normal conformation of intersegmental vessel (ISV) in mock vector-injected embryos at 3 dpf (Fig. 5A), ISV was severely disrupted in ORF119L-expressing embryos (Fig. 5B). In a whole-mount AP staining assay, pronephric ducts (PD), subintestinal vessels (SIV), and posterior cardinal veins (PCV) were normal in the control embryos (Fig. 5C, D, G, and H). However, overexpressing ORF119L led to the absence of SIV and PCV but no effects on PD (Fig. 5E, F, I, and J). In contrast, embryos expressing mutant 119LΔ3ANK did not show any ISV defects (Fig. 5K and L).
FIG 5.
ORF119L overexpression affects angiogenesis in zebrafish embryos. (A and B) The ISV structure was examined in Tg(flk1:GFP) transgenic embryos. ISV development was normal in mock vector-injected embryos (A), whereas disorganized ISV was found after ORF119L overexpression (B) at 3 dpf. (C to J) Whole-mount AP staining showing the vessel development in control (C, D, G, and H) and ORF119L-overexpressing (E, F, I, and J) embryos at 3 dpf. SIV was impaired after ORF119L overexpression. (K and L) ISV structure was normal in 119LΔ3ANK-expressing embryos at 3 dpf. ISV, intersegmental vessel; PCV, posterior cardinal vein; DA, dorsal aorta; PD, pronephric duct; SIV, subintestinal vessel. Scale bars = 500 μm (A, B, C, E, G, I, K, and L) and 200 μm (D, F, H, and J).
ORF119L overexpression induces cardiac defects in zebrafish.
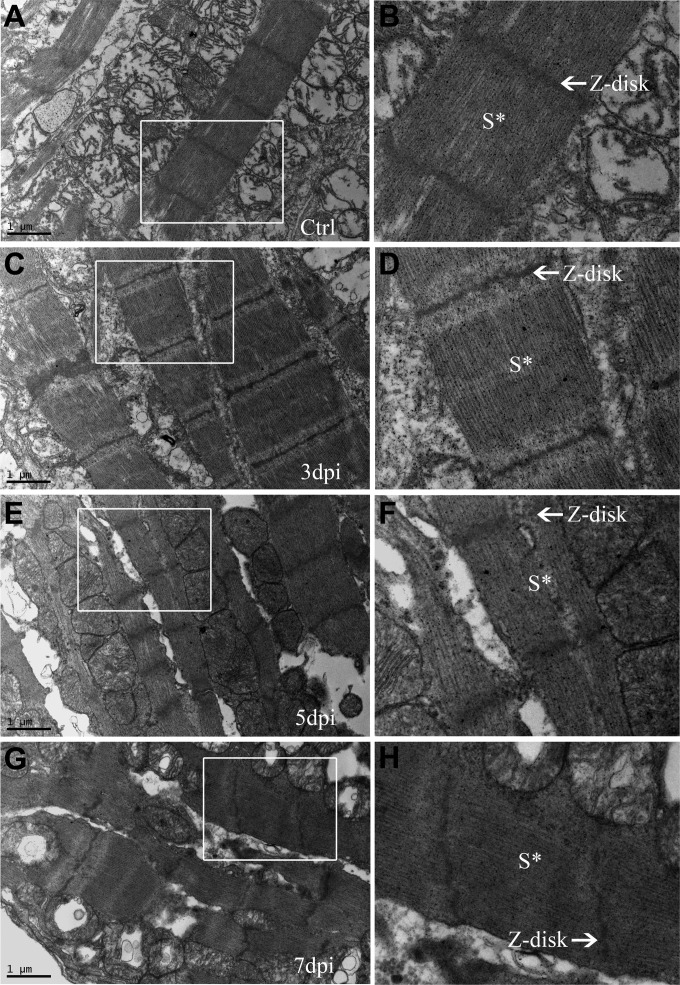
ORF119L overexpressing embryos make evident pericardial edema and stretched heart, which are highly similar to the abnormal phenotype of zebrafish ILKL308P kinase-dead mutant (21, 49). This led us to perform a histological and ultracellular analysis to delineate the detailed cardiac defects in ORF119L embryos. In an H&E staining assay, the structures of ventricle and atrium were normal in mock vector-injected control embryos (Fig. 6A and C). However, stretched hearts (Fig. 6B), enlarged pericardial cavities (Fig. 6D, PC), and thinner ventricular walls (Fig. 6D, arrowheads) were clearly observed in ORF119L-expressing embryos. In a TEM analysis of the embryonic ventricle, sarcomeric structures were clearly found in control embryos (Fig. 6E, S*), whereas they were sparse and immature in ORF119L-expressing embryos (Fig. 6F, S*). Distinct Z disks, A bands, and I bands in the sarcomere were observed in the control embryos (Fig. 6G). In contrast, sarcomeric Z disks disappeared in ORF119L-expressing embryos (Fig. 6H). Intriguingly, a similar disruption of the sarcomeric Z disk was also found in mandarin fish cardiomyocytes 5 days (Fig. 7E and F) and 7 days (Fig. 7G and H) after ISKNV infection.
FIG 6.
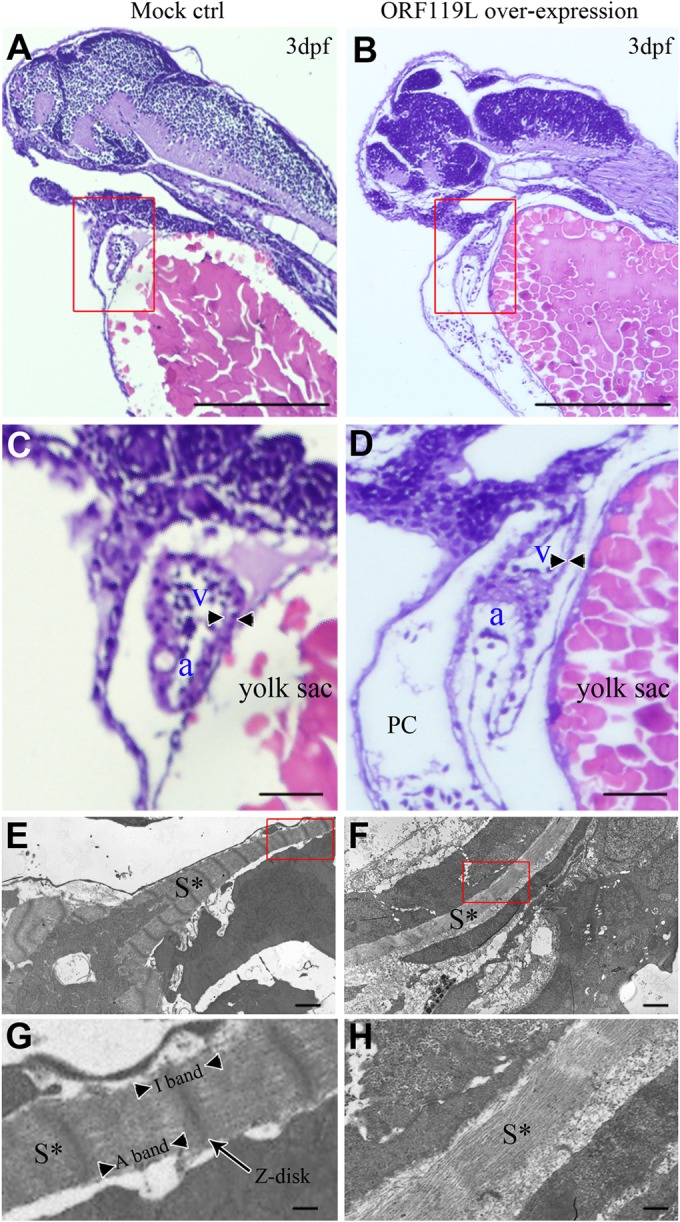
ORF119L overexpression results in cardiac defects in zebrafish. (A to D) Paraffin section and H&E staining in control (A and C) and ORF119L-overexpressing (B and D) embryos at 3 dpf. The arrowheads indicate the ventricular wall. (E to H) TEM assay detecting the ultracellular structure of ventricle muscle in control (E and G) and ORF119L-overexpressing embryos (F and H) at 3 dpf. Images in panels C, D, G, and H showed higher magnifications of the boxed areas in panels A, B, E, and F, respectively. PC, pericardial cavity; v, ventricle; a, atrium; S*, sarcomeric structure. Bars = 500 μm (A and B), 100 μm (C and D), 2 μm (E and F), and 500 nm (G and H).
FIG 7.
ISKNV infection disrupted the sarcomeric Z disks of cardiomyocytes in mandarin fish. The structures of the sarcomeres and Z disks of cardiac muscle in PBS-injected control (A and B) and ISKNV-infected mandarin fish at 3 (C and D), 5 (E and F), and 7 (G and H) days postinfection are shown. Images in panels B, D, F, and H are higher magnifications of the boxed areas in panels A, C, E, and G. S*, sarcomeric structure.
ORF119L overexpression resembles the effect of ILK inhibition.
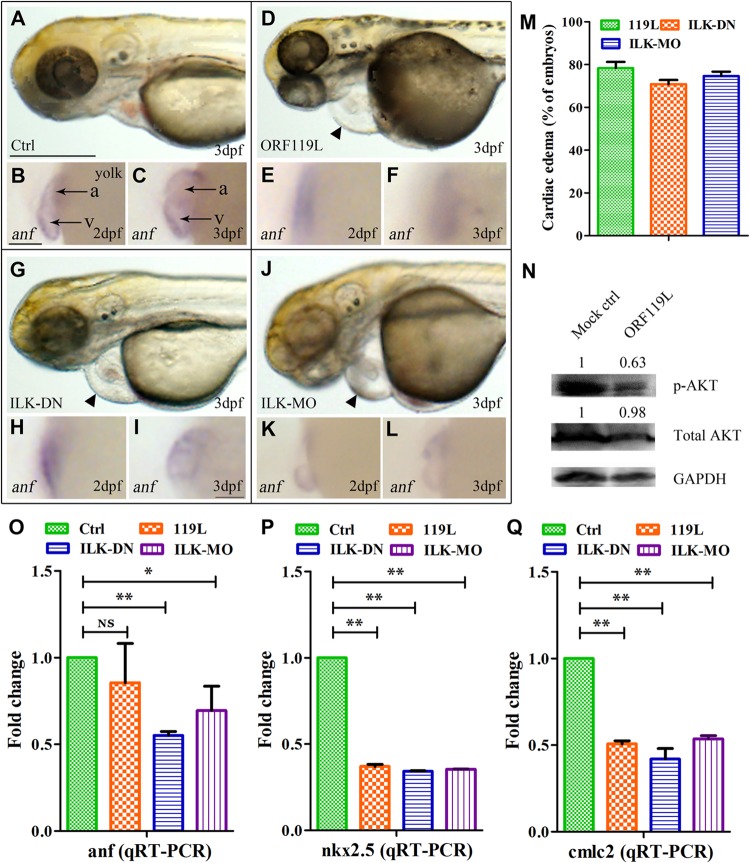
All of the above results prompted us to speculate on a mechanism by which the ISKNV ORF119L-induced abnormal phenotype might contribute to dysfunctional ILK signaling. We blocked endogenous ILK expression, either by expressing the dominant negative form of ILK (ILK-DN) or by injecting an ILK antisense morpholino oligonucleotide (ILK-MO). While the heart development in the standard control MO (Ctrl-MO)-injected embryos (Fig. 8A), as well as the mock vector-injected (control) and 119LΔ3ANK-overexpressing embryos (as shown in Fig. 4A to F and M to R), was relatively normal at 3 dpf, inhibition of endogenous ILK by either ILK-DN overexpression (Fig. 8G, arrowhead) or ILK-MO injection (Fig. 8J, arrowhead) resulted in evident pericardial edema similar to the pattern found in ORF119L-expressing embryos (Fig. 8D, arrowhead, and M). Because the heart beating rate was hard to quantify by using our experimental device, here we used the gene for atrial natriuretic factor (anf) (21) as a marker of heart development in the whole-mount in situ hybridization analysis. While anf transcription was normal in ventricles and atria in Ctrl-MO-injected embryos at 2 dpf (Fig. 8B) and 3 dpf (Fig. 8C), anf transcription was impaired in ORF119L-expressing embryos (Fig. 8E and F), ILK-DN-expressing embryos (Fig. 8H and I), and ILK-MO-expressing embryos (Fig. 8K and L), respectively. In addition, the percentage of cardiac edema was similar among the ORF119L-overexpressing, ILK-DN-overexpressing, and ILK-MO-injected embryos at 3 dpf (Fig. 8M). Furthermore, phosphorylation of AKT, a downstream effector of ILK, was decreased in ORF119L-expressing embryos (Fig. 8N). Compared with the Ctrl-MO-injected embryos, ORF119L-overexpressing, ILK-DN-overexpressing, and ILK-MO-injected embryos showed decreased transcription of the cardiac markers anf (Fig. 8O), nkx2.5 (Fig. 8P), and cmlc2 (Fig. 8Q) by the qRT-PCR assay, demonstrating that ORF119L may affect the cardiac function.
FIG 8.
ORF119L overexpression resembles the effect of ILK inhibition. (A to L) Developmental morphology and whole-mount in situ hybridization of anf in Ctrl-MO control embryos (A to C), ORF119L-overexpressing embryos (D to F), ILK-DN-expressing embryos (G to I), and ILK-MO-injected embryos (J to L). Arrowheads indicate pericardial edema. (M) Percentage of ORF119L-overexpressing, ILK-DN-overexpressing, and ILK-MO-injected embryos showing pericardial edema at 3 dpf. (N) Fifty 3-day-old embryos were collected and lysed by using radioimmunoprecipitation assay (RIPA) lysis buffer containing phosphatase/protease inhibitor cocktail (Merck). Western blotting was performed to examine the phosphorylation of AKT (Ser473) and total AKT in control and ORF119L-overexpressing embryos at 3 dpf. The relative levels of p-AKT and total AKT were calculated by normalizing to their corresponding GAPDH levels (Quantity One 4.6.2 software program). (O to Q) Quantitative RT-PCR showing the change in levels of anf (O), nkx2.5 (P), and cmlc2 (Q) transcription in Ctrl-MO-injected, ORF119L-overexpressing, ILK-DN-overexpressing, and ILK-MO-injected embryos at 3 dpf. v, ventricle; a, atrium. Scale bars = 500 μm (A, D, G, and J) and 100 μm (B, C, E, F, H, I, K, and L). *, P < 0.05; **, P < 0.01.
DISCUSSION
In this study, we identified a three ankyrin-repeat-domain-containing protein (ISKNV ORF119L) from a megalocytivirus. In vitro studies show that ORF119L directly interacts with zebrafish PINCH and affects the binding of PINCH to ILK. In vivo studies show that overexpression of ORF119L in zebrafish predominantly affects the angiogenic and cardiac systems. Phosphorylation of AKT, a downstream effector of ILK, was decreased in ORF119L-overexpressing embryos. Intriguingly, ISKNV infection alters the cardiac sarcomeric structure of mandarin fish. In addition, ORF119L-induced phenotypes are similar to the abnormality derived from the expression of dominant negative ILK kinase-dead mutant in zebrafish.
When a mutant subunit of a multisubunit complex is coexpressed with a functionally related wild-type protein, a dysfunctional complex is formed due to the dominant negative inhibitory (DNI) effect (50, 51). The DNI phenomenon has been observed in mammals, such as inhibition of wild-type p53 by ΔNp73 (52), NF-κB by IκBm (53), C/EBP by CHOP (54), and promyelocytic leukemia zinc finger protein (PLZF) by AML-1/ETO fusion protein (34). Viral DNI factors also have been reported, such as the human cytomegalovirus-derived truncated unique short 3 (US3) isoform (55) and Epstein-Barr virus EBNA-1 (56). In this study, we demonstrate that escalated expression of ISKNV ORF119L attenuates PINCH-ILK interaction, affects cardiomyocyte development in zebrafish, and decreases AKT phosphorylation. In fact, we further show that the 119LΔ3ANK mutant did not bind to PINCH and the escalated expression of 119LΔ3ANK mutant did not show a competitive binding effect on the PINCH-ILK interaction. Moreover, overexpression of the 119LΔ3ANK mutant in vivo did not affect the embryonic morphology or the cardiovascular system, confirming that the ILK-like dominant negative inhibiting effect of ISKNV ORF119L is due to the 3ANK-containing domain. Therefore, we propose that ISKNV ORF119L is a novel ILK-like dominant negative inhibitor, most likely due to the fact that ISKNV ORF119L contains a 3ANK-containing domain and lacks the kinase domain of ILK.
ILK-mediated PINCH/AKT signal transduction functions as the cardiac mechanical stretch sensor machinery and has a role that is crucial for contractility in the zebrafish heart (21, 57). In an ethylnitrosourea (ENU) mutagenesis screening, Bendig and colleagues (21) identified a main squeeze (msq) zebrafish mutant, which contains a point mutation (L308P) in the kinase domain of ILK. The msq ILKL308P mutation leads to reduced ILK kinase activity and cardiac defects in zebrafish embryos (21). In line with these findings, we show that ORF119L is a three-ankyrin-repeat-domain-containing protein without a kinase domain. Overexpression of ORF119L induces a stretched heart in zebrafish. In addition, we observed reduced expression of the cardiac marker anf by whole-mount in situ hybridization (WISH) and qRT-PCR assay (not significant in the qRT-PCR assay because of the variation of the triplicate experiments) in ORF119L-overexpressing embryos. Likewise, additional cardiac markers such as nkx2.5 and cmlc2 were consistently and significantly reduced after ORF119L overexpression, providing evidence that ORF119L may affect the cardiac function. We hypothesized that the cardiac defects may result from disrupting the ILK-PINCH interaction, thereby decreasing the phosphorylation of AKT. In fact, both ILK and PINCH localize at the sarcomeric Z disk in the zebrafish heart and skeletal muscle (21, 57) and are pivotal for the sarcomeric cytoarchitecture and Z-disk integrity (58). In keeping with these observations, we also show the disorganized sarcomere and disintegrated Z disks of cardiomyocytes in ORF119L-expressing embryos. Intriguingly, ISKNV infection leads to disordered sarcomeres and Z disks in mandarin fish cardiomyocytes, providing a potential explanation for low vitality in mandarin fish showing accelerated breathing and dyspnea, as previously described (59). Taken together, our studies on ORF119L might provide novel insights into the pathogenesis of ISKNV infection.
The complex formed by PINCH-ILK-parvin provides crucial physical linkages between integrins and the actin cytoskeleton for transducing diverse signals from the extracellular matrix to intracellular effectors (60). In this study, we demonstrated that ORF119L attenuates the binding of PINCH to ILK and induces cardiomyocytes defects in zebrafish. However, the effects of ORF119L on the function of Parvin and on the function of spleen or kidney necrosis remain to be determined.
ACKNOWLEDGMENTS
We sincerely thank Zi-Liang Wang, Xiao-Peng Xu, Jun-Feng Xie, and Jing Wang for technical assistance, Chuan-Fu Dong for providing the ISKNV virus and fish cell lines, Qiu-Ling Liang for help with cell cultures, and Hai-Bin Liu for careful zebrafish maintenance. We thank Wen-qing Zhang (Southern Medical University, Guangzhou, China) for providing the zebrafish line.
This work was supported by the National Natural Science Foundation of China under grants 31330080, 31322056, and 31370048, the National Basic Research Program of China (973 Program) (no. 2012CB114402), the Guangdong Natural Science Foundation (no. S2013010012161), the Pearl River Nova Program of Guangzhou (no. 2014J2200055), the Foundation for Young Teachers (no. 20130171220009), and the doctoral student innovative talent training projects of Sun Yat-sen University.
REFERENCES
- 1.He JG, Deng M, Weng SP, Li Z, Zhou SY, Long QX, Wang XZ, Chan SM. 2001. Complete genome analysis of the mandarin fish infectious spleen and kidney necrosis iridovirus. Virology 291:126–139. doi: 10.1006/viro.2001.1208. [DOI] [PubMed] [Google Scholar]
- 2.Lu L, Zhou SY, Chen C, Weng SP, Chan SM, He JG. 2005. Complete genome sequence analysis of an iridovirus isolated from the orange-spotted grouper, Epinephelus coioides. Virology 339:81–100. doi: 10.1016/j.virol.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 3.Chen XH, Lin KB, Wang XW. 2003. Outbreaks of an iridovirus disease in maricultured large yellow croaker, Larimichthys crocea (Richardson), in China. J Fish Dis 26:615–619. doi: 10.1046/j.1365-2761.2003.00494.x. [DOI] [PubMed] [Google Scholar]
- 4.Sudthongkong C, Miyata M, Miyazaki T. 2002. Iridovirus disease in two ornamental tropical freshwater fishes: African lampeye and dwarf gourami. Dis Aquat Organ 48:163–173. doi: 10.3354/dao048163. [DOI] [PubMed] [Google Scholar]
- 5.Dong C, Weng S, Shi X, Xu X, Shi N, He J. 2008. Development of a mandarin fish Siniperca chuatsi fry cell line suitable for the study of infectious spleen and kidney necrosis virus (ISKNV). Virus Res 135:273–281. doi: 10.1016/j.virusres.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Wang YQ, Lu L, Weng SP, Huang JN, Chan SM, He JG. 2007. Molecular epidemiology and phylogenetic analysis of a marine fish infectious spleen and kidney necrosis virus-like (ISKNV-like) virus. Arch Virol 152:763–773. doi: 10.1007/s00705-006-0870-4. [DOI] [PubMed] [Google Scholar]
- 7.Werden SJ, Lanchbury J, Shattuck D, Neff C, Dufford M, McFadden G. 2009. The myxoma virus m-t5 ankyrin repeat host range protein is a novel adaptor that coordinately links the cellular signaling pathways mediated by Akt and Skp1 in virus-infected cells. J Virol 83:12068–12083. doi: 10.1128/JVI.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werden SJ, Barrett JW, Wang G, Stanford MM, McFadden G. 2007. M-T5, the ankyrin repeat, host range protein of myxoma virus, activates Akt and can be functionally replaced by cellular PIKE-A. J Virol 81:2340–2348. doi: 10.1128/JVI.01310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanie S, Gelfi J, Bertagnoli S, Camus-Bouclainville C. 2010. MNF, an ankyrin repeat protein of myxoma virus, is part of a native cellular SCF complex during viral infection. Virol J 7:56. doi: 10.1186/1743-422X-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonnberg S, Fleming SB, Mercer AA. 2011. Phylogenetic analysis of the large family of poxvirus ankyrin-repeat proteins reveals orthologue groups within and across chordopoxvirus genera. J Gen Virol 92:2596–2607. doi: 10.1099/vir.0.033654-0. [DOI] [PubMed] [Google Scholar]
- 11.Sonnberg S, Seet BT, Pawson T, Fleming SB, Mercer AA. 2008. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc Natl Acad Sci U S A 105:10955–10960. doi: 10.1073/pnas.0802042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng X, Xiang Y. 2006. Vaccinia virus K1L protein supports viral replication in human and rabbit cells through a cell-type-specific set of its ankyrin repeat residues that are distinct from its binding site for ACAP2. Virology 353:220–233. doi: 10.1016/j.virol.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 13.Guo CJ, Chen WJ, Yuan LQ, Yang LS, Weng SP, Yu XQ, He JG. 2011. The viral ankyrin repeat protein (ORF124L) from infectious spleen and kidney necrosis virus attenuates nuclear factor-kappaB activation and interacts with IkappaB kinase beta. J Gen Virol 92:1561–1570. doi: 10.1099/vir.0.031120-0. [DOI] [PubMed] [Google Scholar]
- 14.Wickstrom SA, Lange A, Montanez E, Fassler R. 2010. The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase! EMBO J 29:281–291. doi: 10.1038/emboj.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legate KR, Montanez E, Kudlacek O, Fassler R. 2006. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat Rev Mol Cell Biol 7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- 16.Chiswell BP, Zhang R, Murphy JW, Boggon TJ, Calderwood DA. 2008. The structural basis of integrin-linked kinase-PINCH interactions. Proc Natl Acad Sci U S A 105:20677–20682. doi: 10.1073/pnas.0811415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Wang X, Hawkins CA, Chen K, Vaynberg J, Mao X, Tu Y, Zuo X, Wang J, Wang YX, Wu C, Tjandra N, Qin J. 2009. Structural basis of focal adhesion localization of LIM-only adaptor PINCH by integrin-linked kinase. J Biol Chem 284:5836–5844. doi: 10.1074/jbc.M805319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu C. 1999. Integrin-linked kinase and PINCH: partners in regulation of cell-extracellular matrix interaction and signal transduction. J Cell Sci 112:4485–4489. [DOI] [PubMed] [Google Scholar]
- 19.Velyvis A, Yang Y, Wu C, Qin J. 2001. Solution structure of the focal adhesion adaptor PINCH LIM1 domain and characterization of its interaction with the integrin-linked kinase ankyrin repeat domain. J Biol Chem 276:4932–4939. doi: 10.1074/jbc.M007632200. [DOI] [PubMed] [Google Scholar]
- 20.Moik D, Bottcher A, Makhina T, Grashoff C, Bulus N, Zent R, Fassler R. 2013. Mutations in the paxillin-binding site of integrin-linked kinase (ILK) destabilize the pseudokinase domain and cause embryonic lethality in mice. J Biol Chem 288:18863–18871. doi: 10.1074/jbc.M113.470476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bendig G, Grimmler M, Huttner IG, Wessels G, Dahme T, Just S, Trano N, Katus HA, Fishman MC, Rottbauer W. 2006. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes Dev 20:2361–2372. doi: 10.1101/gad.1448306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Guo L, Chen K, Wu C. 2002. A critical role of the PINCH-integrin-linked kinase interaction in the regulation of cell shape change and migration. J Biol Chem 277:318–326. doi: 10.1074/jbc.M108257200. [DOI] [PubMed] [Google Scholar]
- 23.Guo L, Wu C. 2002. Regulation of fibronectin matrix deposition and cell proliferation by the PINCH-ILK-CH-ILKBP complex. FASEB J 16:1298–1300. doi: 10.1096/fj.02-0089fje. [DOI] [PubMed] [Google Scholar]
- 24.Ellett F, Lieschke GJ. 2010. Zebrafish as a model for vertebrate hematopoiesis. Curr Opin Pharmacol 10:563–570. doi: 10.1016/j.coph.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Eimon PM, Ashkenazi A. 2010. The zebrafish as a model organism for the study of apoptosis. Apoptosis 15:331–349. doi: 10.1007/s10495-009-0432-9. [DOI] [PubMed] [Google Scholar]
- 26.Ma AC, Chung MI, Liang R, Leung AY. 2010. A DEAB-sensitive aldehyde dehydrogenase regulates hematopoietic stem and progenitor cells development during primitive hematopoiesis in zebrafish embryos. Leukemia 24:2090–2099. doi: 10.1038/leu.2010.206. [DOI] [PubMed] [Google Scholar]
- 27.Wang ZL, Xu XP, He BL, Weng SP, Xiao J, Wang L, Lin T, Liu X, Wang Q, Yu XQ, He JG. 2008. Infectious spleen and kidney necrosis virus ORF48R functions as a new viral vascular endothelial growth factor. J Virol 82:4371–4383. doi: 10.1128/JVI.02027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang Z, Dong C, Qi L, Chen W, Huang L, Li Z, Xia Q, Liu D, Huang M, Weng S, He J. 2010. Characteristics of the interferon regulatory factor pairs zfIRF5/7 and their stimulation expression by ISKNV Infection in zebrafish (Danio rerio). Dev Comp Immunol 34:1263–1273. doi: 10.1016/j.dci.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Xu X, Huang L, Wu J, Lu Q, Xiang Z, Liao J, Weng S, Yu X, He J. 2010. Administration of recombinant IFN1 protects zebrafish (Danio rerio) from ISKNV infection. Fish Shellfish Immunol 29:399–406. doi: 10.1016/j.fsi.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 30.Xu X, Zhang L, Weng S, Huang Z, Lu J, Lan D, Zhong X, Yu X, Xu A, He J. 2008. A zebrafish (Danio rerio) model of infectious spleen and kidney necrosis virus (ISKNV) infection. Virology 376:1–12. doi: 10.1016/j.virol.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 31.He BL, Yuan JM, Yang LY, Xie JF, Weng SP, Yu XQ, He JG. 2012. The viral TRAF protein (ORF111L) from infectious spleen and kidney necrosis virus interacts with TRADD and induces caspase 8-mediated apoptosis. PLoS One 7:e37001. doi: 10.1371/journal.pone.0037001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 34.Desper R, Gascuel O. 2002. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol 9:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- 35.Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res 30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 37.Fukami-Kobayashi K, Saito N. 2002. How to make good use of CLUSTALW. Tanpakushitsu Kakusan Koso 47:1237–1239. (In Japanese.) [PubMed] [Google Scholar]
- 38.Habeck H, Odenthal J, Walderich B, Maischein H, Schulte-Merker S. 2002. Analysis of a zebrafish VEGF receptor mutant reveals specific disruption of angiogenesis. Curr Biol 12:1405–1412. doi: 10.1016/S0960-9822(02)01044-8. [DOI] [PubMed] [Google Scholar]
- 39.Serbedzija GN, Flynn E, Willett CE. 1999. Zebrafish angiogenesis: a new model for drug screening. Angiogenesis 3:353–359. doi: 10.1023/A:1026598300052. [DOI] [PubMed] [Google Scholar]
- 40.Kishi S, Bayliss PE, Uchiyama J, Koshimizu E, Qi J, Nanjappa P, Imamura S, Islam A, Neuberg D, Amsterdam A, Roberts TM. 2008. The identification of zebrafish mutants showing alterations in senescence-associated biomarkers. PLoS Genet 4:e1000152. doi: 10.1371/journal.pgen.1000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo Y, Weng S, Wang Q, Shi X, Dong C, Lu Q, Yu X, He J. 2009. Tiger frog virus can infect zebrafish cells for studying up- or down-regulated genes by proteomics approach. Virus Res 144:171–179. doi: 10.1016/j.virusres.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 42.Yuan S, Sun Z. 2009. Microinjection of mRNA and morpholino antisense oligonucleotides in zebrafish embryos. J Vis Exp 7:1113. doi: 10.3791/1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. 2009. A primer for morpholino use in zebrafish. Zebrafish 6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogel B, Meder B, Just S, Laufer C, Berger I, Weber S, Katus HA, Rottbauer W. 2009. In-vivo characterization of human dilated cardiomyopathy genes in zebrafish. Biochem Biophys Res Commun 390:516–522. doi: 10.1016/j.bbrc.2009.09.129. [DOI] [PubMed] [Google Scholar]
- 45.Zhang R, Yang J, Zhu J, Xu X. 2009. Depletion of zebrafish Tcap leads to muscular dystrophy via disrupting sarcomere-membrane interaction, not sarcomere assembly. Hum Mol Genet 18:4130–4140. doi: 10.1093/hmg/ddp362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang R, Dodd A, Lai D, McNabb WC, Love DR. 2007. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim Biophys Sin (Shanghai) 39:384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wani AA, Jafarnejad SM, Zhou J, Li G. 2011. Integrin-linked kinase regulates melanoma angiogenesis by activating NF-kappaB/interleukin-6 signaling pathway. Oncogene 30:2778–2788. doi: 10.1038/onc.2010.644. [DOI] [PubMed] [Google Scholar]
- 48.Hynes RO, Lively JC, McCarty JH, Taverna D, Francis SE, Hodivala-Dilke K, Xiao Q. 2002. The diverse roles of integrins and their ligands in angiogenesis. Cold Spring Harbor Symp Quant Biol 67:143–153. doi: 10.1101/sqb.2002.67.143. [DOI] [PubMed] [Google Scholar]
- 49.Postel R, Vakeel P, Topczewski J, Knoll R, Bakkers J. 2008. Zebrafish integrin-linked kinase is required in skeletal muscles for strengthening the integrin-ECM adhesion complex. Dev Biol 318:92–101. doi: 10.1016/j.ydbio.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 50.Michaels JE, Schimmel P, Shiba K, Miller WT. 1996. Dominant negative inhibition by fragments of a monomeric enzyme. Proc Natl Acad Sci U S A 93:14452–14455. doi: 10.1073/pnas.93.25.14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melnick A, Carlile GW, McConnell MJ, Polinger A, Hiebert SW, Licht JD. 2000. AML-1/ETO fusion protein is a dominant negative inhibitor of transcriptional repression by the promyelocytic leukemia zinc finger protein. Blood 96:3939–3947. [PubMed] [Google Scholar]
- 52.Zaika AI, Slade N, Erster SH, Sansome C, Joseph TW, Pearl M, Chalas E, Moll UM. 2002. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med 196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou M, Gu L, Zhu N, Woods WG, Findley HW. 2003. Transfection of a dominant-negative mutant NF-kB inhibitor (IkBm) represses p53-dependent apoptosis in acute lymphoblastic leukemia cells: interaction of IkBm and p53. Oncogene 22:8137–8144. doi: 10.1038/sj.onc.1206911. [DOI] [PubMed] [Google Scholar]
- 54.Ron D, Habener JF. 1992. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev 6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 55.Shin J, Park B, Lee S, Kim Y, Biegalke BJ, Kang S, Ahn K. 2006. A short isoform of human cytomegalovirus US3 functions as a dominant negative inhibitor of the full-length form. J Virol 80:5397–5404. doi: 10.1128/JVI.02397-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirchmaier AL, Sugden B. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J Virol 71:1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meder B, Huttner IG, Sedaghat-Hamedani F, Just S, Dahme T, Frese KS, Vogel B, Kohler D, Kloos W, Rudloff J, Marquart S, Katus HA, Rottbauer W. 2011. PINCH proteins regulate cardiac contractility by modulating integrin-linked kinase-protein kinase B signaling. Mol Cell Biol 31:3424–3435. doi: 10.1128/MCB.05269-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perkins AD, Ellis SJ, Asghari P, Shamsian A, Moore ED, Tanentzapf G. 2010. Integrin-mediated adhesion maintains sarcomeric integrity. Dev Biol 338:15–27. doi: 10.1016/j.ydbio.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 59.He J, Zeng K, Weng S, Chan S-M. 2002. Experimental transmission, pathogenicity and physical–chemical properties of infectious spleen and kidney necrosis virus (ISKNV). Aquaculture 204:11–24. doi: 10.1016/S0044-8486(01)00639-1. [DOI] [Google Scholar]
- 60.Wu C. 2004. The PINCH-ILK-parvin complexes: assembly, functions and regulation. Biochim Biophys Acta 1692:55–62. doi: 10.1016/j.bbamcr.2004.01.006. [DOI] [PubMed] [Google Scholar]