ABSTRACT
HIV-1 acquires an impressive number of foreign components during its formation. Despite all previous efforts spent studying the nature and functionality of virus-anchored host molecules, the exact mechanism(s) through which such constituents are acquired by HIV-1 is still unknown. However, in the case of ICAM-1, one of the most extensively studied transmembrane proteins found associated with mature virions, the Pr55Gag precursor polyprotein appears to be a potential interaction partner. We investigated and characterized at the molecular level the process of ICAM-1 incorporation using initially a Pr55Gag-based virus-like particle (VLP) model. Substitution of various domains of Pr55Gag, such as the nucleocapsid, SP2, or p6, had no effect on the acquisition of ICAM-1. We found that the structural matrix protein (MA) is mandatory for ICAM-1 incorporation within VLPs, and we confirmed this novel observation with the replication-competent HIV-1 molecular clone NL4.3. Additional studies suggest that the C-terminal two-thirds of MA, and especially 13 amino acids positioned inside the fifth α-helix, are important. Moreover, based on three-dimensional (3D) modeling of protein-protein interactions (i.e., protein-protein docking) and further validation by a virus capture assay, we found that a series of acidic residues in the MA domain interact with basic amino acids located in the ICAM-1 cytoplasmic tail. Our findings provide new insight into the molecular mechanism governing the acquisition of ICAM-1, a host molecule known to enhance HIV-1 infectivity in a significant manner. Altogether, these observations offer a new avenue for the development of antiviral therapeutics that are directed at a target of host origin.
IMPORTANCE Intercellular adhesion molecule 1 (ICAM-1) is a cell surface host component known to be efficiently inserted within emerging HIV-1 particles. It has been demonstrated that host-derived ICAM-1 molecules act as a strong attachment factor and increase HIV-1 infectivity substantially. Despite previous efforts spent studying virus-associated host molecules, the precise mechanism(s) through which such constituents are inserted within emerging HIV-1 particles still remains obscure. Previous data suggest that the Pr55Gag precursor polyprotein appears as a potential interaction partner with ICAM-1. In the present study, we demonstrate that the HIV-1 matrix domain plays a key role in the ICAM-1 incorporation process. Some observations were confirmed with whole-virus preparations amplified in primary human cells, thereby providing physiological significance to our data.
INTRODUCTION
Nascent HIV-1 particles incorporate a large spectrum of proteins of host origin during assembly and budding processes (1–3). Such host-derived molecules have been found either located inside viral particles or embedded in the virus envelope (ENV), depending on their natural location in target cells (2). For example, different cytoplasmic constituents are acquired by HIV-1, such as actin, actin-binding proteins, and EF-1α (3, 4). Several other intracellular host proteins have also been detected within virions, such as Tsg101, Staufen, INI1, and cyclophilin A, with each of them participating in distinct steps of the viral life cycle (1, 3). HIV-1 also incorporates various transmembrane proteins that are found anchored in the viral ENV, of which ICAM-1 is probably the most extensively studied (5). Despite numerous studies aimed at the identification of virus-anchored host molecules, the exact mechanism(s) through which such cellular constituents are acquired by emerging HIV-1 particles remains obscure.
ICAM-1 is a heavily glycosylated transmembrane protein that contains five distinct extracellular domains, a transmembrane region, and a short cytoplasmic tail of 28 amino acids. The intracellular region interacts with cytoskeletal proteins such as α-actinin, ezrin, and actin (6, 7). Moreover, it has been demonstrated that the cytoplasmic tail of ICAM-1 associates with phosphatidylinositol 4,5-biphosphate [PI(4,5)P2] through a basic region resembling the PI(4,5)P2-binding site. This results in an electrostatic interaction, which facilitates the association between ICAM-1 and the actin-binding protein ezrin (7). The major biological role of ICAM-1 is to mediate cell-to-cell adhesion via interactions with cell surface β2 integrins such as LFA-1 and Mac-1. This type of association is essential for leukocyte transmigration and immunological synapse formation, two phenomena associated with inflammation and the establishment of the immune response. ICAM-1 is continuously expressed on endothelial cells and leukocytes, particularly on CD4+ T cells and macrophages, the two major target cell types of HIV-1.
The efficient incorporation of ICAM-1 within HIV-1 has been demonstrated with different laboratory and clinical isolates of HIV-1 that were produced in established cell lines, primary human cells (i.e., peripheral blood mononuclear cells [PBMCs]), and human lymphoid tissue cultured ex vivo (8–11). Interestingly, it has been firmly established that the insertion of host-derived ICAM-1 within the virus ENV leads to increases in both HIV-1 attachment and entry within CD4+ T cells (12, 13). It has been reported that interactions between virus-associated host ICAM-1 and cell surface LFA-1 allow such virions to attach more firmly and fuse more rapidly with target cells than isogenic viruses lacking the adhesion molecule (8–10, 12). The higher permissiveness of both memory and activated CD4+ T cells to productive infection with ICAM-1-bearing virions is attributable mostly to surface expression of an activated form of LFA-1 (14). Given that virus-associated host ICAM-1 significantly augments virus infectivity (e.g., up to 100-fold), we were interested in studying the structural determinants responsible for ICAM-1 incorporation because this information could serve to develop new classes of compounds that can abolish the insertion of host-derived ICAM-1 in emerging HIV-1 particles.
Pr55Gag is the structural viral constituent acting as the main actor in HIV-1 particle formation as it drives assembly through protein-protein, protein-RNA, and protein-lipid interactions, orchestrating the incorporation of each of the major virus-encoded components into the assembled particle (15–18). Pr55Gag is translated from unspliced viral mRNA on free ribosomes in the cytoplasm and encodes the internal structural components of the virion, i.e., matrix (MA), capsid (CA), and nucleocapsid (NC), along with the C-terminal p6 domain and two spacer peptides, SP1 and SP2 (19). In the absence of any other viral proteins, Pr55Gag can self-assemble into virus-like particles (VLPs) by ordered multimerization of monomers and/or oligomers to produce a spherical shell (20), which forms the structural framework of the immature virus particle (18, 21).
Several regions of the Pr55Gag polyprotein have been reported to be crucial for particle formation. Notably, previous genetic and structural analyses demonstrated that regions responsible for membrane binding are located at the N-terminal region of MA, which encompasses a cluster of basic residues and a myristyl group interacting with acidic phospholipids of the membrane PI(4,5)P2 (22, 23). However, several studies have shown that MA can be largely deleted or even entirely replaced by a heterologous myristyl anchor without compromising the formation of extracellular viruses (24–26). This suggests that the role of MA in assembly is not as important as its role in Pr55Gag trafficking and in the incorporation of virus-encoded ENV glycoproteins (27, 28).
In the current study, we first show that Pr55Gag is sufficient by itself to lead to the insertion of host-derived ICAM-1 molecules in a VLP model devoid of any other HIV-1 constituents. Data from a virus capture assay indicate that functional VLP mutants carrying substitutions of either NC, SP2, or p6 can still acquire host ICAM-1. Experiments with additional mutants, using VLPs (i.e., MAΔ44-132) as well as a molecular clone of HIV-1 (i.e., NL4.3Δ44-132), suggest that a two-thirds portion of MA is responsible for ICAM-1 incorporation. Using a protein structure modeling approach to engineer additional mutants, we further demonstrate that the insertion of host-derived ICAM-1 molecules in emerging virions requires interactions between certain acidic amino acids in the HIV-1 MA domain and basic residues within the ICAM-1 cytoplasmic tail.
MATERIALS AND METHODS
Ethics statement.
Human PBMCs were obtained from anonymous and paid healthy volunteer donors who were specifically solicited for the provision of these samples. Healthy subjects signed an informed consent form approved by the Institutional Review Board (IRB) of the Centre Hospitalier Universitaire de Québec. The current research project was also approved by our IRB.
Cell culture and transfection.
The human embryonic kidney cell line 293T was used to produce our virus preparations (i.e., VLPs and NL4.3-derived virus). This cell line was maintained in Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Burlington, Ontario, Canada) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Burlington, Ontario, Canada) at 37°C under a 5% CO2 atmosphere. A cell line stably expressing ICAM-1 was obtained by cotransfecting 293T cells with a pCDNA-3.1(+) expression vector containing the entire human ICAM-1 cDNA and pCMV-Hygro at a 10:1 ratio. Such stably transfected 293T cells were maintained under selective pressure with hygromycin B (400 mg/ml; Calbiochem) for 3 weeks and were next isolated by fluorescence-activated cell sorter analysis with the use of a phycoerythrin (PE)-labeled anti-ICAM-1 monoclonal antibody (BD Biosciences, Mississauga, Canada) (11). For calcium phosphate transfection, 40 to 50% confluent 293T cells in T75 flasks (BD, Oakville, Canada) were transfected as described previously (9, 29). Briefly, 15 μg of the different plasmids were used to transfect 293T cells. At 6 h posttransfection, the medium was discarded, cells were washed two times with phosphate-buffered saline (PBS), and 10 ml of fresh medium was added. Immature HIV-1 particles were produced by treating the virus producer cell lines with indinavir (20 μM) during transfection experiments (30). PBMCs from healthy donors were isolated by Ficoll-Hypaque density gradient centrifugation and cultured in the presence of the mitogenic agent phytohemagglutinin P (PHA-P; Sigma, St. Louis, MO, USA) (3 μg/ml) and recombinant human interleukin-2 (30 units/ml) for 3 days at 37°C under a 5% CO2 atmosphere prior to virus infection. Acute HIV-1 infection was pursued for 5 days before performance of a virus immunocapture test (see below for more details).
Molecular constructs.
The VLP constructs P6449LD, NCP6378LZLD, and MAΔ44-132 were described previously (31). Additional HIV-1 mutants were derived from the fully infectious pNL4.3 molecular clone. Matrix regions were first PCR amplified from the proviral DNAs. Specifically, we amplified a region of Gag extending from the BssHII to the SphI unique restriction sites, which includes the MA coding region. The PCR products were TA cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA), and the BssHII-SphI Gag fragments were subsequently excised by restriction enzyme digestion (32). The Gag mutant cassettes were then subcloned into the pNL4.3 proviral backbone with the desired deletion or alanine substitution. Full-length human ICAM-1 cDNA was inserted in the pCDNA3.1(+) expression vector (Invitrogen, Burlington, Canada). ICAM-1 mutants were PCR amplified with Phusion high-fidelity polymerase (ThermoFisher Scientific, Ottawa, Ontario, Canada) by using 5′-phosphorylated primers. The PCR products were cloned back into the empty pCDNA3.1(+) backbone with the desired deletion or alanine substitution. Primers used for mutagenesis are summarized in Table 1.
TABLE 1.
Primers used for mutagenesis
| Mutant | Primer sequence |
|
|---|---|---|
| Forwarda | Reverse | |
| NL4.3Δ44-132 | CCTATAGTGCAGAACCTCCAG | TCGTTCTAGCTCCCTGCTTGC |
| NL4.3 Del 102-114 | GCACAGCAAGCAGCAGCT | TAAGGCTTCCTTGGTGTCTTT |
| D102A/E105A/E106A | AAGGAAGCCTTAGCTAAGATAGCAGCAGAGCAAAACAAAAGT | GGTGTCTTTTACATCTATCCTTTGATGCACACAATAGAG |
| D102A/E105A/E106A/E107A | AAGGAAGCCTTAGCTAAGATAGCAGCAGCACAAAACAAAAGT | GGTGTCTTTTACATCTATCCTTTGATGCACACAATAGAG |
| D102A/E105A/E106A/E107A/S111A | GCAGCACAAAACAAAGCTAAGAAAAAGGCACAGCAAGCA | TGCTATCTTAGCTAAGGCTTCCTTGGTGTCTTTTACATC |
| CD54 Del 505-514 | CAACAGGCCCAAAAAGGG | GTTATAGAGGTACGTGCTGAGGCC |
| CD54 R505,507,513A K508,510,511A | TACCTCTATAACGCCCAGGCCGCAATCGCCGCATACGCACTACAACAGGCC | CGTGCTGAGGCCTGCAGTGCCCATTATGACTGCGGCTGCTAC |
Underlined nucleotides correspond to the mutated codons for alanine substitution.
Preparation of virus stocks.
VLPs and NL4.3-derived virus (both wild-type [WT] and mutated forms) that bear host-derived ICAM-1 were produced by calcium phosphate transfection in 293T cells stably expressing ICAM-1, while isogenic virus preparations lacking ICAM-1 were prepared in the parent 293T cell line (which is ICAM-1 negative). At 48 h posttransfection, cell-free supernatants were collected and centrifuged at 4°C for 5 min at 1,000 × g and filtered through 0.22-μm-pore-size cellulose acetate membranes (ThermoFisher Scientific, Ottawa, Canada) to remove cells debris. The cell-free supernatants were then ultracentrifuged through a 2-ml cushion of 20% sucrose in TSE (10 mM Tris hydrochloride [pH 7.5], 100 mM NaCl, and 1 mM EDTA) at 4°C for 45 min at 40,000 rpm (Beckman Ti70 rotor) (2 ml of cushion for 10 ml of supernatants) (26, 31). The pellets were finally resuspended in PBS. The different Pr55Gag-derived VLP preparations were normalized by Western blot quantification using anti-p24 monoclonal antibody 183-H12-5C (see Fig. 2 and 3), whereas NL4.3-derived VLP stocks (see Fig. 4 and 6 to 10) were normalized by using a sensitive in-house sandwich enzyme-linked immunosorbent assay (ELISA) that can detect both p24 and the Pr55Gag precursor, as previously described (8). Finally, samples were aliquoted before storage at −80°C.
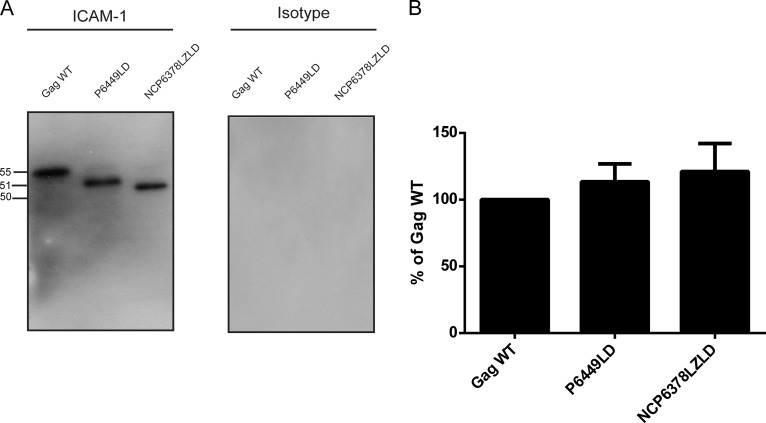
FIG 2.
Immunocapture of VLPs carrying mutations in NC, SP2, and p6 domains. (A) VLPs produced by 293T cells stably expressing ICAM-1 were captured by using magnetic beads coated with either an anti-ICAM-1 (R6.5) or isotype-matched irrelevant biotinylated antibody. Next, VLPs were estimated by Western blotting using an anti-p24 monoclonal antibody (183-H12-5C) recognizing both the Pr55Gag precursor polyprotein and p24. The primary antibody was revealed with a horseradish peroxidase-conjugated anti-mouse secondary antibody. (B) Pr55Gag precursor polyprotein signals were determined by densitometry analyses of Western blot films using ImageJ software. The area density for each band was defined and compared to the area density for the Gag WT (considered 100%). Data shown represent results from three independent experiments for each mutant, and error bars indicate standard deviations.
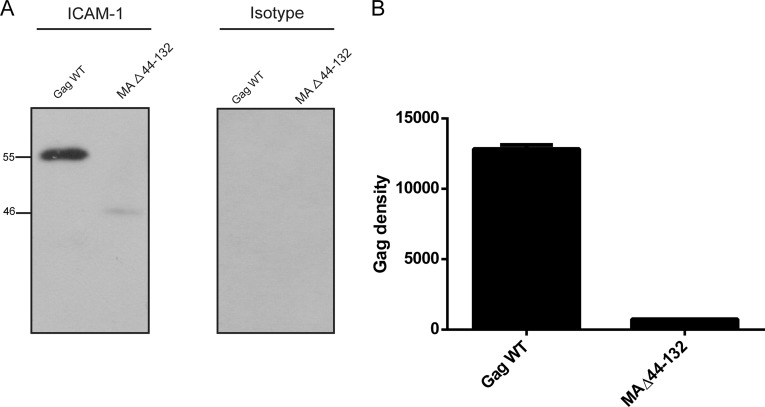
FIG 3.
Immunocapture of VLPs carrying mutations in the MAΔ44-132 region. (A) VLPs produced by 293T cells stably expressing ICAM-1 were captured by using magnetic beads coated with either an anti-ICAM-1 (R6.5) or isotype-matched irrelevant biotinylated antibody. Next, VLPs were estimated by Western blotting using an anti-p24 monoclonal antibody (183-H12-5C) recognizing both the Pr55Gag precursor polyprotein and p24. The primary antibody was revealed with a horseradish peroxidase-conjugated anti-mouse secondary antibody. (B) The Pr55Gag precursor polyprotein signals were determined by densitometry analyses of Western blot films using ImageJ software. Data shown represent results from three independent experiments for each mutant, and error bars indicate standard deviations.
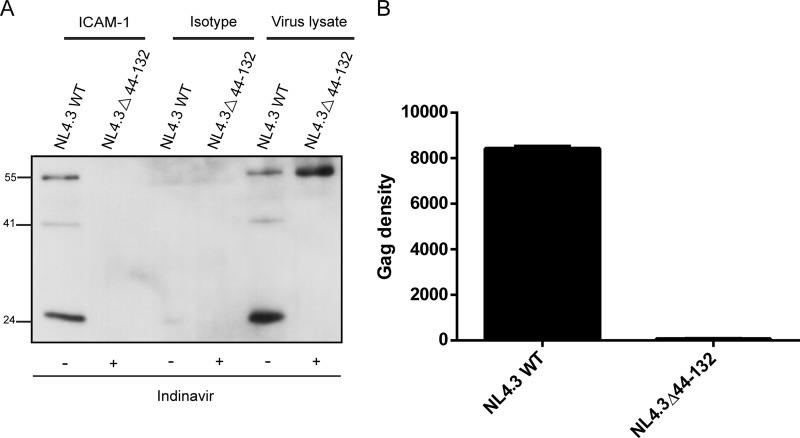
FIG 4.
Immunocapture of NL4.3-derived virus carrying mutations in the MAΔ44-132 region. (A) Complete NL4.3-derived virus particles produced by 293T cells stably expressing ICAM-1 were captured by using magnetic beads coated with either an anti-ICAM-1 (R6.5) or an isotype-matched irrelevant biotinylated antibody. Next, immunocaptured viruses were estimated by Western blotting using an anti-p24 monoclonal antibody (183-H12-5C) recognizing both the Pr55Gag precursor polyprotein and p24. The primary antibody was revealed with a horseradish peroxidase-conjugated anti-mouse secondary antibody. (B) The Pr55Gag precursor polyprotein signals were determined by densitometry analyses of Western blot films using ImageJ software. Data shown represent results from three independent experiments for each mutant, and error bars indicate standard deviations.
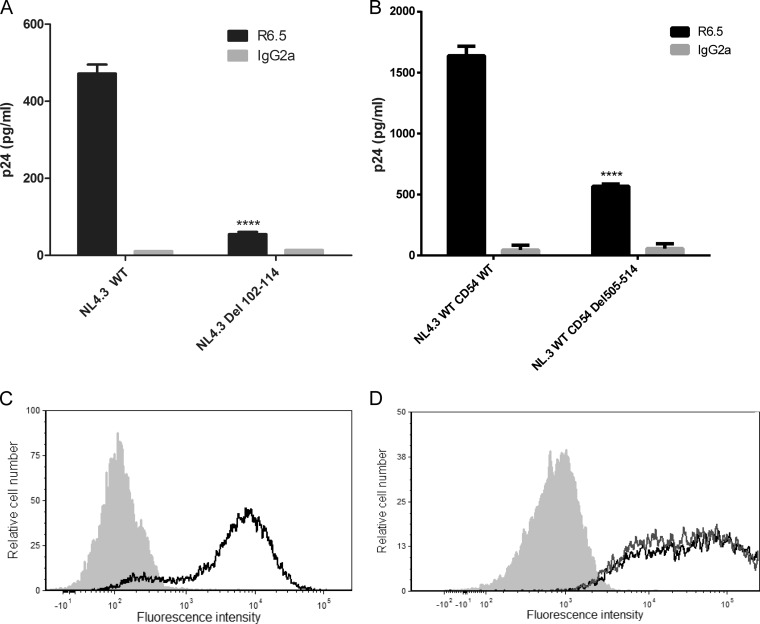
FIG 6.
Immunocapture of NL4.3-derived Del 102-114 mutant virus and NL4.3-derived virus carrying Del 505-514 mutant ICAM-1. (A) NL4.3-derived WT and mutant virus particles were prepared in 293T cells stably expressing wild-type ICAM-1. (B) NL4.3-derived WT viruses were produced in 293T cells transiently expressing either wild-type ICAM-1 or Del 505-514 mutant ICAM-1. Next, virus stocks were captured by using magnetic beads coated with an anti-ICAM-1 antibody (R6.5) or an isotype-matched irrelevant biotinylated antibody (used as a control). Precipitated viral entities were quantified with a homemade p24 ELISA. Data shown represent the means ± standard deviations of data from triplicate samples (i.e., total amounts of virus captured with anti-ICAM-1 divided by total amounts of virus captured with the control antibody) and are representative of three independent experiments. (C and D) Flow cytometry analyses of ICAM-1 surface expression in 293T cells either stably (C) or transiently (D) expressing ICAM-1. Wild-type ICAM-1 is depicted as a solid black line (C and D), Del 505-514 mutant ICAM-1 is shown as a solid gray line (D), and the control is presented as a gray-filled histogram. Data shown represent cell surface ICAM-1 expression after 1 month of selective drug pressure for stable transfectants (C) and 2 days after transfection for transient transfectants (D). Statistical analyses were made by using the Student t test, and asterisks denote a statistically significant difference between the NL4.3 WT or NL4.3 WT CD54 WT and the listed mutants (****, P < 0.0001).
FIG 10.
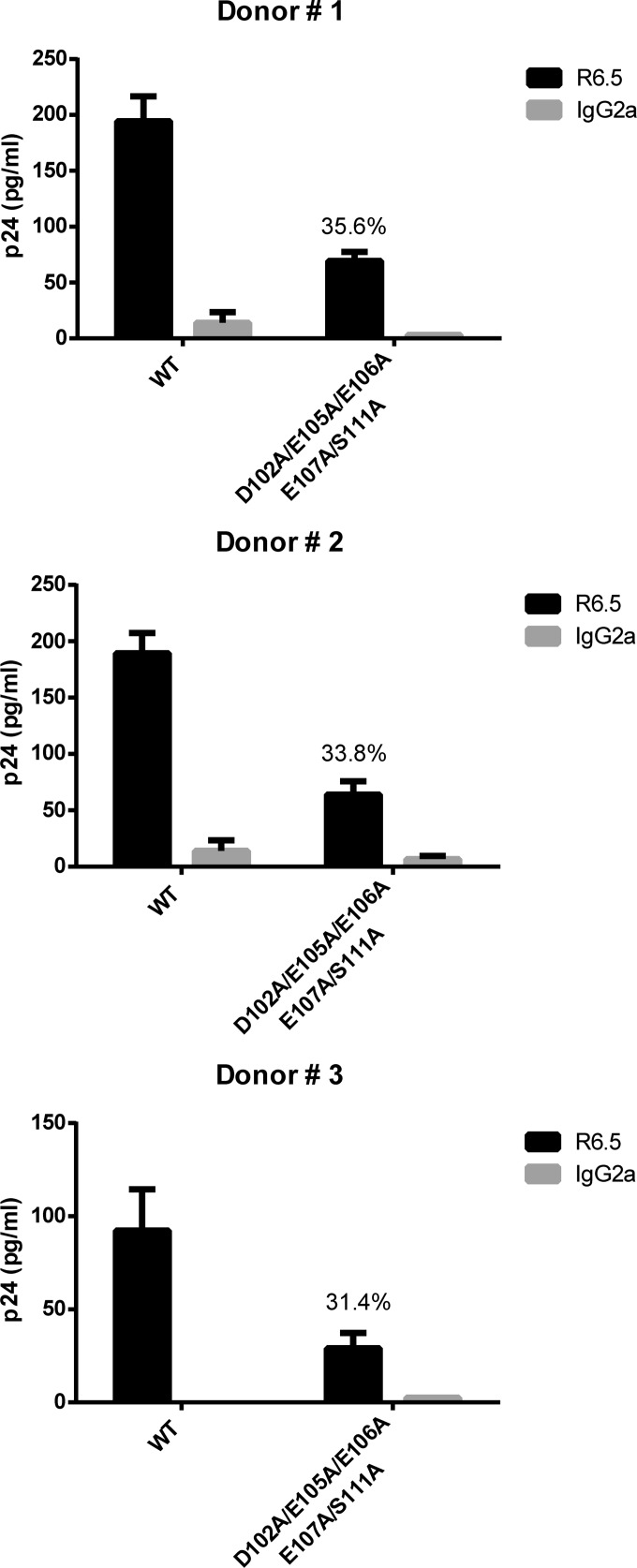
Immunocapture of NL4.3-derived WT and mutant viruses produced in primary human cells. NL4.3-derived WT virions and mutant virus particles carrying the listed alanine substitutions were obtained from mitogen-treated PBMCs from three different healthy donors. Virus infection was allowed to proceed for 5 days before capture of progeny virus with magnetic beads coated with either an anti-ICAM-1 antibody (R6.5) or an isotype-matched irrelevant biotinylated antibody (used as a control). Precipitated viral entities were quantified with a homemade p24 ELISA. Data shown represent the means ± standard deviations of data from triplicate samples and are representative of three independent experiments. The percentage of ICAM-1 incorporation for each individual donor is denoted on each corresponding graphic.
Immunocapture.
The presence of host-derived ICAM-1 within VLPs and NL4.3-derived virus (both WT and mutated forms) was monitored by capturing viruses with immunomagnetic beads using a previously reported procedure, with slight modifications (30). In brief, streptavidin-coated magnetic beads (7.5 × 105 beads) (Dynabeads M280; Invitrogen, Burlington, Canada) were washed once with PBS supplemented with 5% bovine serum albumin (BSA) (binding medium) using a vertical magnetic plate. Next, beads were incubated with a biotinylated anti-ICAM-1 antibody (clone R6.5; IgG2a) for 1 h at room temperature and then washed three times with PBS before use. The immunomagnetic beads were next incubated for 16 h at 4°C with the virus preparations (2 and 40 ng of p24 for virus stocks prepared in 293T cells and PBMCs, respectively) under gentle agitation. The beads were washed four times with 1 volume of binding medium and were finally resuspended in a final volume of 250 μl of PBS. Finally, the amount of immunocaptured VLPs was determined by SDS-PAGE and Western blot experiments against the Pr55Gag precursor, while the amount of immunocaptured NL4.3-derived virus was determined by measuring the viral p24 protein by an ELISA. Beads coated with biotinylated isotype-matched irrelevant antibodies were used as negative controls.
Flow cytometry analysis.
For staining, cells were washed twice with PBS and centrifuged at 300 × g for 5 min at 4°C. Pelleted cells were incubated for 30 min on ice with a saturated concentration of the PE-coupled ICAM-1 antibody, as recommended by the manufacturer (BD Biosciences, Mississauga, Canada). Finally, cells were washed twice in PBS and resuspended in 500 μl of PBS containing 1% paraformaldehyde prior to flow cytometry analysis (FACSCanto II; BD Biosciences, Mississauga, Canada).
Western blots analyses.
After SDS-PAGE migration, proteins were transferred onto Immobilon-P membranes (0.45 μm) (ThermoFisher Scientific, Ottawa, Ontario, Canada) by standard blotting techniques. Proteins were revealed with an anti-p24 antibody (clone 183-H12-5C). Primary antibodies were detected with a horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Lab, Inc.). HIV-1 Gag proteins immunodetected on membranes were quantitated with Image J (1997 to 2012; W. S. Rasband, U.S. National Institutes of Health, Bethesda, MD, USA [http://imagej.nih.gov/ij/]). Graphics were normalized with Gag WT proteins and expressed as a percentage of the WT or as the total Gag density.
3D modeling and protein-peptide docking.
The three-dimensional (3D) model of the cytoplasmic C terminus of ICAM-1 (peptide residues 504 to 532) was modeled de novo by using PepFold (33). The model quality was assessed by analysis of a Ramachandran plot through PROCHECK (34). The peptide spanning residues 504 to 532 (peptide 504-532) was docked into the structure of the N-terminal 283-residue fragment of the HIV-1 Gag polyprotein (PDB accession number 1L6N) by using PatchDock software (35). The latter is based on shape complementarity principles. The 3D complex containing HIV-1 MA and peptide 504-532 was refined by using FlexPepDock (36), which allows full flexibility to the peptide and side-chain flexibility to the receptor. Interactions between peptide 504-532 and MA were computed by PIC (37). Pictures were generated by using PyMol (http://www.pymol.org/).
Statistical analysis.
All experiments were repeated at least three times, and data shown in each figure are the combined results for all different donors unless otherwise specified. Statistically significant differences between groups were determined by using the Student t test when only two groups were evaluated or by one-way analysis of variance (ANOVA) with multiple comparisons using Bonferroni's test. Calculations were made with Prism version 6 (GraphPad Software, Inc., La Jolla, CA, USA). P values of <0.05 were considered statistically significant.
RESULTS
Nucleocapsid, SP2, and p6 Gag domains do not contribute to ICAM-1 incorporation into nascent HIV-1 particles.
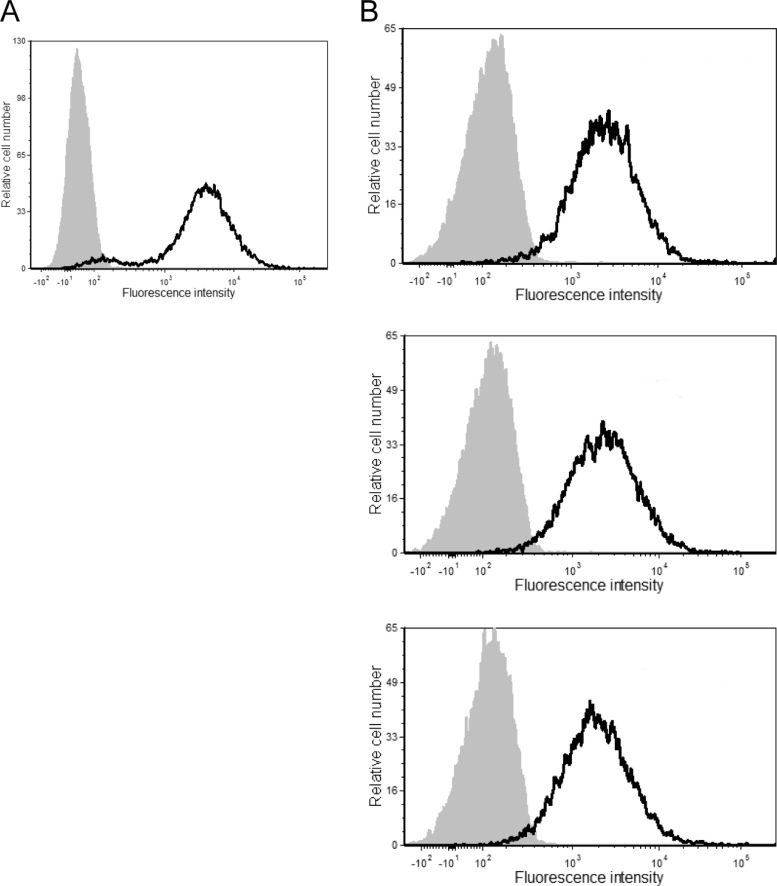
It was established previously that the Pr55Gag precursor polyprotein, which codes for all the structural proteins of HIV-1 (20, 38–41), acts as a dominant actor in the process of acquisition of ICAM-1 (30). Therefore, our initial series of investigations were aimed at corroborating this observation using a simple and pertinent model of VLPs mimicking the immature virion state. For this purpose, only the full-length gag gene or different deletion and substitution Gag mutants were cloned into the pCDNA3.1(+) expression vector. Based on a previous report by our group, in which we characterized different VLP mutants (31), we selected only deletion and substitution mutants giving rise to functional VLPs that can form spherical particles with a morphology similar to that of WT Pr55Gag and be excreted in the extracellular milieu, as shown by transmission electron microscopy. Thus, initial experiments were conducted with the following three molecular constructs: P6449LD, which carries a substitution of the p6 gene by the late p2b domain from Rous sarcoma virus (RSV); NCP6378LZLD, which bears a double substitution of the NC-SP2 and p6 regions by a heterologous leucine zipper domain (bZIP) from the CREB gene and the late domain (LD) from RSV, respectively (42–45); and, finally, MAΔ44-132, which carries a deletion of 89 amino acids in the MA gene. The studied VLP-encoding mammalian expression vectors were transiently transfected in a 293T cellular clone stably expressing a constant level of ICAM-1 on its surface (11). As depicted in Fig. 1, the surface expression level of ICAM-1 on the stable transfectant is slightly higher than that in mitogen-stimulated primary human cells (i.e., PBMCs), which provides physiological relevance to our studies. Next, the cell-free supernatants were harvested 48 h following transfection, and VLPs were concentrated by ultracentrifugation. Next, a virus capture assay based on the immunomagnetic isolation of ICAM-1-bearing VLPs was performed before lysis of the isolated material in hot SDS sample buffer prior to Western blot analysis. As illustrated in Fig. 2A, the vector coding for the complete Pr55Gag precursor polyprotein (called the Gag WT), which is free of any other virus constituent, produces HIV-1-based VLPs that can efficiently acquire host-derived ICAM-1. Host-derived ICAM-1 molecules were similarly incorporated into VLPs obtained after transfection of the P6449LD and NCP6378LZLD molecular constructs. Virus capture assays with an irrelevant isotype-matched control antibody gave no signal. It can be concluded that NC, SP2, and p6 domains are dispensable for ICAM-1 incorporation in this VLP model. In fact, Gag density quantification of the protein bands confirmed that the levels of captured VLPs are comparable for the three VLP constructs tested (Fig. 2B).
FIG 1.
Surface expression levels of ICAM-1. The stable ICAM-1-expressing 293T cell line (A) and PBMCs from three healthy donors (B) were incubated with either an R-phycoerythrin-conjugated mouse anti-human ICAM-1 antibody (black lines) or an isotype-matched irrelevant antibody (used as a control) (gray areas). The results shown are representative of data from 1 out of a total of 3 independent experiments.
The C-terminal region of MA is required for ICAM-1 incorporation.
The third VLP mutant, called MAΔ44-132, was subjected to the same experimental procedure, and in sharp contrast to the two other mutants, we could barely isolate any ICAM-1-bearing VLPs (Fig. 3A). Indeed, only a very faint signal at 46 kDa, corresponding to the MA mutant, was observed, compared to the strong band detected for the Gag WT. This result was confirmed by densitometry analysis, where <10% of the MA mutant signal was detected compared to the Gag WT (Fig. 3B). To our knowledge, this is the first demonstration that a defined domain in Gag (i.e., MA) is involved in the acquisition of host ICAM-1 molecules by HIV-1, at least in a VLP experimental model system. To further characterize and confirm this finding with fully competent HIV-1, we reproduced the Gag deletion in the prototype laboratory proviral clone pNL4.3. Transposing the deletion of 89 amino acids in the MA gene to the protease-positive NL4.3 provirus (called NL4.3Δ44-132), we then faced a constraint with regard to the virus assembly process (27, 46). Obviously, a deletion of 89 amino acids from the C-terminal end of MA might affect the capacity to produce spherical and functional virions, which could be essential for ICAM-1 incorporation. Thus, to circumvent this issue, the cell line producing the NL4.3Δ44-132 mutant was pretreated with a potent protease inhibitor (i.e., indinavir). The rationale for pretreating cells transfected with the NL4.3Δ44-132 mutant was based on our previously reported demonstration that significant increases in the levels of Gag products and ICAM-1–Pr55Gag complexes were seen in the NL4.3 WT upon indinavir treatment (30). We found that VLPs were produced in ICAM-1-expressing cells transfected with the NL4.3Δ44-132 mutant and treated with indinavir (Fig. 4A). Bands migrating at the 55-, 41-, and 24-kDa levels, corresponding to Pr55Gag, the maturation intermediate p41, and the capsid p24, respectively, were detected for the NL4.3 WT, while no signals were seen for the NL4.3Δ44-132 mutant. Quantification of the bands revealed a virtually complete loss of ICAM-1 incorporation (Fig. 4B). In this context, we were able to confirm that the MA region is really involved in the process of ICAM-1 incorporation in both immature and mature HIV-1 particles. For the ensuing experiments using MA mutants bearing either a short deletion or point mutations, which imply less structural and morphological consequences for emerging viruses, a classical virus production system was used based on p24 quantification by ELISA of pelleted viral particles.
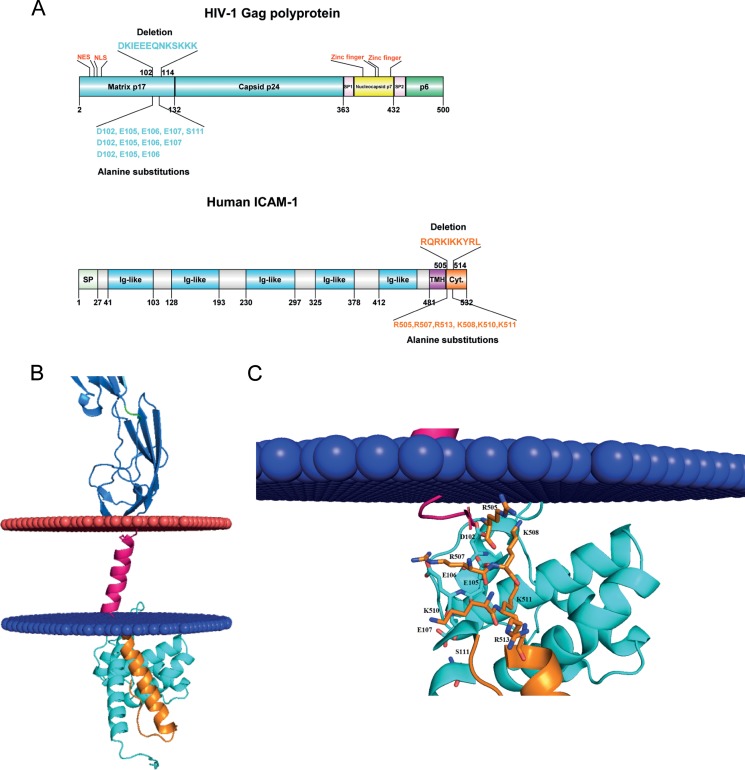
Protein-protein docking reveals a putative direct interaction between HIV-1 MA and human ICAM-1.
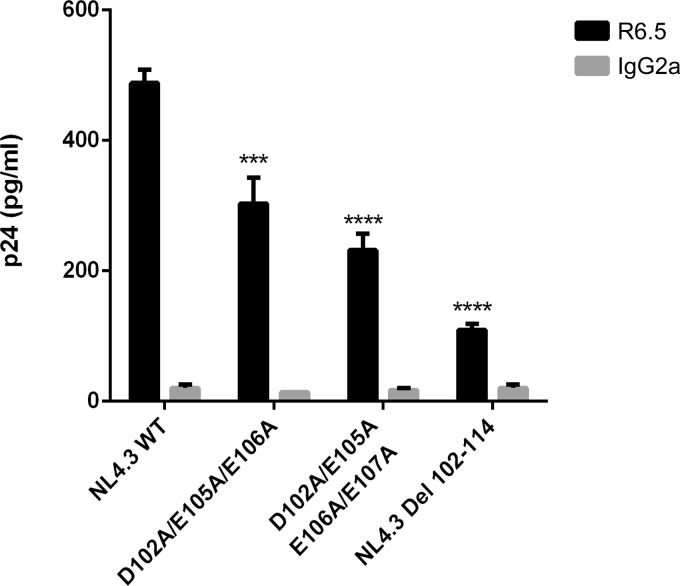
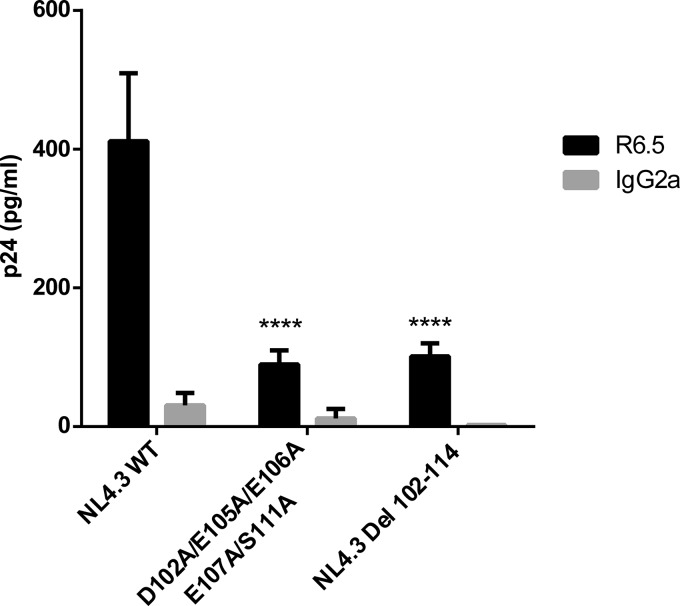
Thereafter, we aimed at refining the molecular relationship between these two proteins of distinct origins (i.e., viral MA and cellular ICAM-1). For this purpose, we investigated any potential interactions between HIV-1 MA and ICAM-1 by a protein-protein docking approach. This method highlighted areas of interest in each molecule (Fig. 5A and B). The putative interacting region in HIV-1 MA appears to be localized in the fifth α-helix and encompasses the following amino acids: 96-DTKEALDKIEEEQNKSKKKAQQAAAD-121 (amino acids of interest are shown in boldface) (Fig. 5C). On the other hand, de novo 3D modeling revealed a stretch of amino acids contained inside the predicted α-helix in the intracytoplasmic tail of ICAM-1 that appears to potentially interact with HIV-1 MA, i.e., 504-NRQRKIKKYRLQQAQKGTP-523 (Fig. 5C). First, to experimentally confirm the computational protein modeling and demonstrate the contribution of these two domains to ICAM-1 incorporation, we engineered the respective deleted mutants in both the HIV-1 MA (i.e., NL4.3 Del 102-114) and human ICAM-1 (i.e., CD54 Del 505-514) genes. As depicted in Fig. 6A, we observed a decrease in ICAM-1 incorporation (75 to 90%) with NL4.3 Del 102-114 compared to the NL4.3 WT. A substantial reduction in the level of ICAM-1 incorporation (65 to 75%) was also detected when the ICAM-1-deleted mutant named CD54 Del 505-514 was tested (Fig. 6B). The difference in ICAM-1 incorporation between these two deleted mutants is not caused by different ICAM-1 expression levels in cells producing the NL4.3 Del 102-114 (Fig. 6C) and CD54 Del 505-514 (Fig. 6D) mutant viruses.
FIG 5.
Molecular docking of putative interactions between HIV-1 MA and host-derived ICAM-1. (A) Diagram representation of the HIV-1 Pr55Gag precursor polyprotein (top) and ICAM-1 (bottom). Matrix and CA are shown in cyan, NC is shown in yellow, p6 is shown in green, and SP1 and SP2 regions are depicted in pink. For ICAM-1, the signal peptide (SP) is illustrated in green, the five-immunoglobulin-like domain is in blue, the hydrophobic transmembrane domain (TMH) is in purple, and the cytoplasmic domain is in orange. NLS, nuclear localization signal; NES, nuclear export signal. (B) Close-up view of negative residues (HIV-1 MA) and positive residues (cytoplasmic region of ICAM-1) that participate in the interaction. Negative and positive residues are shown as sticks. In this 3D model, D102 forms ionic bonds with R507-K508, E105 forms bonds with R507-K508-K510-K511, and E106 forms bonds with R507-K510. (C) Schematic representation of the 3D model of HIV-1 MA (PDB accession number 1L6N) docked to a 3D model of human ICAM-1 anchored in the cytoplasmic membrane (blue and red). The colors used for the model are the same as those described above for panel A.
The negative charges in HIV-1 MA interact with basic residues within the cytoplasmic tail of ICAM-1 and drive its incorporation.
Given that our results suggest that the two predicted domains within HIV-1 MA and human ICAM-1 might play a role in the insertion of this host adhesion molecule in emerging virions, we intended to refine our observation by trying to identify the specific amino acids involved in this process. Based on our predictive model (Fig. 5), we concentrated our efforts on a series of negatively charged amino acids in the HIV-1 MA domain spanning residues 102 to 114 that can potentially associate with positive charges in the cytoplasmic end of ICAM-1. We first tried to substitute aspartate 102 and the two glutamates at residues 105 and 106 for alanine because these residues should engage ionic interactions with the corresponding ICAM-1 amino acids. We then evaluated the ability of this mutant to acquire ICAM-1 by the virus capture assay. We found that such changes resulted in a diminution of 35 to 40% of ICAM-1 incorporation compared to that of WT NL4.3 (Fig. 7). This decrease in ICAM-1 incorporation is less important than the data obtained with the Del 102-114 mutant (i.e., 75 to 90%). Therefore, we hypothesized that the rest of the negative amino acids of the region spanning MA residues 102 to 114 are involved in the interaction and/or that a structural rearrangement involves the other negative residues in the interaction (amino acid compensation) and that we needed to completely remove all other negative charges present in the vicinity of this domain (47). Thus, we included glutamate 107, which immediately follows E105 and E106 in our substitution mutant, and observed a decrease in ICAM-1 incorporation up to 55 to 59%. Although close to that of the Del 102-114 deletant, the diminution is still weaker, and other amino acids might also be involved in the ICAM-1 incorporation phenomenon. Analysis of the sequence revealed the presence of a serine residue at position 111. After phosphoprediction with the NetPhos 2.0 server, we observed that this serine is potentially phosphorylated at 99%. Therefore, we also substituted this serine in order to totally eliminate all potential negative charges in the region. As shown in Fig. 8, this cumulative new substitution mutant, called the D102A/E105A/E106A/E107A/S111A mutant, diminished ICAM-1 incorporation up to 88% (75 to 88%), which is comparable to what was seen with NL4.3 Del 102-114. It can be concluded that the incorporation of host-derived ICAM-1 by nascent HIV-1 particles relies heavily on the participation of only five negatively charged amino acids in the complete HIV-1 MA domain.
FIG 7.
Immunocapture of NL4.3-derived WT and mutant viruses into which MA acidic residues were replaced by alanines. NL4.3-derived WT virions and mutant virus particles carrying the listed alanine substitutions were prepared in 293T cells stably expressing wild-type ICAM-1. Next, virus stocks were captured by using magnetic beads coated with an anti-ICAM-1 antibody (R6.5) or an isotype-matched irrelevant biotinylated antibody (used as a control). Precipitated viral entities were quantified with a homemade p24 ELISA. Data shown represent the means ± standard deviations of data from triplicate samples and are representative of three independent experiments. Statistical analyses were made by using one-way ANOVA, and asterisks denote a statistically significant difference between the NL4.3 WT and the listed mutants (***, P < 0.005; ****, P < 0.0001).
FIG 8.
Immunocapture of NL4.3-derived WT and mutant viruses into which MA acidic residues were replaced by alanines. NL4.3-derived WT virions and mutant virus particles carrying the listed alanine substitutions were prepared in 293T cells stably expressing wild-type ICAM-1. Next, virus stocks were captured by using magnetic beads coated with an anti-ICAM-1 antibody (R6.5) or an isotype-matched irrelevant biotinylated antibody (used as a control). Precipitated viral entities were quantified with an in-house p24 ELISA. Data shown represent the means ± standard deviations of data from triplicate samples and are representative of three independent experiments. Statistical analyses were made by using one-way ANOVA, and asterisks denote a statistically significant difference between the NL4.3 WT and the listed mutants (****, P < 0.0001).
Basic residues in the intracellular helix of ICAM-1 account for its incorporation within HIV-1.
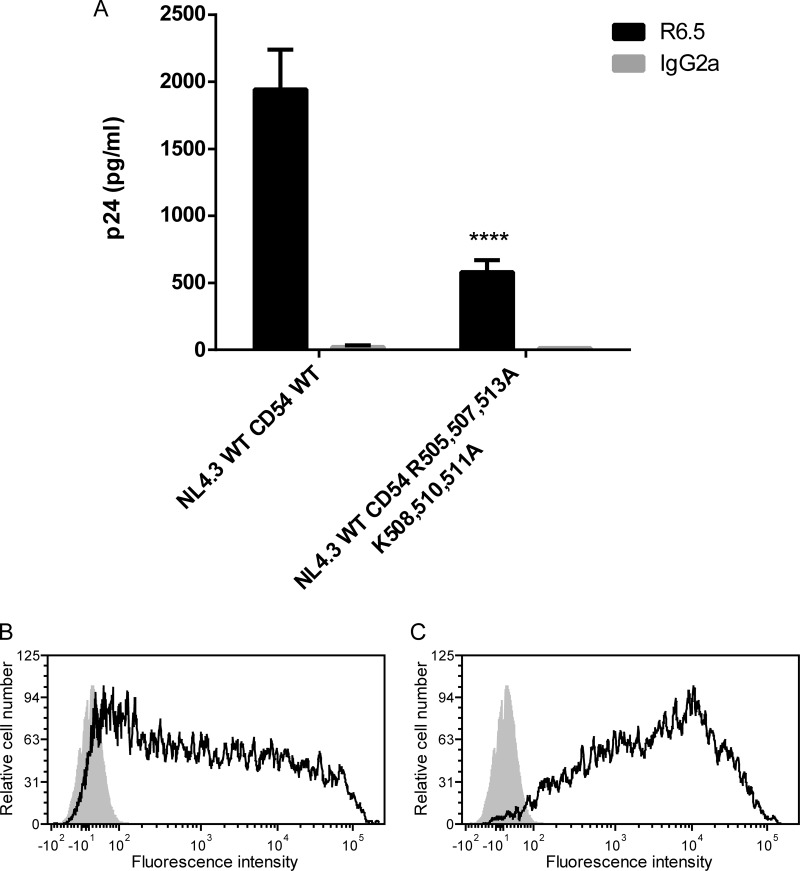
Concerning ICAM-1, there are six basic residues present within its cytoplasmic end, three of which are arginines located at positions 505, 507, and 513 and the other three of which are lysines found at positions 508, 510, and 511. Unfortunately, for each of these amino acids mutated individually or together, we could not detect a significant modulation in ICAM-1 incorporation compared to the WT protein (data not shown). We thought that this could be due to a conformational rearrangement in the predicted α-helix within the cytoplasmic part of ICAM-1 and to a compensation of the remaining basic amino acids for the mutated amino acids. Therefore, we applied an alternative strategy by substituting all basic amino acids with an alanine in the putative α-helix of ICAM-1. The corresponding substitution mutant, called CD54 R505,507,513A/K508,510,511A, was made and tested for the insertion of host ICAM-1 in progeny virus. Using this molecular construct, we are able to detect a significant decrease in ICAM-1 incorporation (65 to 70%) (Fig. 9). Therefore, it can be concluded that these six basic residues in the cytoplasmic tail of ICAM-1 are important for the selective acquisition of this host constituent by HIV-1.
FIG 9.
Immunocapture of NL4.3 WT virus produced in 293T cells transiently expressing wild-type or mutated ICAM-1. (A) NL4.3 WT virus particles were produced in 293T cells transiently expressing either WT or mutated ICAM-1. Next, virus stocks were captured by using magnetic beads coated with an anti-ICAM-1 antibody (R6.5) or an isotype-matched irrelevant biotinylated antibody (used as a control). Precipitated viral entities were quantified with an in-house p24 ELISA. Data shown represent the means ± standard deviations of data from triplicate samples and are representative of three independent experiments. (B and C) Flow cytometry analyses of surface expression of ICAM-1 in 293T cells transiently expressing WT (B) or mutated (C) ICAM-1. WT and mutated ICAM-1 are shown as solid black lines, while negative controls are presented as gray-filled histograms. Data shown represent cell surface expression of ICAM-1 at 2 days following transfection. Statistical analyses were made by using the Student t test, and asterisks denote statistically significant difference between the NL4.3 WT and the listed mutant (****, P < 0.0001).
ICAM-1 incorporation is significantly reduced in HIV-1 MA mutant viruses grown in primary human cells.
Finally, we sought to investigate if the previous findings made with the HIV-1 MA D102A/E105A/E106A/E107A/S111A mutant remained similar for virus preparations made in a more physiological cell system. To this end, mitogen-treated PBMCs from three different donors were inoculated with the NL4.3 WT and the NL4.3 D102A/E105A/E106A/E107A/S111A mutant, and progeny virus obtained after 5 days of acute HIV-1 infection was monitored for the extent of ICAM-1 incorporation. First of all, we observed that the HIV-1 MA D102A/E105A/E106A/E107A/S111A mutant can still replicate in PBMCs but to a lower level (i.e., 28 to 45%) than the NL4.3 WT (data not shown). More importantly, a significant decrease in ICAM-1 incorporation was seen in each instance (Fig. 10). This experiment illustrates that the intimate link between HIV-1 MA and ICAM-1 is not cell type specific and can also occur in progeny virus produced in natural target cells.
DISCUSSION
Previous observations have revealed that the cell surface adhesion molecule ICAM-1 is selectively acquired by HIV-1. This incorporation process involves the Pr55Gag polyprotein precursor, as shown previously (30) and as confirmed in the current experiments. Pr55Gag is sufficient per se for the insertion of host-derived ICAM-1 into emerging virions, whereas ENV and other regulatory/accessory HIV-1 proteins are dispensable in this regard. Studies performed with a series of Pr55Gag-derived VLP mutants revealed that we could easily replace NC, SP2, and P6 regions by heterologous sequences without affecting the ability of ICAM-1 to be efficiently inserted within nascent viruses. We thus conclude that these Pr55Gag domains do not contribute to ICAM-1 incorporation. Moreover, we found that a large deletion of 89 amino acids in the MA gene gave rise to VLPs lacking ICAM-1, and this crucial observation was confirmed by using the laboratory strain NL4.3 (NL4.3Δ44-132). Examining this hot spot in MA in more detail by molecular protein-protein docking and point mutations, we came down to only a few negatively charged relevant residues (i.e., D102, E105, E106, and E107) and phosphorylated serine 111, which account for ∼90% of ICAM-1 incorporation.
On the other hand, the cytoplasmic tail of ICAM-1 was also shown to be a key player in the process of acquisition of this host constituent by HIV-1 (30). Unfortunately, the crystal structure of the cytoplasmic domain of ICAM-1 had never been elucidated due to the well-known difficulty in crystallizing transmembrane proteins. De novo modeling allowed us to generate a new and accurate 3D model of the ICAM-1 intracellular tail. We found a predicted α-helix proximal to the plasma membrane, composed of 19 amino acids (i.e., 504-NRQRKIKKYRLQQAQKGTP-522), which is followed by a flexible tail (i.e., 523-MKPNTQATPP-532). Within the α-helix, basic residues potentially interact with the acidic residues of the MA ligand (shown in boldface type). This interaction relies mainly on electrostatic forces. We found a series of ionic interactions within <10 Å, enough to generate a strong binding of the two molecules within such proximity (48).
The intracellular domain 507-RKIKK-512 of ICAM-1 was previously described to be critical for its spatial and dynamic localization, with functional implications for leukocyte adhesion and transmigration in the fibroblast cell line COS-7 (49). Based on this notion, one may hypothesize that this domain might have a similar role in the HIV-1 context, directing ICAM-1 in specific areas of the membrane where the virus is emerging. However, we initially removed amino acids 507-RKIKK-512 from the intracellular tail, but ICAM-1 incorporation was not affected, as monitored by a virus capture assay (data not shown). In light of this result, we then tested whether the two basic residues surrounding this motif, namely, R505 and R513, might compensate for the loss of positive charges required for the ICAM-1 interaction with MA. For this reason, we decided to replace all these positively charged residues with a neutral alanine. Our results revealed that this domain is deeply involved in the association between host-derived ICAM-1 and the virus-encoded MA region, since its deletion diminished ICAM-1 incorporation up to 70%. According to our experimental model, it is clear that a region rich in basic residues present in the intracytoplasmic tail drives ICAM-1 incorporation. We demonstrate for the first time that a cellular transmembrane molecule interacts in very close proximity with Pr55Gag. Coimmunoprecipitation studies were performed, but convincing evidence of an intimate contact between the MA domain and host-derived ICAM-1 could not be obtained due to technical constraints (i.e., the inability to find a lysis buffer with a suitable strength allowing virus lysis without disrupting the association between MA and ICAM-1 proteins). Therefore, although our work cannot permit us to conclude without coimmunoprecipitation experiments that ICAM-1 incorporation is due to a direct interaction with MA, our experimental model strongly suggests that this might be the case given the theoretical, very intimate, charged interactions between residues on both sides.
The MA protein has been associated with important processes in HIV-1 biology. These functions have so far been almost exclusively mapped in the globular N-terminal portion of the protein, through helices 1 to 4 (50). The most studied and understood of these is its association with the internal leaflet of the cellular membrane for Gag trafficking and particle formation (51, 52). The N-terminal myristoylated residue in conjunction with a group of conserved basic residues form a bipartite domain that has been demonstrated to target the Pr55Gag protein to membranes (23, 53). The basic residues interact with acidic phospholipids, such as PI(4,5)P2. The N-terminal myristoyl moiety is well buried inside the p17 protein, and the interaction between MA and PI(4,5)P2 induces the anchorage of the myristate inside the lipid membrane, especially within the lipid rafts. Residues in the basic region of MA are also involved in the recruitment of the ENV proteins within the virion via the cytoplasmic tail of gp41. However, the exact mechanisms and necessity of this interaction remain poorly understood and need to be defined in more detail. Finally, other interactions with calmodulin, AP-3, TIP47, SOCS1, lyric, EF1-α, and HO3 have been described (52, 54, 55). To the best of our knowledge, we are the first group to suggest a possible association between a cellular factor and the fifth helix of the MA globular head. In a crystal model in which trimeric MA is bound to a lipid leaflet, as first demonstrated by Hill and colleagues (56, 57), helix 5 extends away from the globular head and thus away from the plasma membrane. We thus believe that it is highly plausible that in this configuration, helix 5 would be able to interact with the putative intracellular helix of the ICAM-1 intracellular tail to allow its eventual recruitment to nascent HIV-1 particles.
Our proposed model brings out new insights into the incorporation process of transmembrane molecules acquired by HIV-1. In the case of cell surface ICAM-1, a large part of the MA fifth α-helix appears to be directly involved. Interestingly, sequence alignment of >1,600 MA proteins from HIV-1 group M subtype B in the Los Alamos HIV database reveals a remarkably high degree of conservation of the sequence 102-DKIEEEQNKSKKK-114. Indeed, each amino acid contained in the sequence is highly conserved (i.e., ranging from 89 to 99%), except for residue D102, which is conserved at ∼64.6%. Nevertheless, in this case, aspartate is exchanged for a glutamate residue, which retains the same negative charge at this position, therefore not substantially affecting the overall negative charge of this region. Most importantly, we should mention that all the relevant negatively charged residues (i.e., D102, E105, E106, and E107) and the serine residue at position 111 are all highly conserved. It is tempting to speculate that these key conserved residues might have evolved to maintain ICAM-1 incorporation within HIV-1. Nonetheless, no clear function for the MA fifth α-helix has been described yet, although a role in interacting with CA p24 was suggested (58). In this sense, we cannot rule out that this highly conserved region might play a dominant role in another aspect of HIV-1 biology/pathology. A broader evaluation of the conservation of these residues in other HIV-1 subtypes and variants is needed to draw any further conclusions.
The promising results obtained with virus preparations made in primary human cells (i.e., PBMCs), where the MA mutant achieved a diminution of ICAM-1 incorporation of 66.4%, are in accordance with our observations made with virus stocks produced in 293T cells. This experiment illustrates that the intimate link between HIV-1 MA and ICAM-1 is not cell type specific and can be reproduced in cells that more closely mimic natural targets of HIV-1. It has now been clearly established that ICAM-1 incorporation increases virus infectivity, presumably by offering superior virus adhesion onto target cells, improved fusion kinetics, and cytosolic entry leading to a more productive virus infection (9, 10, 12). This enhancement is even more amplified when the LFA-1 counterligand on susceptible cells is in a high-affinity/avidity state, reaching up to almost 2 logs in some in vitro experiments. Our present study sheds light on the intimate molecular interactions between cellular ICAM-1 and viral Gag proteins. This opens a new avenue in the design of short peptide inhibitors that could circumvent the acquisition of ICAM-1 by HIV-1 and weaken its replicative capacity.
ACKNOWLEDGMENTS
This study was performed by Pascal Jalaguier in partial fulfillment of his Ph.D. degree at the Microbiology and Immunology Program, Faculty of Medicine, Laval University.
This study was supported by an operating grant to M.J.T. from the Canadian Institutes of Health Research (CIHR) (MOP-14438).
M.J.T. holds the CIHR-Canada Research Chair in Human Immuno-Retrovirology (tier 1 level).
We declare that we have no competing financial interests.
REFERENCES
- 1.Cantin R, Méthot S, Tremblay MJ. 2005. Plunder and stowaways: incorporation of cellular proteins by enveloped viruses. J Virol 79:6577–6587. doi: 10.1128/JVI.79.11.6577-6587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ott DE. 2008. Cellular proteins detected in HIV-1. Rev Med Virol 18:159–175. doi: 10.1002/rmv.570. [DOI] [PubMed] [Google Scholar]
- 3.Tremblay MJ, Fortin J-F, Cantin R. 1998. The acquisition of host-encoded proteins by nascent HIV-1. Immunol Today 19:346–351. doi: 10.1016/S0167-5699(98)01286-9. [DOI] [PubMed] [Google Scholar]
- 4.Ott DE, Coren LV, Johnson DG, Kane BP, Sowder RC II, Kim YD, Fisher RJ, Zhou XZ, Lu KP, Henderson LE. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42–51. doi: 10.1006/viro.1999.0075. [DOI] [PubMed] [Google Scholar]
- 5.Beauséjour Y, Tremblay MJ. 2004. Envelope glycoproteins are not required for insertion of host ICAM-1 into human immunodeficiency virus type 1 and ICAM-1-bearing viruses are still infectious despite a suboptimal level of trimeric envelope proteins. Virology 324:165–172. doi: 10.1016/j.virol.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 6.Carpen O, Pallai P, Staunton DE, Springer TA. 1992. Association of intercellular adhesion molecule-1 (ICAM-1) with actin-containing cytoskeleton and alpha-actinin. J Cell Biol 118:1223–1234. doi: 10.1083/jcb.118.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heiska L, Alfthan K, Gronholm M, Vilja P, Vaheri A, Carpen O. 1998. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4,5-bisphosphate. J Biol Chem 273:21893–21900. [DOI] [PubMed] [Google Scholar]
- 8.Bounou S, Leclerc JE, Tremblay MJ. 2002. Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4(+)-T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J Virol 76:1004–1014. doi: 10.1128/JVI.76.3.1004-1014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortin J-F, Cantin R, Lamontagne G, Tremblay M. 1997. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J Virol 71:3588–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortin J-F, Cantin R, Tremblay MJ. 1998. T cells expressing activated LFA-1 are more susceptible to infection with human immunodeficiency virus type 1 particles bearing host-encoded ICAM-1. J Virol 72:2105–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paquette J-S, Fortin J-F, Blanchard L, Tremblay MJ. 1998. Level of ICAM-1 surface expression on virus producer cells influences both the amount of virion-bound host ICAM-1 and human immunodeficiency virus type 1 infectivity. J Virol 72:9329–9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tardif MR, Tremblay MJ. 2003. Presence of host ICAM-1 in human immunodeficiency virus type 1 virions increases productive infection of CD4+ T lymphocytes by favoring cytosolic delivery of viral material. J Virol 77:12299–12309. doi: 10.1128/JVI.77.22.12299-12309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tardif MR, Tremblay MJ. 2005. LFA-1 is a key determinant for preferential infection of memory CD4+ T cells by human immunodeficiency virus type 1. J Virol 79:13714–13724. doi: 10.1128/JVI.79.21.13714-13724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tardif MR, Tremblay MJ. 2005. Regulation of LFA-1 activity through cytoskeleton remodeling and signaling components modulates the efficiency of HIV type-1 entry in activated CD4+ T lymphocytes. J Immunol 175:926–935. doi: 10.4049/jimmunol.175.2.926. [DOI] [PubMed] [Google Scholar]
- 15.Adamson CS, Freed EO. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol 55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 16.Adamson CS, Jones IM. 2004. The molecular basis of HIV capsid assembly—five years of progress. Rev Med Virol 14:107–121. doi: 10.1002/rmv.418. [DOI] [PubMed] [Google Scholar]
- 17.Cimarelli A, Darlix JL. 2002. Assembling the human immunodeficiency virus type 1. Cell Mol Life Sci 59:1166–1184. doi: 10.1007/s00018-002-8495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganser-Pornillos BK, Yeager M, Sundquist WI. 2008. The structural biology of HIV assembly. Curr Opin Struct Biol 18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wills JW, Craven RC. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 21.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. 2004. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 22.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J Virol 82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W, Parent LJ, Wills JW, Resh MD. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol 68:2556–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borsetti A, Ohagen A, Gottlinger HG. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol 72:9313–9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reil H, Bukovsky AA, Gelderblom HR, Gottlinger HG. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J 17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang CT, Lai HY, Li JJ. 1998. Analysis of minimal human immunodeficiency virus type 1 gag coding sequences capable of virus-like particle assembly and release. J Virol 72:7950–7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol 68:5311–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottlinger HG, Sodroski JG, Haseltine WA. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantin R, Fortin J-F, Lamontagne G, Tremblay M. 1997. The presence of host-derived HLA-DR1 on human immunodeficiency virus type 1 increases viral infectivity. J Virol 71:1922–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beauséjour Y, Tremblay MJ. 2004. Interaction between the cytoplasmic domain of ICAM-1 and Pr55Gag leads to acquisition of host ICAM-1 by human immunodeficiency virus type 1. J Virol 78:11916–11925. doi: 10.1128/JVI.78.21.11916-11925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jalaguier P, Turcotte K, Danylo A, Cantin R, Tremblay MJ. 2011. Efficient production of HIV-1 virus-like particles from a mammalian expression vector requires the N-terminal capsid domain. PLoS One 6:e28314. doi: 10.1371/journal.pone.0028314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhatia AK, Campbell N, Panganiban A, Ratner L. 2007. Characterization of replication defects induced by mutations in the basic domain and C-terminus of HIV-1 matrix. Virology 369:47–54. doi: 10.1016/j.virol.2007.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thevenet P, Shen Y, Maupetit J, Guyon F, Derreumaux P, Tuffery P. 2012. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res 40:W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 35.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. 2005. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raveh B, London N, Schueler-Furman O. 2010. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins 78:2029–2040. doi: 10.1002/prot.22716. [DOI] [PubMed] [Google Scholar]
- 37.Tina KG, Bhadra R, Srinivasan N. 2007. PIC: Protein Interactions Calculator. Nucleic Acids Res 35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jouvenet N, Bieniasz PD, Simon SM. 2008. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jouvenet N, Simon SM, Bieniasz PD. 2009. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A 106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jouvenet N, Simon SM, Bieniasz PD. 2011. Visualizing HIV-1 assembly. J Mol Biol 410:501–511. doi: 10.1016/j.jmb.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crist RM, Datta SA, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J Virol 83:2216–2225. doi: 10.1128/JVI.02031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ott DE, Coren LV, Gagliardi TD, Nagashima K. 2005. Heterologous late-domain sequences have various abilities to promote budding of human immunodeficiency virus type 1. J Virol 79:9038–9045. doi: 10.1128/JVI.79.14.9038-9045.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strack B, Calistri A, Gottlinger HG. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J Virol 76:5472–5479. doi: 10.1128/JVI.76.11.5472-5479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J Virol 72:1782–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finzi A, Orthwein A, Mercier J, Cohen EA. 2007. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J Virol 81:7476–7490. doi: 10.1128/JVI.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark AR, Naylor CE, Bagneris C, Keep NH, Slingsby C. 2011. Crystal structure of R120G disease mutant of human alphaB-crystallin domain dimer shows closure of a groove. J Mol Biol 408:118–134. doi: 10.1016/j.jmb.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, Witham S, Alexov E. 2011. On the role of electrostatics in protein-protein interactions. Phys Biol 8:035001. doi: 10.1088/1478-3975/8/3/035001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh HM, Lee S, Na BR, Wee H, Kim SH, Choi SC, Lee KM, Jun CD. 2007. RKIKK motif in the intracellular domain is critical for spatial and dynamic organization of ICAM-1: functional implication for the leukocyte adhesion and transmigration. Mol Biol Cell 18:2322–2335. doi: 10.1091/mbc.E06-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hearps AC, Jans DA. 2007. Regulating the functions of the HIV-1 matrix protein. AIDS Res Hum Retroviruses 23:341–346. doi: 10.1089/aid.2006.0108. [DOI] [PubMed] [Google Scholar]
- 51.Chukkapalli V, Ono A. 2011. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J Mol Biol 410:512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghanam RH, Samal AB, Fernandez TF, Saad JS. 2012. Role of the HIV-1 matrix protein in Gag intracellular trafficking and targeting to the plasma membrane for virus assembly. Front Microbiol 3:55. doi: 10.3389/fmicb.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan X, Yu X, Lee TH, Essex M. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol 67:6387–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cimarelli A, Luban J. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol 73:5388–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lama J, Trono D. 1998. Human immunodeficiency virus type 1 matrix protein interacts with cellular protein HO3. J Virol 72:1671–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci U S A 93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol 244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 58.Verli H, Calazans A, Brindeiro R, Tanuri A, Guimaraes JA. 2007. Molecular dynamics analysis of HIV-1 matrix protein: clarifying differences between crystallographic and solution structures. J Mol Graph Model 26:62–68. doi: 10.1016/j.jmgm.2006.09.009. [DOI] [PubMed] [Google Scholar]